
In silico analysis of maize and wheat miRNAs as potential regulators of human gene expression
Highlight box
Key findings
• Corn and wheat microRNAs (miRNAs) have a potential to interact with messenger RNA (mRNA) of human genes.
What is known and what is new?
• Human consumption of food and herbal medicines supplies the body with not only nutrients but also associated biologically active compounds, such as exogenic miRNAs. Numerous miRNAs from maize and wheat might have effective binding sites (BSs) in the mRNA of human genes. Many zma-miRNAs and tae-miRNAs can bind to mRNAs of target genes of various human diseases. Given the high conservation of some miRNAs in many plant species, one should expect edible plants to influence the expression of human genes.
What is the implication, and what should change now?
• As a result, after experimental verification of our in silico results the diets of both healthy and sick people should be changed to accommodate the knowledge on impact of edible plants miRNAs on the expression of human genes.
Introduction
Plant messenger RNA (mRNA) inhibitory RNAs (miRNAs) are nanoscale noncoding RNAs 19–24 nucleotides long that regulate the expression of genes in animals (1) and plants (2,3) at the posttranscriptional level. These molecules can regulate the expression of target genes by cleaving mRNA, or they can prevent translation (4,5). Many plant miRNAs are crucial in the control of plant growth, development, and tolerance to biotic and abiotic stresses. In both plants and animals, miRNAs are key posttranscriptional regulators of gene expression.
Since a number of publications describe the entry of plant miRNAs into the human body through food intake (6), it is important to determine the possibilities of interaction between plant miRNAs and human genes. It was shown that that exogenous plant miRNA ingested by mammals can survive through the gastrointestinal system, and enter the bloodstream and various organs of mice (7). Liang et al. detected a variety of fruit miRNAs in the human plasma after feeding recruited volunteers with watermelon juice or mixed fruits (8). Liu et al. provided a novel information for the research and application of soybean-derived miRNAs in cancer occurrence (9). A recent study showed that olive small RNA is functionally homologous to human miRNA34 in cross-kingdom regulation of tumorigenesis (10). It has been demonstrated that plant small RNAs from Moringa oleifera may restore normalcy in immune system and reduce replication of human immunodeficiency virus (HIV) infection (11). Such small non-coding RNAs have also recently been used as tumor suppressor RNAs to play a role in tumor progression (12). The available information on the possible effect of plant miRNAs on human physiological processes is not sufficient for the target-based utilization of certain miRNAs for specific genes (13,14). Plant miR159 was previously shown to inhibit cancer growth in mammals (15). According to Zhang et al., rice-derived miRNA-168a regulates gene expression effectively and has been shown to target LDLRAP1 protein, a gene involved in cholesterol metabolism (16). It has been demonstrated that plant miRNAs have a regulatory effect on human genes involved in lipid metabolism (17). Plant mir156a has been shown to have an atheroprotective effect on inflamed human endothelial cells (18). Plant mir167e-5p can inhibit enterocyte proliferation by acting on β-catenin (19-21). The role of plant miRNAs as an immunomodulator in humans and animals has been revealed (22). Cavalieri et al. from the same study suggest that plant miRNAs can prevent chronic-inflammation related diseases (22).
The cross-kingdom regulation of miRNAs has become an attractive topic in nutrition science due to their ingestion with food (23-27). Plant miRNAs have the capacity to be stored in the gastrointestinal tract (28), enter the bloodstream, and control the production of protein synthesis via interactions with mRNA (1,23,24). Exogenous miRNAs can diffuse through body fluid, allowing cross-kingdom delivery (7,16,29,30). After surviving digestion, miRNAs use extracellular vesicles (31), e.g., exosomes (32-34), vesicles (35), specific lipoproteins (36), some viruses and Argonaute-containing bodies (37), to enter the bloodstream. “Trancytosis” is another process by which miRNAs can enter body fluids and involves the formation of a vesicle from the intestinal wall (38). Immune system cells (39) and specific transporters (40,41) may also help in this transportation. Previous studies have shown the slight reduction in miRNA content that occurs during storage, processing, and preparation of plant-based foods is one factor impacting the entry of plant miRNA into the human body (16,42). The potential of exogenous plant miRNAs to control target genes both in animals and humans opens up new possibilities for the treatment of human diseases using ex-miRNAs during therapy (43-45). Yang et al. have shown that dietary sRNAs could survive circulation and are excreted in urine (30). miR172, the most abundant miRNA in cabbage, was found in mice stomachs, serum, intestines, and feces (23). This study showed that ingested plant miRNAs may be involved in metabolism comparable to endogenous miRNAs, and with continuous feeding, the miRNA concentration can be consistent for few days (8,42). Following the consumption of watermelon juice, ten exogenous plant miRNAs were discovered in human plasma (8,46). After feeding maize for seven days, plant miRNAs were detected in pig tissues and blood (42). Human sera has been found to contain the plant-derived miR159, which was shown to target the TCF7 gene and prevent the development of breast cancer (15). The research revealed that orally administered plant miRNAs have the potential to be found in human sera and could affect the development of cancer in mammals in vitro. It was discovered that the exogenous plant miR168a was present in animal and human plasma and that miR168a regulated the expression of the human LDLRAP1 gene (16). miR156, one of the most conserved miRNAs in plants, is highly expressed in various crops, including wheat (47) and maize (48). In 2018, Hou et al., revealed that exogenous miR156a from green vegetables could be detected in human serum and showed the involvement of plant miR156a in cardiovascular diseases (18). Broccoli-derived miR156a has been shown to have therapeutic potential for the therapy of nasopharyngeal cancer patients (49). This result is confirmed by MirTarget program in the present work. Other findings indicate that miR156 inhibits intestinal cell proliferation by targeting Wnt10b (20).
The diversity of miRNAs in each plant species is great, however, in each of them there are identical key miRNAs that determine the regulation of plant growth and development (50,51). Previous evolutionary analyses have reported the functional and evolutionary significance of miR156 in land plants (52). Yang et al. proposed a work model for Arabidopsis leaf development and drought tolerance mediated by miR160 and miR165/166 interactions (53). Recently, bioinformatics approaches have been successfully used to predict the effect of ex-miRNAs on the mRNA of human genes, which reduces the amount of work and materials needed to determine how miRNAs interact with mRNAs (54-56). Computational approaches have been assessed for Hypericum perforatum flower miRNAs in relation to cancer. It was demonstrated that some of these miRNAs potentially have a significant and critical tumor-suppressive role for prostate cancer (57). In silico results indicated the role of miR156, miR414 and miR5021 in essential oil biosynthesis by the modulation of terpenoid backbone synthesis pathways (58). Although plant miRNAs have the potential to regulate gene expression, it is important to correctly find the target genes and prevent negative impacts (59). A number of studies have shown that miRNAs of different plants can enter the human body and circulate in the blood together with endogenous miRNAs, but the target genes of these miRNAs were not specified. The above data on the detection of plant miRNAs in the human body and their possible effect on gene expression indicate the need for studies on the effect of plant miRNAs on human genes. The aim of our research was to examine the interaction of human genes with Zea mays miRNAs (zma-miRNAs) and Triticum aestivum miRNAs (tae-miRNAs), which are some of the most valuable and high-yielding cereals cultivated on all continents and are the staple food for approximately one-third of the world’s population. These plant miRNAs might be used in future medicine as therapeutic agents in the regulation of the expression of target genes involved in a variety of biological processes in human body (60-62).
Methods
GenBank database (http://www.ncbi.nlm.nih.gov) was used to collect 17,508 human mRNA nucleotide sequences. Mature plant miRNA sequences were obtained from miRBase repository (v.22) (http://www.mirbase.org). The WebLogo (63,64) program was used to assess nucleotide sequence variability (https://weblogo.berkeley.edu/logo.cgi). The MirTarget software was used to search for the target genes of zma-miRNAs and tae-miRNAs (65,66). The MirTarget program determines the following binding characteristics: the start of the plant miRNA binding site (BS) of mRNA; the localization of plant miRNA BSs [5'-untranslated region (5'UTR); coding domain sequence (CDS) and 3'-untranslated region (3'UTR)]; the nucleotide interaction schemes between plant miRNAs and mRNAs and interaction free energy (∆G, kJ/mol). The ∆G value depends on GC-content and the number of nucleotides in miRNA. For each BS, the ΔG/ΔGm (%) ratio was found, where ΔGm is equal to the free energy binding of plant miRNA with its full complementary nucleotide sequence. The MirTarget software identifies hydrogen bonds between adenine (A) and uracil (U), guanine (G) and cytosine (C), G and U, and A and C. The distances between A and C (1.04 nm) and G and U (1.02 nm) bonds are comparable to the 1.03 nanometer distance between A and U, G and C (59-61). The number of hydrogen bonds in the G-C, A-U, G-U and A-C interactions has been found to be 3, 2, 1 and 1, respectively (67-70). The adequacy of the MirTarget program in terms of finding BSs has been confirmed in several publications (71-73).
To support the selection of the MirTarget software, we analyzed the existing tools for searching for BSs between plant miRNA and mRNA. We used TAPIR (74), psRNATarget (75), and RNA 22 (76) to show the effectiveness of the MirTarget compared to other programs (Table S1). To determine whether predicted plant miRNA target genes are involved in the development of human diseases, the DisGeNET (http://www.disgenet.org) platform was used (77). DisGeNET combines information from catalogs, scientific journals, and archives that have been expertly curated. The functional analyses were generated through the use of Ingenuity Pathway Analysis (QIAGEN Inc., Venlo, The Netherlands; https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/) (78).
Statistical methods
A total of 17,500 genes were searched—assuming 2,000 nt of sequence per transcript as an approximation that is about 35 million base pairs of sequence. The probability if having a particular 21-nt sequence is 421, i.e., 4.4×1,012. It can be noted, however, that our identified motifs either 9 or 10 perfectly conserved bases and a number of others that are partially conserved. As these number varies from sequence to sequence, the common expectation value is not the same for all these sequences. Instead, following a referee suggestion we assume the 14-mer as commonly found number of identities. The probability of finding certain n nucleotides in certain positions in the of k-mer sequence is equal to: probability of choosing certain n positions in k-mer sequence multiplied by the probability of finding correct nucleotides in these positions .
which for the 14-mer evaluates to .
Results
Characteristics of single zma-miRNA interactions with mRNAs of human target genes
We used NCBI GenBank data and novel bioinformatics tool to define miRNA binding parameters. Study of the interaction of 325 maize miRNAs with mRNAs of 17,508 protein coding genes indicated only 31 human target genes for nine maize miRNAs chosen based on the ∆G/∆Gm ratio (∆G, the free energy of the interaction between miRNA and the mRNA; ∆Gm equals the free energy of the miRNA binding with its fully complementary nucleotide sequence) equal to 90% and greater. The results attained are presented in Table 1. zma-miR162-5p, zma-miR529-5p, and zma-miR827-3p were found to have only one target gene. Many of the discovered target genes for maize-derived miR529-3p perform a variety of functions in cells and are essential for controlling numerous processes in the human organism (table available at https://cdn.amegroups.cn/static/public/exrna-23-4-1.xlsx). All identified interactions of zma-miRNAs with mRNA of human genes had a free binding energy ranged from −96 to −106 kJ/mol (Table 1). The ∆G/∆Gm values were also more than 90%, which indicates that the interaction of miRNA nucleotides and human gene BSs occurs mainly due to canonical nucleotide pairs.
Table 1
| zma-miRNA | Gene | ∆G, kJ/mol | ∆G/∆Gm, % | Length, nt |
|---|---|---|---|---|
| miR162-5p | ZNF853 | −104 | 91 | 21 |
| miR162-3p | DNAH3, PLXNA3, RIMS3 | −98 to −96 | 92 to 94 | 20 |
| miR482-5p | CDKN1C, FAM168A, FGF17, KCNK16, ROBO4, SLC44A4 | −98 to −96 | 94 to 96 | 19 |
| miR482-3p | ATP2A3, CSF1, LARP1, LMBRD2, PTER, SLC35A2, STK32A, TMCO5A, TTC25 | −98 to −96 | 92 to 94 | 20 |
| miR529-5p | PPIE | −102 | 92 | 21 |
| miR529-3p | ATP6V0A4, CHD2, LMO7, LRTOMT, LTB4R, MLL, NUDC, PDE4B, POMC, SOX9 | −106 to −102 | 91 to 94 | 21 |
| miR827-3p | AKAP11 | −100 | 94 | 21 |
mRNA, messenger RNA; zma, Zea mays; miRNA, mRNA-inhibitory RNA; ∆G, free energy of miRNA binding; ∆Gm, free energy of miRNA binding to a fully complementary nucleotide sequence; nt, nucleotide.
The ΔG/ΔGm ratio for maize miR482-5p BSs ranged between 94% and 96%, which shows that the interactions of zma-miR482-5p with mRNAs of human target genes have a high degree of complementarity (Table 1). The functions of the identified human target genes may have a role in the development of many diseases, including abdominal carcinoma, nonhematologic malignant neoplasm, thyroid carcinoma, etc. (table available at https://cdn.amegroups.cn/static/public/exrna-23-4-1.xlsx).
A wide range of interactions between maize-derived miRNAs and a number of human target genes may indicate a possible preventative effect of plant exogenous miRNAs on changes in human gene expression levels associated with various diseases. Particularly, the study of the therapeutic effect of medicinal plant miRNAs is being actively studied (60-62). The effect of both 5p and 3p strands of miRNAs on human target genes is one of the features of plant miRNAs. According to Table 1, both strands of maize-derived miR162, miR482, and miR529 can bind to the mRNA of several target genes. The human miRNA-5p and miRNA-3p from the same miRNA rarely have a similar number of target genes (71).
Interaction characteristics of zma-miRNA families with mRNAs of human target genes
Many plant miRNA sequences vary by only one or two nucleotides at the miR-5p or miR-3p, which is the basis for grouping plant miRNAs into families. Lettered suffixes are assigned to the name to indicate closely related mature miRNAs expressed from different precursors or genomic loci and signify miRNA family members (e.g., zma-miR160e-5p and zma-miR160g-5p) (79). With the most regular length of plant miRNAs (21–22 nt), the family members are 90% identical. The characteristics of the interaction of members of the zma-miRNA families with human mRNA genes are shown in Table 2. miRNAs from the same family had similar free binding energies with mRNA of human genes, since the length of their nucleotide sequences usually differed by only one nucleotide. Highly complementary interactions of zma-miRNAs with mRNA of target genes had miR169q-3p (∆G/∆Gm was 94–98%), miR319a,c-5p (∆G/∆Gm was 92–96%), miR399j-5p and miR528a,b-3p (∆G/∆Gm was 91–96%).
Table 2
| zma-miRNA | Gene | ∆G, kJ/mol | ∆G/∆Gm, % | Length, nt |
|---|---|---|---|---|
| miR156j-5p | AP2A2 | −104 | 94 | 21 |
| miR156i-3p | SGSM1 | −110 | 95 | 22 |
| miR159e-5p | BZRAP1, FEN1, LZTS1, PDE6B, PRCD | −106 to −104 | 91 to 93 | 21 |
| miR159h,i-3p | ATF6B, DEAF1, DGCR8, DOK6, GPSM3, TMEM229B, TTN, URM1 | −104 to −102 | 91 to 92 | 21 |
| miR160a-e,g-5p | DAB2 | −108 | 93 | 21 |
| miR164f-5p | ASXL1 | −110 | 95 | 21 |
| miR160f-3p | ALKBH5, BCL9L | −110 | 91 | 21 |
| miR164b-3p | DYSF, EXOC7, HPD, ITGA7, PTPRF, ZNF37A | −102 to −100 | 92 to 94 | 20 |
| miR166b-i-3p | CHRM1 | −102 | 92 | 20 |
| miR166m-5p | CCNY, HGS, INSM1, SGIP1, TNFSF13, TSNARE1 | −110 to −106 | 91 to 95 | 21 |
| miR167e-j-5p | HSF1, FAM43B, PRICKLE2 | −106 to −102 | 91 to 94 | 21 |
| miR167j-3p | FAM57A, FMNL2, GPR107 | −98 to −93 | 92 to 96 | 20 |
| miR168a-3p | FLAD1 | −102 | 92 | 20 |
| miR169i,j,k-5p | GRHL3, NCOA6 | −104 | 92 | 21 |
| miR169q-3p | ADRA1D, CA6, CYP1A1, DUOX1, EPHB6, MAP3K12, MARCH10, OXSR1, PIK3C2B, TRMT2A, TUBA3C, WNT16 | −104 to −100 | 94 to 98 | 19 |
| miR171d,e-5p | NFATC2, SIRT7 | −106 to −104 | 91 to 93 | 21 |
| miR171g-3p | BRD3 | −104 | 91 | 21 |
| miR172a-d-3p | GPR31, LEF1, TECTA, TUFM | −96 to −93 | 92 to 94 | 20 |
| miR172c-5p | GON4L, LASP1, SLC30A8, YY1AP1 | −100 to −98 | 92 to 94 | 20 |
| miR2118g-3p | DYRK1A, XPO6 | −106 to −104 | 91 to 93 | 22 |
| miR2275a-3p | GPR22, SACS | −102 to −100 | 92 to 94 | 22 |
| miR2275d-5p | AHCYL2, CER1, FAM205A, SPRR1B | −100 to −98 | 92 to 94 | 21 |
| miR319a,c-5p | PLCD4, RNF157 | −104 to −100 | 92 to 96 | 20 |
| miR319a-d-3p | DMAP1, IL4I1 | −102 | 92 | 20 |
| miR390a,b-5p | ASPSCR1, HFE | −108 to −106 | 91 to 93 | 21 |
| miR390a,b-3p | PDAP1 | −102 | 91 | 21 |
| miR393a,c-5p | TMEM136 | −106 | 93 | 22 |
| miR393c-3p | LRP1B, TP53I3 | −106 | 93 | 22 |
| miR394a,b-5p | BIN2, GRIK4, HAVCR2, HNF1B, IGSF3, MICAL3, PGAP1, REXO4, RXFP1 | −102 to −100 | 92 to 94 | 20 |
| miR394a,b-3p | ALDH4A1, CIB3, PURG | −100 | 92 | 20 |
| miR395a-j,n,p-3p | CD22, EDN1, TEAD2 | −108 to −102 | 91 to 96 | 21 |
| miR395k-5p | CCDC117, MYOM1 | −106 to −104 | 92 to 94 | 22 |
| miR396a,b-5p | SCAMP5, SYVN1, VPS13B | −102 to −100 | 92 to 94 | 21 |
| miR396g-3p | EVC2 | −100 | 92 | 21 |
| miR397a,b-5p | PKP3 | −102 | 91 | 21 |
| miR397b-3p | C6orf223 | −106 | 93 | 21 |
| miR398a,b-3p | EPS8, NUAK1, PCDHGA12 | −108 | 91 | 21 |
| miR398b-5p | BAHCC1, NUP62 | −108 | 91 | 21 |
| miR399d-3p | APBB1, CDK18, DDX11, JAGN1, OSTM1 | −108 to −106 | 91 to 93 | 21 |
| miR399j-5p | AMMECR1L, CHTF18, EDN3, IKZF3, LARP4B, LDLRAD2, LGI4, SIAH3, SPSB3, SOX18, SPOCK2, TRAK1 | −115 to −108 | 91 to 96 | 21 |
| miR408a,b-3p | ADARB2, ADCY6, ALPK3, ANO4, EDEM1, ERICH1, FNIP2, KIRREL3, NGB, PAQR6, RBMS2, RLBP1, SMARCC2, TJP3, UBE2K, UNG | −113 to −108 | 91 to 95 | 21 |
| miR408b-5p | ADCY1, ARHGAP30, CPNE6, DYRK1B, GCGR, GPR124, IL6R, MAFK, MAP3K6, NELL2, PLXNA4, PPM1F, PXN, RAB37, TMEM91, TNIP1, WDR46, ZNF132 | −110 to −108 | 91 to 93 | 21 |
| miR444a,b-3p | CEP250, CSRNP1, MPDZ | −102 to −100 | 92 to 94 | 21 |
| miR528a,b-5p | CACNB1, CD276, DNAJB6, GSK3B, GTF3C1, PPP1R26, PROB1 | −108 to −106 | 91 to 93 | 21 |
| miR528a,b-3p | EXOC7, FREM2, LSM4, MC1R, MGAT3, NFE2L1, RANBP1, RNF14, SPAG5, UIMC1 | −110 to −104 | 91 to 96 | 21 |
mRNA, messenger RNA; zma, Zea mays; miRNA, mRNA-inhibitory RNA; ∆G, free energy of miRNA binding; ∆Gm, free energy of miRNA binding to a fully complementary nucleotide sequence; nt, nucleotide.
The results obtained show that some of maize-derived miRNA families have from one up to 18 (miR408b-5p) human target genes, table available at https://cdn.amegroups.cn/static/public/exrna-23-4-1.xlsx lists the human target genes of maize miRNAs that might be involved in the development of deafness—ADCY1; lung cancer—ARHGAP30; glioblastoma multiforme—CPNE6; diabetes—GCGR; ischemic stroke—GPR124; mental depression—IL6R; liver injury—MAFK; gastric cancer—MAP3K6; morphological development for neuronal polarization and axon growth—NELL2; Alzheimer’s disease—PLXNA4; hepatocellular carcinoma—PPM1F; cervical cancer—PXN; nasopharyngeal carcinoma—RAB37; aspirin-exacerbated respiratory disease—WDR46; and esophageal squamous cell carcinoma—ZNF132, among others. The presented list of target genes involved in numerous human diseases demonstrates the potential of maize miRNAs in the regulation of the pathogenesis of the indicated diseases. The majority of the human target genes have a role in the development of oncological, cardiovascular and neurogenerative diseases (table available at https://cdn.amegroups.cn/static/public/exrna-23-4-1.xlsx).
The interaction schemes of nucleotides in these molecules are the most important characteristic in determining how plant-derived miRNAs interact with the mRNA of their target genes. The interactions of zma-miR482-5p, zma-miR169n-3p and zma-miR166l,m-3p with the mRNA of ROBO4, TRMT2A, and XAB2 human target genes are shown schematically in Figure 1. The figure shows that noncanonical A-C and G-U pairs enhance the free energy of interaction between plant miRNAs and human mRNAs. Furthermore, they maintain stacking interactions between nucleotides in each strand of the RNA helix (67,68).

Table S2 demonstrates the binding characteristics of maize miR529-3p to human mRNA genes, which show where miRNA BSs are located in the 5'UTR, CDS, and 3'UTR. It is essential for plant miRNA to have BSs in any of these target mRNA regions (Table S3).
WebLogo (63,64) schemes of the nucleotide sequence variability in the regions of human mRNA genes containing the BSs for maize miRNAs and wheat miRNAs show conserved BSs of several nucleotides at the 5- and 3-end. This demonstrates the significance of the 5- and 3-end in plant miRNAs binding to human mRNAs with higher GC content, which allows the binding of miRNAs with the highest free energy interaction (Figure 2).
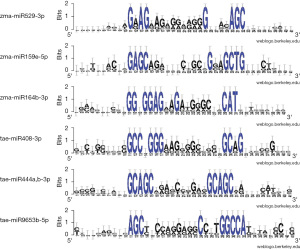
Therefore, determining the BS based on only a few nucleotides from the 5-end of the “seed” is insufficient. The study of the interaction of maize-derived miR159e-5p (Tables S4,S5) and miR164b-3p (Tables S6,S7) with human mRNA genes revealed similar results.
Characteristics of single tae-miRNA interactions with mRNAs of human target genes
Among the wheat miRNAs demonstrated in Table 3, miR156 (80,81), miR160 (82), miR164 (82), miR319 (83), miR396 (84), miR397 (85), miR398 (85), miR399 (83), miR408 (85,86), miR5048, miR9654b-miR9657b, miR9676-miR9679, miR9661, miR5384, miR9662a,b, and miR9652-5p (50) are found in many plants and are conserved. The free energy of interaction of miR169-3p, miR1119-3p, miR1129-5p, and miR9780-3p with mRNAs of the corresponding genes ranged from −113 to −129 kJ/mol, which indicates a strong interaction of these molecules (Table 3).
Table 3
| tae-miRNA | Gene | ∆G, kJ/mol | ∆G/∆Gm, % | Length, nt |
|---|---|---|---|---|
| miR156-5p | AP2A2, ZNF652, F11R (JAMA) | −104 to −102 | 90 to 94 | 21 |
| miR160-5p | C11orf16, DAB2 | −108 to −106 | 91 to 93 | 21 |
| miR164-5p | ASXL1, RASL10B, UGT1A7, UGT1A9 | −108 to −106 | 91 to 93 | 21 |
| miR169-3p | AARS2, GTPBP3, LYNX1 | −119 to −113 | 90 to 95 | 22 |
| miR319-3p | C15orf55, IL4I1, PTPMT1 | −104 | 91 | 21 |
| miR396-5p | AIM1 | −104 | 91 | 21 |
| miR397-3p | ING1 | −108 | 93 | 21 |
| miR398-3p | EPS8, NUAK1, PCDHGA12 | −108 | 91 | 21 |
| miR399-3p | HSPG2, JAGN1, U2AF2 | −98 to −96 | 94 to 96 | 19 |
| miR408-3p | ALPK3, RBMS2, EDEM1, NGB, PAQR6, UNG | −113 to −108 | 91 to 95 | 21 |
| miR531-5p | SLC5A10 | −113 | 91 | 21 |
| miR1118-5p | DCLK1, HECTD3, MDP1, OR51E2 | −110 | 93 | 23 |
| miR1119-3p | HPDL, KCNA1, SLC27A3 | −121 | 90 | 24 |
| miR1124-3p | AGRP, ARVCF, CPN1, FNTA, SPRN, URB1 | −110 | 90 | 22 |
| miR1129-5p | CDAN1, ENTPD2, LMO3, SLC47A1, RAX, RERE, WDR45 | −129 | 92 | 24 |
| miR1134-3p | CCND2, NIPAL4, NOTCH2, TCF7L2, TMEM178B | −108 | 91 | 24 |
| miR1138-3p | C21orf2, MFI2, MOG | −104 | 91 | 23 |
| miR1139-5p | CDON | −98 | 90 | 22 |
| miR1847-5p | ATRIP, HSD3B1 | −104 | 91 | 21 |
| miR5048-5p | FLCN | −100 | 90 | 22 |
| miR5050-5p | MIER1 | −102 | 91 | 21 |
| miR5084-3p | RYR2 | −110 | 90 | 24 |
| miR5085-5p | TGOLN2, ZFHX3 | −100 | 92 | 21 |
| miR5086-5p | DHX30, HBP1, LTBP4, MYO1G, SERPING1, SPTAN1 | −104 to −102 | 91 to 92 | 21 |
| miR5200-3p | LYST, TGFBR3 | −102 to −100 | 92 to 94 | 21 |
| miR5384-3p | MAF, PAX1 | −113 to −110 | 91 to 93 | 21 |
| miR9652-5p | LZTFL1, PTCHD1, STX6 | −100 to −98 | 90 to 92 | 22 |
| miR9655-3p | CIB2, GIPR, GPR123, GPR55, TACR1, ZNF488 | −102 | 91 | 21 |
| miR9656-3p | C9orf50 | −106 | 93 | 21 |
| miR9661-5p | DNAH14, GSE1, NUP50, SHANK1 | −104 to −102 | 91 to 92 | 21 |
| miR9664-3p | ORAOV1, SIK2 | −102 | 91 | 21 |
| miR9667-5p | REEP3 | −96 | 94 | 21 |
| miR9670-3p | TPM3, USP14 | −100 to −98 | 92 to 94 | 21 |
| miR9772-5p | C7 | −100 | 92 | 21 |
| miR9773-3p | CCDC178, CFLAR, INO80D, LRRC34, RHBDD1 | −100 | 90 | 24 |
| miR9774-3p | NPFF | −100 | 90 | 22 |
| miR9776-5p | SPRY4, TRAK2 | −106 to −102 | 91 to 94 | 21 |
| miR9777-3p | PDK2 | −93 | 92 | 20 |
| miR9778-5p | SORL1 | −102 | 91 | 21 |
| miR9779-3p | ASRGL1 | −100 | 96 | 20 |
| miR9780-3p | HES4, MEX3C, UBE2K | −119 to −117 | 92 to 93 | 21 |
| miR9781-3p | LRP8, SLC38A2, PCLO, RNASE3 | −91 to −89 | 91 to 93 | 21 |
| miR10520-5p | TCFL5, TSPYL6 | −98 to −96 | 92 to 94 | 20 |
mRNA, messenger RNA; tae, Triticum aestivum; miRNA, mRNA-inhibitory RNA; ∆G, free energy of miRNA binding; ∆Gm, free energy of miRNA binding to a fully complementary nucleotide sequence; nt, nucleotide.
The discovered wheat miRNA target genes perform multiple functions in cells, and many of them are important in the regulation of crucial processes in the human body and the development of different diseases (table available at https://cdn.amegroups.cn/static/public/exrna-23-4-1.xlsx).
Interaction characteristics of tae-miRNA families with mRNAs of human target genes
The wheat miRNA families consist of only 2–3 tae-miRNAs that target one to nine human genes, as demonstrated in Table 4. The ∆G/∆Gm value varies from 91% to 98%, which indicates a high degree of complementarity between miRNA and mRNA nucleotides. The G/Gm ratio varied from 91% to 98%, which shows that the nucleotides of miRNA and mRNA are highly complementary to each other. The values of free energy of interaction between wheat miRNAs and human mRNAs were higher than −90 kJ/mol.
Table 4
| tae-miRNA | Gene | ∆G, kJ/mol | ∆G/∆Gm, % | Length, nt |
|---|---|---|---|---|
| miR159a,b-3p | KCNJ15, PKHD1 | −102 | 92 | 21 |
| miR167a-c-5p | ARHGEF10, EME1, HSF1, PRICKLE2 | −108 to −102 | 91 to 96 | 21 |
| miR171b-3p | LTB4R2 | −102 | 91 | 21 |
| miR395a-3p | CD22, EDN1, TEAD2 | −108 to −102 | 91 to 96 | 21 |
| miR444a,b-3p | AP1B1, COL6A3, DOCK1, HIC1, KRT6A, KRT6B, KRT6C, RHBDF1, RUNDC1 | −108 to −104 | 91 to 94 | 21 |
| miR1120b-3p | SEMA3A, FAM177A1 | −98 | 92 | 21 |
| miR1120c-5p | EPHA4 | −91 | 91 | 21 |
| miR1122c-3p | KCNAB3 | −100 | 92 | 21 |
| miR1127b-3p | SEMA6A | −102 | 92 | 21 |
| miR1137a-3p | INO80D | −96 | 96 | 20 |
| miR9653a-3p | ARHGEF6, CLCN7, ENDOD1, H1FOO, KDM6B, KIF7, PHLDB2 | −106 to −102 | 91 to 94 | 21 |
| miR9653b-5p | AOC3, ITIH4, RBM19, TEAD2, TEAD3, TMEM184A | −113 to −104 | 91 to 98 | 21 |
| miR9654b-3p | NEBL | −104 | 91 | 22 |
| miR9657b-5p | RNF168 | −104 | 92 | 21 |
| miR9662a,b-3p | DNMT3B, FAM124A, GLI2, HMX3 | −110 to −104 | 91 | 21 |
| miR9666b-3p | HMX3 | −110 | 91 | 22 |
| miR9674b-5p | ROBO3 | −102 | 92 | 21 |
| miR9677a-3p | AKT1, RBFOX3, STAC | −108 | 91 | 22 |
| miR9677b-5p | ABCA3, CASKIN1, ENC1, GALR1, LRRFIP1, NLGN2, SNX8, SQSTM1 | −117 to −113 | 91 to 95 | 21 |
mRNA, messenger RNA; tae, Triticum aestivum; miRNA, mRNA-inhibitory RNA; ∆G, free energy of miRNA binding; ∆Gm, free energy of miRNA binding to a fully complementary nucleotide sequence; nt, nucleotide.
Table S8 shows the binding characteristics of wheat miR408-3p interaction with six human genes, indicating their effective binding. The sequences of regions of human mRNA genes containing wheat miR408-3p BSs reveal (Table S9) conserved nucleotides at the 5- and 3-end (Figure 2). Wheat miR444a,b-3p, which interacts with the CDS of nine mRNA genes, also had similar results (Table S10). Based on the nucleotides of the wheat miR444a,b-3p BSs (Table S11), the created WebLogo scheme demonstrates high conservation of some nucleotide sequences, which is identical at the 5- and 3-end of the BSs (Figure 2). Four G-C pairs with three hydrogen bonds each provide high free energy of interaction for wheat miR444a,b-3p with the mRNA of nine human genes. The characteristics of the interaction of wheat miR9653b-5p with the mRNA of seven human genes also show a high interaction efficiency (Table S12) and the conservation of flanking nucleotide sequences (Table S13 and Figure 2).
Discussion
The results show that many corn and wheat miRNAs have the potential to effectively interact with mRNA of human target genes. Similar studies done with rice miRNA are discussed in previous publication (87). Fully complementary interactions of plant miRNAs with mRNA genes have been previously established in many plant species, which may be important in the control of gene expression (80,82-84). Such an interaction between miRNAs and mRNAs has been already studied in animals (71,88,89). In the present work, the range of known examples was extended by studying the interaction between wheat and maize miRNAs and human mRNAs. Further studies can be done to determine which miRNAs of other plants can interact with mRNAs of human and animal genes.
The miRNAs are found in many plant tissues and organs’ cells in relatively large quantities, which enables the determination of their functional importance Many plant miRNAs have the potential to regulate gene expression in a cross-kingdom manner (90-94). Along with juices, fruits, and vegetables a large number of plant miRNAs enter the human body (8).
The findings of in silico study contribute to the understanding of a potential function of plant miRNAs in the control of human gene expression. In silico analysis can be considered a first step for gene repression regulation analysis. There are obstacles and challenges that dietary miRNAs need to surpass to exert biological effects, including digestion. The literature describes that digestion can be an important part of possible function of miRNAs both in mammalian dietary miRNAs (95) and plant-derived miRNAs (96). Transport of dietary miRNAs within exosomes could help reduce the degradation of dietary miRNAs (97).
The interaction of plant miRNAs and human mRNA genes depend on many factors: for example, on the concentration of miRNAs in cells of various human organs, on the concentration of target genes in the cells, since a low concentration of plant miRNAs cannot significantly change the expression of the target gene. Both positive and negative effects of plant miRNAs may be shown on the human genome expression. For instance, some plant miRNAs can block the expression of oncogenes and suppress oncogenesis, while other plant miRNAs might promote tumor development by inhibiting oncosuppressors. As a result, in order to use plant miRNAs in medicine, extensive research needs to be done to understand the impact of these miRNAs on the function of various human genes. It will also allow to create synthetic analogs of plant miRNAs for therapeutic purposes (98).
Conclusions
Analysis of the possibilities of interaction of maize and wheat miRNAs with mRNAs of human target genes demonstrated that many of these plant miRNAs have potential to bind to human mRNA genes. zma-miRNAs and tae-miRNAs interacting with mRNA of human genes include the most well-known plant miRNAs important for the regulation of plant growth and development, as well as miRNAs that respond to plant stress. Many zma-miRNAs and tae-miRNAs can bind to mRNAs of target genes of various human diseases. Given the high conservation of some miRNAs in many plant species, one should expect edible plants to influence the expression of human genes. The results obtained in this work are consistent with the published data of our (72,74-76,80,82) and other researchers (8,20,21,50). Further study of such plant miRNAs may contribute to their medical applications.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://exrna.amegroups.com/article/view/10.21037/exrna-23-4/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-23-4/coif). PZ serves as an unpaid editorial board member of ExRNA. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab (Lond) 2018;15:68. [Crossref] [PubMed]
- Samad AFA, Sajad M, Nazaruddin N, et al. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front Plant Sci 2017;8:565. [Crossref] [PubMed]
- Nair SK, Wang N, Turuspekov Y, et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci U S A 2010;107:490-5. [Crossref] [PubMed]
- Biswas K, Jolly MK, Ghosh A. First passage time properties of miRNA-mediated protein translation. J Theor Biol 2021;529:110863. [Crossref] [PubMed]
- Sedaghat N, Fathy M, Modarressi MH, et al. Identifying functional cancer-specific miRNA-mRNA interactions in testicular germ cell tumor. J Theor Biol 2016;404:82-96. [Crossref] [PubMed]
- Del Pozo-Acebo L, López de Las Hazas MC, Margollés A, et al. Eating microRNAs: pharmacological opportunities for cross-kingdom regulation and implications in host gene and gut microbiota modulation. Br J Pharmacol 2021;178:2218-45. [Crossref] [PubMed]
- Liang G, Zhu Y, Sun B, et al. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr 2014;2:380-8. [Crossref] [PubMed]
- Liang H, Zhang S, Fu Z, et al. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J Nutr Biochem 2015;26:505-12. [Crossref] [PubMed]
- Liu J, Wang F, Weng Z, et al. Soybean-derived miRNAs specifically inhibit proliferation and stimulate apoptosis of human colonic Caco-2 cancer cells but not normal mucosal cells in culture. Genomics 2020;112:2949-58. [Crossref] [PubMed]
- Minutolo A, Potestà M, Gismondi A, et al. Olea europaea small RNA with functional homology to human miR34a in cross-kingdom interaction of anti-tumoral response. Sci Rep 2018;8:12413. [Crossref] [PubMed]
- Minutolo A, Potestà M, Roglia V, et al. Plant microRNAs from Moringa oleifera Regulate Immune Response and HIV Infection. Front Pharmacol 2020;11:620038. [Crossref] [PubMed]
- Potestà M, Roglia V, Fanelli M, et al. Effect of microvesicles from Moringa oleifera containing miRNA on proliferation and apoptosis in tumor cell lines. Cell Death Discov 2020;6:43. [Crossref] [PubMed]
- Chen X, Rechavi O. Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol 2022;23:185-203. [Crossref] [PubMed]
- Schuh CMAP, Cuenca J, Alcayaga-Miranda F, et al. Exosomes on the border of species and kingdom intercommunication. Transl Res 2019;210:80-98. [Crossref] [PubMed]
- Chin AR, Fong MY, Somlo G, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 2016;26:217-28. [Crossref] [PubMed]
- Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012;22:107-26. [Crossref] [PubMed]
- Roglia V, Potestà M, Minchella A, et al. Exogenous miRNAs from Moringa oleifera Lam. recover a dysregulated lipid metabolism. Front Mol Biosci 2022;9:1012359. [Crossref] [PubMed]
- Hou D, He F, Ma L, et al. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J Nutr Biochem 2018;57:197-205. [Crossref] [PubMed]
- Li M, Chen T, He JJ, et al. Plant MIR167e-5p Inhibits Enterocyte Proliferation by Targeting β-Catenin. Cells 2019;8:1385. [Crossref] [PubMed]
- Li M, Chen T, Wang R, et al. Plant MIR156 regulates intestinal growth in mammals by targeting the Wnt/β-catenin pathway. Am J Physiol Cell Physiol 2019;317:C434-48. [Crossref] [PubMed]
- Zempleni J, Baier SR, Howard KM, et al. Gene regulation by dietary microRNAs. Can J Physiol Pharmacol 2015;93:1097-102. [Crossref] [PubMed]
- Cavalieri D, Rizzetto L, Tocci N, et al. Plant microRNAs as novel immunomodulatory agents. Sci Rep 2016;6:25761. [Crossref] [PubMed]
- Samad AFA, Kamaroddin MF, Sajad M. Cross-Kingdom Regulation by Plant microRNAs Provides Novel Insight into Gene Regulation. Adv Nutr 2021;12:197-211. [Crossref] [PubMed]
- Zhang L, Chen T, Yin Y, et al. Dietary microRNA-A Novel Functional Component of Food. Adv Nutr 2019;10:711-21. [Crossref] [PubMed]
- Zhao Q, Liu Y, Zhang N, et al. Evidence for plant-derived xenomiRs based on a large-scale analysis of public small RNA sequencing data from human samples. PLoS One 2018;13:e0187519. [Crossref] [PubMed]
- Marzano F, Caratozzolo MF, Consiglio A, et al. Plant miRNAs Reduce Cancer Cell Proliferation by Targeting MALAT1 and NEAT1: A Beneficial Cross-Kingdom Interaction. Front Genet 2020;11:552490. [Crossref] [PubMed]
- He JJ, Ye R, Chen T, et al. Exploration of miRNAs in Neolamarckia cadamba and their potential cross-kingdom functions. ExRNA 2020;2:6. [Crossref]
- Spinler JK, Oezguen N, Runge JK, et al. Dietary impact of a plant-derived microRNA on the gut microbiome. ExRNA 2020;2:11. [Crossref] [PubMed]
- Jimenez-Jimenez S, Hashimoto K, Santana O, et al. Emerging roles of tetraspanins in plant inter-cellular and inter-kingdom communication. Plant Signal Behav 2019;14:e1581559. [Crossref] [PubMed]
- Yang J, Farmer LM, Agyekum AA, et al. Detection of an Abundant Plant-Based Small RNA in Healthy Consumers. PLoS One 2015;10:e0137516. [Crossref] [PubMed]
- Zhao C, Sun X, Li L. Biogenesis and function of extracellular miRNAs. ExRNA 2019;1:38. [Crossref]
- Yarmarkovich M, Hirschi KD. Digesting dietary miRNA therapeutics. Oncotarget 2015;6:13848-9. [Crossref] [PubMed]
- Melnik BC, Weiskirchen R, Schmitz G. Milk exosomal microRNAs: friend or foe?—a narrative review. ExRNA 2022;4:22. [Crossref]
- Mutai E, Ramer-Tait AE, Zempleni J. MicroRNAs in bovine milk exosomes are bioavailable in humans but do not elicit a robust pro-inflammatory cytokine response. ExRNA 2020;2:2. [Crossref]
- Li L, Wang J. Roles of extracellular microRNAs in central nervous system. ExRNA 2019;1:13. [Crossref]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol 2010;3:478-81. [Crossref] [PubMed]
- Witwer KW, Hirschi KD. Transfer and functional consequences of dietary microRNAs in vertebrates: concepts in search of corroboration: negative results challenge the hypothesis that dietary xenomiRs cross the gut and regulate genes in ingesting vertebrates, but important questions persist. Bioessays 2014;36:394-406. [Crossref] [PubMed]
- Chen Q, Zhang F, Dong L, et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res 2021;31:247-58. [Crossref] [PubMed]
- Witwer KW. XenomiRs and miRNA homeostasis in health and disease: evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol 2012;9:1147-54. [Crossref] [PubMed]
- Luo Y, Wang P, Wang X, et al. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci Rep 2017;7:645. [Crossref] [PubMed]
- Cui J, Zhou B, Ross SA, et al. Nutrition, microRNAs, and Human Health. Adv Nutr 2017;8:105-12. [Crossref] [PubMed]
- Lukasik A, Zielenkiewicz P. Plant MicroRNAs-Novel Players in Natural Medicine? Int J Mol Sci 2016;18:9. [Crossref] [PubMed]
- Kasarello K, Köhling I, Kosowska A, et al. The Anti-Inflammatory Effect of Cabbage Leaves Explained by the Influence of bol-miRNA172a on FAN Expression. Front Pharmacol 2022;13:846830. [Crossref] [PubMed]
- Snow JW, Hale AE, Isaacs SK, et al. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol 2013;10:1107-16. [Crossref] [PubMed]
- Sun F, Guo G, Du J, et al. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol 2014;14:142. [Crossref] [PubMed]
- Ding D, Wang Y, Han M, et al. MicroRNA transcriptomic analysis of heterosis during maize seed germination. PLoS One 2012;7:e39578. [Crossref] [PubMed]
- Tian Y, Cai L, Tian Y, et al. miR156a Mimic Represses the Epithelial-Mesenchymal Transition of Human Nasopharyngeal Cancer Cells by Targeting Junctional Adhesion Molecule A. PLoS One 2016;11:e0157686. [Crossref] [PubMed]
- Han R, Jian C, Lv J, et al. Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.). BMC Genomics 2014;15:289. [Crossref] [PubMed]
- Lukasik A, Pietrykowska H, Paczek L, et al. High-throughput sequencing identification of novel and conserved miRNAs in the Brassica oleracea leaves. BMC Genomics 2013;14:801. [Crossref] [PubMed]
- Morea EG, da Silva EM, Silva GF, et al. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol 2016;16:40. [Crossref] [PubMed]
- Yang T, Wang Y, Teotia S, et al. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci Rep 2019;9:2832. [Crossref] [PubMed]
- Pirrò S, Minutolo A, Galgani A, et al. Bioinformatics Prediction and Experimental Validation of MicroRNAs Involved in Cross-Kingdom Interaction. J Comput Biol 2016;23:976-89. [Crossref] [PubMed]
- Dai X, Zhuang Z, Zhao PX. Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform 2011;12:115-21. [Crossref] [PubMed]
- Zhang B, Pan X, Wang Q, et al. Computational identification of microRNAs and their targets. Comput Biol Chem 2006;30:395-407. [Crossref] [PubMed]
- Ergün S. Cross-Kingdom Gene regulation via miRNAs of Hypericum perforatum (St. John’s wort) flower dietetically absorbed: An in silico approach to define potential biomarkers for prostate cancer. Comput Biol Chem 2019;80:16-22. [Crossref] [PubMed]
- Singh N, Srivastava S, Shasany AK, et al. Identification of miRNAs and their targets involved in the secondary metabolic pathways of Mentha spp. Comput Biol Chem 2016;64:154-62. [Crossref] [PubMed]
- Li J, Zhang Y. Current experimental strategies for intracellular target identification of microRNA. ExRNA 2019;1:6. [Crossref]
- Tan ZL, Li JF, Luo HM, et al. Plant extracellular vesicles: A novel bioactive nanoparticle for tumor therapy. Front Pharmacol 2022;13:1006299. [Crossref] [PubMed]
- Mohanty JN, Sahoo S, Routray SP, et al. Does the diverse source of miRNAs affect human health? An approach towards diagnosis and therapeutic management. Gene Rep 2022;28:101656. [Crossref]
- Saiyed AN, Vasavada AR, Johar SRK. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Futur J Pharm Sci 2022;8:24. [Crossref] [PubMed]
- Crooks GE, Hon G, Chandonia JM, et al. WebLogo: a sequence logo generator. Genome Res 2004;14:1188-90. [Crossref] [PubMed]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 1990;18:6097-100. [Crossref] [PubMed]
- Ivashchenko A, Berillo O, Pyrkova A, et al. MiR-3960 binding sites with mRNA of human genes. Bioinformation 2014;10:423-7. [Crossref] [PubMed]
- Ivashchenko A, Pyrkova A, Niyazova R, et al. Prediction of miRNA binding sites in mRNA. Bioinformation 2016;12:237-40. [Crossref]
- Garg A, Heinemann U. A novel form of RNA double helix based on G·U and C·A(+) wobble base pairing. RNA 2018;24:209-18. [Crossref] [PubMed]
- Kool ET. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu Rev Biophys Biomol Struct 2001;30:1-22. [Crossref] [PubMed]
- Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 2002;30:3497-531. [Crossref] [PubMed]
- Lemieux S, Major F. RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire. Nucleic Acids Res 2002;30:4250-63. [Crossref] [PubMed]
- Yurikova OY, Aisina DE, Niyazova RE, et al. The Interaction of miRNA-5p and miRNA-3p with the mRNAs of Orthologous Genes. Mol Biol (Mosk) 2019;53:692-704. [PubMed]
- Davis E, Caiment F, Tordoir X, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol 2005;15:743-9. [Crossref] [PubMed]
- Koyama T, Sato F, Ohme-Takagi M. Roles of miR319 and TCP Transcription Factors in Leaf Development. Plant Physiol 2017;175:874-85. [Crossref] [PubMed]
- Bonnet E, He Y, Billiau K, et al. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010;26:1566-8. [Crossref] [PubMed]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 2018;46:W49-54. [Crossref] [PubMed]
- Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006;126:1203-17. [Crossref] [PubMed]
- Piñero J, Saüch J, Sanz F, et al. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J 2021;19:2960-7. [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA 2003;9:277-9. [Crossref] [PubMed]
- Bari A, Orazova S, Ivashchenko A. miR156- and miR171-binding sites in the protein-coding sequences of several plant genes. Biomed Res Int 2013;2013:307145. [Crossref] [PubMed]
- Zhang LL, Li Y, Zheng YP, et al. Expressing a Target Mimic of miR156fhl-3p Enhances Rice Blast Disease Resistance Without Yield Penalty by Improving SPL14 Expression. Front Genet 2020;11:327. [Crossref] [PubMed]
- Rakhmetullina A, Zielenkiewicz P, Pyrkova A, et al. Prediction of characteristics of interactions of miRNA with mRNA of GRAS, ERF, C2H2 genes of A. thaliana, O. sativa and Z. mays. Curr Plant Biol 2021;28:100224. [Crossref]
- Rakhmetullina AK, Pyrkova AY, Goncharova AV, et al. Predicting characteristics of the potentially binding sites for miRNA in the mRNA of the TCP transcription factor genes of plants. Russ J Plant Physiol 2020;67:606-17. [Crossref]
- Bari AA, Sagaidak AI, Pinskii IV, et al. Binding of miR396 to mRNA of genes encoding growth-regulating transcription factors of plants. Russ J Plant Physiol 2014;61:807-10. [Crossref]
- Zhao Y, Xu K, Liu G, et al. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci Rep 2020;10:15812. [Crossref] [PubMed]
- Zhao XY, Hong P, Wu JY, et al. The tae-miR408-Mediated Control of TaTOC1 Genes Transcription Is Required for the Regulation of Heading Time in Wheat. Plant Physiol 2016;170:1578-94. [Crossref] [PubMed]
- Rakhmetullina A, Pyrkova A, Aisina D, et al. In silico prediction of human genes as potential targets for rice miRNAs. Comput Biol Chem 2020; Epub ahead of print. [Crossref] [PubMed]
- Atambayeva S, Niyazova R, Ivashchenko A, et al. The Binding Sites of miR-619-5p in the mRNAs of Human and Orthologous Genes. BMC Genomics 2017;18:428. [Crossref] [PubMed]
- Myrzabekova M, Labeit S, Niyazova R, et al. Identification of Bovine miRNAs with the Potential to Affect Human Gene Expression. Front Genet 2021;12:705350. [Crossref] [PubMed]
- Ito Y, Taniguchi K, Kuranaga Y, et al. Uptake of MicroRNAs from Exosome-Like Nanovesicles of Edible Plant Juice by Rat Enterocytes. Int J Mol Sci 2021;22:3749. [Crossref] [PubMed]
- Li C, Wong AYP, Wang S, et al. miRNA-Mediated Interactions in and between Plants and Insects. Int J Mol Sci 2018;19:3239. [Crossref] [PubMed]
- Ji H, Zhang K, Pan G, et al. Deoxyelephantopin Induces Apoptosis and Enhances Chemosensitivity of Colon Cancer via miR-205/Bcl2 Axis. Int J Mol Sci 2022;23:5051. [Crossref] [PubMed]
- Woith E, Fuhrmann G, Melzig MF. Extracellular Vesicles-Connecting Kingdoms. Int J Mol Sci 2019;20:5695. [Crossref] [PubMed]
- Lukasik A, Brzozowska I, Zielenkiewicz U, et al. Detection of Plant miRNAs Abundance in Human Breast Milk. Int J Mol Sci 2017;19:37. [Crossref] [PubMed]
- López de Las Hazas MC, Del Pozo-Acebo L, Hansen MS, et al. Dietary bovine milk miRNAs transported in extracellular vesicles are partially stable during GI digestion, are bioavailable and reach target tissues but need a minimum dose to impact on gene expression. Eur J Nutr 2022;61:1043-56. [Crossref] [PubMed]
- Chapado LA, Martín-Hernández R, Hernández de la Red S, et al. Connection between miRNA Mediation and the Bioactive Effects of Broccoli (Brassica oleracea var. italica): Exogenous miRNA Resistance to Food Processing and GI Digestion. J Agric Food Chem 2021;69:9326-37. [Crossref] [PubMed]
- Del Pozo-Acebo L, López de Las Hazas MC, Tomé-Carneiro J, et al. Therapeutic potential of broccoli-derived extracellular vesicles as nanocarriers of exogenous miRNAs. Pharmacol Res 2022;185:106472. [Crossref] [PubMed]
- Fu Y, Chen J, Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019;1:24. [Crossref]
Cite this article as: Rakhmetullina A, Ivashchenko A, Pyrkova A, Uteulin K, Zielenkiewicz P. In silico analysis of maize and wheat miRNAs as potential regulators of human gene expression. ExRNA 2023;5:4.


