MED12 in hematopoietic stem cells—cell specific function despite ubiquitous expression
All cells contain several multi-component molecular machines for executing steps in the fundamental cellular processes of transcription and translation. Paradoxically, despite their ubiquitous expression, individual components of these protein conglomerates are responsible for extraordinarily cell-specific functions.
One of these fundamental molecular complexes is Mediator. Mediator is expressed and required in cells where genes are actively being expressed. Its major role is to act as a molecular bridge anchoring two regions of otherwise unconnected DNA (promoter and enhancer), bringing together the constellation of promoter and enhancer associated transcription factors required for expression of genes transcribed by RNA polymerase II. It also plays associated roles in architecting chromatin organisation, transcription pre- and re-initiation, elongation and pausing [reviewed in Allen and Taatjes, 2015 (1)].
MED12 is one component of the four-part kinase module of Mediator along with MED13, cyclin-dependent kinase 8 (CDK8) and cyclin C (CCNC), which regulates the function of Mediator. Mutations in MED12 were originally associated with intellectual disability syndromes including Lujan (2), Opitz-Kaveggia (3), and Ohdo syndromes (4). Since then, strong association of acquired somatic mutations in MED12 with uterine leiomyoma has surfaced (5). An essential, specific hematopoietic role for Med12 in myelopoiesis and T cell development was evident from analysis of the syrah zebrafish mutant that resulted from a forward genetic screen to identify as yet unrecognised factors important for myeloid development (6). Somatic mutations in MED12 have since been associated with or identified as driver mutations in T- and NK-cell post-transplant lymphoproliferative disorders (7) and pediatric T-cell acute lymphoblastic leukemia (8), and are a poor prognosis marker in chronic lymphocytic leukemia (9). How ubiquitously expressed components of a universally required macromolecular machine can generate cell-specific functional outcomes is an intriguing mechanistic question that has been elegantly explored by Aranda-Orgilles et al. in their recent paper published in Cell Stem Cell using MED12 and its role in hematopoiesis as an exemplar (10).
Two different conditional knockout approaches allowing specific deletion of MED12 in the hematopoietic system of both neonates and adults enabled a requirement for MED12 in maintaining hematopoietic stem cell (HSC) number and function to be demonstrated. HSCs and progenitors were largely absent in MED12 deleted animals and failed to engraft. Even after permitting equivalent engraftment by transplanting prior to inducing Cre recombinase expression, transplanted HSCs with subsequent deletion of MED12 failed to reconstitute blood lineages in competitive bone marrow transplant assays, pointing to a failure in de novo hematopoiesis. This requirement for MED12 was conserved in human CD34+ peripheral blood stem cells depleted of MED12 using an shRNA knockdown approach.
The next question was whether this requirement was universal for all cells or specific to HSCs. Med12 was deleted from immortalised Med12fl mouse embryonic fibroblasts in vitro and mouse embryonic stem cells were obtained from germline MED12-deficient mice. Both showed no changes in growth. Med12fl mice were also crossed on to a CreER-T2 strain to achieve global Med12 deletion. While these mice showed decreased bone marrow hematopoietic stem and progenitor cells (HSPCs), histological analysis of other tissues revealed no significant differences. Taken together, these observations indicated that although ubiquitously expressed, the requirement for MED12 in cell growth, differentiation and function was not universal, but rather affected HSCs preferentially.
This presents a paradox. How could such a specific functional consequence in just one cell type be effected by loss of a general component of the transcriptional machinery that is present in all cell types?
Since MED12 is one of four components comprising the regulatory kinase module of Mediator, the HSC requirement for MED12 might have depended on the kinase activity of this module. However, systematic deletion of each of the other kinase module components (MED13, CDK8 and CCNC) did not recapitulate the quantitative and qualitative disruption of HSCs seen with just deficiency of the MED12 component. This led to the hypothesis that MED12 could have an intrinsic function separate from the kinase module and Mediator.
To explore this comprehensively, RNAseq transcriptome profiling was performed in MED12-deficient HSPCs, uncovering a decrease in expression of transcription factor gene networks associated with HSC/HSPC growth and survival. Conversely, gene expression signatures for proliferation and apoptosis were both enriched and functionally validated, suggesting disruption of the carefully balanced pathways that simultaneously maintain homeostasis while avoiding transformation. The authors suggested that increased proliferation may occur in a bid to compensate for increased apoptosis, but this has yet to be experimentally tested.
To mechanistically understand the basis for these changes, ChIPseq was performed in human CD34+ HSPCs, identifying that >80% of MED12-associated DNA sequences occurred outside promoters. Comparison of MED12 ChIPseq profiling with epigenetic profiling of CD34+ cells showed that the genomic regions associated with MED12 binding occurred within active chromatin (flagged by H3K4me1 and H3K27ac) and not within actively transcribed gene bodies, and that these corresponded to enhancers as defined by their epigenetic signatures. These enhancer regions contained binding sites for hematopoietic transcription factors including FLI1, RUNX1 and GATA1, correlating with decreased expression of these gene networks in MED12 deficient mice. Remarkably, a c-kit enhancer containing a MED12 element that is active only in CD34+ cells was identified, strongly making the point that MED12 is capable of marking cell-type specific active enhancers. Epigenetic analysis together with 4C-seq suggested that MED12 loss resulted in enhancer inactivation. CRISPR/Cas9 deletion of the c-kit MED12 element confirmed its identity as a functional element whose loss decreased c-kit expression and increased apoptosis.
How could the association of MED12 with these regions be regulating expression of key hematopoietic transcription factors? Immunoprecipitation of MED12 in HPC-7 cells followed by mass spectrometry was employed to identify other proteins that could be interacting with MED12 in order to execute its regulator function at enhancers. Both p300 and CBP, the two main acetylases responsible for targeting H3K27, were found to interact with MED12. Bioinformatics analysis demonstrated >80% overlap between sites occupied by p300 and MED12, which included hematopoiesis-relevant enhancers. Occupation of these enhancers by p300 was depleted in the absence of MED12, including the c-kit enhancer, but not those of many other non-hematopoietic genes. This points to a mechanism whereby in the absence of MED12, p300 is reduced at these specific sites, thereby decreasing acetylation and resulting in inactivation of these enhancers in HSPCs.
Table 1 charts the insights into MED12 function gained from animal models and in human disease syndromes. This highlights that despite its ubiquitous expression, MED12 loss of function can have quite specific phenotypic consequences. The insights of Aranda-Orgilles et al. provide a mechanism for how MED12 can act as a hub to integrate and regulate different gene networks that are dependent on cellular context. A model of Mediator function is emerging in which it comprises a “plug and play” series of modules where transcription factors bind to their cognate Mediator subunit and induce structural changes that exert a domino effect through the complex to produce a specific transcriptional outcome. The Mediator complex, by virtue of its multiple subunits, size and plasticity, is particularly suited to this role as a switchboard for integrating cell signalling circuits. Consistent with this picture, it was previously found that MED1 deficiency in mouse leads to a defect in GATA-1-mediated transactivation, which is due in part to Mediator components at GATA-1-occupied enhancer sites, and results in failure of erythropoiesis (21). Now, Aranda-Orgilles et al. have demonstrated that another subunit, MED12, helps to maintain open chromatin by its interaction with p300 at enhancers engaged by hematopoietic transcription factors, thereby controlling key HSC gene regulatory networks.
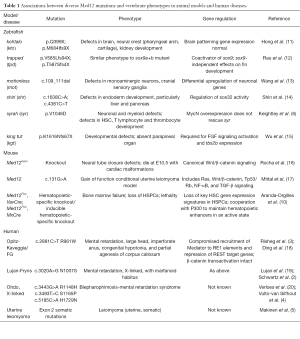
Full table
This study provides an example of a Mediator component that has an intrinsic tissue-specific role in hematopoietic enhancer recruitment independent of its previously known general role in modulating Mediator through activation of CDK8 kinase. The mechanisms by which these large multicomponent, ubiquitously expressed complexes achieve specificity of function in individual cell types are likely to be diverse and reflect nuanced tissue-specific roles of individual components. The spliceosome is another ubiquitous molecular machine; it is responsible for editing out a specific configuration of introns from primary RNA transcripts to generate correctly assembled transcripts for expression of fully functional proteins [reviewed in Matera and Wang (22)]. This “housekeeping” function is required in all cells synthesising protein. Mutations affecting some spliceosome components also result in paradoxes where despite their ubiquitous expression, a disease phenotype is surprisingly tissue-specific. The core spliceosomal component PRPF8 provides one such example. Congenital C-terminal PRPF8 mutations specifically affect pigmented retinal epithelial cells, resulting in an autosomal dominant form of the disease retinitis pigmentosa that causes blindness, despite the mutation being present in all cells of affected individuals (23). Adding another dimension to this paradox, acquired somatic mutations in other regions of PRPF8 are associated with myelodysplastic syndrome (MDS) and secondary acute leukemia (24), as are mutations of a number of other splicing factors [including SRSF2, SF3B1, U2AF1 (25)]. Studies exploring the mechanistic basis of such paradoxes are likely to reveal new functional roles for these particular components independent of their contribution to their complex’s primary function.
As was the case for MED12 in the study of Aranda-Orgilles et al., unravelling just how individual components of general macromolecular complexes function to transduce cell-specific signals into targeted rather than general transcriptional outcomes promises to be a rewarding area of further investigation.
Acknowledgements
GJ Lieschke, SK Nilsson and MC Keightley are supported by the National Health and Medical Research Council (1070687, 1044754, 1081933, 1084990). SK Nilsson is a CSIRO OCE Science Leader. The Australian Regenerative Medicine Institute is supported by funds from the State Government of Victoria and the Australian Federal Government.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015;16:155-66. [Crossref] [PubMed]
- Schwartz CE, Tarpey PS, Lubs HA, et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet 2007;44:472-7. [Crossref] [PubMed]
- Risheg H, Graham JM Jr, Clark RD, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet 2007;39:451-3. [Crossref] [PubMed]
- Vulto-van Silfhout AT, de Vries BB, van Bon BW, et al. Mutations in MED12 cause X-linked Ohdo syndrome. Am J Hum Genet 2013;92:401-6. [Crossref] [PubMed]
- Mäkinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011;334:252-5. [Crossref] [PubMed]
- Keightley MC, Layton JE, Hayman JW, et al. Mediator subunit 12 is required for neutrophil development in zebrafish. PLoS One 2011;6:e23845. [Crossref] [PubMed]
- Margolskee E, Jobanputra V, Jain P, et al. Genetic landscape of T- and NK-cell post-transplant lymphoproliferative disorders. Oncotarget 2016;7:37636-48. [PubMed]
- Spinella JF, Cassart P, Richer C, et al. Genomic characterization of pediatric T-cell acute lymphoblastic leukemia reveals novel recurrent driver mutations. Oncotarget 2016;7:65485-65503. [PubMed]
- Kämpjärvi K, Järvinen TM, Heikkinen T, et al. Somatic MED12 mutations are associated with poor prognosis markers in chronic lymphocytic leukemia. Oncotarget 2015;6:1884-8. [Crossref] [PubMed]
- Aranda-Orgilles B, Saldaña-Meyer R, Wang E, et al. MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis. Cell Stem Cell 2016;19:784-799. [Crossref] [PubMed]
- Hong SK, Haldin CE, Lawson ND, et al. The zebrafish kohtalo/trap230 gene is required for the development of the brain, neural crest, and pronephric kidney. Proc Natl Acad Sci U S A 2005;102:18473-8. [Crossref] [PubMed]
- Rau MJ, Fischer S, Neumann CJ. Zebrafish Trap230/Med12 is required as a coactivator for Sox9-dependent neural crest, cartilage and ear development. Dev Biol 2006;296:83-93. [Crossref] [PubMed]
- Wang X, Yang N, Uno E, et al. A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci U S A 2006;103:17284-9. [Crossref] [PubMed]
- Shin CH, Chung WS, Hong SK, et al. Multiple roles for Med12 in vertebrate endoderm development. Dev Biol 2008;317:467-79. [Crossref] [PubMed]
- Wu SY, de Borsetti NH, Bain EJ, et al. Mediator subunit 12 coordinates intrinsic and extrinsic control of epithalamic development. Dev Biol 2014;385:13-22. [Crossref] [PubMed]
- Rocha PP, Scholze M, Bleiss W, et al. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 2010;137:2723-31. [Crossref] [PubMed]
- Mittal P, Shin YH, Yatsenko SA, et al. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest 2015;125:3280-4. [Crossref] [PubMed]
- Ding N, Zhou H, Esteve PO, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell 2008;31:347-59. [Crossref] [PubMed]
- Lujan JE, Carlin ME, Lubs HA. A form of X-linked mental retardation with marfanoid habitus. Am J Med Genet 1984;17:311-22. [Crossref] [PubMed]
- Verloes A, Bremond-Gignac D, Isidor B, et al. Blepharophimosis-mental retardation (BMR) syndromes: A proposed clinical classification of the so-called Ohdo syndrome, and delineation of two new BMR syndromes, one X-linked and one autosomal recessive. Am J Med Genet A 2006;140:1285-96. [Crossref] [PubMed]
- Stumpf M, Waskow C, Krötschel M, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A 2006;103:18504-9. [Crossref] [PubMed]
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014;15:108-21. [Crossref] [PubMed]
- McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 2001;10:1555-62. [Crossref] [PubMed]
- Kurtovic-Kozaric A, Przychodzen B, Singh J, et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia 2015;29:126-36. [Crossref] [PubMed]
- Boultwood J, Dolatshad H, Varanasi SS, et al. The role of splicing factor mutations in the pathogenesis of the myelodysplastic syndromes. Adv Biol Regul 2014;54:153-61. [Crossref] [PubMed]
Cite this article as: Keightley MC, Nilsson SK, Lieschke GJ. MED12 in hematopoietic stem cells—cell specific function despite ubiquitous expression. Stem Cell Investig 2017;4:3.


