Accurate and Rapid Antigen Assay for COVID-19 Diagnostics Using Saliva
Article Information
Camille Troup1, Debnath Mukhopadhyay1, Tania Chakrabarty1, Anup Madan1, Sri Satyanarayana1, Vel Murugan2, and Su Dwarakanath1*
1Kaya17, Livermore CA, USA
2 Biodesign Institute, Phoenix AZ, USA
*Corresponding author: Su Dwarakanath, Kaya17, Livermore CA, USA.
Received: 15 February 2022; Accepted: 22 February 2022; Published: 07 March 2023
Citation: Camille Troup, Debnath Mukhopadhyay, Tania Chakrabarty, Anup Madan, Sri Satyanarayana, Vel Murugan and Su Dwarakanath. Development of an Accurate and Rapid Antigen Assay for COVID-19 Diagnostics Using Saliva. Archives of Clinical and Biomedical Research 7 (2023): 133-146.
View / Download Pdf Share at FacebookAbstract
The global outbreak of COVID-19 highlighted the need for rapid and accurate diagnostic testing to control the spread of this highly contagious disease [1-5]. Here, we describe the nCoVega COVID-19 antigen rapid test (~ 15min) that can detect the presence of the SARS-COV-2 virus particles from saliva sample on a portable device. The portable reader instrument, the Vega-200, has a small footprint and is designed for use at point of care settings. The test detects the fluorescence signal using wide-field illumination from antigen-antibody complexes captured on a special filter matrix ([6]. Results of this clinical evaluation of 183 subjects demonstrates that the nCoVega COVID-19 test performs at par with qRT-PCR tests [7] (gold standard) for both symptomatic and asymptomatic patients with a strong inverse correlation between RFU (relative fluorescence units) and Ct counts (from RT-PCR). The test has an analytical performance of 15.3 TCID50/mL, and 100% specificity for COVID-19 as compared to other human respiratory viruses, including other human coronaviruses. The working principle of this assay and test system can be used for developing other rapid, inexpensive antigen assays and it can offer an end-to-end, point-of-care solution to meet the continuous demand in tackling existing and emerging infectious diseases across the globe.
Keywords
Antigen; COVID-19; immuno-fluorescence; Point of care; SARS-COV-2; Saliva
Antigen articles; COVID-19 articles; immuno-fluorescence articles; Point of care articles; SARS-COV-2 articles; Saliva articles
Article Details
1. Introduction
The nCoVega COVID-19 Antigen test, developed by Kaya 17, Inc., is an antigen-capture assay intended for the qualitative detection of spike protein antigen from SARS-CoV-2 in saliva from individuals, as viral antigen is generally detectable in saliva during the acute phase of infection. Positive results indicate the presence of viral antigens. Our antigen test can be performed by trained non-laboratory or laboratory technicians for point of care use or for general use in clinical labs and hospital settings. The test works on a simple portable instrument called Vega-200 using a custom cartridge, assay reagents, and Vega software. The workflow uses a QR code reader and printer for tracking patient information. The vast majority of COVID-19 antigen tests utilize lateral flow technology, which has inherent limitations in terms of dynamic range, sensitivity and specificity [8-12]. The current rapid antigen tests are approved for nasal or NP swab. Other approaches include magnetic force assisted ELISA, microfluidic ELISA and digital chromatographic immunoassays, all of which require costly and complex custom manufacturing. The nCoVega test utilizes selective antigen-antibody complex capture via filtration and wide field fluorescence imaging on an inexpensive and easy to manufacture disposable cartridge, resulting in sensitivity and specificity comparable to the qRT-PCR based gold standard (ref). The developed tests therefore can be easily deployed for large-scale population testing under community health settings, hospitals, and at the point of care with minimal operator training. Our test, which is both rapid and inexpensive, can help with COVID-19 diagnosis and controlling the spread of the global pandemic. The test can also be used to support decisions on infection control strategies such as detecting and isolating asymptomatic cases that could spread the virus as we return to normal lifestyles. The novel nCoVega COVID19 Antigen test is a direct dual probe antigen capture test using two fluorescently labeled primary antibodies (S1 and S2 spike protein recombinant IgG) that bind to SARS-CoV-2 spike protein on viral particles present in a saliva sample.
The workflow for our end-to-end solution including the test and the system is shown below in figure 1.
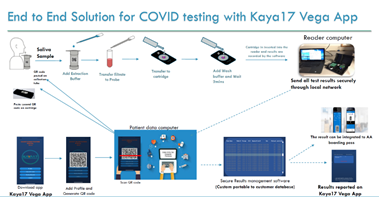
Figure 1: Schematic diagram showing the workflow for the nCoVega Test on the Vega-200.
2. Materials and Methods
Sample collection and processing: A saliva sample (100ul or more) is collected at the point of care testing by directly spitting into a vial that is placed in a syringe fitted with a filter. 1mL of the proprietary sample processing reagent is added. A plunger is pushed to allow the treated saliva sample to pass through the filter and is collected in a second tube for antibody labeling. The filtrate is incubated with 30ul of fluorescently-labeled S1 and S2 antibodies for 10 minutes at room temperature. The mixture is then passed through a disposable vertical flow cartridge fitted with a proprietary capture matrix. The fluorescence signal from the captured antigen-antibody complex is recorded using a portable device equipped with wide field illumination. Antibodies and conjugation: The S1 and S2 antibodies used in our test are purchased from GeneTex, TX and conjugated with a Biotium Mix-n-Stain CF 405L fluorophore (Biotium Inc, Fremont, CA, USA). The fluorophore used in our test has a large Stokes Shift with an excitation peak at 395nm and emission peak around 545 nm. S1 and S2 antibodies are conjugated using the Mix-n-Stain protocol following the manufacturer’s protocol, which consists of direct labeling of the antibodies to the fluorophore via an NHS-ester covalent bond formation. The conjugated antibodies are then purified via spin column centrifugation and buffer exchanged into a PBS-based storage buffer. The CF dyes were carefully chosen for their superior brightness, photostability, large Stokes Shift, and resistance to hydrolysis offering superior performance over most dyes. Instrument and data collection: The patient saliva sample labeled with fluorescent antibodies is placed on the cartridge in a well lined at the bottom with a capture matrix. An absorbent pad inside the cartridge holder to remove excess liquid and the unbound antibody. The bound antibody/antigen complex is selectively retained on the capture matrix via size exclusion mechanism. 500ul of wash buffer is added to the cartridge and allowed to wick through. Finally, 500ul of proprietary read buffer is added to the cartridge and the cartridge is immediately placed on the Vega-200 instrument for readout. The Vega-200 instrument records the fluorescence from the captured complex on the cartridge and reports a qualitative result (Positive or Negative). The system portable Vega-200 instrument is a fluorescence reader (U.S. Patent Application Serial Number: 16/690,589). The Vega analysis software, Patient ID software, an ID scanner with a QR code printer were all run on a standard PC. In addition, data reporting via a mobile device is also available.
2.1 Data Availability
Detailed data directly used to generate each figure or table of this study are available within the article, Supplementary Information and source data are also provided with this paper.
2.2 Method used to Determine LoD
Concentrations ranging from 1.6 x 105 TCID50/mL to 1.0 x 101 TCID50/mL Heat killed Zeptometrix SARS-CoV-2 (Cat. No. 0810587CFHI Lot No.324930) was spiked into artificial saliva matrix from Pickering laboratory (1700-0316). Limit of Detection (LoD) for Kaya17 nCoVega test was determined by performing serial dilution (Table 1). A 10-fold serial dilution of was performed from 10,000, 1,000, 100, 10 and 0 of Heat Killed SARS CoV 2 from Zeptometrix in triplicate in Artificial saliva matrix from Pickering laboratory (1700-0316) (Figure 2a). From this, the LoD was estimated as 15.3 TCID50 per ml by Probit analysis (Figure 2b). Raw data is included in the supplementary material section (Table 1, supplementary materials section).
|
ID |
Bgd_Rdg |
Thr_Rdg |
Raw_Avg |
Test_Results |
Sample_Name |
|
C1 |
-2.362 |
0.3 |
0.298 |
NEG |
NEG Control |
|
C2 |
-2.362 |
0.3 |
0.347 |
POS |
POS control |
|
S3 |
-2.362 |
0.3 |
0.337 |
POS |
20 |
|
S4 |
-2.362 |
0.3 |
0.333 |
POS |
20 |
|
S5 |
-2.362 |
0.3 |
0.336 |
POS |
20 |
|
S6 |
-2.362 |
0.3 |
0.338 |
POS |
20 |
|
S7 |
-2.362 |
0.3 |
0.331 |
POS |
20 |
|
S8 |
-2.362 |
0.3 |
0.334 |
POS |
20 |
|
S9 |
-2.362 |
0.3 |
0.333 |
POS |
20 |
|
S10 |
-2.362 |
0.3 |
0.335 |
POS |
20 |
|
S11 |
-2.362 |
0.3 |
0.334 |
POS |
20 |
|
S12 |
-2.362 |
0.3 |
0.337 |
POS |
20 |
|
S13 |
-2.362 |
0.3 |
0.338 |
POS |
20 |
|
S14 |
-2.362 |
0.3 |
0.333 |
POS |
20 |
|
S15 |
-2.362 |
0.3 |
0.338 |
POS |
20 |
|
S16 |
-2.362 |
0.3 |
0.336 |
POS |
20 |
|
S17 |
-2.362 |
0.3 |
0.335 |
POS |
20 |
|
S18 |
-2.362 |
0.3 |
0.336 |
POS |
20 |
|
S19 |
-2.362 |
0.3 |
0.331 |
POS |
20 |
|
S20 |
-2.362 |
0.3 |
0.332 |
POS |
20 |
|
S21 |
-2.362 |
0.3 |
0.331 |
POS |
20 |
|
S22 |
-2.362 |
0.3 |
0.335 |
POS |
20 |
|
S23 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S24 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S25 |
-2.362 |
0.3 |
0.299 |
NEG |
0 |
|
S26 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S27 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S28 |
-2.362 |
0.3 |
0.299 |
NEG |
0 |
|
S29 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S30 |
-2.362 |
0.3 |
0.298 |
NEG |
0 |
|
S31 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S32 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S33 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S34 |
-2.362 |
0.3 |
0.298 |
NEG |
0 |
|
S35 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S36 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S37 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S38 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S39 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S40 |
-2.362 |
0.3 |
0.297 |
NEG |
0 |
|
S41 |
-2.362 |
0.3 |
0.3 |
NEG |
0 |
|
S42 |
-2.362 |
0.3 |
0.299 |
NEG |
0 |
Table 1: LOD LOB testing raw data.
Operator: Arsh
ReaderID: R6
|
Sample |
N Gene |
IC |
ORF1ab |
PE RESULT |
Kaya17 (RFU) |
Kaya17 RESULT |
Operator |
|
NP-1 |
UND |
36.77 |
UND |
NOT DETECTED |
0.275 |
NEG |
1 |
|
NP-2 |
30.05 |
33.47 |
30.07 |
DETECTED |
0.392 |
POS |
|
|
NP-3 |
UND |
33.84 |
UND |
NOT DETECTED |
0.266 |
NEG |
|
|
NP-4 |
UND |
32.56 |
UND |
NOT DETECTED |
0.268 |
NEG |
|
|
NP-5 |
UND |
33.47 |
UND |
NOT DETECTED |
0.289 |
NEG |
|
|
NP-6 |
UND |
34.90 |
UND |
NOT DETECTED |
0.292 |
NEG |
|
|
NP-7 |
UND |
33.59 |
UND |
NOT DETECTED |
0.284 |
NEG |
|
|
NP-8 |
22.87 |
33.53 |
22.67 |
DETECTED |
0.64 |
POS |
|
|
NP-9 |
UND |
33.64 |
UND |
NOT DETECTED |
0.281 |
NEG |
|
|
NP-10 |
30.59 |
35.99 |
31.96 |
DETECTED |
0.293 |
NEG |
|
|
NP-11 |
31.49 |
33.79 |
UND |
DETECTED |
0.385 |
POS |
|
|
NP-12 |
33.10 |
33.56 |
35.57 |
DETECTED |
0.382 |
POS |
|
|
NP-13 |
24.81 |
33.01 |
25.53 |
DETECTED |
0.562 |
POS |
|
|
NP-14 |
29.64 |
34.02 |
29.71 |
DETECTED |
0.396 |
POS |
|
|
NP-15 |
19.83 |
32.21 |
20.71 |
DETECTED |
0.767 |
POS |
|
|
NP-16 |
UND |
31.14 |
UND |
NOT DETECTED |
0.279 |
NEG |
|
|
NP-17 |
UND |
33.67 |
UND |
NOT DETECTED |
0.284 |
NEG |
|
|
NP-18 |
UND |
33.15 |
UND |
NOT DETECTED |
0.278 |
NEG |
|
|
NP-19 |
27.92 |
32.49 |
23.68 |
DETECTED |
0.446 |
POS |
|
|
NP-20 |
UND |
33.75 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-21 |
32.94 |
33.16 |
34.98 |
DETECTED |
0.383 |
POS |
|
|
NP-22 |
UND |
32.56 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-23 |
17.86 |
33.58 |
17.55 |
DETECTED |
0.845 |
POS |
|
|
NP-24 |
UND |
33.63 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-25 |
UND |
32.89 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-26 |
UND |
34.45 |
UND |
NOT DETECTED |
0.276 |
NEG |
|
|
NP-27 |
27.42 |
33.36 |
26.99 |
DETECTED |
0.448 |
POS |
|
|
NP-28 |
UND |
33.31 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-29 |
32.99 |
32.23 |
34.33 |
DETECTED |
0.388 |
POS |
|
|
NP-30 |
19.55 |
33.49 |
19.75 |
DETECTED |
0.763 |
POS |
|
|
NP-31 |
UND |
31.48 |
UND |
NOT DETECTED |
0.323 |
POS |
|
|
NP-32 |
UND |
32.89 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-33 |
UND |
33.02 |
UND |
NOT DETECTED |
0.278 |
NEG |
|
|
NP-34 |
28.52 |
33.94 |
28.70 |
DETECTED |
0.38 |
POS |
|
|
NP-35 |
33.95 |
33.58 |
34.86 |
DETECTED |
0.376 |
POS |
|
|
NP-36 |
UND |
34.46 |
UND |
NOT DETECTED |
0.278 |
NEG |
2 |
|
NP-37 |
UND |
33.92 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-38 |
UND |
32.53 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-39 |
28.82 |
35.67 |
28.30 |
DETECTED |
0.401 |
POS |
|
|
NP-40 |
UND |
36.41 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-41 |
UND |
33.23 |
UND |
NOT DETECTED |
0.298 |
NEG |
|
|
NP-42 |
UND |
32.34 |
UND |
NOT DETECTED |
0.272 |
NEG |
|
|
NP-43 |
UND |
33.63 |
UND |
NOT DETECTED |
0.284 |
NEG |
|
|
NP-44 |
UND |
32.22 |
UND |
NOT DETECTED |
0.281 |
NEG |
|
|
NP-45 |
UND |
34.12 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-46 |
UND |
33.41 |
UND |
NOT DETECTED |
0.285 |
NEG |
|
|
NP-47 |
UND |
33.30 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-48 |
UND |
33.20 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-49 |
UND |
33.46 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-50 |
UND |
34.85 |
UND |
NOT DETECTED |
0.281 |
NEG |
|
|
NP-51 |
UND |
33.56 |
UND |
NOT DETECTED |
0.283 |
NEG |
|
|
NP-52 |
UND |
32.66 |
UND |
NOT DETECTED |
0.289 |
NEG |
|
|
NP-53 |
UND |
33.96 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-54 |
34.32 |
32.80 |
34.16 |
DETECTED |
0.375 |
POS |
|
|
NP-55 |
UND |
33.93 |
UND |
NOT DETECTED |
0.279 |
NEG |
|
|
NP-56 |
35.10 |
32.53 |
35.91 |
DETECTED |
0.379 |
POS |
|
|
NP-57 |
18.76 |
33.67 |
19.82 |
DETECTED |
0.817 |
POS |
|
|
NP-58 |
34.40 |
32.93 |
33.86 |
DETECTED |
0.387 |
POS |
|
|
NP-59 |
UND |
32.79 |
UND |
NOT DETECTED |
0.286 |
NEG |
|
|
NP-60 |
UND |
34.54 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-61 |
UND |
33.64 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-62 |
UND |
35.73 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-63 |
UND |
33.16 |
UND |
NOT DETECTED |
0.291 |
NEG |
|
|
NP-64 |
UND |
35.99 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-65 |
UND |
32.35 |
UND |
NOT DETECTED |
0.279 |
NEG |
|
|
NP-66 |
UND |
33.90 |
UND |
NOT DETECTED |
0.289 |
NEG |
3 |
|
NP-67 |
UND |
34.65 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-68 |
23.55 |
33.55 |
24.67 |
DETECTED |
0.675 |
POS |
|
|
NP-69 |
34.74 |
32.82 |
35.92 |
DETECTED |
0.377 |
POS |
|
|
NP-70 |
25.66 |
33.78 |
UND |
DETECTED |
0.555 |
POS |
|
|
NP-71 |
26.11 |
33.00 |
28.00 |
DETECTED |
0.479 |
POS |
|
|
NP-72 |
UND |
34.75 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-73 |
UND |
35.03 |
UND |
NOT DETECTED |
0.284 |
NEG |
|
|
NP-74 |
35.16 |
33.87 |
36.84 |
DETECTED |
0.368 |
POS |
|
|
NP-75 |
35.30 |
33.85 |
33.48 |
DETECTED |
0.379 |
POS |
|
|
NP-76 |
UND |
35.00 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-77 |
UND |
34.78 |
UND |
NOT DETECTED |
0.292 |
NEG |
|
|
NP-78 |
UND |
33.98 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-79 |
UND |
35.67 |
UND |
NOT DETECTED |
0.289 |
NEG |
|
|
NP-80 |
UND |
34.79 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-81 |
39.23 |
33.18 |
UND |
DETECTED |
0.329 |
POS |
|
|
NP-82 |
35.63 |
33.12 |
34.82 |
DETECTED |
0.366 |
POS |
|
|
NP-83 |
UND |
34.63 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-84 |
UND |
34.56 |
UND |
NOT DETECTED |
0.276 |
NEG |
|
|
NP-85 |
UND |
33.78 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-86 |
18.87 |
34.01 |
19.05 |
DETECTED |
0.842 |
POS |
|
|
NP-87 |
25.12 |
33.43 |
24.93 |
DETECTED |
0.528 |
POS |
|
|
NP-88 |
26.76 |
33.72 |
26.40 |
DETECTED |
0.492 |
POS |
|
|
NP-89 |
UND |
32.91 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-90 |
UND |
33.54 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-91 |
33.74 |
31.96 |
34.55 |
DETECTED |
0.39 |
POS |
|
|
NP-92 |
28.52 |
33.60 |
29.88 |
DETECTED |
0.421 |
POS |
|
|
NP-93 |
35.47 |
33.66 |
35.45 |
DETECTED |
0.376 |
POS |
|
|
NP-94 |
22.45 |
33.41 |
21.77 |
DETECTED |
0.674 |
POS |
|
|
NP-95 |
UND |
33.03 |
UND |
NOT DETECTED |
0.286 |
NEG |
|
|
NP-96 |
UND |
32.91 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-97 |
UND |
35.30 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-98 |
UND |
33.84 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-99 |
UND |
34.77 |
UND |
NOT DETECTED |
0.293 |
NEG |
4 |
|
NP-100 |
34.33 |
32.07 |
34.50 |
DETECTED |
0.374 |
POS |
|
|
NP-101 |
UND |
32.10 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-102 |
UND |
32.21 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-103 |
UND |
32.42 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-104 |
34.46 |
32.29 |
34.85 |
DETECTED |
0.371 |
POS |
|
|
NP-105 |
37.67 |
32.55 |
38.11 |
DETECTED |
0.368 |
POS |
|
|
NP-106 |
UND |
31.77 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-107 |
UND |
34.54 |
UND |
NOT DETECTED |
0.279 |
NEG |
|
|
NP-108 |
UND |
33.32 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-109 |
UND |
33.65 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-110 |
35.98 |
32.73 |
36.01 |
DETECTED |
0.366 |
POS |
|
|
NP-111 |
34.66 |
33.21 |
34.97 |
DETECTED |
0.37 |
POS |
|
|
NP-112 |
26.43 |
34.88 |
27.98 |
DETECTED |
0.487 |
POS |
|
|
NP-113 |
30.12 |
31.44 |
UND |
DETECTED |
0.396 |
POS |
|
|
NP-114 |
UND |
32.28 |
UND |
NOT DETECTED |
0.285 |
NEG |
|
|
NP-115 |
UND |
32.69 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-116 |
UND |
34.55 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-117 |
UND |
33.99 |
UND |
NOT DETECTED |
0.329 |
POS |
|
|
NP-118 |
UND |
33.24 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-119 |
UND |
33.40 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-120 |
UND |
32.59 |
UND |
NOT DETECTED |
0.289 |
NEG |
|
|
NP-121 |
31.54 |
32.73 |
30.98 |
DETECTED |
0.387 |
POS |
|
|
NP-122 |
33.39 |
33.11 |
33.28 |
DETECTED |
0.379 |
POS |
|
|
NP-123 |
UND |
36.46 |
UND |
NOT DETECTED |
0.297 |
NEG |
|
|
NP-124 |
22.85 |
33.26 |
22.37 |
DETECTED |
0.646 |
POS |
|
|
NP-125 |
UND |
33.55 |
UND |
NOT DETECTED |
0.288 |
NEG |
|
|
NP-126 |
UND |
31.67 |
UND |
NOT DETECTED |
0.285 |
NEG |
|
|
NP-127 |
UND |
34.21 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-128 |
UND |
33.44 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-129 |
UND |
32.45 |
UND |
NOT DETECTED |
0.292 |
NEG |
|
|
NP-130 |
14.73 |
32.65 |
15.10 |
DETECTED |
1.272 |
POS |
|
|
NP-131 |
21.58 |
33.32 |
21.99 |
DETECTED |
0.683 |
POS |
|
|
NP-132 |
33.97 |
33.48 |
35.56 |
DETECTED |
0.384 |
POS |
|
|
NP-133 |
35.25 |
33.77 |
36.95 |
DETECTED |
0.368 |
POS |
|
|
NP-134 |
32.89 |
34.25 |
34.22 |
DETECTED |
0.381 |
POS |
|
|
NP-135 |
35.33 |
33.92 |
35.02 |
DETECTED |
0.374 |
POS |
|
|
NP-136 |
37.74 |
34.67 |
38.08 |
DETECTED |
0.361 |
POS |
5 |
|
NP-137 |
15.76 |
32.69 |
15.59 |
DETECTED |
0.921 |
POS |
|
|
NP-138 |
UND |
32.29 |
UND |
NOT DETECTED |
0.285 |
NEG |
|
|
NP-139 |
UND |
31.90 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-140 |
UND |
32.65 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-141 |
UND |
32.98 |
UND |
NOT DETECTED |
0.297 |
NEG |
|
|
NP-142 |
35.17 |
32.28 |
34.66 |
DETECTED |
0.327 |
POS |
|
|
NP-143 |
UND |
33.58 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-144 |
33.75 |
33.88 |
33.21 |
DETECTED |
0.384 |
POS |
|
|
NP-145 |
33.76 |
32.55 |
34.86 |
DETECTED |
0.388 |
pos |
|
|
NP-146 |
29.58 |
33.75 |
29.57 |
DETECTED |
0.397 |
POS |
|
|
NP-147 |
21.65 |
33.45 |
21.50 |
DETECTED |
0.683 |
POS |
|
|
NP-148 |
39.00 |
32.71 |
37.06 |
DETECTED |
0.328 |
POS |
|
|
NP-149 |
UND |
31.90 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-150 |
34.79 |
32.56 |
36.85 |
DETECTED |
0.374 |
POS |
|
|
NP-151 |
35.02 |
32.32 |
34.62 |
DETECTED |
0.377 |
POS |
|
|
NP-152 |
UND |
34.56 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-153 |
UND |
33.67 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-154 |
UND |
33.84 |
UND |
NOT DETECTED |
0.287 |
NEG |
|
|
NP-155 |
35.22 |
32.18 |
36.37 |
DETECTED |
0.374 |
POS |
|
|
NP-156 |
34.82 |
33.00 |
34.67 |
DETECTED |
0.375 |
POS |
|
|
NP-157 |
34.48 |
32.91 |
34.44 |
DETECTED |
0.372 |
POS |
|
|
NP-158 |
UND |
33.10 |
UND |
NOT DETECTED |
0.296 |
NEG |
|
|
NP-159 |
UND |
33.87 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-160 |
UND |
33.04 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-161 |
UND |
32.29 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-162 |
UND |
33.46 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-163 |
UND |
33.85 |
UND |
NOT DETECTED |
0.297 |
NEG |
|
|
NP-164 |
UND |
34.06 |
UND |
NOT DETECTED |
0.294 |
NEG |
|
|
NP-165 |
UND |
31.54 |
UND |
NOT DETECTED |
0.295 |
NEG |
|
|
NP-166 |
36.03 |
33.01 |
34.70 |
DETECTED |
0.365 |
POS |
|
|
NP-167 |
UND |
30.97 |
38.06 |
DETECTED |
0.328 |
POS |
|
|
NP-168 |
UND |
33.15 |
38.87 |
DETECTED |
0.325 |
POS |
|
|
NP-169 |
35.31 |
32.80 |
35.16 |
DETECTED |
0.376 |
POS |
|
|
NP-170 |
37.82 |
33.47 |
38.92 |
DETECTED |
0.365 |
POS |
|
|
NP-171 |
30.79 |
34.47 |
29.69 |
DETECTED |
0.392 |
POS |
|
|
NP-172 |
31.62 |
33.25 |
32.08 |
DETECTED |
0.383 |
POS |
|
|
NP-173 |
UND |
36.19 |
UND |
NOT DETECTED |
0.297 |
NEG |
|
|
NP-174 |
26.75 |
34.21 |
26.73 |
DETECTED |
0.482 |
POS |
|
|
NP-175 |
27.52 |
33.68 |
27.59 |
DETECTED |
0.447 |
POS |
|
|
NP-176 |
29.30 |
34.07 |
29.27 |
DETECTED |
0.398 |
POS |
|
|
NP-177 |
UND |
33.13 |
UND |
NOT DETECTED |
0.293 |
NEG |
|
|
NP-178 |
33.56 |
33.41 |
36.53 |
DETECTED |
0.38 |
POS |
|
|
NP-179 |
35.30 |
32.72 |
35.66 |
DETECTED |
0.372 |
POS |
|
|
NP-180 |
34.86 |
33.66 |
36.34 |
DETECTED |
0.369 |
POS |
|
|
NP-181 |
35.80 |
33.63 |
37.02 |
DETECTED |
0.364 |
POS |
|
|
NP-182 |
UND |
34.56 |
UND |
NOT DETECTED |
0.292 |
NEG |
|
|
NP-183 |
UND |
33.55 |
UND |
NOT DETECTED |
0.297 |
NEG |
Table 2: Test Results for PE Nucleic Acid Detection Kit and Kaya17 nCoVega Test.
2.3 Cross Reactivity (Analytical Specificity)
Each organism was tested in the absence or presence of Heat Killed SARS-CoV-2 from Zeptometrix at 3 x LoD in Artificial saliva matrix from Pickering laboratory (1700-0316). The final concentration of the organisms is in the Table 3 below (the concentrations of 106 CFU/mL or higher for bacteria and 105 PFU/mL or higher for viruses is recommended). For some of the microorganisms, the stock concentration was lower than or equal to the recommended testing concentration. In such cases, these microorganisms were used at the stock concentration.
2.4 Microbial Interference Studies
Each organism was tested in the absence or presence of Heat Killed SARS-CoV-2 from Zeptometrix at 3 x LoD in Artificial saliva matrix from Pickering laboratory (1700-0316). The final concentration of the organisms is in the Table 4 below (the concentrations of 106 CFU/mL or higher for bacteria and 105 PFU/mL or higher for viruses is recommended). For some of the microorganisms, the stock concentration was lower than or equal to the recommended testing concentration. In such cases, these microorganisms were used at the stock concentration.
2.5 Endogenous Interference Studies
Each substance was tested in triplicate in the absence or presence of Heat Killed SARS-CoV-2 from Zeptometrix at 3 x LOD in Artificial saliva matrix from Pickering laboratory (1700-0316). Substances for testing were selected based on the respiratory specimen’s guidance in http://www.accessdata.fda.gov/cdrh_docs/reviews/K112177.pdf.
2.6 Hook Effect Testing
To determine if the Kaya17 nCoVega Test suffers from any high dose Hook effect, increasing concentrations of Heat Killed SARS-CoV-2 from Zeptometrix at 3 x LoD, were tested up to a concentration of 1.6 x 105 TCID50/mL. The starting material was spiked into a pooled negative saliva matrix obtained from healthy donors and confirmed negative for SARS-CoV-2. At each dilution, 500μL samples were added to Extraction buffer and tested using the nCoVega test.
2.7 Clinical Evaluation
A prospective, single center, comparative study for validation of a rapid point-of-care test for diagnosis of SARS-COV-2 infection was conducted to evaluate the clinical use of the Kaya17 nCoVega COVID-19 Antigen test for use with saliva specimens collected neat. The Kaya17 nCoVega COVID-19 Antigen test was clinically validated by Infinity BiologiX (IBX) using 183 paired saliva and swab samples (79 positives and 104 negatives) collected by St. Rose Hospital, Hayward, CA as per KAYA-PROTOPOCT-007 Prospective Clinical Study Protocol. The Western IRB approval number for the clinical trial was IRB protocol #20204097. Informed consents were collected from each subject along with basic medical history. The samples utilized in the study were analyzed using an RT PCR assay (Control), PerkinElmer New Coronavirus Nucleic Acid Detection Kit. The primary objective of the study was to demonstrate efficacy of the Kaya17 nCoVega COVID-19 Antigen test in a non-laboratory setting by verifying the sensitivity and specificity of the Kaya17 nCoVega COVID-19 Antigen test in detecting SARS-COV-2 in positive saliva samples (yields a “positive” result) and not detecting SARS-COV-2 in negative saliva samples (yields a “negative” result) when performed by non-laboratory personnel. The secondary objective in the same protocol was to demonstrate that non-laboratory healthcare providers can perform the Kaya17 nCoVega COVID-19 Antigen test accurately in the intended use environment to Support Point of Care (POC) Use. (Point of Care study)
The data generated from the clinical study were statistically analyzed as per the following techniques
- Positive percent agreement (PPA),
- Negative percent agreement (NPA),
- Overall percent agreement (OPA),
- Associated 95% Wilson score confidence intervals
- Concordance analysis of the Kaya17 nCoVega Antigen test RFU vs Ct values of PCR
Subject status (negative/positive) was determined using a receiver operating characteristic (ROC) curve cutoff analysis. The evaluation metrics included positive percent agreement (PPA), negative percent agreement (NPA), overall percent agreement (OPA), Youden index, distance to (0,1), and the absolute value of the sensitivity minus the specificity. Before testing any of the samples, sample cartridges with positive and negative controls were inserted on the Vega 200 instrument that recorded fluorescence from these samples in RFU. This allowed the Vega analysis software to decide the cut-off value to use when deciding on positive and negative COVID-19 calls for sample testing. Details of the result interpretation are cited below in their respective sections. Raw data for the plots are also included in the supplementary information section. All test controls were examined prior to interpretation of results. If the controls were not valid, the samples were not analyzed using the kit/instrument. Additional kits or instruments are required to be validated prior to running samples. The test result was determined by cutoff values based on fluorescence measurements on the Vega-200 instrument (excitation at 395 nm and emission at 545 nm). A “POSITIVE” result is called by the Vega software if the fluorescence measurement of the sample is above baseline fluorescence that has been determined by running the negative control multiple times. If the fluorescence value is at or below baseline, a “NEGATIVE” result is called by the Vega software. The results are stored in a CSV file on the Vega Reader computer. Further details are provided in the supplementary section.
3. Results
For laboratory validation of the nCoVega COVID19 Antigen test, a number of different studies were conducted such as analytical performance testing, LOD, cross-reactivity, inclusion and exclusion panel testing and hook effect testing. The details of each of those tests served to establish the sensitivity and specificity of the assay and the results are presented below.
3.1 Analytical Performance
3.1.1 Limit of Detection (LoD) Determination: LoD studies were performed to determine the lowest detectable concentration of SARS-CoV-2 at which approximately 95% of all (true positive) replicates test positive.
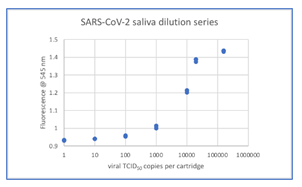
Figure 2 (a): LoD determination and dynamic range based on data from Table 1 (see supplementary section).
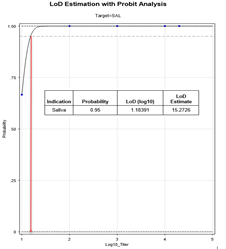
Figure 2 (b): Probit Analysis to determine LoD using data from Table 1.
From the ten-fold dilution series results in Table 1, the LoD can be determined as at least 0.95 (100 TCID50/mL) since all samples with concentrations from 100000 TCID50/mL down to 100 TCID50/mL were positive based on the 0.94 cutoff. The ability to detect SARS-CoV-2 with Kaya17 nCoVega test at 20 TCID50 per ml was further confirmed by testing 20 replicates (Table 1 in supplementary section). Limit of Blank (LoB) experiments were performed by running 20 samples at 0 TCID50 per ml LOB (Table 1 in supplementary section).
3.1.2 Assay Cutoff Determination: Determining the assay cutoff was a key part of the data analysis. The assay cutoff determination for positives and negatives is determined using the Currie’s method11 and CLSI EP17 guidance12 for our qualitative purposes as follows.
From the guidance, the LoB’ is defined as:
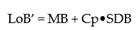
where M refers to the mean of “blank” samples and Cp is the multiplier and SDB is the Standard Deviation of the Blank Samples. As the data are very precise, in this case using a two-sided 5% significance level, Cp is defined as
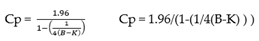
where B is the total number of blank replicates and K is the number of blank samples.
Therefore, from the data in the spreadsheet (see supplementary section), the LOB’ is calculated as:

and SDL = 0.00266 is the standard deviation of the positive low concentration samples with appropriate provision for degrees of freedom depending on the number of samples used to obtain the total replicates. Therefore, if the LoB’ is taken as the cutoff (11), then we can establish a conservative equivocal zone between the LoB’ and the LoD’ as between 0.936 and 0.942 mean RR with the actual cutoff as 0.939 (as determined by ROC curve methodology in clinical evaluation (12) ). Therefore, if a patient were to test in this equivocal zone, their result should be assigned as INDETERMINATE and a retest is performed.
3.1.3 Cross-Reactivity (Analytical Specificity): We performed cross-reactivity studies for Kaya17 nCoVega test using a panel of related pathogens, normal or pathogenic flora that are reasonably found in the clinical specimen. Also, the high prevalence disease agents likely to be encountered in the clinical specimen were tested for specificity of the test and the results showed no potential cross-reactivity with the Kaya17 nCoVega test with these agents, including various microorganisms and pathogens.
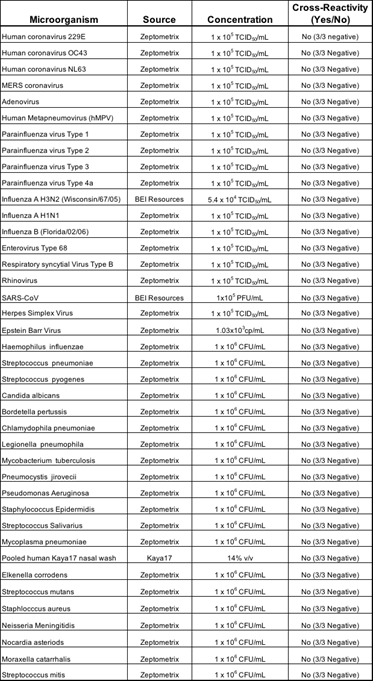
Table 3: Results of Cross-Reactivity Study
3.1.4 Microbial Interference Studies: Microbial Interference for Kaya17 nCoVega test was evaluated by using a panel of related pathogens, normal or pathogenic flora that are reasonably found in the clinical specimen. Also, high prevalence disease agents were tested for specificity to demonstrate that false negatives do not occur when SARS-CoV-2 is present in a specimen with other microorganisms.
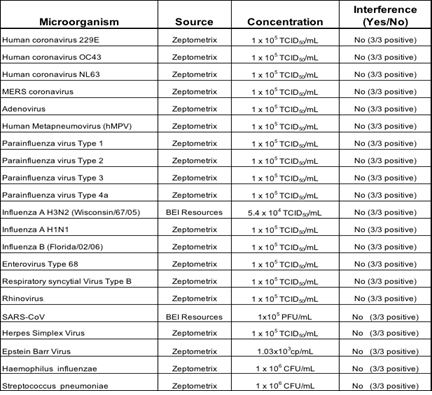
Table 4: Results of Microbial Interference studies
3.1.5 Endogenous Interference Substances Studies: The following study was conducted to investigate whether potentially interfering substances, which may be found in the mouth and throat of symptomatic subjects (including over-the-counter medications), cross-react or interfere with the detection of SARS-CoV-2 using the Kaya17 nCoVega test. There was no interference with any of the tested materials in the study as described in Table 5.
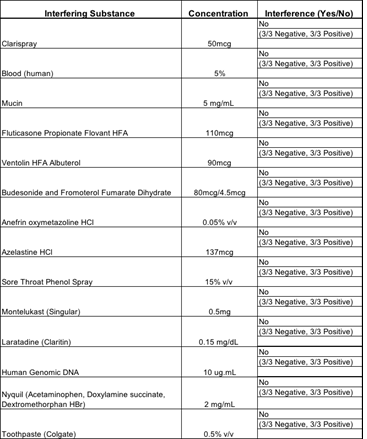
Table 5: Results of Interference Studies.
3.2 High-dose Hook Effect:
The Weibull statistical model was used for the data analysis (x axis=log10 TCID50 copies; y axis =Raw Average). The analysis showed that at the top end of the assay (1.6 x 105 TCID50 copies) there is a plateau. This provides evidence that a Hook effect does not exist for this assay up to 1.6 x 105 TCID50 copies. The model fits, as measured by the correlation and coefficient of determination, were each about 0.99 (closer to 1.00 is desired). Specific model information is shown in the figure 3.
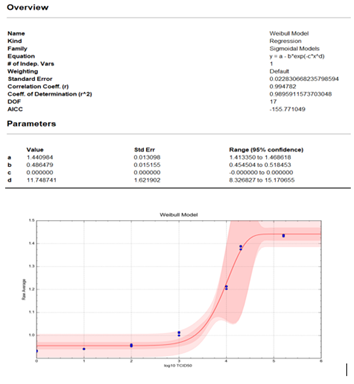
Figure 3: Weibull statistical analysis of Hook effect data.
3.3 Clinical Evaluation
A total of n=183 samples were collected during the clinical evaluation. A total of n=183 test samples utilizing the subject device resulted in the same result (i.e., positive or negative) in comparison to the control, except two (n=2) test sample rendered a positive result when using the subject device and a negative result when using the control. One (n=1) test samples rendered a negative result when using the subject device and a positive result when using the control. After conducting statistical analysis, a 98.4% degree of accuracy is established based on the data set (Table 6).
3.3.1 Accuracy in Overall Patient Population: The test results include 79 True Positives (RT-PCR Positives) and 104 True negatives (RT-PCR Negatives). When we analyzed the performance of the Kaya17 assay in overall population, the PPA value is calculated to be 98.7% and at 95% Confidence Interval, values are found to be at (93.2% Lower bound, 99.8% Upper bound).
|
PCR Reference Result |
Agreement |
||||||
|
Comparator Assay |
Test Result |
Positive |
Negative |
Total |
Measure [a] |
% (n/N) |
95% CI [b] |
|
Kaya17 (Overall) |
Positive |
78 |
2 |
80 |
PPA |
98.7 (78/79) |
(93.2, 99.8) |
|
Negative |
1 |
102 |
103 |
NPA |
98.1 (102/104) |
(93.3, 99.5) |
|
|
Total |
79 |
104 |
183 |
OPA |
98.4 (180/183) |
(95.3, 99.4) |
|
Table 6: Accuracy in Overall Patient Population.
[a] PPA=Positive Percent Agreement, NPA=Negative Percent Agreement, OPA=Overall Percent Agreement.
[b] Wilson Score Confidence Interval.
3.3.2 Accuracy in Symptomatic Patient Population: In the Symptomatic population (Table 7), there are 73 RT-PCR positives (True Positives) and 45 RT-PCR negatives (True Negatives). When we analyzed the performance of the Kaya17 assay in symptomatic population, the PPA value is calculated to be 98.6% and at 95% Confidence Interval, values are found to be at (92.60% Lower bound, 99.8% Upper bound). It meets the FDA's requirement for symptomatic population that positive percent agreement (PPA) of at least 80% with 70% at the lower bound of the two-sided 95% confidence interval, in symptomatic patients suspected of COVID-19 infection by their healthcare providers.
|
PCR Reference Result |
Agreement |
||||||
|
Comparator Assay |
Test Result |
Positive |
Negative |
Total |
Measure [a] |
% (n/N) |
95% CI [b] |
|
Kaya17 (Symptomatic) |
Positive |
72 |
1 |
73 |
PPA |
98.6 (72/73) |
(92.6, 99.8) |
|
Negative |
1 |
44 |
45 |
NPA |
97.8 (44/45) |
(88.4, 99.6) |
|
|
Total |
73 |
45 |
118 |
OPA |
98.3 (116/118) |
(94.0, 99.5) |
|
Table 7: Accuracy in Symptomatic Patient Population.
[a] PPA=Positive Percent Agreement, NPA=Negative Percent Agreement, OPA=Overall Percent Agreement.
[b] Wilson Score Confidence Interval.
3.3.3 Accuracy in Asymptomatic Patient Population: In the Asymptomatic population (Table 8), there are 6 RT-PCR positives (True Positives) and 59 RT-PCR negatives (True Negatives). When we analyzed the performance of the Kaya17 assay in asymptomatic population, the PPA value is calculated to be 100% and at 95% Confidence Interval, values are found to be at (61% Lower bound, 100% Upper bound).
|
PCR Reference Result |
Agreement |
|||||||
|
Comparator Assay |
Test Result |
Positive |
Negative |
Total |
Measure [a] |
% (n/N) |
95% CI [b] |
|
|
Kaya17 (Asymptomatic) |
Positive |
6 |
1 |
7 |
PPA |
100 (6/6) |
(61.0, 100.0) |
|
|
Negative |
0 |
58 |
58 |
NPA |
98.3 (58/59) |
(91.0, 99.7) |
||
|
Total |
6 |
59 |
65 |
OPA |
98.5 (64/65) |
(91.8, 99.7) |
||
Table 8: Accuracy in Asymptomatic Patient Population.
[a] PPA=Positive Percent Agreement, NPA=Negative Percent Agreement, OPA=Overall Percent Agreement.
[b] Wilson Score Confidence Interval.
Out of 183 clinical study samples (Table 9), 34 of them are N gene low positives and 31 of them are ORF low positives (calculated from the results of comparator test. i.e., PerkinElmer (PE) New Coronavirus Nucleic Acid Detection Kit RT-PCR). The PE RT-PCR LoD for N gene is 35.8 and the LoD for ORF is 37 as per the IFU of PE Nucleic Acid Detection Kit (provided as Annexure A to clinical protocol). Hence, we considered the Ct value range of 32.8 – 35.8 for N gene and the range of 34 – 37 for ORF to calculate the percentage of low positives from the overall clinical samples as recommended by FDA (Section J10 of FDA Template for Developers of Antigen Tests under EUA dated 6th October 2021) that low positives are defined as samples in which any gene target is within 3 cycle thresholds (Ct’s) of the mean Ct count at the comparator test’s LoD. The low positive values from the RT-PCR test results are highlighted in blue color in Table 7. The percentage of low positive samples are determined to be 18.58 % for N gene & 16.94% for ORF of the total sample size (183) which meets the FDA requirement of having approximately 10-20% of the clinical samples should be low positives in the clinical samples.
|
Total |
N Gene Low Positives (Ct Value range: 32.8 - 35.8) |
ORF Low Positives (Ct Value range: 34 - 37) |
|
34 |
31 |
|
|
Percentage w.r.t Overall Sample size (183) |
18.58% |
16.94% |
Table 9: Low Positives in RT-PCR Results.
The operator specific accuracy is measured to support the POC Study to demonstrate that the Kaya17 nCoVega COVID-19 Antigen test can be performed by the untrained operator following the Quick Reference Guide without any training. Each untrained operator has achieved the 95% accuracy. Please refer to the Table 10.
|
S.No |
Operator |
Total No. Samples Tested |
Correctly measured w.r.t Control |
Accuracy (%) |
|
1 |
Operator 1 |
41 |
39 |
95.1 |
|
2 |
Operator 2 |
36 |
36 |
100 |
|
3 |
Operator 3 |
39 |
39 |
100 |
|
4 |
Operator 4 |
43 |
42 |
97.7 |
|
5 |
Operator 5 |
54 |
54 |
100 |
Table 10: It is also observed that there is no reader instrument variability while interpreting the results when the samples are tested. All the RT-PCR comparative results correlate with the Kaya17 results when 5 different reader instruments were used for testing by 5 different untrained operators.
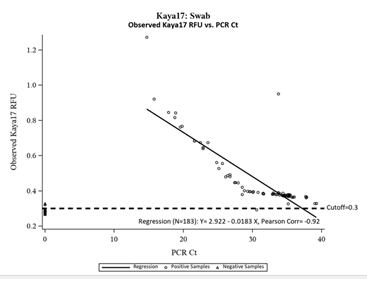
Figure 4: Inverse Relationship between Kaya17 RFUs vs Ct values from RT-PCR.
Figure 4 shows the inverse linear relationship between the observed Kaya17 RFUs versus the mean values of the N and ORF1ab Gene cycle thresholds (n=183, Pearson Correlation= -0.92). The figure shows that all of the negative samples by PCR were also Negative in Kaya17 nCoVega test, except for two samples (Sample Number 31and 117), all positive samples by PCR were also positive by Kaya17 nCoVega test, except for one sample (Sample Number 10).
4. Discussion
The Kaya17 Vega-200 system and nCoVega antigen test is a simple-to-use, rapid assay that is also highly sensitive and specific. The system can process 30-50 samples in one hour with minimal hands-on time. It can also detect very low levels of SARS-CoV-2 as seen in the high correlation with RT-PCR across all ranges of viral loads, all the way down to Ct counts of 38. The reader instrument is light, portable and runs off USB-power from its connected laptop, so it does not need external power source. Our end-to-end solution can be deployed in mobile units, at schools, offices and events and also in remote areas, with its small footprint. Easily scalable manufacturing processes llow this test to meet any surge in demand for COVID-19 testing as we open our schools, businesses and global travel and new variants emerge. In the future, this test system can be productized for self-testing purposes and for remote monitoring via a mobile app. The nCoVega test can address the unmet need for rapid, accurate and inexpensive COVID-19 testing and it is suitable for broader public dissemination.
Author Contributions
SD and VM conceived the study and cross-platform validation experiments at ASU. SS led the design and development of the Vega-200 system. SD, CBT, TC, AM, DM developed and optimized the nCoVega SARS-CoV-2 assay. DM and SD ran the analytical study. SD, CBT ran the clinical validation study at ASU. JC, JM and CBT analyzed the data. TC, SM and SD drafted the manuscript. CBT revised the manuscript. All authors read and approved the final manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Western IRB (IRB protocol #20204097; Kaya17 protocol #KAYA-PROTOPOCT-007 Rev002) on 01/06/2021.
Supplementary Information
Download the supplementary information from https://www.fortunejournals.com/supply/ACBR_7871.pdf
References
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17 (2019): 181-192.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (2020): 270-273.
- Contini C, Di Nuzzo M, Barp N, et al. The novel zoonotic COVID-19 pandemic: An expected global health concern. J Infect Dev Ctries 14 (2020): 254-264.
- Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 26 (2020): 506-510.
- Kissler SM, Tedijanto C, Goldstein E, et al. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science (2020): eabb5793.
- Dwarakanath S, Satyanarayana S, John Bruno J, et al, Ultrasensitive Fluorescent Nanoparticle-Based Binding Assays for Foodborne and Waterborne Pathogens of Clinical Interest. J. of Clinical Ligand Assay, Fall 29 (2006): 3.
- Altamimi, AM, Obeid, DA, Alaifan, TA, et al. Assessment of 12 Qualitative RT-PCR commercial kits for the detection of SARS-CoV-2. Journal of medical virology (2021).
- Eshghifar N, Busheri A, Shrestha R, et al. Evaluation of Analytical Performance of Seven Rapid Antigen Detection Kits for Detection of SARS-CoV-2 Virus. International journal of general medicine 14 (2021): 435-440.
- Mak GC, Lau SS, Wong KK, et al. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J Clin Virol (2020).
- Kohmer N, Toptan T, Pallas C, et al. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity in Vitro. J Clin Med 10 (2021): 328.
- Currie L (1968). Limits for qualitative detection and quantitative determination. Application to radiochemistry. Anal. Chem 40 (1 968): 586-593.
- Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline—Second Edition. CLSI document EP17-A2. Wayne, PA: Clinical and Laboratory Standards Institute; (2012).


 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 