
Cancer exosomes in cerebrospinal fluid
Introduction
It is well documented that there are over 100 distinct histological types of primary central nervous system (CNS) tumours (1,2). However, one of the most prevalent CNS based type tumours, known as a glioma, had an estimated 23,000 new cases and 16,000 deaths reported in the USA in 2016 (3). Gliomas are derived from glial cells and include a spectrum of tumour types including glioblastomas (GBM), astrocytomas, oligodendrogliomas, ependymomas and mixed gliomas. The CBTRUS Statistical report by Ostrom et al. (4), indicates that the broad category of glioma represents approximately 24.7% of all primary brain and CNS tumours, but overall they account for a majority (74.6%) of malignant CNS tumours. In addition, GBM and astrocytomas account for about 74.9% of all gliomas (55.4% and 19.5% respectively of these registered cases in the USA) (4).
The 2016 World Health Organization (WHO) Classification of tumours of the CNS is an integration of the phenotypic or histological classification of brain tumours utilized for the majority of the last 100 years, and has now been incorporated with data from studies over the last 20 years examining the molecular parameters which may contribute to the additional sub-classification of some tumours (5,6). Nonetheless, it is understood that the lower grade astrocytomas are known to be slow growing, less aggressive tumours, whilst the higher grade, GBM is the most aggressive with an extremely dismal prognosis. Despite the current standard clinical combinatorial treatment approach, which involves maximal safe surgical resection and post-operative fractionated radiation therapy coupled with concomitant chemotherapy with temozolomide (7), the median survival for GBM patients is only 14.6 months (8) and the 5-year survival rate is only 5.5% (4). An Australian study examining 542 GBM patients over a 12-year period had a shorter median overall survival of 7.7 months (9).
Diagnostic and therapeutic monitoring strategies that are employed include magnetic resonance imaging (MRI) and repeat brain biopsies. As a result of limitations in the resolution of MRI leading to potential delays in disease diagnosis or detection of treatment resistance (10-12) and the risks of significant morbidity with highly invasive multiple post-treatment treatment tumour biopsies (13,14), there is an urgent requirement for alternative monitoring techniques for following disease progression and response to treatment. Serum based tumour markers have been proposed as an alternative for providing a diagnostic, disease progression or treatment response profile of the patient (15-20). However, due to the variations in the permeability of the blood-brain barrier that can influence or prevent the released amount of biomarker into the bloodstream (21), there have been efforts in identifying alternate clinical sampling avenues. Cerebrospinal fluid (CSF) has been investigated as a potential source of brain tumour markers.
CSF
CSF is a clear fluid that is primarily produced in the ventricular choroid plexus with a portion also generated by capillaries in the brain parenchyma (22) and is subsequently distributed within the ventricular system and the subarachnoid space. Up to 500 mL of CSF can be produced daily, even though only about 100–150 mL is present in the ventricular system and subarachnoid space at any one time, and production being influenced by a number of factors including venous pressure and blood hydrostatic pressure (23-25). It can serve a number of functions, with the most important being a neuroprotective agent in the form of a protective cushion, ultimately dispersing force away from the delicate structures of the brain and spinal cord (26). Additionally, CSF can assist in the maintenance of normal intracranial pressure, regulate brain blood flow and provide buoyancy for the brain (22). Importantly, with CSF being renewed about four times every 24 hours (27), a critical regulatory function is to maintain a homeostatic environment via the transfer between the CSF and bloodstream of various biologic/pathologic products, as well as acting as a medium for nutrient delivery (28,29).
CSF is an ideal source for viable biomarkers, as it exists in a state of equilibrium, while it interacts directly with the extracellular space and neighbouring microenvironment within the brain and potentially can reflect changes in both biological and pathological systems (30-36). Therefore as a result, CSF analysis is widely used for both diagnostic and prognostic purposes to provide critical information for numerous conditions. CSF is routinely obtained by a lumbar puncture (37) or during intra-operative sampling of the ventricles (38). Serial sampling of CSF is also achievable with up to 20 mL being collected at any one time by lumbar puncture due to the high production and turnover rates. It is less complex in composition than serum, which would allow for a more straightforward detection of tumour-specific markers. Given the continuous proximity of CSF with the CNS, it is possible that cancer cells from primary CNS cancers can infiltrate the CSF directly, allowing for the examination of CSF under the microscope to detect the presence of cancer cells. However, the cytological analysis of CSF samples is problematic due to the low sensitivity in detecting tumour cells, even though tumour progression has been observed and ultimately does not provide for quantification or molecular analysis of tumour cells that are identified (39-41).
Evolution in technological advances and techniques in the laboratory have allowed laboratory studies to focus on potential biomarker changes at the molecular level preceding observable macroscopic tumour alterations (40). These include proteomic profiling (42-44), immunocytochemistry (43,45), mass spectrometry (46) and the detection of a number of conventional serum based markers such as epidermal growth factor receptor (EGFR) (47) and β-human chorionic gonadotropin (48,49). However, due to variations in sensitivities and specificities, none are currently clinically validated. Several studies have also reported the detection of microRNAs (miRNAs) in biological fluids including CSF from CNS malignancies (50-52). As miRNAs have been observed to be either oncogenic (oncomirs) or tumour suppressive with different types of brain cancer possessing distinct miRNA signatures (51,53-55), whilst also being undetectable in normal brain (56), there has been an increased interest in investigating if there are distinctive brain tumour miRNA signatures present in biofluids such as CSF. Laboratory studies have shown that miR-21, miR-125b, miR-223, miR-451 and miR-771 have been detected in the CSF of patients with CNS malignancies (50,57).
Extracellular vesicles (EVs)
EVs, which were discovered over 30 years ago (58), are a very heterogeneous and molecularly complex group of membrane-bound cell secreted vesicles that can range from 30–2,000 nm in size and are secreted by a variety of cell types including normal and cancer cells (59-63). Numerous laboratory studies have demonstrated the ability to isolate EVs from body fluids such as urine (64), plasma (65), saliva (66), breast milk (67), ascites (68), semen (69) and CSF (57), indicating their potential in acting as circulating biomarkers for the diagnosis and progression of pathologic diseases.
EVs have been categorized into a number of different classes based on size and their subcellular origin including: (I) exosomes (40–120 nm in diameter; originating from the endocytic recycling pathway); (II) microvesicles (50–1,000 nm in diameter; originating directly from the plasma membrane) and (III) apoptotic bodies (1,000–5,000 nm in diameter; released from the plasma membrane as blebs during apoptosis). EVpedia (70,71), ExoCarta (72,73) and Vesiclepedia (74) are comprehensive public online databases that list currently identified cargos of EVs. As gliomas account for the majority of all CNS tumours (of which over 50% are GBMs) (3,4) and recent studies have shown that GBM cells secrete EVs that contain tumour-specific genetic material such as DNA, mRNA and miRNA (75-79), we will focus on glioma based studies. Also, as the field of EV research has been rapidly expanding over the last 30 years, this review will include a brief overview on the available literature focussing on the role of exosomes in glioma and the relationship with CSF.
Exosomes
Intracellular crosstalk occurs between both neighbouring and distant tumour cells, immune and stromal cells in the tumour microenvironment and the surrounding normal tissue that can ultimately contribute to various aspects of tumour development including the proliferative, migration and invasive phenotypes (76,78-80). Whilst it is known that this can occur via secreted cellular factors into the surrounding microenvironment or direct cell to cell contact, there is an increased focus on exosomes acting as important mediators of cell to cell communication. Exosomes are known as spherical to cup-shaped vesicles that possess a distinct lipid bi-layer and are of a diameter between 40–120 nm (81), are of endocytic origin and sediment at approximately 100,000 g in a sucrose gradient of 1.13–1.19 g/mL (81). Exosomes are also known to be highly heterogeneous in their composition (82). The lipid bilayer can contain transmembrane proteins whilst their aqueous core may also contain numerous proteins, mRNA and small non-coding RNA including miRNA and DNA that are a reflective fingerprint of the donor cell (80,83,84) (Figure 1).
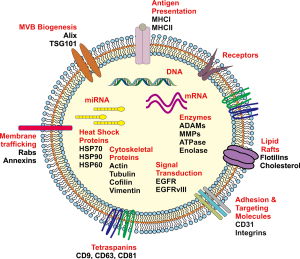
Exosomes can influence various stages of tumour biology or progression through the protected transfer of their bioactive cargo by the lipid bilayer in travelling between donor and recipient cells (61,85,86). Release of their cargo into the recipient cells, in particular mRNA and microRNA, can ultimately lead to the modulation of gene expression through translational regulation of target mRNAs, altering the transcriptome and signalling activity which can induce phenotypic changes in the recipient cells (87-89). The recognition of exosomes as important mediators in the communication between cells in both physiological and pathological processes can be seen in Figure 2, which shows the number of manuscripts published each year since the initial few in 1987, as determined by a PubMed search. There has been an increasing trajectory of exosome research that has been published within the last decade.

Whilst the biogenesis of exosomes is a multifaceted process and incompletely understood, current evidence indicates that they are derived from the release of intraluminal vesicles (ILVs) through the fusion of multivesicular bodies (MVB) with the plasma membrane that may occur via a number of highly structured pathways. These include ESCRT (Endosomal Sorting Complex Required for Transport) and ESCRT-independent pathways (90). ESCRT-dependent pathways function primarily by acting on MVBs, allowing for the sorting of ubiquitinylated proteins or receptors into ILVs, which in turn may be degraded during the fusion of MVBs with lysosomes, or exocytosed (90). The ESCRT machinery is required for a number of different processes and is comprised of a number of peripheral membrane sub complexes including ESCRT-0, -I, -II, -III and the ALIX homodimer. It has also been proposed that soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are required for the fusion of exosomes with the plasma membrane, as they form the anchor-complex between the vesicular and plasma membrane SNARES (91).
Sorting of ubiquitinylated proteins in the MVB pathway is mediated by the ESCRT0 subcomplex, via the binding and clustering of this cargo for delivery into MVBs, coupled with the recruitment of ubiquitin ligases, deubiquitinating enzymes and clathrin (90). ESCRT0 can then bind to the ESCRTI complex, allowing the ESCRTI complex to assemble with its crucial partners, the ESCRTII subunits, resulting in MVB biogenesis and bud formation (92). ESCRTII subsequently facilitates the association of the ubiquitin-bound ESCRT complexes with the ESCRTIII complex, leading to the production of free vesicles as the result of cleavage of the nascent ILVs. The ESCRTIII complex, as the central membrane scission mechanism, is inactive in the cytoplasm and only once it polymerizes on a membrane can it be activated, permitting the fusion of the MVB with the peripheral membrane and ensuing release of exosomes into the extracellular space (92). This membrane bound ESCRTIII complex undergoes a process of disassembly allowing for the return of ESCRTIII subunits to the cytoplasm.
Whilst the ESCRT system is understood to govern the majority of cargo incorporation into exosomes, the ESCRT-independent pathway, aided by tetraspanins (CD63, CD81, CD9) (93) also plays a role for many proteins (94). In general exosomes are enriched not only in tetraspanins, but also export molecules such as (Rab27a/b, TSG101 and ALIX), heat shock proteins (HSP90, HSP70, HSP60), proteases (ADAM10, MMP-2), integrins, receptor tyrosine kinases, phospholipids and immunomodulating molecules (95). Therefore exosomal protein constituents not only reflect molecules that are central to the exosome biogenesis pathway, but there will also exist proteins that were present in the cell of origin, which may provide an insight into the biological function of the released exosomes. Furthermore, there has been an emphasis on identifying the lipid composition of exosomes as it is recognized that lipids are essential in providing exosomal membrane rigidity and stability (96). A number of different lipids have been identified to occur in the lipid bilayer of exosomes including sphingomyelin, phosphatidylserine, phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, cholesterol, ganglioside, GM3 and prostaglandins (97,98).
Exosomes and normal brain
Whilst this review is focussing on cancer derived exosomes in the CSF, it is important to briefly outline what is understood about exosomes in normal brain. Homeostasis within the brain relies upon the coordinated interactions of a number of cell types (including neurons, microglia, astrocytes and oligodendrocytes) via an efficient cell-cell communication system. The modulation of synaptic plasticity has been linked to the secretion of neuronal based exosomes (99), especially as their secretion at synapses is known to be internalized by neighbouring cells through endocytosis (100). It has been postulated that exosomes might be involved in the physiological regulation of the synapse and this study showed that glutamatergic synaptic activity modulated exosomal release given that they carry the GluR2/3 subunit of the AMPA receptor. Exosomes from oligodendrocytes function in an autocrine fashion through a second messenger cascade involving a number of molecules, but not the reinternalization of the exosomes that can inhibit myelin formation by these cells (101). Neurons can also internalize oligodendroglial exosomes to regulate their own internal cargo as a way of on-going cell maintenance (102).
Astrocytes are involved in neuronal growth and survival, but also form the blood brain barrier (BBB). The exosomal cargo of astrocytes is varied but includes growth factors, angiogenic factors, MMPs and lipids (79), however to maintain a homeostatic environment in response to heat or oxidative stress, exosomes from astrocytes are also known to contain heat shock proteins which can provide a neuroprotective effect to neurons (103). Microglial exosomes are involved in the execution of immune functions through antigen presentation in a pathologic situation, however, in a physiological environment, the exosomes can contain the lactate transporter and glycolytic enzymes, allowing for the delivery of energy substrates to neurons (104). The exosomes released by brain cancer cells, in particular gliomas, carry cargo which may be oncogenic in nature, but also facilitate angiogenesis. Donor cells can then internalize these vesicles to promote cell transformation or the tubular growth of endothelial cells, ultimately producing an environment that is ideal for tumour growth, invasion and suppression of anti-tumour immune responses (75,76). The field of exosome function in normal brain is still within its infancy and has not been thoroughly elucidated at a mechanistic level.
Exosomes and glioma—diagnostic biomarkers
One hallmark of all cancers, including glioma, is tumour heterogeneity. This can have a significant impact on patient outcome due to difficulty in providing a clear diagnosis, as well as reducing the effect of current clinical treatments or designing efficacious therapies to target the mixed population of cancer cells within tumours (intra-tumour heterogeneity) and between tumours from different patients (inter-tumour heterogeneity). The degree of heterogeneity can also lead to variations in proliferation rate, level of aberrant vascularization within tumours, and extent of diffuse invasion of the infiltrative cells, contributing to the difficulty in defining response to therapeutic intervention and patient outcome. As mentioned earlier, determining the clinical status of glioma patients involves diagnostic tools such as clinical manifestations, computed tomography (CT), MRI and histological examination of surgical biopsies. At the present, histological analysis remains the gold standard for obtaining tumour diagnosis, however there are inherent risks involved with this invasive method including the potential risk of neurological damage and brain haemorrhaging, in particular when serial biopsies are involved. The ability to monitor the disease normally only appears after sufficient tumour burden has occurred to alter the neurologic examination in defining the overall clinical manifestation. Importantly, the resolution limit of MRI is only 2–3 mm (105), with a specificity of between 50–80% of correctly identifying GBM tumours from other intracranial lesions or changes that have been induced by treatments such as radiation and temozolomide (106,107). Therefore, an urgent need exists for a clinical application which is potentially less invasive and has a lower risk of morbidity, but is able to allow in a timely manner for the detection of an ideal biomarker which may have diagnostic, disease progression and/or predictive capabilities.
Even though tumour biopsy and histologic analysis has been the mainstay of disease diagnosis, it only provides a snapshot of the heterogeneous tumour at any one time. But it is known that, in addition to the numerous growth factors, cytokines and metabolites that can be secreted by glioma cells (108), these cells have also been shown to release various types of EVs that can support many critical biologic and pathological processes of a tumour cell including proliferation and invasion (80). EVs (including exosomes) are released into the surrounding extracellular environment and can potentially travel across various anatomical compartments to enter the systemic blood circulation or the CSF (75,109). As a consequence of the detection of EVs in these biological fluids and their capacity to act as carriers of a bioactive cargo (such as RNA, DNA, proteins and lipids) that can be a ‘fingerprint’ representation of the donor cell, it has been proposed that CSF or serum can act as a ‘liquid biopsy’, providing a potential prospect of an avenue for diagnostic markers, monitoring glioma progression and also response to therapy [see review by Lin et al. presenting an overview of exosomes as novel biomarkers for clinical diagnosis in a number of cancers including GBM (110)]. EGFR mutations, rearrangements or amplifications occur in up to 70% of GBM tumours (111,112) and around 50% of patients that harbour EGFR amplification also possess an EGFRvIII mutation, resulting from an in-frame deletion of exons 2–7, leading to constitutive and ligand independent receptor activity (113). A qRT-PCR based study investigating the EGFRvIII status of exosomes derived from GBM patient sera identified that EGFRvIII mRNA was found in the exosomes of patients with EGFRvIII expressing tumours (75). Interestingly, EGFRvIII positive exosomes were undetectable after tumour resection. This has also been confirmed in a study by Nilsson et al. (114) highlighting the diagnostic potential of GBM derived exosomes. As EGFRvIII has been linked with the ‘classical’ molecular subtype of GBM (115), this may have great potential in the application of a personalized therapeutic approach for these patients based on their exosomal profile. Significantly, a correlation between the levels of wild-type EGFR in glioma patient CSF-derived exosomes and chemotherapeutic response has also been observed (112), reinforcing the relevance of CSF derived exosomes as a clinical liquid biopsy.
By comparison, the isolation of CSF based exosomes is ideal compared to serum as (I) the BBB is not a concentration-limiting factor as it would be with serum derived exosomes and (II) CSF would provide a ‘cleaner’ source of glioma based exosomes compared to serum which would contain exosomes from normal (non-tumour) based processes (for example—maturation of reticulocytes and platelet activation). Furthermore, isocitrate dehydrogenase 1 (IDH1) is a citric acid cycle enzyme with mutations in this enzyme occurring in approximately 10% of all GBMS and 80% of secondary GBMs (116). This has allowed for the IDH1 mutation status to be utilized in the detection of a secondary tumour arising from a lower grade tumour versus a primary tumour that has arisen de novo. A novel study by Chen et al. (117) involving the exosomal RNA analysis of human CSF and serum samples using BEAMing RT-PCR, reliably detected and quantified mutant and wild-type IDH1 RNA transcripts in CSF-based exosomes from glioma patients (5 out of 8 patients). The authors were not able to detect the mutant IDH1 transcript in exosomes isolated from the sera of patients that possessed tumours positive for this IDH1 mutation. Significantly, the levels of mutant IDH1 transcript showed a strong correlation with tumour volume, signifying the strong possibility of CSF-based exosomes predicting glioma disease burden. Additionally, it has also been shown that IDH1 mutations may appear to be a significant marker of a positive chemosensitivity response of secondary GBM to TMZ (118). Therefore, CSF-derived exosomes with IDH1 mutant transcripts may provide clinical information on two levels: (I) the presence of a secondary GBM and (II) predicting the response to TMZ treatment. An important consideration is malignant tumour cells (including GBM) exhibit altered cellular energetics that can influence the metabolism of tumour cells, promoting aerobic glycolysis and ATP generation (119). In terms of IDH1 and the mutations which can occur in GBM (120), this is noteworthy as independent studies in other systems have shown that exosomes can possess the ability to aerobically synthesize ATP (121,122). Exosomes isolated from urine (121) and mesenchymal stem cells (122) were able to carry out oxidative phosphorylation to synthesize ATP and consume oxygen. It has been suggested that since mitochondrial redox chain proteins and ATP synthase are ectopically expressed on the surface of exosomes that they can transfer their oxidative phosphorylation machinery and ultimately rescue aerobic respiration (123). IDH1 catalyzes the oxidative decarboxylation of isocitrate into α-ketoglutarate (120), whereas IDH1 mutations in some gliomas convert α-ketoglutarate to 2-hydroxyglutarate (124). The presence of these mutations may be of significance in terms of treatment response (radiotherapy and chemotherapy), as there is some evidence suggesting increased sensitivity to treatment (125). However, there are also conflicting reports indicating that the mutations do not correlate with an improved treatment response (126). Therefore the role of exosomes in mediating the aerobic synthesis of ATP in cells must also be considered.
In recent years, there has also been a focus on exosome associated miRNAs, since they are small, non-coding RNAs that regulate gene expression by binding to specific target mRNAs and inhibiting their translation. This is highly relevant as exosomes can protect this donor cell ‘tumour-specific’ material from the action of RNAses in the extracellular environment, and have this transferred to a recipient cell. Ultimately, the presence of miRNAs within exosomes in a biofluid such as CSF may also allow us to understand the spectrum of disease progression or response to therapy. A meta-analysis of data from different studies and databases investigating the potential diagnostic value of miRNA in primary CNS lymphoma [PCNSL—a rapidly growing tumour that has a median survival of around 3 months once diagnosed (127)] and glioma was carried out by Wei and colleagues (128). This analysis invovled 23 studies, 299 CNS patients and 418 control subjects involving the qRT-PCR analysis of blood and CSF samples to determine the level of detection and sensitivity of the assays. This study determined that CSF-based miRNA assays yielded more reproducible and accurate results than those from blood-based samples which further supports the use of studies based on CSF-isolated exosomes.
Akers et al. (129) examined the miRNA contents of exosomes isolated from the CSF and plasma of GBM patients and glioblastoma cell lines. In exosomes derived from GBM patient plasma or glioblastoma cell lines, the relative abundance of miRNA was highly variable with miRNA species alternating between exosomes and microvesicles in different specimens. However, they were able to demonstrate that CSF-derived exosomes were enriched for miRNAs relative to CSF microvesicles which indicates that CSF exosomes are the major compartment that can harbour miRNAs. Utilizing a TaqMan OpenArray human microRNA panel, their analysis revealed 46 miRNAs that were detected in the CSF exosomes (including miR-21, miR-24, miR-103, and miR-125) not in the microvesicles. In addition, 6–8 of the miRNAs that were found to be expressed in both exosomes and microvesicles were up to 150-fold higher in the exosomes relative to the microvesicles implying that miRNAs are enriched in the CSF-derived exosomes. A critical follow-up study was also carried out by Akers et al. (130) for maximizing the exosome related miRNA derived from clinical CSF samples. They proposed the clinical CSF samples should firstly be aliquoted and stored at −80 °C at the volume that will be required for laboratory analysis, as one single freeze thaw cycle does not affect the content of the CSF exosomes or their associated miRNAs. Importantly, they also determined that CSF can be safely stored and transported at room temperature for up to seven days, an important factor to consider if samples are going to be transported from smaller hospitals to specialized laboratories for exosome isolation and miRNA analysis. A study by Teplyuk et al. (52) investigating miRNA profiles of CSF miRNA as a means of discriminating between metastatic brain cancers and GBM revealed that by using the combined analysis of a group of seven cancer-related miRNA, they were able to discriminate between these two types of brain cancers with 90% accuracy. MiR-21 and miR-10b expression levels were elevated in the CSF of GBM patients compared to the metastatic brain cancer patients and CSF from non-neoplastic tissue, whilst the miR-200 family was detected only in the CSF of the metastatic brain cancer patients. This allowed them to distinguish between GBM and metastatic cancers. MiR-21, known to be a promising biomarker for the progression of GBM patients, was also found to be overexpressed in CSF exosomes in the Akers study (129), further emphasizing the relevance of utilizing CSF-derived exosomes.
Exosomes and glioma—disease progression
Exosomes contain various membrane-associated proteins that facilitate many different cellular functions, such as the tetraspanins, which include CD151. The tetraspanins are protein superfamilies that aid the organization of membrane microdomains, known as tetraspanin-enriched microdomains, by establishing clusters and also interacting with numerous transmembrane and cytoplasmic signalling proteins (131). CD151 has been shown to contribute to host matrix remodelling due to exosomal-integrin and tetraspanin-protease associations (131), with one of these associations regulating cell motility via protease activity (132). CD151 induces matrix metalloproteinase-9 (MMP-9) expression via CD151-associated integrin signalling (133) and it also associates with MMP14 to regulate ADAM10/ADAM17 (A Disintegrin and Metalloproteinases) activity (134). Secreted proteases such as the matrix metalloproteinases (MMPs) and the ADAMS with thrombospondin motifs (ADAMTS) cleave various extracellular matrix (ECM) components and cell surface proteases, and also regulate stimulatory autocrine/paracrine factors in glioma cells to facilitate an invasive phenotype (135). The critical characteristic of all gliomas is their extensive infiltration which thwarts efforts to completely remove or ablate malignant cells at the time of surgery (136).
A number of these proteases have also been observed to exert their proteolytic matrix remodelling function through the aid of finger-like membrane structures in glioma cells known as invadopodia (137,138). Many types of tumour cells, including glioma, can form these actin-rich dynamic protrusions which aid invasion through the involvement of numerous adaptor, signalling, adhesion and proteolytic proteins, including those linked to CD151. The association between exosomes and invadopodia is strengthened by the involvement of TSG101. It is known that TSG101 as an ESCRT pathway protein plays a key role in ESCRT-1 function by binding to ubiquitinated receptors and facilitating interactions with other ESCRT complexes. Importantly, TSG101 can also mediate the translocation of active Src from endosomes to assist in tumour cell invasion through the promotion of functional invadopodia (139).
Tks5 (also known as SH3PXD2A) is an adaptor protein that facilitates invadopodia activity and we have demonstrated that it has a potential prognostic role in glioma, with increased Tks5 expression resulting in significantly reduced survival among glioma patients (137). Importantly, Tks5 is also known as a Src substrate (140,141) and through Src mediated phosphorylation, and subsequent association directly with the SH3-SH2 domain adaptor proteins Nck1/Nck2, we identified a Src-Tks5-Nck pathway in the activity of matrix degrading invadopodia (140). Therefore it is emerging that TSG101 (and potentially ESCRT complexes), as a mediator of Src translocation, may be vital in coordinating the dynamic trafficking of Src from endosomes to its sight of function in invadopodia, thereby facilitating the remodelling or degradation of the matrix surrounding the tumour cells. Significantly, this identifies that exosomes possess a vital role in promoting glioma cell invasion through a functional relationship linking exosome secretion and invadopodia activity.
Through a complex network-based communication system between cellular components, secreted factors (such as growth factors and cytokines), and the interaction with the surrounding cells and environment, cancer cells including glioma can promote a plethora of processes such as proliferation, ECM degradation, invasion, angiogenesis and evasion of immune-surveillance to drive disease progression. It is recognized that glioma derived exosomes with their multifunctional cargo can facilitate these processes (142). This can be achieved through a number of avenues. Exosomes isolated from primary GBM cells which contain EGFRvIII can stimulate the in vitro proliferation of human glioma cells (75). Proliferation of glioma cells has also been observed to be driven by the miRNA, miR-21 (51,55). This oncomir (miR-21) has been detected in CSF derived exosomal fractions (along with miR-24, miR-103, and miR-125) when compared directly with the corresponding CSF derived microvesicles. Therefore, it has been proposed that CSF derived exosomes, and not microvesicles, are the major EV compartment that harbour miRNAs which can potentially drive the increased proliferation of exosome recipient cells (129). It has also been demonstrated that the miRNA cargo of GBM derived exosomes can also include miR-29a and miR-30e, which are known to be drivers of angiogenesis (77). MiR-29a through the targeting of PTEN, which is coupled to the activation of Akt, results in the activation of endothelial cells leading to their migration as a part of the angiogenesis process (143). Alternatively, another stimulator of angiogenesis, VEGF-C, is upregulated as a result of the enhanced expression of NF-κB-regulated genes which have been linked to the miR-30e induced hyperactivation of NF-κB (144).
A study by Valadi et al. (87) in 2007 showed that exosomes isolated from human mast cells contained over 1,330 mRNA and 120 non-coding RNA/miRNA, which has been updated as further research into exosome biogenesis in normal and tumour cells is conducted. With this aspect of mRNA/miRNA in mind, we undertook an analysis of glioma based studies within the online Oncomine® platform for datasets that contained mRNA expression levels of genes that are involved in the biogenesis of exosomes (Table 1). Oncomine (version 4.5—www.oncomine.org, Compendia BioscienceTM, Ann Arbor, MI, USA, Thermo Fisher) is an online tool that contains 715 mRNA and copy number expression datasets from 86,733 cancer and normal tissue samples (12,764 samples are normal tissue samples) (145). A total of 62 datasets (5,571 samples) are brain/CNS related datasets. Our data mining of the 62 brain/CNS datasets deposited in the Oncomine Compendium examined the relative mRNA levels of exosomal related gene markers in both glioma and normal brain tissue. The exosomal markers which were differentially expressed between normal brain and glioma tissue included Alix, CD9, CD63, CD81, CD151 and TSG101. As can be observed by the data presented in Table 1, there was an elevation of mRNA expression levels in glioma tissue relative to normal brain. Approximately 47% of the exosome markers in the listed studies were ranked in the top 10% of differentially expressed genes between glioma and normal tissue.
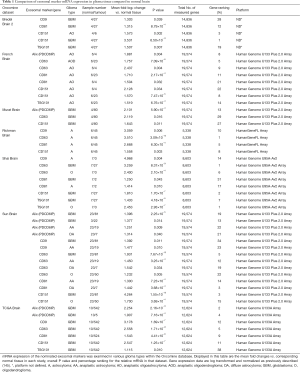
Full table
The potential importance of exosomal markers as a diagnostic or prognostic tool in glioma is further enhanced by the fact that up to 90% of the exosome markers examined here were ranked in the top 20% of differentially expressed genes. In addition, it can also be observed that the mRNA expression levels of multiple exosomal markers were elevated in the same tissue grade in a number of the studies. Interestingly, the datasets which contained exosomal mRNA expression data from GBM patient samples (Bredel Brain 2, Murat, Shai, Sun and TCGA), CD151 and TSG101 were the two exosomal markers with mRNA expression levels that were elevated in GBM relative to normal brain in all five studies. A recent study by Shi et al. (57) identified that miR-21, which is known to be upregulated in the tissue of higher grade glioma (grade II and grade IV), was significantly higher in CSF derived exosomes in glioma patients than in control samples isolated from patients without tumours and the exosomal marker CD63 was also observed to be present.
Exosomes and glioma—therapeutic capacity?
Despite the considerable progress achieved over the years with technological advances in neuroimaging, surgery and post-operative treatments, the prognosis for patients with glioma, in particular GBM tumours, remains poor (4,146). Consequently, there is a continual effort to develop novel therapeutic approaches for the treatment of glioma. Exosomes are emerging as an attractive strategy for potentially providing a flexible drug delivery system. Exosomes, as spherical structures with a well-defined lipid bilayer and an aqueous core, have the ability to house both hydrophilic and hydrophobic molecules, but importantly can function as ‘protectors’ of their cargo while they cross biological barriers to reach specific cellular targets. Importantly, they possess a long circulating half-life, are taken up by recipient cells leading to changes in biological processes within these cells, do not activate acute immune rejection upon being administered (147), and their exosomal cargo or their membrane can be tailored for tumour-specific targets or patient personalized treatment. Yet one of the most critical features of exosomes is that they can cross the BBB, making them an ideal courier for drugs or therapeutic molecules in treating glioma. The BBB is partly permeable due to a weakened association between endothelial cells and astrocytes (148), permitting for an easier passage of the exosomes to the tumour cells, which is normally highly impermeable to many chemotherapeutic drugs. In addition, the phospholipid bilayers allow direct delivery of the cargo into the cytoplasm of the recipient tumour cells as a result of fusion with the cell membrane (149) and their small size assists them in bypassing rapid phagocytosis from the circulation by mononuclear phagocytic mechanisms.
A laboratory based study by Yang et al. (150) using a zebrafish model employing U87 glioma cells demonstrated that exosomes derived from brain endothelial cells utilized as drug carriers facilitated the delivery of paclitaxel and doxorubicin across the BBB resulted in toxicity. This could not be achieved in the absence of exosome-drug cargo loading, with the drugs remaining within the vasculature, unable to cross the BBB. Exosome transfer across the BBB has also been demonstrated in additional laboratory based models. Delivery of siRNA via systemically injected exosomes allowed for the transport into brain tissue, specifically neurons, microglia and oligodendrocytes to efficiently target RNA expression in these cells (151). Dendritic cells have been genetically engineered to release exosomes which expressed the protein Lamp2b fused to the neuron peptide RVG (targeting neural cells) on the exosome membrane, allowing for the effective delivery of siRNA cargo to the neurons (151).
The successful delivery of exosome-encapsulated anti-inflammatory drugs across the BBB via intranasal administration in treating brain inflammatory diseases further indicates their suitability as cargo carriers through the BBB (152). Since exosomes can function as cargo carriers to deliver drugs into the cells, it is also logical that secreted vesicles can potentially function as facilitators of chemoresistance through the expression of genes driving vesicle secretion in removing cytotoxic drugs from within the cell. This was demonstrated by Munoz et al. (153) to revert TMZ resistance in GBM cells through the exosomal delivery of anti-miR-9 from mesenchymal stem cells (MSCs). MiR-9 levels are normally elevated in GBM cells which in turns lead to the increased expression of the drug efflux transporter, P-glycoprotein. Exosomal-mediated delivery of the anti-miR-9 to the GBM cells resulted in a reduction in the expression levels of miR-9 leading to a restoration in TMZ chemosensitivity through increased apoptosis and caspase activity. Whilst, the interaction between the GBM cells and the donor MSCs may have been the result of a gap-junction (contact dependent) mechanism between the two cell types, the authors were able to demonstrate that the anti-miR-9 transfer occurred via a contact independent mechanism mediated via exosomes. Overall, it appears that exosomes have considerable promise as cargo delivery agents for the treatment of glioma. However, the studies are based on in vitro and in vivo models in the laboratory and a greater understanding of the distribution of administered exosomes clearance and tumour-targeting properties are required for the successful application of exosomal-mediated therapeutics in the treatment of glioma. However, it must be noted that CSF sampling may be limited to ventricular/lumbar collection at the time of primary or repeat surgery for glioma patients.
Conclusions
Research in the field of EVs, including exosomes, is moving forward at an increasing rate. Coupled with the ongoing evolution in isolation and characterization techniques, of various EVs isolated from normal cell and tumour cell populations, there has been a significant increase in the number of EV related publications over the last decade. The information presented in this review highlights the relevance of glioma-derived exosomes as a ‘liquid biopsy’ to potentially develop avenues for diagnostic and prognostic markers to monitor disease progression, personalize treatment through the stratification of patients based on the profile of the exosomal biomarkers, and potentially lead to an improvement in glioma patient outcome. Whilst serum has been proposed as a liquid biopsy for exosomes, there is the complicating factor that it also contains exosomes from a greater variety of normal cells and processes in the body. This emphasizes the importance of utilizing CSF as a purer source of glioma-derived exosomes, since it interacts directly with the extracellular space and neighbouring environment around the tumour, ultimately reflecting gradual changes that may be occurring over time as the disease progresses or as the tumour cells respond to treatment. Whilst current evidence is promising, this field of research is still in its early stages. However, continued development of exosome composition and biomarker analytics being linked to technologies such as miRNA arrays and qRT-PCR assays have potential for CSF-derived glioma exosomes being incorporated into the clinical management of glioma patients in the future.
Acknowledgments
Funding: The authors would like to acknowledge support from The Royal Melbourne Hospital Neuroscience Foundation.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Edward R. Sauter) for the series “Body Fluid Exosomes and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.31). The series “Body Fluid Exosomes and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol 2015;17:iv1-iv62. [Crossref] [PubMed]
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ostrom QT, Gittleman H, Xu J, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro Oncol 2016;18:v1-v75. [Crossref] [PubMed]
- Louis DN, Perry A, Burger P, et al. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014;24:429-35. [Crossref] [PubMed]
- Louis DN. The next step in brain tumor classification: "Let us now praise famous men"... or molecules? Acta Neuropathol 2012;124:761-2. [Crossref] [PubMed]
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Ammirati M, Chotai S, Newton H, et al. Hypofractionated intensity modulated radiotherapy with temozolomide in newly diagnosed glioblastoma multiforme. J Clin Neurosci 2014;21:633-7. [Crossref] [PubMed]
- Field KM, Drummond KJ, Yilmaz M, et al. Clinical trial participation and outcome for patients with glioblastoma: multivariate analysis from a comprehensive dataset. J Clin Neurosci 2013;20:783-9. [Crossref] [PubMed]
- Sorensen AG, Batchelor TT, Wen PY, et al. Response criteria for glioma. N at Clin Pract Oncol 2008;5:634-44.
- Yu Y, Lee DH, Peng SL, et al. Assessment of Glioma Response to Radiotherapy Using Multiple MRI Biomarkers with Manual and Semiautomated Segmentation Algorithms. J Neuroimaging 2016;26:626-34. [Crossref] [PubMed]
- Wiestler B, Kluge A, Lukas M, et al. Multiparametric MRI-based differentiation of WHO grade II/III glioma and WHO grade IV glioblastoma. Sci Rep 2016;6:35142. [Crossref] [PubMed]
- Livermore LJ, Ma R, Bojanic S, et al. Yield and complications of frame-based and frameless stereotactic brain biopsy--the value of intra-operative histological analysis. Br J Neurosurg 2014;28:637-44. [Crossref] [PubMed]
- Woodworth GF, McGirt MJ, Samdani A, et al. Frameless image-guided stereotactic brain biopsy procedure: diagnostic yield, surgical morbidity, and comparison with the frame-based technique. J Neurosurg 2006;104:233-7. [Crossref] [PubMed]
- Short SC. Science in Focus: MicroRNA in Glioma - Potential as Biomarkers and Therapeutic Targets. Clin Oncol (R Coll Radiol) 2016;28:543-6. [Crossref] [PubMed]
- Yu X, Li Z. Serum microRNAs as potential noninvasive biomarkers for glioma. Tumour Biol 2016;37:1407-10. [Crossref] [PubMed]
- Tabouret E, Boudouresque F, Farina P, et al. MMP2 and MMP9 as candidate biomarkers to monitor bevacizumab therapy in high-grade glioma. Neuro Oncol 2015;17:1174-6. [Crossref] [PubMed]
- Kros JM, Mustafa DM, Dekker LJ, et al. Circulating glioma biomarkers. Neuro Oncol 2015;17:343-60. [PubMed]
- Hochberg FH, Atai NA, Gonda D, et al. Glioma diagnostics and biomarkers: an ongoing challenge in the field of medicine and science. Expert Rev Mol Diagn 2014;14:439-52. [Crossref] [PubMed]
- Tumilson CA, Lea RW, Alder JE, et al. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol 2014;50:545-58. [Crossref] [PubMed]
- Holdhoff M, Yovino SG, Boadu O, et al. Blood-based biomarkers for malignant gliomas. J Neurooncol 2013;113:345-52. [Crossref] [PubMed]
- Bradley WG Jr. CSF Flow in the Brain in the Context of Normal Pressure Hydrocephalus. AJNR Am J Neuroradiol 2015;36:831-8. [Crossref] [PubMed]
- Kittisupamongkol W. Cerebrospinal fluid production rate. Clin Exp Ophthalmol 2009;37:827; author reply 827-8.
- Czosnyka M, Czosnyka Z, Schmidt EA, et al. Cerebrospinal fluid production. J Neurosurg 2003;99:206-7; author reply 207. [PubMed]
- May C, Kaye JA, Atack JR, et al. Cerebrospinal fluid production is reduced in healthy aging. Neurology 1990;40:500-3. [Crossref] [PubMed]
- Wright BL, Lai JT, Sinclair AJ. Cerebrospinal fluid and lumbar puncture: a practical review. J Neurol 2012;259:1530-45. [Crossref] [PubMed]
- Sakka L, Coll G, Chazal J. Anatomy and physiology of cerebrospinal fluid. Eur Ann Otorhinolaryngol Head Neck Dis 2011;128:309-16. [Crossref] [PubMed]
- Stoicea N, Du A, Lakis DC, et al. The MiRNA Journey from Theory to Practice as a CNS Biomarker. Front Genet 2016;7:11. [Crossref] [PubMed]
- Shalaby T, Grotzer MA. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int J Mol Sci 2015;16:29103-19. [Crossref] [PubMed]
- Kang JH, Mollenhauer B, Coffey CS, et al. CSF biomarkers associated with disease heterogeneity in early Parkinson's disease: the Parkinson's Progression Markers Initiative study. Acta Neuropathol 2016;131:935-49. [Crossref] [PubMed]
- Shekhar R, Rao JR, Devi KA, et al. CSF Proteins as Discreminatory Markers of Tubercular and Pyogenic Meningitis. J Clin Diagn Res 2013;7:1586-8. [PubMed]
- Preusser M, Hainfellner JA. CSF and laboratory analysis (tumor markers). Handb Clin Neurol 2012;104:143-8. [Crossref] [PubMed]
- Sussmuth SD, Sperfeld AD, Hinz A, et al. CSF glial markers correlate with survival in amyotrophic lateral sclerosis. Neurology 2010;74:982-7. [Crossref] [PubMed]
- Alexander GM, Perreault MJ, Reichenberger ER, et al. Changes in immune and glial markers in the CSF of patients with Complex Regional Pain Syndrome. Brain Behav Immun 2007;21:668-76. [Crossref] [PubMed]
- Teunissen CE, Dijkstra C, Polman C. Biological markers in CSF and blood for axonal degeneration in multiple sclerosis. Lancet Neurol 2005;4:32-41. [Crossref] [PubMed]
- Martinez-Rodriguez JE, Santamaria J. CSF markers in sleep neurobiology. Clin Chim Acta 2005;362:12-25. [Crossref] [PubMed]
- Seehusen DA, Reeves MM, Fomin DA. Cerebrospinal fluid analysis. Am Fam Physician 2003;68:1103-8. [PubMed]
- Souweidane MM, Morgenstern PF, Christos PJ, et al. Intraoperative arachnoid and cerebrospinal fluid sampling in children with posterior fossa brain tumors. Neurosurgery 2009;65:72-8; discussion 78. [Crossref] [PubMed]
- Grewal J, Saria MG, Kesari S. Novel approaches to treating leptomeningeal metastases. J Neurooncol 2012;106:225-34. [Crossref] [PubMed]
- Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: current methods and future directions. Fluids Barriers CNS 2011;8:14. [Crossref] [PubMed]
- Patel AS, Allen JE, Dicker DT, et al. Identification and enumeration of circulating tumor cells in the cerebrospinal fluid of breast cancer patients with central nervous system metastases. Oncotarget 2011;2:752-60. [Crossref] [PubMed]
- Samuel N, Remke M, Rutka JT, et al. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J Neurooncol 2014;118:225-38. [Crossref] [PubMed]
- Albulescu R, Codrici E, Popescu ID, et al. Cytokine patterns in brain tumour progression. Mediators Inflamm 2013;2013:979748 [PubMed]
- Saratsis AM, Yadavilli S, Magge S, et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol 2012;14:547-60. [Crossref] [PubMed]
- Rubenstein JL, Wong VS, Kadoch C, et al. CXCL13 plus interleukin 10 is highly specific for the diagnosis of CNS lymphoma. Blood 2013;121:4740-8. [Crossref] [PubMed]
- Dekker LJ, Boogerd W, Stockhammer G, et al. MALDI-TOF mass spectrometry analysis of cerebrospinal fluid tryptic peptide profiles to diagnose leptomeningeal metastases in patients with breast cancer. Mol Cell Proteomics 2005;4:1341-9. [Crossref] [PubMed]
- Zhao J, Ye X, Xu Y, et al. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol 2016;78:1305-10. [Crossref] [PubMed]
- Allen J, Chacko J, Donahue B, et al. Diagnostic sensitivity of serum and lumbar CSF bHCG in newly diagnosed CNS germinoma. Pediatr Blood Cancer 2012;59:1180-2. [Crossref] [PubMed]
- Qaddoumi I, Sane M, Li S, et al. Diagnostic utility and correlation of tumor markers in the serum and cerebrospinal fluid of children with intracranial germ cell tumors. Childs Nerv Syst 2012;28:1017-24. [Crossref] [PubMed]
- Drusco A, Bottoni A, Lagana A, et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015;6:20829-39. [Crossref] [PubMed]
- Qu K, Lin T, Pang Q, et al. Extracellular miRNA-21 as a novel biomarker in glioma: Evidence from meta-analysis, clinical validation and experimental investigations. Oncotarget 2016;7:33994-4010. [Crossref] [PubMed]
- Teplyuk NM, Mollenhauer B, Gabriely G, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 2012;14:689-700. [Crossref] [PubMed]
- Rolle K. miRNA Multiplayers in glioma. From bench to bedside. Acta Biochim Pol 2015;62:353-65. [Crossref] [PubMed]
- Liu S, Yin F, Zhang J, et al. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J Cell Biochem 2014;115:1368-80. [Crossref] [PubMed]
- Li Y, Xu J, Chen H, et al. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res 2013;41:e203 [Crossref] [PubMed]
- . Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Shi R, Wang PY, Li XY, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 2015;6:26971-81. [Crossref] [PubMed]
- Trams EG, Lauter CJ, Salem N Jr, et al. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63-70. [Crossref] [PubMed]
- Takahashi RU, Prieto-Vila M, Hironaka A, et al. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med 2017;55:648-56. [Crossref] [PubMed]
- Zha QB, Yao YF, Ren ZJ, et al. Extracellular vesicles: An overview of biogenesis, function, and role in breast cancer. Tumour Biol 2017;39:1010428317691182 [Crossref] [PubMed]
- Becker A, Thakur BK, Weiss JM, et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016;30:836-48. [Crossref] [PubMed]
- Wendler F, Favicchio R, Simon T, et al. Extracellular vesicles swarm the cancer microenvironment: from tumor-stroma communication to drug intervention. Oncogene 2017;36:877-84. [Crossref] [PubMed]
- Kosaka N, Yoshioka Y, Fujita Y, et al. Versatile roles of extracellular vesicles in cancer. J Clin Invest 2016;126:1163-72. [Crossref] [PubMed]
- Street JM, Koritzinsky EH, Glispie DM, et al. Urine Exosomes: An Emerging Trove of Biomarkers. Adv Clin Chem 2017;78:103-22. [Crossref] [PubMed]
- Hong CS, Funk S, Muller L, et al. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles 2016;5:29289. [Crossref] [PubMed]
- Zlotogorski-Hurvitz A, Dayan D, Chaushu G, et al. Human saliva-derived exosomes: comparing methods of isolation. J Histochem Cytochem 2015;63:181-9. [Crossref] [PubMed]
- Zhou Q, Li M, Wang X, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci 2012;8:118-23. [Crossref] [PubMed]
- Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep 2011;25:749-62. [PubMed]
- Vojtech L, Woo S, Hughes S, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014;42:7290-304. [Crossref] [PubMed]
- Kim DK, Lee J, Kim SR, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 2015;31:933-9. [Crossref] [PubMed]
- Kim DK, Kang B, Kim OY, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles 2013;2. [PubMed]
- Keerthikumar S, Chisanga D, Ariyaratne D, et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol 2016;428:688-92. [Crossref] [PubMed]
- Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009;9:4997-5000. [Crossref] [PubMed]
- Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012;10:e1001450 [Crossref] [PubMed]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- van der Vos KE, Balaj L, Skog J, et al. Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol Neurobiol 2011;31:949-59. [Crossref] [PubMed]
- Li CC, Eaton SA, Young PE, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol 2013;10:1333-44. [Crossref] [PubMed]
- Godlewski J, Krichevsky AM, Johnson MD, et al. Belonging to a network--microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro Oncol 2015;17:652-62. [Crossref] [PubMed]
- Guescini M, Genedani S, Stocchi V, et al. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna) 2010;117:1-4. [Crossref] [PubMed]
- Redzic JS, Ung TH, Graner MW. Glioblastoma extracellular vesicles: reservoirs of potential biomarkers. Pharmgenomics Pers Med 2014;7:65-77. [PubMed]
- Xu R, Greening DW, Zhu HJ, et al. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest 2016;126:1152-62. [Crossref] [PubMed]
- Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 2016;113:E968-77. [Crossref] [PubMed]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [PubMed]
- Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta 2012;1820:940-8. [Crossref] [PubMed]
- Keller S, Ridinger J, Rupp AK, et al. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med 2011;9:86. [Crossref] [PubMed]
- Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014;3. [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci 2012;37:460-5. [Crossref] [PubMed]
- Zomer A, Vendrig T, Hopmans ES, et al. Exosomes: Fit to deliver small RNA. Commun Integr Biol 2010;3:447-50. [Crossref] [PubMed]
- Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 2013;126:5553-65. [Crossref] [PubMed]
- Gross JC, Chaudhary V, Bartscherer K, et al. Active Wnt proteins are secreted on exosomes. Nat Cell Biol 2012;14:1036-45. [Crossref] [PubMed]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010;464:864-9. [Crossref] [PubMed]
- van Niel G, Charrin S, Simoes S, et al. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 2011;21:708-21. [Crossref] [PubMed]
- Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 2011;23:452-7. [Crossref] [PubMed]
- Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014;30:255-89. [Crossref] [PubMed]
- Zoller M. Exosomes in Cancer Disease. Methods Mol Biol 2016;1381:111-49. [Crossref] [PubMed]
- Subra C, Grand D, Laulagnier K, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res 2010;51:2105-20. [Crossref] [PubMed]
- Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res 2017;66:30-41. [Crossref] [PubMed]
- Lachenal G, Pernet-Gallay K, Chivet M, et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 2011;46:409-18. [Crossref] [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Bakhti M, Winter C, Simons M. Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J Biol Chem 2011;286:787-96. [Crossref] [PubMed]
- Nave KA. Myelination and the trophic support of long axons. Nat Rev Neurosci 2010;11:275-83. [Crossref] [PubMed]
- Taylor AR, Robinson MB, Gifondorwa DJ, et al. Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev Neurobiol 2007;67:1815-29. [Crossref] [PubMed]
- Potolicchio I, Carven GJ, Xu X, et al. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol 2005;175:2237-43. [Crossref] [PubMed]
- Weber MA, Zoubaa S, Schlieter M, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology 2006;66:1899-906. [Crossref] [PubMed]
- Chen CC, Taniguchi T, D'Andrea A. The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J Mol Med (Berl) 2007;85:497-509. [Crossref] [PubMed]
- Chittiboina P, Connor DE Jr, Caldito G, et al. Occult tumors presenting with negative imaging: analysis of the literature. J Neurosurg 2012;116:1195-203. [Crossref] [PubMed]
- Hoelzinger DB, Demuth T, Berens ME. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J Natl Cancer Inst 2007;99:1583-93. [Crossref] [PubMed]
- Street JM, Barran PE, Mackay CL, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 2012;10:5. [Crossref] [PubMed]
- Lin J, Li J, Huang B, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal 2015;2015:657086 [PubMed]
- Furnari FB, Cloughesy TF, Cavenee WK, et al. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer 2015;15:302-10. [Crossref] [PubMed]
- Taylor TE, Furnari FB, Cavenee WK. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets 2012;12:197-209. [Crossref] [PubMed]
- Del Vecchio CA, Giacomini CP, Vogel H, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene 2013;32:2670-81. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [Crossref] [PubMed]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Chen WW, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids 2013;2:e109 [Crossref] [PubMed]
- SongTao Q. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 2012;103:269-73. [Crossref] [PubMed]
- Dang CV. Links between metabolism and cancer. Genes Dev 2012;26:877-90. [Crossref] [PubMed]
- Rossetto M, Ciccarino P, Boisselier B, et al. Metabolism of glioma and IDH1/IDH2 mutations. Rev Neurol (Paris) 2011;167:699-703. [Crossref] [PubMed]
- Bruschi M, Santucci L, Ravera S, et al. Human urinary exosome proteome unveils its aerobic respiratory ability. J Proteomics 2016;136:25-34. [Crossref] [PubMed]
- Panfoli I, Ravera S, Podesta M, et al. Exosomes from human mesenchymal stem cells conduct aerobic metabolism in term and preterm newborn infants. FASEB J 2016;30:1416-24. [Crossref] [PubMed]
- Arslan F, Lai RC, Smeets MB, et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res 2013;10:301-12. [Crossref] [PubMed]
- Ohba S, Hirose Y. Biological Significance of Mutant Isocitrate Dehydrogenase 1 and 2 in Gliomagenesis. Neurol Med Chir (Tokyo) 2016;56:170-9. [Crossref] [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [Crossref] [PubMed]
- van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res 2010;16:1597-604. [Crossref] [PubMed]
- Schafer N, Glas M, Herrlinger U. Primary CNS lymphoma: a clinician's guide. Expert Rev Neurother 2012;12:1197-206. [Crossref] [PubMed]
- Wei D, Wan Q, Li L, et al. MicroRNAs as Potential Biomarkers for Diagnosing Cancers of Central Nervous System: a Meta-analysis. Mol Neurobiol 2015;51:1452-61. [Crossref] [PubMed]
- Akers JC, Ramakrishnan V, Kim R, et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J Neurooncol 2015;123:205-16. [Crossref] [PubMed]
- Akers JC, Ramakrishnan V, Yang I, et al. Optimizing preservation of extracellular vesicular miRNAs derived from clinical cerebrospinal fluid. Cancer Biomark 2016;17:125-32. [Crossref] [PubMed]
- Yue S, Mu W, Erb U, et al. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2015;6:2366-84. [Crossref] [PubMed]
- Yanez-Mo M, Gutierrez-Lopez MD, Cabanas C. Functional interplay between tetraspanins and proteases. Cell Mol Life Sci 2011;68:3323-35. [Crossref] [PubMed]
- Shi GM, Ke AW, Zhou J, et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology 2010;52:183-96. [Crossref] [PubMed]
- Yanez-Mo M, Barreiro O, Gonzalo P, et al. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood 2008;112:3217-26. [Crossref] [PubMed]
- Mentlein R, Hattermann K, Held-Feindt J. Lost in disruption: role of proteases in glioma invasion and progression. Biochim Biophys Acta 2012;1825:178-85. [PubMed]
- Giese A, Westphal M. Treatment of malignant glioma: a problem beyond the margins of resection. J Cancer Res Clin Oncol 2001;127:217-25. [Crossref] [PubMed]
- Stylli SS. Prognostic significance of Tks5 expression in gliomas. J Clin Neurosci 2012;19:436-42. [Crossref] [PubMed]
- Stylli SS, Kaye AH, Lock P. Invadopodia: at the cutting edge of tumour invasion. J Clin Neurosci 2008;15:725-37. [Crossref] [PubMed]
- Tu C, Ahmad G, Mohapatra B, et al. ESCRT proteins: Double-edged regulators of cellular signaling. Bioarchitecture 2011;1:45-8. [Crossref] [PubMed]
- Stylli SS, Stacey TT, Verhagen AM, et al. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J Cell Sci 2009;122:2727-40. [Crossref] [PubMed]
- Lock P, Abram CL, Gibson T, et al. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J 1998;17:4346-57. [Crossref] [PubMed]
- Giusti I, Di Francesco M, Dolo V. Extracellular Vesicles in Glioblastoma: Role in Biological Processes and in Therapeutic Applications. Curr Cancer Drug Targets 2017;17:221-35. [Crossref] [PubMed]
- Wang J, Wang Y, Wang Y, et al. Transforming growth factor beta-regulated microRNA-29a promotes angiogenesis through targeting the phosphatase and tensin homolog in endothelium. J Biol Chem 2013;288:10418-26. [Crossref] [PubMed]
- Jiang L, Lin C, Song L, et al. MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-kappaB/IkappaBalpha negative feedback loop. J Clin Invest 2012;122:33-47. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chen TS, Arslan F, Yin Y, et al. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. J Transl Med 2011;9:47. [Crossref] [PubMed]
- Nduom EK, Yang C, Merrill MJ, et al. Characterization of the blood-brain barrier of metastatic and primary malignant neoplasms. J Neurosurg 2013;119:427-33. [Crossref] [PubMed]
- Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014;35:2383-90. [Crossref] [PubMed]
- Yang T, Martin P, Fogarty B, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res 2015;32:2003-14. [Crossref] [PubMed]
- Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011;29:341-5. [Crossref] [PubMed]
- Zhuang X, Xiang X, Grizzle W, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther 2011;19:1769-79. [Crossref] [PubMed]
- Munoz JL, Bliss SA, Greco SJ, et al. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids 2013;2:e126 [Crossref] [PubMed]


