The Clinicopathological Features of Epithelioid Undifferentiated Sarcoma with TFE3 Amplification with One Case Report
Article Information
Yuejiao Lang1#, Xiaojuan Li2#, Shaoyu Chen3, Pei Xiang4, Anjia Han1*
#Authors contributed equally to this work
1Department of Pathology, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China
2Department of Pathology, Yuncheng Central Hospital, Yuncheng, China
3Guangzhou LBP medical Technology Co., Ltd., Guangzhou, China
4Department of Medical Imaging, The First Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China
*Corresponding Author: Anjia Han, MD, PhD, Department of Pathology, The First Affiliated Hospital, Sun Yat-sen University, 58, Zhongshan Road II, Guangzhou 510080, China
Received: 17 December 2020; Accepted: 08 January 2021; Published: 14 January 2021
Citation: Yuejiao Lang, Xiaojuan Li, Shaoyu Chen, Pei Xiang, Anjia Han. The Clinicopathological Features of Epithelioid Undifferentiated Sarcoma with TFE3 Amplification with One Case Report. Archives of Clinical and Medical Case Reports 5 (2021): 129-136.
View / Download Pdf Share at FacebookAbstract
Herein, we first reported a case of undifferentiated sarcoma with epithelioid features harboring TFE3 amplification. A 66-year-old woman with a history of chronic lymphocytic leukemia and chemotherapy presented with a 4 cm palpable nodule in the left lower leg. Microscopically, the large epithelioid tumor cells with remarkable pleomorphism and the small round tumor cells intermingled with each other in a diffuse sheet or a hemangiopericytoma-like vascular growth pattern. Atypical mitotic figures and lymph node metastasis were found while tumor necrosis was absent. Immunohistochemically, the tumor was positive for vimentin, TFE3, and CD34. TFE3 gene amplification was identified by fluorescence in situ hybridization. The patient was alive and well without recurrence or metastasis for 12 months after tumor resection. The tumor should differentiate from PEComa, alveolar soft part sarcoma, epithelioid sarcoma, epithelioid angiosarcoma, and epithelioid rhabdomyosarcoma.
Keywords
Epithelioid undifferentiated sarcoma; TFE3; Gene amplification
Article Details
1. Introduction
As a number of microphthalmia/transcription factor E (MiT/TFE) family, the transcription factor E3 (TFE3) protein plays an important role in tumorigenesis [1-2]. Strong nuclear expression of TFE3 protein is characteristically identified in several tumors including alveolar soft part sarcoma (ASPS), perivascular epithelioid cell tumor (PEComas), Xp11.2 renal cell carcinomas, granular cell tumor, and epithelioid haemangioendothelioma [3-7]. The genetic or epigenetic alterations involving TFE3 expression are complicated and have not been fully elucidated. Apart from gene translocation, TFE3 gene amplification has been proven to be an additional, novel mechanism leading to increased TFE3 protein expression in several cases of PEComas [8-10] and renal cell carcinoma [11]. In this article, we first presented a case of undifferentiated sarcoma with epithelioid features harboring TFE3 amplification.
2. Case Presentation
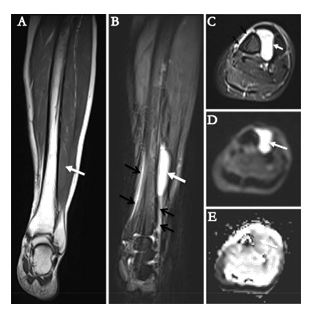
A 66-year-old woman presented with a 4 cm palpable and painful nodule in the left lower leg that was detected approximately 1 month before consultation. The patient had a history of chronic lymphocytic leukemia and had been treated with chemotherapy 8 years ago. The therapy was effective and completed 2 years ago. The familial history was uneventful. Magnetic resonance imaging revealed an oval juxtacortical lesion with a well-defined margin to the anterolateral left tibia, measuring 4.9 × 2.4 × 1.2 cm. Periosteal reaction of the tibia was identified, indicating its aggressiveness. Neither cortical erosion nor marrow infiltration was observed (Figure 1A-E). Tumor resection was performed.
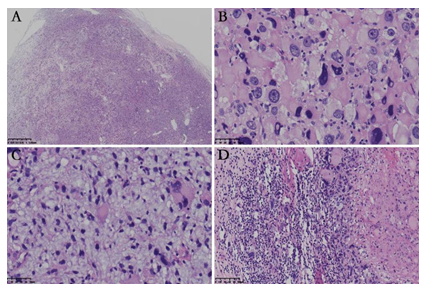
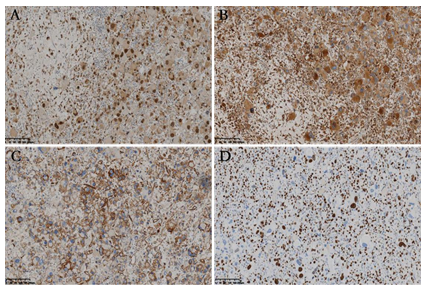
Grossly, the resected specimen consisted of multiple fragments with gray or white on cut surface, measuring 6 × 3 × 2 cm in size. Microscopically, the well-circumscribed but unencapsulated lesion displayed variable cellularity. Two morphologically distinct types of tumor cells intimately intermingled with each other and were arranged in a diffuse sheet or a hemangiopericytoma-like vascular pattern (Figure 2A). One type of tumor cells was the large epithelioid cells with remarkable cytologic atypia. The abundant eosinophilic cytoplasm was homogeneous, granular, or vacuolated. The appearance of large hyperchromatic nuclei ranged from oval to polygonal and centric to eccentric. Multinucleated giant cells with up to three to four nuclei per cell, were not uncommon. Prominent eosinophilic nucleoli and intranuclear inclusions were observed (Figure 2B). The other type of tumor cells was the small round cells with an oval or irregular nucleus. The cytoplasm was scant without a distinct cellular boundary. Myxoid stromal change was evident focally, imparting a relative hypocellular appearance (Figure 2C). Small- to medium-size blood vessels were scattered throughout the lesion. Lymphocytic infiltration was identified and was prominent along the perivascular spaces. Atypical mitotic figures were abundant in the large epithelioid tumor cells but less remarkable in the small round cells (Figure 2B). Tumor necrosis was absent. Tumor metastasis was present in one lymph node (Figure 2D). Immunohistochemically, both the components were diffusely positive for vimentin, TFE3 (Figure 3A), and CD68 (Figure 3B), partially positive for CD34 (Figure 3C), and retained the nuclear expression of INI1. The tumor cells were negative for CK, desmin, myogenin, MyoD1, SMA, HMB45, MelanA, MiTF, STAT6, CDK4, MDM2, S-100 protein, CD31, CD163, FLI-1 and MPO. The Ki-67 index was up to 60% in the hot spot area (Figure 3D).
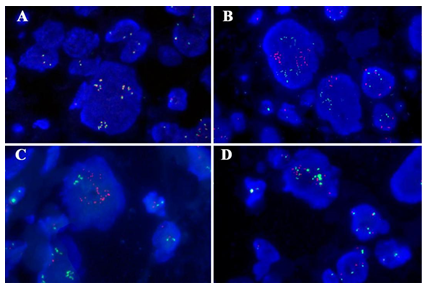
Fluorescence in situ hybridization (FISH) analysis with TFE3 Probe was breakapart designed to detect gene rearrangement and could be also used for the detection of polyploidy or target gene amplification. In normal cells of the present case (female), the copy of TFE3 gene is 2. But in tumor cells, TFE3 Probe show more copies, and the average copy number in each cell is 7. Of which, the number of Green probe and Orange probe is equal in each amplified cell. One hundred consecutive nuclei were counted, 1R1G1F accounted for 1%, and the polyploid proportion was 68% (Figure 4A). Gene fusion of TFE3 with PRCC, ASPSCR1 or NONO was not identified by FISH with gene fusion probe. One hundred consecutive nuclei were counted, 1R1G1F accounted for 1%, 2% and 3%, the polyploid proportion was 68%, 65% and 61%, respectively (Figure 4B-D). Therefore, no TFE3 rearrangement but TFE3 gene amplification was identified in the present case. We made the diagnosis of undifferentiated sarcoma with epithelioid features harboring TFE3 amplification. The patient was alive and well at 12 month follow-up information after surgery by radiological examination.
Figure 1: Radiological characteristics illustrated by magnetic resonance imaging. The tumor (white arrows) appeared iso-intense on coronal T1WI (A), hyper-intense on coronal (B) and axial (C) T2WI with fat-suppression. It appeared hyper-intense on diffusion-weighted image (b=800 s/mm2) (D) with corresponding decreased apparent diffusion coefficient values on apparent diffusion coefficient map (E), indicating diffusion restriction. The periosteal reaction of tibia was noted on T2WI with fat-suppression (black arrows on B and C).
Figure 2: Histological characteristics of the tumor. (A) The well-circumscribed lesion was unencapsulated with variable cellularity; (B) Large epithelioid tumor cells with marked nuclear pleomorphism arranged in a diffuse sheet pattern. Atypical mitotic figures were identified; (C) Small bland cells bear oval or irregular nuclei and scant cytoplasm, with large epithelioid cells in between. Myxoid stromal change was prominent; (D) Metastasis was identified in one lymph node.
Figure 4: Genetic characteristics of the tumor. (A) TFE3 break-apart probe showed no TFE3 rearrangement but TFE3 gene amplification by FISH. The TFE3 probe contains a 627 kb probe labeled in green, which lies proximal to the TFE3 gene that located at Xp11.2 region, and a 637 kb probe labeled in orange extends distally from the TFE3 gene. (B-D) Gene fusion of TFE3 with PRCC(B), ASPSCR1(C) or NONO(D) was not identified by FISH with gene fusion probe. Green, the 5′- terminal region of the TFE3 gene; orange, the 3′- terminal region of the PRCC/ASPSCR1/NONO gene, respectively.
3. Discussion
Undifferentiated sarcoma with epithelioid features harboring TFE3 amplification is very rare. The tumor should differentiate from several tumors. Based on the morphological and genetic features, PEComa is a strong consideration for the diagnosis. PEComas can be composed of epithelioid and spindle cells arranged in a hemangiopericytoma-like vascular pattern. TFE3 is positively expressed in a number of PEComas resulting from gene fusion [12]. TFE3 gene amplification, a characteristic finding in the present case, has also been documented in three cases of PEComas; all of these PEComas were found in adults and showed predominant or pure epithelioid morphology. Among these, two cases displayed marked atypical large cells with abundant eosinophilic cytoplasm, nuclear anaplasia, and multipolar mitotic figures, sharing histological overlaps with the present case. However, tumor cells in PEComas commonly are positive for melanocytic and smooth muscle markers; all the three cases were positive for Melan-A and actin/SMA, two were positive for HMB45 [8-10], whereas none of Melan-A, HMB45 and actin/SMA were expressed in the present case. Therefore, evidence for classifying the tumor as PEComa is insufficient.
Several sarcomas including ASPS, epithelioid sarcoma, epithelioid angiosarcoma, and epithelioid rhabdomyosarcoma should be differentiated from our diagnosis. ASPS typically is composed of large eosinophilic cells in nest or pseudoalveolar growth pattern delineated by fibrous septa. Increased cytologic pleomorphism and nuclear pleomorphism are uncommon but have been described [13]. ASPS demonstrates nuclear expression of TFE3, resulting from the oncogenic fusion or rearrangement of TFE3 with ASPSCR1 on chromosome 17 [5]. However, such characteristic chromosomal translocation was not detected by FISH in the present tumor. The intracytoplasmic accumulation of rod-shaped crystals, which is another specific character of ASPS, could be found in a majority of the cases. Both epithelioid sarcoma and epithelioid angiosarcoma are composed of diffusely arranged large tumor cells with abundant eosinophilic cytoplasm, vesicular nuclei, and prominent nucleoli. As for epithelioid sarcoma, especially in the proximal-type, pleomorphism is prominent [14]. It is one of the few mesenchymal neoplasms that metastasize to lymph nodes. However, epithelioid sarcoma is positive for CK and EMA, and usually loss of nuclear INI1 expression [15]. Epithelioid angiosarcoma presents irregular vascular spaces lined by protuberant epithelioid tumor cells which usually positive for endothelial markers such as CD31, CD34, ERG and FLI-1 [13]. In contrast to the obvious cytologic pleomorphism in the present case, cells and nuclei in epithelioid rhabdomyosarcoma tend be relatively uniform in size. Skeletal muscle differentiation should be proven based on immunostaining for myogenin and myoD1 [16].
The most common chromosomal abnormalities involving TFE3 are translocations and rearrangements, and are among the earliest reported gene fusion in tumorigenesis [17-18]. TFE3 gene fusion can occur with different partners, such as PRCC, ASPSCR1, SFPQ, and NONO, all of which provide active gene promoters, and therefore, lead to higher levels of TFE3 fusion proteins than wild-type TFE3 [5, 19-20]. Apart from PEComas, it has also been confirmed in four cases of renal cell carcinomas with moderate to strong nuclear expression of TFE3 [11]. An interesting finding is that it might be associated with aggressive biological behavior [8-11]. As for renal cell carcinoma, patients with TFE3 amplification exhibited a significantly poorer cancer-specific survival rate than those with translocation [11]. As for PEComas, distant metastasis and recurrence were documented [8-10]. In the present case, tumor aggressiveness has been suggested by radiological examination, and one lymph node metastasis was identified.
4. Conclusion
In conclusion, we present a case of epithelioid undifferentiated sarcoma with characteristic genetic alteration. Even comprehensive immunohistochemistry and molecular examination could not help in classifying the tumor under any of the existing soft tissue tumor classification. Therefore, we designated it as epithelioid undifferentiated sarcoma with TFE3 amplification. Our report indicated TFE3 amplification as an additional chromosomal abnormality might induce increased expression of TFE3 protein, and suggested the possible relationship between the genetic alteration and aggressive biological behavior of tumor.
Disclosure of Conflict of Interest
None.
References
- Hemesath TJ, Steingr í msson E, McGill G, et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes and development 8 (1994): 2770-2780.
- Haq R, Fisher DE. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 29 (2011): 3474-3482.
- Sandberg A, Bridge J. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: alveolar soft part sarcoma. Cancer Genet Cytogenet 136 (2002): 1-9.
- Folpe AL, Mentzel T, Lehr HA, et al. Perivascular epithelioid cell neoplasms of soft tissue and gynecologic origin: a clinicopathologic study of 26 cases and review of the literature. Am J Surg Pathol 29 (2005): 1558-1575.
- Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal 8 cell carcinomas of children and adolescents. Am J Pathol 159 (2001): 179-192.
- Liu Y, Zheng Q, Wang C, et al. Granular cell tumors overexpress TFE3 without gene rearrangement: Evaluation of immunohistochemistry and break-apart FISH in 45 cases. Oncol Lett 18 (2019): 6355-6360.
- Mohamed AD, Tremblay AM, Murray GI, et al. The Hippo signal transduction pathway in soft tissue sarcomas. Biochim Biophys Acta 1856 (2015): 121-129.
- Argani P, Aulmann S, Illei PB, et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol 34 (2010): 1395-1406.
- Jimbo N, Nishigami T, Noguchi M, et al. Hepatic angiomyolipomas may overexpress TFE3, but have no relevant genetic alterations. Hum Pathol 61 (2017): 41-48.
- Wang H, Zhan H, Yao Z, et al. Malignant renal epithelioid angiomyolipoma with TFE3 gene amplification mimicking renal carcinoma. Clin Nephrol Case Stud 6 (2018): 11-15.
- Macher-Goeppinger S, Roth W, Wagener N, et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod Pathol 25 (2012): 308-315.
- Thway K, Fisher C. PEComa: morphology and genetics of a complex tumor family. Ann Diagn Pathol 19 (2015): 359-368.
- Matthew R, Lindberg DL, Jerad M Gardner, et al. Diagnostic Pathology: Soft Tissue Tumors. 2nd Canada: Elsevier (2016).
- Guillou L, Wadden C, Coindre JM, et al. “Proximal-type” epithelioid sarcoma, a distinctive aggressive neoplasm showing rhabdoid features. Clinicopathologic, immunohistochemical, and ultrastructural study of a series. Am J Surg Pathol 21 (1997): 130-146.
- Hornick JL, Dal Cin P, Fletcher CD. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am J Surg Pathol 33 (2009): 542-550.
- Jo VY, Marino-Enriquez A, Fletcher CD. Epithelioid rhabdomyosarcoma: clinicopathologic analysis of 16 cases of a morphologically distinct variant of rhabdomyosarcoma. Am J Surg Pathol 35 (2011): 1523-1530.
- Shipley JM, Birdsall S, Clark J, et al. Mapping the X chromosome breakpoint in two papillary 9 renal cell carcinoma cell lines with a t(X;1)(p11.2;q21.2) and the first report of a female case. Cytogenetics and cell genetics 71 (1995): 280-284.
- Sidhar SK, Clark J, Gill S, et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet 5 (1996): 1333-1338.
- Clark J, Lu YJ, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 15 (1997): 2233-2239.
- Weterman MA, Wilbrink M, Geurts van Kessel A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proceedings of the National Academy of Sciences of the United States of America 93 (1996): 15294-15298.


 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
