Design, Molecular Docking, and in Silico Analysis of Analogues of Chloroquine, and Hydroxychloroquine Against Sars-Cov-2 Target (6W63.pdb)
Article Information
Yakubu SN*, Poyi CO, Afolabi EO
Department of Pharmaceutical and Medicinal Chemistry, Faculty of Pharmaceutical Sciences, University of Jos, Nigeria.
*Corresponding author: Yakubu Solomon Naruka, Department of Pharmaceutical and Medicinal Chemistry, Faculty of Pharmaceutical Sciences, University of Jos, Nigeria.
Received: 29 November 2021; Accepted: 12 December 2021; Published: 21 December 2021
Citation: Yakubu SN, Poyi CO, Afolabi EO. Design, Molecular Docking, and in Silico Analysis of Analogues of Chloroquine, and Hydroxychloroquine Against Sars-Cov-2 Target (6W63.pdb). Archives of Clinical and Biomedical Research 5 (2021): 1004-1017.
View / Download Pdf Share at FacebookAbstract
Computer-aided drug design has been an effective strategy and approach to discover, develop, analyze, accelerate, and economize design, and development of drugs and biologically active molecules. A total of twelve analogues of Chloroquine (CQ) and Hydroxychloroquine (HCQ) were designed and virtually analyzed using PyRx software, Molinspiration, Swiss ADME, Swiss-Target Prediction software, and ProTox-II-Prediction of toxicity platform. Based on the docking studies carried out using Autodock vina, five analogues; H-368 (-6.0Kcal/mol), H-372 (-6.0Kcal/mol), H-156 (-5.9Kcal/mol), H-139 (-5.7Kcal/mol), C-136 (-5.7Kcal/mol) exhibited higher binding affinity compared to HCQ (-5.5Kcal/mol), while all twelve analogues exhibited higher binding affinity compared to CQ (-4.5Kcal/mol). In silico analysis of toxicity profile of these analogues shows a lower potential to toxicity and a comparable activity on some major isoforms of cytochrome P450. But unlike the parent molecules, both H-139 and H-156 are substrates of P-glycoproteins (P-gp) which implies that these analogues possess high clearance and less pharmacokinetic-related drug-drug interactions compared to the parent molecules. Herein we propose these analogues as potential inhibitors or lead compounds against SARS- CoV-2 with a view of synthesizing them, conducting more molecular dynamic simulations, and conducting In vitro studies on them.
Keywords
Chloroquine; COVID-19; Docking; Hydroxychloroquine; SARS-CoV-2
Article Details
1. Introduction
The world today is faced with skyrocketing costs for drug design and development of new biologically active molecules hence researchers are currently looking for ways to repurpose older drugs and possibly, even some that failed in initial trials. With the aid of computer-aided drug- design and development, researchers can find countless new tricks for old drugs [1]. According to Atul Butte (2012), a bioinformatician at the University of California, San Francisco, drug repositioning is a complement to the discovery of new molecules, rather than an alternative. More so, modern medicine is becoming better at figuring out that each disease is actually made up of five or ten different diseases and there are simply not enough companies out there to develop new drugs to treat them all [2]. For researchers in the academia and industries, taking drugs that have been developed for one disorder and repositioning them-with little or no modification to tackle another disorder/disease, is an increasingly important strategy for researchers. These efforts are been inspired from numerous classic success stories as documented in many literatures [3].
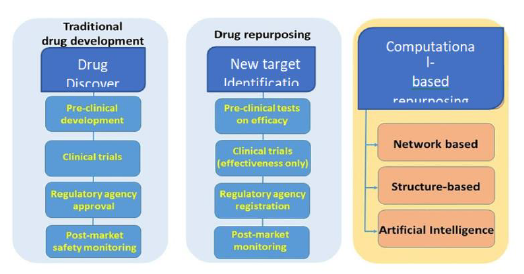
Drug repurposing (also known as drug repositioning) involves the identification of new applications for existing drugs at a lower cost and in a shorter time. Currently, there are different computational drug-repurposing strategies and some of these approaches have been applied to the Coronavirus Disease 2019 (COVID-19) pandemic. Computational drug-repositioning approaches applied to COVID-19 can be broadly categorized into (i) Network-Based models, (ii) Structure-Based approaches and (iii) artificial intelligence (AI) approaches as summarized in Figure 1. Network-based approaches are sub-divided into two: network-based clustering approaches and network-based propagation approaches. Both of them allowed to annotate some important patterns, to identify proteins that are functionally associated with COVID-19 and to discover novel drug–disease or drug–target relationships useful for new therapies. Structure-based approaches allowed to identify small chemical compounds able to bind macromolecular targets to evaluate how a chemical compound can interact with the biological counterpart, trying to find new applications for existing drugs. AI-based networks appear, at the moment, less relevant since they need more data for their application [4]. Drug repositioning plays a very important role in drug discovery in that old drugs are being rescued from the shelves and patency extended while they are being used to treat new diseases. This fact also contributes to the reason why drug repositioning is quite attractive to many scientists and multinational companies around the world [5-8]. Till date, the most notable repurposed drugs have been discovered either through serendipity, based on specific pharmacological insights or using experimental screening platforms [9].
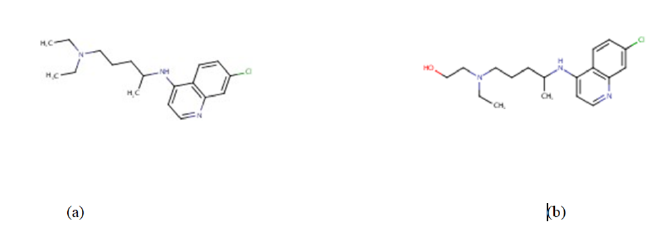
Presently, vaccines such as Pfizer/BioNTech, Moderna, Johnson & Johnson’s Janssen, AstraZeneca are authorized and recommended for COVID-19 infection prevention [10] and have been approved by the world health organization-WHO to help curb the spread of the corona virus disease [10,11]. Quite a number of antiviral drugs, formerly discovered and used in the management of malaria, MERS, and SARS are being tested as potential treatment for COVID-19 and some of them are being used in clinical trial treatments for COVID-19 infection [11]. In the same light, drugs like chloroquine and hydroxychloroquine (Figure 2) are the focus of our research here.
The serendipity responsible for the earlier discovery of drugs like chloroquine (Figure 2a) for the prevention and treatment of malaria and the discovery of hydroxychloroquine (Figure 2b) for the treatment of rheumatoid arthritis, lupus and porphyria cutanea cannot be overemphasized [12]. This is so because both chloroquine and hydroxychloroquine are currently being studied in order to be repositioned for the treatment of COVID-19 [13]. Chloroquine (CQ) is an old antimalarial agent with some pharmacodynamic properties including anti-inflammatory and immunomodulatory properties. It has gained significant interest as a potential therapeutic option for the management and treatment of COVID-19 based on the research conducted in China. Wang et al., 2021 demonstrated potent in vitro activity of chloroquine against SARS-CoV-2 with an EC50 at 48 hours of 1.13 μM in Vero E6 cells. Hydroxychloroquine (HQ) on the other hand, is a compound that differs from CQ only by a single hydroxyl group which may be responsible for its tolerability and its long-term usage in rheumatological disorders compared to CQ [12]. These data were consistent with previous data for chloroquine’s inhibitory activity against SARS-CoV-1 and MERS-CoV in various cell lines, where EC50 values of 1–8.8 and 3.0 μM were demonstrated, respectively.Previous studies reported that CQ/HCQ possess a broad spectrum of antiviral effects on a variety of viruses as diverse as (HIV) Marburg virus, Zika virus, dengue virus, Ebolavirus, and SARS-CoV-1. CQ and HCQ can interfere with the binding of viral particles to their cellular cell surface receptor or the pH-dependent endosome-mediated viral entry of enveloped viruses to inhibit the viral cycle. They can also interfere with the posttranslational modification of viral proteins or impair the proper maturation of viral protein by pH modulation. In addition, CQ and HCQ can regulate the immune system by affecting cell signaling and the production of proinflammatory cytokines [14]. Moreover, CQ on the growth of SARS-CoV-2 in vitro and an early clinical trial conducted in COVID-19 Chinese patients showed a significant effect, in terms of both clinical outcome and viral clearance. Chinese experts recommend that patients diagnosed with mild, moderate, and severe cases of COVID19 pneumonia and without contraindications to it be treated with 500 mg chloroquine twice per day for ten-day treatment duration. HCQ (an analog of chloroquine) has been demonstrated to have an anti-SARS-CoV activity in vitro. Its clinical safety profile is better than that of CQ (during long-term use) and allows higher daily dose and has fewer concerns about drug-drug interactions. HCQ/CQ alone and in combination with azithromycin was highly effective in clearing viral nasopharyngeal carriage within six days in COVID-19 subjects [15]. However, according to Mittra and Mieler 2013 [16], these drugs molecules (including chloroquine and hydroxychloroquine) are not without some serious side effects (seizures, muscle damage, problems with vision, low blood count, etc.) that can possibly limit their use and apparently their potential application(s) in other disease conditions. Consequently, this research work is taking advantage of computational chemistry to design twelve (12) analogues of CQ and HCQ in order to potentiate their safety, efficacy and overall potency against the dreaded COVID-19. And possibly, to reduce some of the serious side effects that accompanies them. There is an unprecedent response by both the medical and scientific communities to tackle the COVID-19 disease due to the rapid spread of SARS-CoV-2 Virus (Fig.3) and the growing number of morbidity and deaths it has catalyzed. Many communities seek to completely understand its epidemiology and the mechanisms of its druggable protein targets, resolve its molecular structures, identify effective therapeutic agents and developing vaccines to prevent the virus spread [15].
Identifying key protein targets for drug development is one of the first tasks to be addressed. Once a druggable protein model or structure is available, numerous molecular modelling methods allow us to identify drugs with high specificity and efficacy. While these methods involve de novo design strategies, the urgency of the current situation makes computational- based repurposing approaches applied to COVID-19 (Figure 1) one of the most economic and efficient therapeutic strategies to pursue. We can also leverage on the vast reservoir of knowledge about agents currently known to be effective against SARS-CoV. Virtual screening methods including docking and pharmacophore modelling are ideal to identify and rank- prioritize lead candidates for further investigation and possible optimization. Worthy of note is the fact that relying on physical experimentation alone is not economically sustainable in today’s rapidly evolving COVID-19 environment.
2. Materials and Methods
2.1 Hardware
All the computational analysis/screening were done using x64-based PC, windows 10 Pro, 4 compute cores 2C+2G, 2 CORES, 4GB memory, and 32-Bit operating system. Protein and Ligand Library 12 analogues of CQ and HCQ were designed in PubChem Sketcher V2.4 [17] and downloaded as an MDL file. The structures were optimized and converted to .sdf in discovery studio 4.5 visualizer [18]. The crystal structure of the SARS- CoV-2 target (6W63.pdb) was downloaded from the protein data bank [19], its original ligands and water were eliminated using discovery studio 4.5 visualizer [18]. Molecular Docking Ligands and Protein target for molecular docking were prepared in Autodock Tools using PyRx 0.8 package [20](27), a grid box (x: -2.3200, y: 19.1496, z: -26.3281, dimensions (Angstrom); x:y:z: = 25.0000) was employed, and docking simulations of bioactive conformations was done using Autodock Vina [20]. The results obtained were analyzed using PyMol [21] and discovery studio visualizer [18].
2.2 Method
The 3D structures of the receptors were downloaded from protein databank (www.pdb.org) and subsequently prepared; first by removing the ligands which were in complexed with the downloaded receptors and the water molecules with the help of using Discovery Studio. More so, all the prepared receptors were saved in pdbqt format, which is the required format when using Audodock vina or Autodock docking tool for docking. The 3D structures of the ligands were downloaded from pubchem (www.ncbi.nlm-nih.gov/pubchem), and then saved in sdf format. The prepared 3D structures of the receptors were loaded onto the PyRx platform, which has both the Autodock vina and Autodock. After loading the structures, the structures were then converted into macromolecules and then the receptors were then selected to get the binding pockets so as to generate the grid box. and they were all converted to pdbqt formats and the energy minimized, the ligands were then docked into the various receptors.
2.3 In Silico Pharmacokinetic Studies
All the structures were drawn using Swiss ADME platform, the SMILES were generated and loaded onto the molinspiration platform where pharmacokinetic properties, bioactivity scores and other parameters were being obtained.
All the molecules were also loaded into the ProTox-II virtual lab in the form of a ‘MOLfile’ with the aid of Advanced Chemistry Development, Inc.-ACD/Labs, ACD/ChemSketch software [11-22]. The organ toxicity (hepatotoxicity), toxicity end point (carcinogenicity, immunotoxicity, mutagenicity, cytotoxicity), LD50, and the toxicity class of all the compounds were determined.
3. Results and Discussion
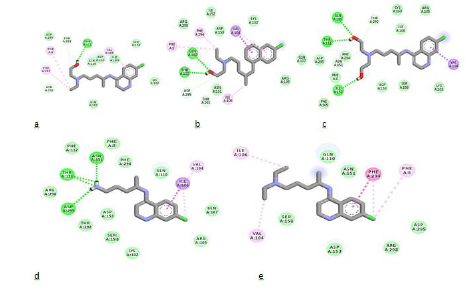
Understanding binding affinity is key in appreciating the intermolecular interactions driving biological processes, structural biology and structure-function relationships. It is also measured as part of the drug discovery process to help design drugs that bind their targets selectively and specifically. Binding affinity is the strength of the binding interaction between a single biomolecule (e.g., protein or DNA) to its ligand/binding partner-drug or inhibitor. Binding affinity is typically measured and reported by the dissociation constant-KD, the smaller the KD value, the greater the binding affinity of the ligand for its target. [23]. When a drug molecule associates with a target, it results in a lowered energy which compensates for any transformation of the ligand from its energy minimum to its bound conformation with the protein [24,25]. With respect to the Structure-Activity Relationship (SAR) of these molecules, as summarized in Table 1, the removal of one ethyl group from the terminal nitrogen atom of CQ increased the binding affinity of the molecule with a characteristic binding energy of -4.5 Kcal/mol (CQ) to - 4.7 Kcal/mol (C-383). Further hydroxylation of the ethyl group led to a further increase in binding affinity with a corresponding binding energy of -5.7 Kcal/mol (H-139). This can also be seen in the binding interaction of C-136 (Figure 3d), where the hydrogens of the terminal amino group participated in hydrogen-bonding. The 3D view of binding conformation of H-372, H-156, and C-136 to the active site residues of SARS-CoV-2-6W63.pdb showing hydrogen-bond interactions is shown in Fig. 4. The complete conversion of the two alkyl groups attached to the terminal nitrogen of HCQ to alcoholic groups also led to an increase in binding affinity with a binding energy of -5.9Kcal/mol (H-156). Also, the removal of C-11 along with the amino group attached to C-9 and C-10 led to an increase in binding affinity of the molecule H-372 (- 6.0Kcal/mol). All the designed molecules have a synthetic accessibility-SA of less than 3. A compound’s SA is a very important aspect of computer-aided drug design since in some cases computer-designed compounds/molecules cannot be synthesized [26]. It is often reported within the range of 1 (very easy to synthesize) and 10 (Difficult to synthesize).
Table1: Molecular properties and Binding Energy of Compounds
Figure 4: 2D binding interactions of HCQ (a), H-372 (b), H-156 (c), C-136 (d) and CQ (e) to the active site residues of SARS-CoV-2 (6W63.pdb). Ligands are shown in stick forms while amino acid residues are shown in disc forms. Hydrogen- bond interaction with amino acid main chain is indicated by green discontinuous lines, green colored discs show van der waal's interaction while purple discs show pi-sigma interactions.
The LogP values (Table 2) of all the molecules are within the range of 2 and less than 5 which indicate compounds of intermediate polarity, good balance between aqueous and lipid solubility, good absorption and distribution. The logP value of a compound, which is the logarithm of its partition coefficient between n-octanol and water i.e., log (Coctanol/Cwater), is a well-established measure of the compound’s hydrophilicity. Low hydrophilicity, and therefore high logP, poor absorption or permeation. Compounds have been shown to have a reasonable probability of being well absorbed, their logP values should not be greater than 5.0 [27]. The designed molecules possess up to 60% activity on the G-protein-coupled receptors (GPCRs) (Table 2) specifically on the family A (rhodopsin-like receptors) as obtained on the Swiss-Target platform. Presently, there are over four hundred (400) drug molecules i.e., approximately 34% of all FDA approved drugs, that act on more than 100 unique targets of GPCRs. Generally, GPCRs are among the most numerous groups of transmembrane proteins of the mammalian genome. Till date, about 800 of these proteins have been identified in humans [28]. The relevance of their manifold functions has made them therapeutically attractive as shown by the fact that they are the targets of over 30% of United States Food and Drug Administration-approved drugs [29]. Two analogues of HCQ (H-139 and H-156) are substrates of the permeability-glycoprotein (P- gp) which implies that these molecules will undergo less pharmacokinetic-related drug-drug interactions and will also be easily cleared from the human system. The knowledge about compounds being substrate or non-substrate of P-gp is key to appraise active efflux through biological membranes, for instance from the gut wall to the lumen or from the brain [30]. One important role of the P-gp is to protect the CNS from xenobiotics [31].
|
S/N |
Name |
LogP |
B.A |
G.I |
BBB |
GPCRs (%) |
P-gp |
LD50 |
T.C |
|
Score |
Absorption |
(mg/Kg) |
|||||||
|
1 |
CQ |
4.15 |
0.55 |
High |
Yes |
60 |
No |
311 |
4 |
|
2 |
HCQ |
3.37 |
0.55 |
High |
Yes |
53.3 |
No |
1240 |
4 |
|
3 |
C-136 |
2.83 |
0.55 |
High |
Yes |
60 |
No |
750 |
4 |
|
4 |
H-139 |
2.77 |
0.55 |
High |
Yes |
53.3 |
Yes |
1240 |
4 |
|
5 |
H-156 |
2.88 |
0.55 |
High |
Yes |
53.3 |
Yes |
1240 |
4 |
|
6 |
C-189 |
3.49 |
0.55 |
High |
Yes |
60 |
No |
311 |
4 |
|
7 |
H-140 |
3.36 |
0.55 |
High |
Yes |
60 |
No |
750 |
4 |
|
8 |
C-383 |
3.57 |
0.55 |
High |
Yes |
53.3 |
No |
311 |
4 |
|
9 |
H-7715 |
3.67 |
0.55 |
High |
Yes |
60 |
No |
750 |
4 |
|
10 |
H-97 |
3.36 |
0.55 |
High |
Yes |
53.3 |
No |
750 |
4 |
|
11 |
H-368 |
3.32 |
0.55 |
High |
Yes |
26.7 |
No |
200 |
3 |
|
12 |
H-372 |
4.8 |
0.55 |
High |
Yes |
40 |
No |
416 |
4 |
|
13 |
H-369 |
2.95 |
0.55 |
High |
Yes |
53.3 |
No |
750 |
4 |
|
14 |
H-347 |
3.12 |
0.55 |
High |
Yes |
53.3 |
No |
750 |
4 |
Table2: Pharmacokinetic properties and Toxicity Prediction.
The interaction of the molecules with CYP450 isoforms and kinase is as presented in Table 3. The HCQ analogue (H-156) stands out as it inhibits only one isoform of CYP450 i.e., CYP2D6 and as earlier mentioned, H-156 is also a substrate of P-gp. The knowledge about interaction of molecules with CYP450 is essential because it plays a major role in drug elimination through metabolic biotransformation. It’s being documented that both CYP and P-gp can synergistically process small molecules to enhance tissue protection and most therapeutic molecules are the substrate of five major isoforms (Table 3). Inhibition of these major isoforms is certainly one major cause of pharmacokinetic-related drug-drug interactions leading to toxic or other unwanted adverse effects due to lower clearance and accumulation of the drug or its metabolite. It is therefore important for drug discovery to predict the propensity with which the molecule will cause significant drug interactions through inhibition of CYPs and to determine which isoforms are affected. [32,33]
|
CYP450 Isoforms Inhibitors |
|||||||
|
S/N |
Name |
Kinase |
CYP1A2 |
CYP2C19 |
CYP2C9 |
CYP2D6 |
CYP3A4 |
|
Inhibitor |
|||||||
|
1 |
CQ |
0.38 |
Yes |
No |
No |
Yes |
Yes |
|
2 |
HCQ |
0.44 |
Yes |
No |
No |
Yes |
No |
|
3 |
C-136 |
0.46 |
Yes |
Yes |
No |
Yes |
Yes |
|
4 |
H-139 |
0.46 |
Yes |
No |
No |
Yes |
Yes |
|
5 |
H-156 |
0.43 |
No |
No |
No |
Yes |
No |
|
6 |
C-189 |
0.41 |
Yes |
No |
No |
Yes |
Yes |
|
7 |
H-140 |
0.33 |
Yes |
No |
No |
Yes |
No |
|
8 |
C-383 |
0.4 |
Yes |
Yes |
No |
Yes |
Yes |
|
9 |
H-7715 |
0.44 |
Yes |
No |
No |
Yes |
Yes |
|
10 |
H-97 |
0.52 |
Yes |
No |
No |
Yes |
Yes |
|
11 |
H-368 |
0.04 |
Yes |
No |
No |
Yes |
Yes |
|
12 |
H-372 |
-0.01 |
Yes |
Yes |
No |
Yes |
Yes |
|
13 |
H-369 |
0.55 |
Yes |
No |
No |
Yes |
Yes |
|
14 |
H-347 |
0.49 |
Yes |
No |
No |
Yes |
Yes |
Table 3: Enzyme activity.
4. Conclusion
In this study, twelve analogues of CQ and HCQ were designed and subjected to virtual screening, five lead molecules showed better binding affinity and strong interactions with active site residues of SARS-CoV-2 target (6W63.pdb) as compared to both chloroquine-CQ and hydroxychloroquine-HCQ. More so, all the designed analogues exhibited superior binding affinity compared to CQ. They were seen to have a number of non-covalent interactions which included, hydrogen bonding, van der waal’s forces and hydrophobic interactions. Based on the predicted synthetic accessibility, all the compounds can easily be synthesized and In vitro testing can also be carried out against the SARS-CoV-2 Vero cell lines as well as comparative toxicity studies using drosophila model prior to subsequent clinical evaluations. These twelve molecules therefore, can serve as potential leads to the development of potent and effective drug molecules against the dreaded Covid-19 pandemic.
References
- Scanndl JW, Blanckly A, Boldon H, et al. Diagnosing the decline in pharmaceutical R&D efficiency. Nature Rev. Drug Discov 11 (2012): 191-200
- Lekka E, Deftereos SN, Persidis A, et al. Drug Discov. Today Ther. Strateg 8 (2011): 103-108.
- Singh N, Halliday A, Thomas J, et al. A safe lithium mimetic for bipolar disorder. Nat Commun 4 (2013): 1332.
- Coronavirus Disease 2019 (COVID-19): Situation Report, WHO, Geneva, Switzerland (2020).
- Adhikari SP,Meng S, Wu YJ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infectious Diseases of Poverty 9 (2020): 29.
- Velavan TP, Meyer CG. The COVID-19 epidemic,” 2020. CDC. About COVID-19 (2020).
- Hopkins J. Coronavirus Resource Center. Im Internet (2020)
- Monti S, Delvino P, Bellis E, et al. Clinical course of COVID-19 in a series of patients with chronic arthritis treated with immunosuppressive targeted therapies. Annals of the Rheumatic Diseases 79 (2020): 667-668.
- Mahase E. Covid-19: where are we on vaccines and variants?. British Medical Journal 372 (2021): 7.
- Khan Z, Karatas Y, Rahman H. Anti COVID-19 drugs: need for more clinical evidence and global action. Advances in Herapy 37 (2020): 2575-2579.
- McCreary EK, Pogue JM. Coronavirus disease 2019 treatment: a review of early and emerging options. Open Forum Infectious Diseases 7 (2020).
- Cortegiani A, Ingoglia G, Ippolito M, et al. A systematic review on the afficacy and safety of chloroquine for the treatment of COVID-19 (2020).
- W Zhang, Y Zhao, F Zhang, et al. The use of anti-inflammatory drugs in the treatment of people with severe corona virus disease 2019 (COVID-19): the perspectives of clinical immunologists from China. Clinical Immunology 214 (2020).
- Otuechere C Repurposing of chloroquine and its derivative, hydroxychloroquine for COVID-19: implications for people living with HIV in Africa. Preprints 328 (2020).
- Mittra RA, Mieler WF. Chapter 89-Drug Toxicity of the Posterior Segment”. Retina (Fifth ed.). W.B. Saunders (2013): 1532-1554.
- Ihlenfeldt WD, Bolton EE, Bryant SH. The PubChem chemical structure sketcher. Journal of cheminformatics 1 (2009): 1-20.
- Dassault Systemes BIOVIA (2016) Discovery Studio Modeling Environment, Release 2017, San Diego.
- Burley SK, Helen Chen L, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy Nucleic Acids Research 47 (2019): 464-474.
- Trott O, Olson AJ. Autoduck Vina: improving the speed and accuracy of ducking with a new scoring function, efficient optimization and multithreading. journal of computational chemistry 31 (2010): 455-461
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
- ACD/Structure Elucidator, version 2018.1, Advanced Chemistry Development, Inc., Toronto, ON, Canada, www.acdlabs.com, 2019.
- Baron R, Setny P, Mc Cammon JA. Water in cavity-ligand recognition. Journal of the American Chemical Society 132 (2010): 12091-12097.
- Wienken CJ, Baaske P, Rothbauer U, et al. Protein-binding assays in biological liquids using microsale thermophoresis.” Nature Communications 1 (2010): 100-101.
- Foscato M, Jensen VR. Automated in Silico Design of Homogeneous Catalysts.ACS Catalysis 10 (2020): 2354-2377.
- Vorsilak M, Nonpher DS. Computational method for design of hard-to-synthesize structures. Journal of Cheminformatics 9 (2017).
- Parker MA, Kurasch DM, Nichols DE. The role of lipophilicity in determining binding affinity and functional activity for 5-HT2A receptor ligands. Biorg Med Chem 16 (2008): 4661-4669.
- Fredriksson R, Lagerstrom MC, Lundin LG, et al. The G-protein-coupled-receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and finger Mol Pharmacol 63 (2003): 1256-1272.
- Hauser AS, Attwood MM, Rask-Andersen M, et al. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov 16 (2017): 829-842.
- Szakacs G, Váradi A, Ozvegy-Laczka C, et al. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox). Drug Discov Today 13 (2008): 379-393.
- Huang SM, Strong JM, Zhang L, et al.. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J Clin.Pharmacol 48 (2008): 662-670.
- Veith H, Southalla N, Huang R, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nature Biotechnol 27 (2009): 1050-1055.
- Mittra RA, Mieler WF. Chapter 89-Drug Toxicity of the Posterior Segment. Retina (fifth ed.) (2013): 1532-1554
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal friendliness of small molecules. Scientific Reports 7 (2017).
- Shoichet BK, Kobilka BK. Structure-based drug screening for G-protein coupled receptors. Trends Pharmacol Sci 33 (2012): 268-272.
- Niswender CM, Conn PJ. Mtebotropic glutamate receptors: physiology, pharmacology and disease. Annu Rev Pharmacol Toxicol 50 (2010): 295-322.
- Tian S, Wang J, Li Y, et al.. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev 86 (2015): 2-10.
- Hay M, Thomas DW, Craighead JL, et al. Clinical development success rates for investigational drugs. Nature Biotechnol 32 (2014): 40-51.
- Dahlin JL, Inglese J, Walters MA. Mitigating risk in academic preclinical drug discovery. Nature Rev Drug Discov 14 (2015): 279-294.
- Mishra NK, Agarwal S, Raghava GP. Prediction of cytochrome P450 isoform responsible for metabolizing a drug molecule. BMC Pharmacol 10 (2010): 8.



 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 