
Adipose tissue fibrosis assessed by high resolution ex vivo MRI as a hallmark of tissue alteration in morbid obesity
Introduction
Obesity was defined by the World Health Organization (WHO) as a disproportionate adipose tissue (AT) growth that impairs health. In 2016, more than 650 million adults aged 18 years and older were obese, representing 13% of the world’s adult population (1). Obesity is associated with increased risks of type 2 diabetes mellitus, cardiovascular and liver diseases as well as cancer, osteoarthritis, and chronic kidney disease, among others (2,3).
Low grade inflammation and fibrosis accumulation in different organs such as the liver (4), heart (5) and AT are factors related to comorbidity development. The discovery of the biological relevance of AT fibrosis is recent (6). Subcutaneous AT (SAT) fibrosis is increased during obesity and higher levels of collagen gene expression and protein turnover are found in patients with deleterious profiles of AT distribution i.e. more visceral depots (7). In addition, fibrosis was shown to impair AT homeostasis and function (8). In obesity, the relative increase in SAT extracellular matrix (ECM) reduces tissue elasticity and is associated with adipocyte dysfunction, ectopic fat storage and metabolic disorders (8,9). Importantly, such changes to the ECM, and consequent fibrosis in human SAT, are strongly linked to insulin resistance and type 2 diabetes mellitus (10-13).
Scoring of SAT fibrosis based on tissue biopsy was recently proposed (14) through methods based on surgical samples analyzed by picrosirius red staining in histology, which are considered as the gold standard to detect and quantify tissue fibrosis (15,16). Others aimed to quantify adipose tissue fibrosis using different modalities such as immunohistochemistry (11) and polarized light microscopy/histomorphometry (10). These methods are nevertheless invasive for patients, especially for disease follow-up since non-negligible risks are associated with surgical biopsy (e.g., bleeding, hematoma, infection). Moreover, histological analysis suffers from high sampling variability as biopsy samples represent only a very limited part of the whole tissue of interest. Finally, such analysis is also time consuming and subject to inter-individual variability due to the manual delineation of the picrosirius red for subsequent total quantification (9) which is the reason why a semi-quantitative scoring was developed and proved accurate and less time consuming than the manual delineation of picrosirius red (14).
SAT fibrosis imaging is currently becoming of great clinical interest to examine tissue alteration and remodeling and could be used as a proxy for systemic fibrosis evaluation since it is easier to access than liver or kidney tissue. Non-invasive methods to evaluate SAT fibrosis in obese patients were recently proposed. Shear wave elastography has been proposed to quantify tissue stiffness (FibroScan®) (17) and was mainly applied to the liver with subsequent adaptation to SAT (AdipoScan®) characterization revealing positive correlations to histological SAT fibrosis and to metabolic comorbidities (18). These techniques remain new and have not reached widespread validation and availability. Questions related to repeated measurements, reproducibility and clinical utility in the management of non-alcoholic fatty liver disease have been raised recently (19), and such questions have not yet been addressed in the SAT characterization.
MRI allows for non-invasive and targeted assessment of whole body AT thanks to the specific resonance of fat using Dixon-based methods which is a chemical-shift-encoded imaging technique. The major output of this sequence is water and fat separated images, and consequently in-phase and out-of-phase data, as well as fat fraction maps. Recently, MRI T1 mapping has generated considerable interest as it grants tissue characterization of the cellular and extracellular tissue components non-invasively. Indeed, at a macroscopic level, native and post-contrast T1 values and derived parameters such as extracellular volume (ECV), albeit non-specific, have been found to be sensitive markers of the fibrosis presence and degree (20).
The aim of our work was to study the feasibility of detecting and measuring human SAT fibrosis ex vivo in severely obese patients by MR spectroscopy (MRS) and MRI in order to subsequently stratify obese patients in vivo and to ultimately predict their weight-loss response after bariatric surgery and prognosis.
Materials and methods
Patients and tissue samples
This study was approved by the institutional review board (clinicaltrials.org: NCT01655017). Informed consent was obtained from all patients. SAT samples (n=10) were collected by surgical biopsy from obese participants during bariatric surgery performed at la Pitié-Salpêtrière Hospital recruited from a dedicated hospital-wide research and treatment program on severe obesity. The superficial SAT samples were collected under local anesthesia from a small incision made lateral and perpendicular to the central line, horizontal to the umbilicus. The volume of biopsies was around 1 cm3 to avoid scars. Only five samples had histological fibrosis assessment. Samples were immediately fixed in 4% Paraformaldehyde for ex vivo MRI. Patients were included if they met the following bariatric surgery criteria: body mass index (BMI) >40 kg/m2 or BMI >35 kg/m2 with obesity related comorbidities, and age ≥18 years. Patients’ clinical characteristics are displayed in Table 1. None of the participants had acute illnesses, intestinal inflammatory diseases or malignancies. Patients taking medications that could possibly affect adiposity were excluded. Body composition was estimated by whole-body dual-energy X-ray absorptiometry (DXA) scanning.
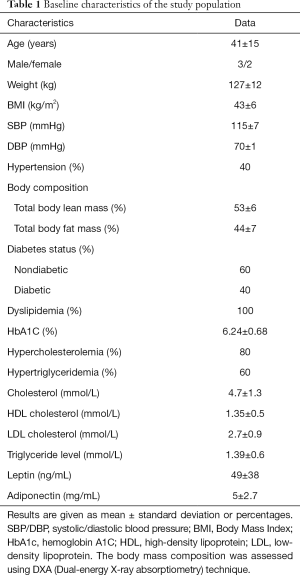
Full table
MR spectroscopy
A laboratory-assembled 4.7 T scanner (30 cm diameter bore) and a gradient system (12 cm diameter, peak gradient amplitude =400 mT/m) was used for MR spectroscopy. Samples were examined using a 16 mm diameter loop-gap resonator (maximum B1=1,174 µT) to provide higher sensitivity supplied by a pulsed amplifier (TOMCO, 1 kW maximum power). The dwell time was equal to 200 µs, the number of points was equal to 1,024 and the number of averages was equal to 4. Spectra were acquired after a 90° hard RF pulse without any gradients with immediate reception of the signal by the RF coil. MRS signal was collected from the entire sample volume. The two main peaks were referenced to water and the dominant methylene lipids (-CH2 and -CH3). The water fraction percentage, which may reflect the fibrosis and edema percentage, was defined as follows: WA/(FA+WA)×100, where WA is the area under the water peak and FA is the area under the fat peak.
MR imaging
MR images were acquired on a 4.0 T scanner (Bruker, Germany) supplied with a gradient insert and a 2.8 cm diameter coil. AT samples were immersed in fluorinertâ FC-770 (Sigma-Aldrich, Germany) to eliminate susceptibility artifacts and placed in tubes.
T1 maps were generated from a 2D RARE (Rapid Acquisition with Relaxation Enhancement) sequence at different TRs (repetition time) using a mono-exponential decay and taking into account saturation efficacy with the following parameters: TE (ms) = 11.4, TR delays (s): 0.1; 0.2; 0.4; 0.7; 1; 2; 4; 7, 3 slices, slice thickness (mm): 1, matrix =128×96 (zero fill to 128), FOV (cm) =2.56×2.56, in-plane spatial resolution (µm) =200, echo train length =2.
A gradient echo (Fast Low-Angle Shot: FLASH) sequence with three echo readout was performed using the following parameters: FOV (cm) =2.56×2.56, TE (ms) =3.414; 4.267; 5.12, TR (ms) =250, slice thickness: 1 mm, flip angle: 30°, 15 slices, number of excitations =8, bandwidth =592 Hz per pixel, matrix: 128×128, spatial resolution (µm) =200. All MRI data were analyzed using Matlab (The Mathworks, Natick, MA, USA). Single-slice and total (over three contiguous slices) fibrosis fractions were measured from T1 maps computed from the RARE sequence using the k-means classification method (21). Two classes were used to identify clusters corresponding to fibrosis and fat. MRI water-content fractions were quantified for each slice and then summed over all slices (3 slices) resulting in a water-content volume fraction. Such total water volume over the entire sample might help minimizing effect of sampling mismatch between MRI and histology, caused by differences between slice thicknesses.
As previously suggested (22), fat maps were reconstructed from in-phase (IP) and out-of-phase (OP) magnitude images acquired with the FLASH sequence. Fat maps were used for total visual assessment of water-content in SAT samples.
Histological analysis
Samples were fixed in PFA*m+ttoem 4% at room temperature. After MRI, SAT samples were embedded in paraffin. They were cut in 5 µm-thick sections and stained with picrosirius red for histological evaluation of fibrosis. Slices were digitized using a high quality resolution technique with an ECLIPSE TiE inverted microscope (Nikon Corporation, Japan). All scanned slices were magnified with an ×10 objective lens.
Histological analysis was performed using HistoLab (Microvision Instruments, France). Semi-automated segmentation through a thresholding method based on the color and illumination contents was performed. Such analysis comprised manual delineation of each tissue component as well as objects of non-interest within the selected area.
Total fibrosis was defined in percentage as the ratio of the respective total fibrosis surface area divided by the total tissue surface area multiplied by 100.
Statistical analysis
Statistical analysis was performed using PRISM 6 (GraphPad Inc., Canada). Fibrosis fractions are presented as percentage values according to the total surface of slices for histological and MRI analysis. Spearman’s correlation coefficients were used to report the relationship between MRI fibrosis fractions and histology and between MRI fibrosis fractions and native T1 values (two-tailed). The significance level was fixed to P<0.05. Bland-Altman analysis was used to assess agreement between single-slice and total MRI quantitative fibrosis fractions and mean bias as well as corresponding 95% confidence intervals (±1.96 SD) were computed.
Results
Spectral analysis
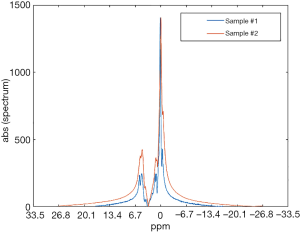
Representative ex vivo spectra obtained from human SAT samples are reported in Figure 1 where several proton resonances attributable to different chemical components can be distinguished. The water proton resonance is visible in the SAT tissue. The fat signal corresponds to the CH2 methylene protons of the lipid chain, and represents the bigger component in the triglyceride spectrum. The water peak broadening is due to the field inhomogeneities. Fibrotic samples showed a higher and larger water peak than less fibrotic samples (Figure 1). Mean fibrosis fraction in all samples was equal to 9.1%±6.4% [1–20%].
MRI and histological quantitative water fraction
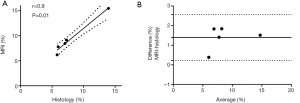
Mean T1 water class was equal to 1,048±182 ms and mean T1 fat class was equal to 394±37 ms. Figure 2 shows a good correlation (r=0.9, P=0.01) between total water fraction quantified from T1 mapping-MRI and fibrosis fraction quantified from histology. The Bland-Altman analysis showed good agreement between the two measurements [mean bias =1.3%, 95% limits of agreement (0.2–2.5%)].
MR imaging
Figure 3A shows a T1 map of a SAT sample highlighting fibrosis infiltration (red) within adipose tissue (yellow). The total fat map of a highly fibrotic sample presented more water-filled pores than a less fibrotic adipose tissue sample (Figure 3B).

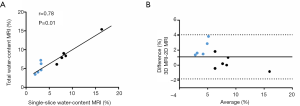
Total water fractions from MRI ranged between 6% and 15% and correlated with single-slice MRI water fractions (r=0.78, P=0.01) (Figure 4A). Bland-Altman plot (Figure 4B) showed that single-slice MRI water fractions were underestimated compared to total MRI water fractions [mean bias =1.1%, 95% limits of agreement (−1–4%)].
Discussion
Fibrosis accumulation in SAT is an important pathological alteration associated with comorbidities in obesity and is considered as an important index for risk stratification. Adipose tissue fibrosis is found to be negatively associated with post bariatric surgery weight loss (11). These findings highlight the potential prognostic importance of fibrosis quantification in obese patients relative to therapeutic strategy.
MR spectroscopy was performed at 4.7 T, two main peaks in the analyzed SAT samples were considered: fat peak and water peak which were spaced by approximately 700 Hz. Water fraction in our samples was consistent with previous findings and may partially represent fibrosis.
Fibrotic SAT samples presented a higher water peak amplitude and a broader fat peak. Indeed, when rigid collagen structures are associated to fat macromolecules, a slow molecular motion is induced resulting in peak broadening. Spectroscopy is the simplest magnetic resonance method to detect the increased amount of collagen in SAT samples however this technique does not provide any information on the spatial distribution of fibrosis within the tissue. The MRS peaks magnitude are related to overall signal within the tissue sample of interest. To avoid this bias in comparing samples, we computed the water fraction as a relative measure of fat/water to allow comparison between samples.
Very good correlation was observed in term of water fractions from MRI and fibrosis fraction from histology in the analyzed SAT samples. Our results show that it is possible to quantify fibrosis in human adipose tissue samples using single-slice and 3D MRI. For 3D MRI, a volumetric method, fibrosis fraction varied between 5% and 11% (7%±2%) in agreement with collagen accumulation percentage reported in invasive studies based on SAT biopsies (11,12).
This work has limitations, primarily a modest sample size. However, our primary goal was to study the feasibility of characterizing the subcutaneous adipose tissue and to validate the MR methodology. This pilot proof-of-concept study paves the way for translation of this approach to quantify SAT fibrosis in animal models to test new drugs which may ameliorate adipose tissue fibrosis, inflammation and glucose intolerance. Assessing this extracellular matrix environment may provide possible targets for pharmacological intervention for future potential treatments of metabolic disorders. Subsequently, a 3D MRI approach may help to stratify patients beyond their BMI according to the relative fibrosis amount in different adipose tissue types (subcutaneous adipose tissue, visceral adipose tissue) without requiring biopsy.
Acknowledgments
We would like to warmly thank Geneviève Guillot for her help during data acquisitions.
Funding: This work was funded by IHU ICAN.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-879). The authors have no conflicts of interest to declare.
Ethical Statement: The study was conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee (CPP Ile-de-France 1). All participants gave signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Available online: http://www.who.int/mediacentre/factsheets/fs311/en/. Access date: 02/06/2020.
- Ng M, Fleming T, Robinson M, Thomson B, Graetz N. Global, regional and national prevalence of overweight and obesity in children and adults 1980-2013: A systematic analysis. Lancet 2014;384:766-81. [Crossref] [PubMed]
- Kopelman PG. Obesity as a medical problem. Nature 2000;404:635-43. [Crossref] [PubMed]
- Abdennour M, Reggio S, Le Naour G, Liu Y, Poitou C, Aron-Wisnewsky J, Charlotte F, Bouillot JL, Torcivia A, Sasso M, Miette V, Zucker JD, Bedossa P, Tordjman J, Clement K. Association of adipose tissue and liver fibrosis with tissue stiffness in morbid obesity: links with diabetes and BMI loss after gastric bypass. J Clin Endocrinol Metab 2014;99:898-907. [Crossref] [PubMed]
- Venteclef N, Guglielmi V, Balse E, Gaborit B, Cotillard A, Atassi F, Amour J, Leprince P, Dutour A, Clément K, Hatem SN. Human epicardial adipose tissue induces fibrosis of the atrial myocardium through the secretion of adipo-fibrokines. Eur Heart J 2015;36:795-805a. [Crossref] [PubMed]
- Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, Zhang BB, Bonaldo P, Chua S, Scherer PE. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol Cell Biol 2009;29:1575-91. [Crossref] [PubMed]
- Pasarica M, Gowronska-Kozak B, Burk D, Remedios I, Hymel D, Gimble J, Ravussin E, Bray GA, Smith SR. Adipose tissue collagen VI in obesity. J Clin Endocrinol Metab 2009;94:5155-62. [Crossref] [PubMed]
- Sun K, Tordjman J, Clément K, Scherer PE. Fibrosis and adipose tissue dysfunction. Cell Metab 2013;18:470-7. [Crossref] [PubMed]
- Khan RS, Newsome PN. Non-alcoholic fatty liver disease and liver transplantation. Metabolism 2016;65:1208-23. [Crossref] [PubMed]
- Muir LA, Neeley CK, Meyer KA, Baker NA, Brosius AM, Washabaugh AR, Varban OA, Finks JF, Zamarron BF, Flesher CG, Chang JS, DelProposto JB, Geletka L, Martinez-Santibanez G, Kaciroti N, Lumeng CN, O'Rourke RW. Adipose tissue fibrosis, hypertrophy, and hyperplasia: Correlations with diabetes in human obesity. Obesity (Silver Spring) 2016;24:597-605. [Crossref] [PubMed]
- Divoux A, Tordjman J, Lacasa D, Veyrie N, Hugol D, Aissat A, Basdevant A, Guerre-Millo M, Poitou C, Zucker JD, Bedossa P, Clément K. Fibrosis in human adipose tissue: composition, distribution, and link with lipid metabolism and fat mass loss. Diabetes 2010;59:2817-25. [Crossref] [PubMed]
- Henegar C, Tordjman J, Achard V, Lacasa D, Cremer I, Guerre-Millo M, Poitou C, Basdevant A, Stich V, Viguerie N, Langin D, Bedossa P, Zucker JD, Clement K. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol 2008;9:R14. [Crossref] [PubMed]
- Lackey DE, Burk DH, Ali MR, Mostaedi R, Smith WH, Park J, Scherer PE, Seay SA, McCoin CS, Bonaldo P, Adams SH. Contributions of adipose tissue architectural and tensile properties toward defining healthy and unhealthy obesity. Am J Physiol Endocrinol Metab 2014;306:E233-46. [Crossref] [PubMed]
- Bel Lassen P, Charlotte F, Liu Y, Bedossa P, Le Naour G, Tordjman J, Poitou C, Bouillot JL, Genser L, Zucker JD, Sokolovska N, Aron-Wisnewsky J, Clément K. The FAT Score, a Fibrosis Score of Adipose Tissue: Predicting Weight-Loss Outcome After Gastric Bypass. J Clin Endocrinol Metab 2017;102:2443-53. [Crossref] [PubMed]
- Hui AY, Liew CT, Go MY, Chim AM, Chan HL, Leung NW, Sung JJ. Quantitative assessment of fibrosis in liver biopsies from patients with chronic hepatitis B. Liver Int 2004;24:611-8. [Crossref] [PubMed]
- Grimm PC, Nickerson P, Gough J, McKenna R, Stern E, Jeffery J, Rush DN. Computerized image analysis of Sirius Red-stained renal allograft biopsies as a surrogate marker to predict long-term allograft function. J Am Soc Nephrol 2003;14:1662-8. [Crossref] [PubMed]
- Cassinotto C, Lapuyade B, Mouries A, Hiriart JB, Vergniol J, Gaye D, Castain C, Le Bail B, Chermak F, Foucher J, Laurent F, Montaudon M, De Ledinghen V. Non-invasive assessment of liver fibrosis with impulse elastography: comparison of Supersonic Shear Imaging with ARFI and FibroScan®. J Hepatol 2014;61:550-7. [Crossref] [PubMed]
- Sasso M, Liu Y, Aron-Wisnewsky J, Bouillot JL, Abdennour M, Clet M, Sandrin L, le Naour G, Bedossa P, Tordjman J, Clément K, Miette V. AdipoScan: A Novel Transient Elastography-Based Tool Used to Non-Invasively Assess Subcutaneous Adipose Tissue Shear Wave Speed in Obesity. Ultrasound Med Biol 2016;42:2401-13. [Crossref] [PubMed]
- Wilder J, Patel K. The clinical utility of FibroScan ® as a noninvasive diagnostic test for liver disease. Med Devices (Auckl) 2014;7:107-14. [PubMed]
- Ambale-Venkatesh B, Lima J. Cardiac MRI: a central prognostic tool in myocardial fibrosis. Nat Rev Cardiol 2015;12:18-29. [Crossref] [PubMed]
- Bouazizi K, Rahhal A, Kusmia S, Evin M, Defrance C, Cluzel P, Berthet M, Atassi F, Leprince P, Lebreton G, Kachenoura N, Hatem SN, Redheuil A. Differentiation and quantification of fibrosis, fat and fatty fibrosis in human left atrial myocardium using ex vivo MRI. PLoS One 2018;13:e0205104. [Crossref] [PubMed]
- Liu CY, Redheuil A, Ouwerkerk R, Lima J, Bluemke D. Myocardial Fat Quantification in Humans: Evaluation by Two- Point Water-Fat Imaging and Localized Proton Spectroscopy. Magn Reson Med 2010;63:892-901. [Crossref] [PubMed]


