Detection of Antibiotic Resistance genes of Multidrug Resistance Enterobacter Cloacae and Enterobacter Aerogenes Isolated from the Patients of Dhaka Medical College Hospital
Article Information
Azmeri Haque1, Nafisa Jabin Mishu2, H M Khaleduzzaman*, 3, S M Shamsuzzaman4, Avizit Sarker5, MD Zabed Ahmed Mitu6, Rizwana Zaman7, Rubaiya Binte Kabir8, MD Asaduzzaman9
1MBBS, M phil (Microbiology), Lecturer, Department of microbiology, Dhaka Medical College, Bangladesh
2MBBS, M phil (Microbiology), Assistant professor, Microbiology, Army Medical College Bogura, Bangladesh
3MBBS, FCPS(Medicine), Classified medicine specialist CMH Bogura, Bangladesh
4MBBS, Mphil (Microbiology), PHD, Prof of microbiology, Department of microbiology Dhaka Medical College Dhaka, Bangladesh
5MBBS (DMC), MD (Microbiology), Medical Officer, Department of Microbiology Dhaka Medical College, Bangladesh
6Associate Professor, Department of Microbiology, Jahurul Islam medical College, Bangladesh
7Assistant Professor (c.c), Department of Microbiology, Dhaka Medical College, Bangladesh
8Lecturer, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
9MBBS, M. Phil, Lecturer, Dhaka Medical College, Bangladesh
*Corresponding author: H M Khaleduzzaman, MBBS, FCPS(Medicine), Classified medicine specialist CMH Bogura, Bangladesh
Received: 24 July 2023 Accepted: 01 August 2023 Published: 10 August 2023
Citation: Azmeri Haque, Nafisa Jabin Mishu, H M Khaleduzzaman, S M Shamsuzzaman, Avizit Sarker, MD Zabed Ahmed Mitu, Rizwana Zaman, Rubaiya Binte Kabir, MD Asaduzzaman. Detection of Antibiotic Resistance genes of Multidrug Resistance Enterobacter Cloacae and Enterobacter Aerogenes Isolated from the Patients of Dhaka Medical College Hospital. Archives of Microbiology and Immunology. 7 (2023): 143-149
View / Download Pdf Share at FacebookAbstract
Emergence of multidrug resistant strains of Enterobacter spp. has a potential threat because these strains were responsible for high morbidity and mortality in recent years due to nosocomial infection in health care settings. Due to shortage of new effective antibiotic, old drug like fosfomycin recently reintroduced in human medicine for the treatment of multi drug resistant (MDR) gram negative bacteria. Sample size was 360. We collected different samples like urine, wound swab, endotracheal aspirate and blood from microbiology lab of Dhaka medical college. Organisms were isolated and identified by culture, gram staining and biochemical tests. Further tests were done only on common Enterobacter species. Antibiotic susceptibility tests were performed by disc diffusion technique. Polymerase chain reaction (PCR) was used to identify different colistin resistance genes (mcr-1 and mcr-2,) fosfomycin resistance genes (fosA, fosA3, fosA4) and carbapenem resistance genes (blaKPC, blaIMP, blaVIM and blaOXA-48 / blaOXA181). Sequencing of fosA4 gene was done. Present study observed the presence of fosfomycin resistance gene of multidrug resistant (MDR) Enterobacter spp. Among the 360 samples 66.11% yielded culture positive results and out of the culture positive samples 10.53% were Enterobacter spp. Among Enterobacter spp. 82.76% E. cloacae and 17.24% E. aerogenes were identified by biochemical tests. Among the 17.24% isolated E. aerogenes, 100% were resistant to amoxiclav and cefoxitin and ceftazidime, 80% were resistant to ciprofloxacin and amikacin, 60% were resistant to piperaicillin-tazobactam, cefuroxime and ceftriaxone, 40% were resistant to aztreonum, cefepime and fosfomycin, 20% were resistant to imipenem and colistin but none were resistant to tigecycline. Among the fosfomycin resistant Enterobacter spp. 80% were positive for fosA gene, 50% were positive for fosA4 gene, 50% were positive for fosA5gene. Sequencing of PCR product of fosA4 gene showed multiple point mutations. So, Antimicrobial susceptibility testing must be done before prescribing antibiotics due to the high rates of resistance of Enterobacter spp. to multiple antibiotics.
Keywords
Enterobacter cloacae, Enterobacter aerogenes, fosfomycin, antibiotic resistance, genes, MDR, Polymerase chain reaction; Agar dilution method
Enterobacter cloacae articles, Enterobacter aerogenes articles, fosfomycin articles, antibiotic resistance articles, genes articles, MDR articles, Polymerase chain reaction articles; Agar dilution method articles
Article Details
1. Introduction
The genus Enterobacter is associated with hospital acquired infection that has been ranked as the third most frequent isolate following Escherichia coli and Klebsiella species [1]. Enterobacter cloacae and Enterobacter aerogenes are the most commonly isolated species among the Enterobacter species [2].
National Nosocomial Infection Surveillance System (NNIS) study found that Enterobacter accounts for 5 to 11% of all nosocomially transmitted blood, wound, respiratory tract and urinary tract infections. Enterobacter spp. caused 11.2% of pneumonia cases in all types of ICUs, ranking third after Staphylococcus aureus (18.1%) and P. aeruginosa (17%). The corresponding rates among patients in pediatric ICUs were 9.8% for pneumonia, 6.8% for bloodstream infections, and 9.5% for UTIs [3]. Enterobacter spp. appears well adapted for survival and threats to cause immense mortality & morbidity by the proliferation of highly drug resistant strains both in the community and hospital environment. These organisms seem to have innate resistance to older anti-microbial agents and have the propensity to rapidly develop resistance to newer anti-microbial agents [4] Treatment of infections with Enterobacter spp. is difficult because of resistance to third generation cephalosporins, penicillin and quinolones is an increasing problem [5].
Enterobacter cloacae can produce chromosome mediated AmpC β-lactamase and are resistant to ampicillin, amoxicillin/clavulanic, cephamycin and first and second generation cephalosporin [6]. Due to the clinical significance of their AmpC-β-lactamase production, these pathogens are a part of the “SPICE”(Serratia, Pseudomonas, indole-positive Proteus, Citrobacter and Enterobacter) of bacteria [7] which is usually associated with complicated UTI (associated with catheters, functional or anatomical abnormalities of genitourinary tract). The carbapenem non-susceptible phenotypes are attributed to production of carbapenemases or more likely, production of extended-spectrum β lactamase (ESBL) plus AmpC β-lactamase with dysfunctional entry routes (i.e., porin change) of carbapenems, integrons and insertion sequence common region 1 (ISCR1) carrying various resistance genes, and/or efflux pumps etc [8]. Also, the mechanism of combinations of either ESBL or AmpC and mutation of porins may hold a certain proportion [9]. Therefore, it is no surprise that the carbapenem resistant Enterobacter spp. is a part of the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.). The purpose of this study was to identify the patterns of antibiotic resistance in Enterobacter species isolated from various clinical cases. Additionally, common Enterobacter species that cause a variety of diseases were found. In addition to reducing treatment failure in hospitalized patients, this would offer crucial information about the empiric management of Enterobacter infections.
2. Materials and Methods
Total of 29 Enterobacter isolates were gathered from various samples of urine, wound swab, endotracheal aspirate and blood during the course of a year (June 2019 to July 2020) in the microbiology department at Dhaka Medical College. The Enterobacter isolates were tested for antibiotic susceptibility using the Kirby Bauer disc diffusion method with Mueller Hinton agar and commercially available antibiotic discs (Oxoid Ltd, UK). Amoxycillin /clavulanic acid (20/10μg), Cefepime (30μg), Ceftazidime (30μg), Cefuroxime sodium (30μg), Cefoxitin (30μg), Amikacin (30μg), Ceftriaxone (30μg), Ciprofloxacin (30μg), Piperacillin/Tazobactam (110/10μg), Imipenam (10μg), Tigecycline (15μg), Gentamycin (30μg) were used. Colistin and fosfomycin (Beximco Pharma Limited) susceptibility was tested by was determined by agar dilution method of MIC. The disc content and the zone of inhibition were used as recommended by the Clinical Laboratory Standards Institute (CLSI, 2020). Criteria of the United States Food and Drug Administration (2010) were used for the interpretation of zone of inhibition of Tigecyclin. Antibiotic discs were obtained from commercial source (Oxoid Ltd.UK). For inoculums preparation & inference of MIC 0.5 McFarland turbidity was used as standard (contains 1×108 cfu/ml) [10] 10 times dilution of the test inoculums was done to achieve 1× 107 cfu/ml. All the inoculated plates were incubated aerobically at 37ºC overnight and the lowest concentration of antibiotic impregnated Mueller-Hinton agar which showing no visible growth on agar media was considered as MIC of the drug of that strain of bacteria E coli ATCC 25922 was used for quality control.
Two species of Enterobacter, E.cloacae and E.aerogenes were identified using several biochemical tests including sugar fermentation test for Adonitol, D-sorbitol, L-rhamnose and Esculin and by two decarboxylation reactions- Arginine dihydrolase test and Lysine decarboxylase test. Species other than these two were categorized as others. Polymerase chain reaction (PCR) was done for the detection of MDR genes in Enterobacter spp. Bacterial pellet formation was done by a loop full of bacterial colonies from Mueller Hinton agar (MHA) media was inoculated into a microcentrifuge tube having sterile TSB and incubated overnight at 37ºC. Centrifugation of the incubated tube was done at 4000g for 10 minutes. Supernatant part was discarded and tubes containing bacterial pellets were kept at -20ºC for the purpose of DNA extraction. To extract the DNA three hundred microlitre of sterile distilled water was added to microcentrifuge tubes having pellets and vortexed until mixed well. Then the mixture was heated at 100° C for 10 minutes in a heat block. After heating, tubes were immediately placed on ice for 5 minutes and centrifuged at 14000 g for 6 minutes at 4ºC. Finally, the supernatant was taken into another microcentrifuge tube. This extracted DNA was preserved at 4ºC for 7-10 days and -20ºC for a long time. Mixing of mastermix with primer and DNA template PCR was performed in a final reaction volume 25 µl in a PCR tube, containing 12.5 µl of master mix (mixture of dNTP, taq polymerase, MgCl2 and PCR buffer), 2 µl forward primer, 2 µl reverse primer (Promega Corporation, USA), 2 µl of extracted DNA and 6.5 µl of nuclease free water. After a brief vortex, the tubes were centrifuged. Amplification in thermal cycler (Gene Atlas, Master cycler gradient, Japan, Model482) PCR assays was performed in a DNA thermal cycler. After amplification products were processed for gel documentation or kept at -20ºC till tested (Gene Atlas, Master cycler gradient, Japan, Model 482).
Agarose gel electrophoresis and visualization PCR products were detected by electrophoresis on 1.5% agarose gel. Gel was prepared with 1 X TBE buffer (Tris EDTA). For 1.5% agarose gel preparation, 0.18 gram agarose powder (LE, analytic grade, Promega, Madison, USA) was mixed with a 1.25 ml TBE buffer. A comb was placed in a gel tray, the gel was poured. After solidification, 1 µl of loading dye and 5 µl of amplicon was mixed on parafilm and was loaded in agarose well. Similarly, 2 µl of 100bp DNA ladder was mixed with 1µl loading dye and was loaded. Gel electrophoresis was done in 230 voltages for 30 minutes. After electrophoresis, the gel was stained with ethidium bromide (20µl ethidium bromide in 200 ml distilled water). The gel was observed under UV transilluminator (Gel Doc, Major Science, Taiwan) for DNA bands. The DNA bands were identified according to their molecular size by comparing with the molecular weight marker (100bp DNA ladder) loaded in a separated lane. For sequencing of bacterial DNA, purification of amplified PCR products was done by using DNA purification kits (FAVOGEN, Biotech Corp). Purified PCR products of Proteus mirabilis were sent to 1st Base Laboratories, Malaysia for sequencing by capillary method (ABI PRISM 3500). BLAST analysis was performed to search for homologous sequences into the Gene Bank at www.ncbi.nlm.nih.gov. Primers used in this study (27, 28, 29, 30, 31):
|
Gene |
Sequence (5’ to 3’) |
Size (bp) |
|
|
mcr 1 |
F |
CGG TCA GTC CGT TTG TTC |
309 |
|
R |
CTT GGT CGG TCT GTA GGG |
||
|
mcr 2 |
F |
TGTTGCTTGTGCCGATTGGA |
567 |
|
R |
AGATGGTATTGTTGGTTGCTG |
||
|
fos A |
F |
ATC TGT GGG TCT GCC TGT CGT |
271 |
|
R |
ATG CCC GCA TAG GGC TTC T |
||
|
fosA3 |
F |
CCT GGC ATT TTA TCA GCA GT |
221 |
|
R |
CGG TTA TCT TTC CAT ACC TCA G |
||
|
fosA4 |
F |
CTG GCG TTT TAT CAG CGG TT |
230 |
|
R |
CTT CGC TGC GGT TGT CTT T |
||
|
fosA5 |
F |
TAT TAG CGA AGC CGA TTT TGC T |
177 |
|
R |
CCC CTT ATA CGG CTG CTC G |
||
|
fosB |
F |
CAG AGA TAT TTT AGG GGC TGA CA |
312 |
|
R |
CTC AAT CTA TCT TCT AAA CTT CCTG |
||
|
fosB2 |
F |
CCT GGC CGA GAA AGA GAT GAG |
392 |
|
R |
AAC CGG TTT TGC AAA GTG CC |
||
|
fosC |
F |
CCT TGC TCA CTG GGG ATC TG |
354 |
|
R |
TAC AAG ACC CGA CGC ACT TC |
||
|
fosC2 |
F |
TGG AGG CTA CTT GGA TTT G |
209 |
|
R |
AGG CTA CCG CTA TGG ATT T |
||
|
fosX |
F |
TGT CCC TCA CCT TCG ACT CT |
|
|
R |
TTG CTG GTC TGT GGA TTTGC |
||
|
blaIMP-1 |
F |
ACCGCAGCAGAGTCTTTGCC |
587 |
|
R |
ACAACCAGTTTTGCCTTACC |
||
|
blaNDM-1 |
F |
GGTTTGGCGATCTGGTTTTC |
621 |
|
R |
CGGAATGGCTCATCACGATC |
||
|
blaVIM |
F |
GATGGTGTTTGGTCGCATA |
390 |
|
R |
CGAATGCGCAGCACCAG |
||
|
blaKPC |
F |
CGTCTAGTTCTGCTGTCTTG |
789 |
|
R |
CTTGTCATCCTTGTTAGGCG |
||
|
blaOXA-48 |
F |
GCGTGGTTAAGGATGAACAC |
438 |
|
R |
CATCAAGTTCAACCCAACCG |
||
3. Result
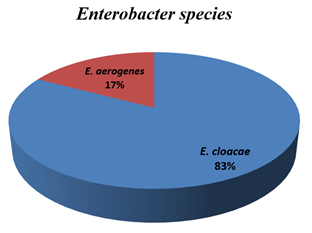
Among the 360 samples 66.11% yielded culture positive results and out of the culture positive samples 10.53% were Enterobacter spp. Figure-1 showed the distribution of isolated Enterobacter into different species. Out of 29 Enterobacter isolates, 24(82.76%) were E. cloacae and 5(17.24%) were E. aerogenes.
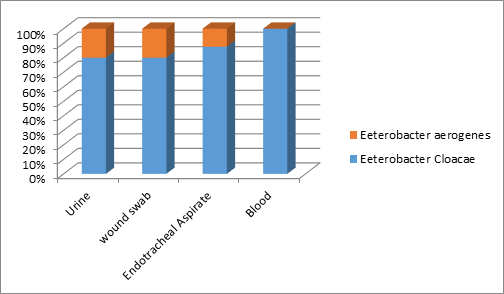
Fig-2 showed among 128 urine sample, 8 (6.25%) were E. cloacae and 2 (1.56%) were E. aerogenes, among wound swab 06.96% were E. cloacae and 1.74% was E. aerogenes. among blood 1.75% were E. cloacae but none was E. aerogenes. and among 60 endotracheal aspirates, 7(11.66%) were E. cloacae and one (1.66%) was E. aerogenes.
Table 1 shows among the 5 isolated E. aerogenes, 100% were resistant to amoxiclav and cefoxitin and ceftazidime, 80% were resistant to ciprofloxacin and amikacin, 60% were resistant to piperacillin-tazobactam, cefuroxime and ceftriaxone, 40% were resistant to aztreonum, cefepime and fosfomycin, 20% were resistant to imipenem and colistin but none were resistant to tiger cycline.
Table 1: Antibiotic resistance pattern of isolated E. cloacae and E. aerogenes (N=29)
|
Antimicrobial drugs |
E. cloacae |
E. aerogenes |
Total N=29 |
|
N=24 n (%) |
N=5 n (%) |
n (%) |
|
|
Amoxiclav |
24 (100.00) |
5 (100.00) |
29 (100.00) |
|
Cefoxitin |
24 (100.00) |
5 (100.00) |
29 (100.00) |
|
Ciprofloxacin |
21 (87.50) |
4 (80.00) |
25 (86.21) |
|
Cefuroxime |
24 (100.00) |
3 (60.00) |
27 (93.10) |
|
Ceftriaxone |
22 (91.67) |
3 (60.00) |
25 (86.21) |
|
Ceftazidime |
17 (70.83) |
5 (100.00) |
22 (75.86) |
|
Aztreonam |
17 (70.83) |
2 (40.00) |
19 (65.52) |
|
Amikacin |
12(50.00) |
4 (80.00) |
16 (55.17) |
|
Imipenem |
12 (50.00) |
1 (20.00) |
13 (44.83) |
|
Piperacilin-tazobactam |
11 (45.83) |
3(60.00) |
14 (48.27) |
|
*Fosfomycin |
8 (33.33) |
2 (40.00) |
10 (34.48) |
|
Cefepime |
7 (29.17) |
2 (40.00) |
9 (31.03) |
|
*Colistin |
7(29.17) |
1 (20.00) |
8 (27.59) |
|
Tigecycline |
4 (16.66) |
0 (0.00) |
4 (13.79) |
N= Total number of isolated bacteria n =Total number of resistant bacteria
*Colistin and Fosfomycin resistance was determined by MIC by agar dilution method.
Table 2 shows distribution of multidrug resistant Enterobacter spp. isolated from different samples. Out of 29 isolated E. cloacae and E. aerogenes, 17 multidrug resistant strains were detected. Of which, 5 (50%) were detected from urine, 6 (60%) from wound swab, 5 (62.50%) from endotracheal aspirates, one (100%) from blood sample.
Table 2: Distribution of multidrug resistant (MDR) E. cloacae and E. aerogenes isolated from different samples
|
Sample |
Total isolates |
MDR isolates |
|
N=29 |
n (%) |
|
|
Urine |
10 |
5 (50.00) |
|
Wound swab |
10 |
6 (60.00) |
|
ETA |
8 |
5 (62.5) |
|
Blood |
1 |
1 (100.00) |
|
Total |
29 |
17 (58.62) |
ETA= Endotracheal aspirate
N= Total number of isolated E. cloacae and E. aerogenes
n = Total number of MDR isolates.
Table 3: shows antimicrobial resistance pattern of the isolated MDR Enterobacter spp. Among the 17 isolated E. cloacae and E. aerogenes, all were resistant to amoxiclav, amikacin and cefoxitin, 88.23 % were resistant to ciprofloxacin, 94.12 % were resistant to ceftriaxone, cefuroxime and ceftazidime, 70.59% were resistant to piperacillin-tazobactam, 76.47% were resistant to imipenem, 47.06 % were resistant to fosfomycin, 41.17% were resistant to colistin and 23.53% were resistant to tigecycline. Among the 14 isolated MDR E. cloacae, 100% were resistant to amoxiclav, cefoxitin, ciprofloxacin, cefuroxime, ceftriaxone, ceftazidime and amikacin, 71.43% were resistant to piperacillin-tazobactam, 85.71% were resistant to imipenem, 50% were resistant to fosfomycin, 42.85% were resistant to colistin and 28.57% were resistant to tigecycline. Among the 3 isolated MDR E. aerogenes, all were resistant to amoxiclav, amikacin and cefoxitin, 66.67% were resistant to cefuroxime, ceftriaxone, ceftazidime and piperacillin-tazobactam, 33.33% were resistant to imipenem, ciprofloxacin, colistin and fosfomycin and no isolated E. aerogenes were resistant to tigecycline.
Table 3: Antibiotic resistance pattern of isolated multidrug resistant (MDR) E. cloacae and E. aerogenes (N=17)
|
Antimicrobial drugs |
E. cloacae |
E. aerogenes |
Total |
|
N=14 n (%) |
N=3 n (%) |
N=17 n (%) |
|
|
Amoxiclav |
14 (100.00) |
3 (100.00) |
17 (100.00) |
|
Cefoxitin |
14 (100.00) |
3 (100.00) |
17 (100.00) |
|
Ciprofloxacin |
14 (100.00) |
1 (33.33) |
15 (88.23) |
|
Cefuroxime |
14 (100.00) |
2 (66.67) |
16 (94.12) |
|
Ceftriaxone |
14 (100.00) |
2 (66.67) |
16 (94.12) |
|
Ceftazidime |
14 (100.00) |
2 (66.67) |
16 (94.12) |
|
Aztreonam |
11 (78.5 7) |
1 (33.33) |
12 (70.59) |
|
Amikacin |
14 (100.00) |
3 (100.00) |
17 (100.00) |
|
Imipenem |
12 (85.71) |
1 (33.33) |
13 (76.47) |
|
Piperacillin- tazobactam |
10 (71.43) |
2 (66.67) |
12 (70.59) |
|
Fosfomycin |
7 (50.00) |
1(33.33) |
8 (47.06) |
|
Cefepime |
5 (35.71 |
1 (33.33) |
6 (35.29) |
|
Colistin |
6(42.85) |
1 (33.33) |
7(41.18) |
|
Tigecycline |
4 (28.57) |
0 (0.00) |
4 (23.53) |
Table-4 shows detection of fosA, fosA3, fosA4, fosA5, fosB, fosB2, fosC, fosC2and fosX genes among fosfomycin resistant E. cloacae and E. aerogenes by PCR. Among 10 fosfomycin resistant Enterobacter spp. 7 (70) were positive for fosA, 5 (50) were positive for fosA4, and 4 (40) were positive for fosA5. No fosA3, fosB, fosC, fosC2 and fosX gene were detected. No fosA, fosA3, fosA4, fosA5, fosB, fosB2, fosC, fosC2 and fosX were detected from blood sample. This table also shows detection of mcr-1 and mcr-2 genes from different isolates among colistin resistant E. cloacae and E. aerogenes by PCR. Among 7 colistin resistant isolates no mcr-1 and mcr-2 genes were detected. No mcr-1 and mcr-2 gene detected from blood sample. This table also shows detection of blaNDM-1, blaKPC, blaIMP, blaVIM and blaOXA-48 / blaOXA181 among imipenem resistant Enterobacter species by PCR. Among 9 imipenem resistant isolates. No blaNDM-1, blaKPC, blaIMP, blaVIM andblaOXA- 48 / blaOXA181 genes were detected. No mcr-1 and mcr-2 gene detected from blood sample.
Table 4: Detection of fosA, fosA3, fosA4, fosA5, fosB, fosB2, fosC, fosC2 and fosX genes among fosfomycin resistant E. cloacae and E. aerogenes by PCR (N=10), mcr-1 and mcr-2 genes from different isolates among colistin resistant E. cloacae and E. aerogenes by PCR (N=8) & blaNDM-1, blaKPC, blaIMP, blaVIM and blaOXA-48/blaOXA181 among imipenem resistant Enterobacter species (N=9)
|
Genes |
Urine |
Wound swab |
ETA |
Total |
|
n (%) |
n (%) |
n (%) |
n (%) |
|
|
fosA |
5(50.00) |
2(10.00) |
1(10.00) |
8 (80.00) |
|
fosA3 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
fosA4 |
2(20.00) |
3(10.00) |
0 (0.00) |
5(50.00) |
|
fosA5 |
4 (40.00) |
1 (10.00) |
0 (0.00) |
4(50.00) |
|
fosB |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
fosB2 |
0(0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
fosC |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
fosC2 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
fosX |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
mcr-1 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
mcr-2 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
blaNDM-1 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
blaKPC |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
blaIMP |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
blaVIM |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
blaOXA-48 |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
/181 |
N=Total number of fosfomycin resistant or colistin resistant or imipenem resistant E. cloacae and E. aerogenes.
n= Total number of fosfomycin resistant or colistin resistant or imipenem resistant gene in different samples.
ETA= Endotracheal aspirates.
Figure-3 shows Comparison of DNA sequence of amplified PCR product of fosA4 gene of Enterobacter cloacae which is 81% identical with Escherichia coli strain CRE1540 chromosome, complete genome available in gene bank (Accession number- gi|1320768276|CP019051.1) fosA4 had point mutation at 169, 170, 171, 180, 192, 195, 197, 203, 214, 218, 219, 222, 228, 238, 244, 250 positions and addition at 128 and 214 position.
Multidrug resistance is the most important problem in antibiotic resistance in treating microorganisms and there is exponential increase in MDR over the last decade [11] The emergence of multidrug-resistant Enterobacter spp. isolates has a negative impact on the clinical outcome of infected patients and increasing mortality rates [12]. In the present study, out of 360 samples, 228 (66.11 %) samples were culture positive of which 29 (12.1 8%) were Enterobacter spp. These finding are in agreement with the recent studies in DMCH reported that 65.14% samples (urine, wound swab, ETA, blood) and 63.20% of samples (urine, wound swab, ETA, blood, sputum, pus) were culture positive [13, 14]. Enterobacter accounts for 11% of all nosocomially acquired blood, wound, respiratory tract and urinary tract infection [15]. These findings are nearly close to the present findings. In the present study, among 29 isolated Enterobacter spp. 24 (82.76 %) were identified as Enterobacter cloacae and 5 (17.24 %) were identified as Enterobacter aerogenes by biochemical tests. These findings were similar to the recent study in DMCH found that 22 (78.57%) were identified as Enterobacter cloacae and 6 (21.43%) were identified as Enterobacter aerogenes [13].
Moreover, a study in India reported that 77.94% were E. cloacae and 22.05% were E. aerogenes [16]. This similarity may be attributed to the fact that these two studies were conducted in same geographic areas. In the present study, 55.17% Enterobacter spp. were resistant to amikacin. Study in Iran reported 48.60% Enterobacter resistant to amikacin which was lower than the present findings [17]. The reason behind the higher resistance rate in the present study might be due to the fact that, there was a significant irrational use of antibiotic.In the present study, imipenem resistance in Enterobacter spp. was 44.83%. Imipenem resistance among the species varies widely in different parts of the world. Study in India reported that 53.8% imipenem resistance where as study in Palestine showed resistance rates as low as 12.2% [18, 19]. Studied from DMCH noted 28% and 42.86% imipenem resistance among Enterobacter spp [20, 13].
The frequency of imipenem resistant Enterobacter spp. is increasing in Bangladesh which is reflected by these studies. Among the isolated Enterobacter spp. 8 (27.59%) colistin resistant Enterobacter spp. were identified. A study in Korea reported that 16% of Enterobacter spp.were resistant to colistin [21]. Another study reported 13.9%- 20.1% colistin resistances in Enterobacter spp [22]. Now a day, the use of colistin has increased in Enterobacter which might be the reason for emergence of resistance of this reserve drug. In the present study, among the 29 isolated Enterobacter spp. 34.48 % were resistant to fosfomycin. A study in India reported that 40% Enterobacter spp. were resistant to fosfomycin which were in agreement with the present study [23]. All the resistant strains were isolated from urine sample which is close to the present study. In the present study, 60% of Enterobacter spp. resistant to fosfomycin were isolated from urine.
The reason behind such finding in present study might be due to horizontal transfer of resistance genes between different species. Plasmids containing ESBL and fos genes may facilitate the dissemination of antibiotic resistance. Recent studies indicate that the recombination of plasmid-encoding carbapenemase and fosfomycinase occurs via mobile elements, thus presenting new treatment challenges [24]. Clinical use of fosfomycin in Bangladesh is rare and there is no data regarding fosfomycin resistance. In the present study, among the fosfomycin resistant Enterobacter spp. 80% were positive for fosA, 50% were positive for fosA4 and 40% were positive for fosA5. A study in China reported that 80% Enterobacter isolates were positive for fosA and 10% E. cloacae were positive for fosA4 [25]. A recent study in DMCH reported resistance rate of fosB2 genes are 40% among fosfomycin resistant Enterobacter spp [13]. Acquisition of fosfomycin resistance by antibiotics modifying enzyme that shows a higher incidence in multidrug resistant strains. The multidrug resistance plasmid, pKP46 carries nine gene (fosA among them) conferred resistance to several antibiotics including penicillins, cephalosporins, fosfomycin, aminoglycosides, quinolones [26]. This multi resistance plasmid might be the reason behind increasing fosfomycin resistance among MDR Enterobacter spp.
The identification of fosA4 gene in Enterobacter cloacae was also further validated by sequencing. In the present study, DNA sequence of amplified PCR product of fosA4 gene detected in E. cloacae which was 81% identical with Esherichia coli which is available in gene bank (Accession no-gi|1320768276|CP019051.1) suggested that the fosA4 gene found in E. cloacae in the present study might have been transferred from Esherichia coli through plasmid. In this study, DNA sequence of amplified PCR product and translated nucleotide base sequence of fosA4 showed point mutations at multiple positions. Comparison of DNA sequence of amplified PCR product of fosA4 gene of Enterobacter cloacae which is 81% identical with Escherichia coli strain CRE1540 chromosome, complete genome available in gene bank (Accession number- gi|1320768276|CP019051.1) fosA4 had point mutation at 169, 170, 171, 180, 192, 195, 197, 203, 214, 218, 219, 222, 228, 238, 244, 250 positions and addition at 128 and 214 positions.
5. Conclusion
cloacae and E. aerogenes have been increasingly associated with multidrug resistance and one of the common threats for hospital patients of critical care unit. E. cloacae and E. aerogenes found to be resistant to most commonly used antibiotics, of which 58.62% were MDR. Among the fosfomycin resistant Enterobacter spp. 70% were positive for fosA gene, 50% were positive for fosA4 & 40% were positive for fosA5. So antimicrobial susceptibility testing must be done before prescribing antibiotics due to the high rates of resistance of Enterobacter spp. to multiple antibiotics.
References
- Kim T, Hong SI, Park SY, Jung J, Chong YP, Kim SH, et al. Clinical features and outcomes of spontaneous bacterial peritonitis caused by streptococcus pneumoniae: A matched Case-control study. Medicine 95 (2016): e3796.
- Vishwanath S, Swetha PS, SuShMa M. Extended-Spectrum β-lactamase Production among Enterobacter cloacae and Enterobacter aerogenes at a Tertiary Care Center in Coastal Karnataka. National Journal of Laboratory Medicine 3 (2016).
- System NN. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2003, issued August 2003. American Journal of Infection Control 31 (2003): 481-98.
- Chavda KD, Chen L, Fouts DE, Sutton G, Brinkac L, Jenkins SG, et al. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. MBio 30 (2016): e02093-16.
- Preston KE, Radomski CC, Venezia RA. Nucleotide sequence of the chromosomal ampC gene of Enterobacter aerogenes. Antimicrobial agents and chemotherapy 44 (2000): 3158-62.
- Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clinical microbiology reviews 31 (2018): e00079-17.
- Rui Y, Lu W, Li S, Cheng C, Sun J, Yang Q. Integrons and insertion sequence common region 1 (ISCR1) of carbapenem-non-susceptible Gram-negative bacilli in fecal specimens from 5000 patients in southern China. International journal of antimicrobial agents 52 (2018): 571-6.
- Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrobial agents and chemotherapy 54 (2010): 573-7.10.
- Cienfuegos-Gallet AV, Ocampo AM, Chavda K, Chen L, Kreiswirth BN, Jiménez JN. Molecular epidemiology of carbapenem-resistant Enterobacter cloacae complex infections uncovers high frequency of non-carbapenemase-producers in five tertiary care hospitals from Colombia. bioRxiv. 2018 Dec 12:494807.
- Ramos JR, Lletí MS. Fosfomycin in infections caused by multidrug-resistant Gram-negative pathogens. Revista Española de Quimioterapia 32 (2019): 45.
- Liu CP, Wang NY, Lee CM, Weng LC, Tseng HK, Liu CW, et al. Nosocomial and community-acquired Enterobacter cloacae bloodstream infection: risk factors for and prevalence of SHV-12 in multiresistant isolates in a medical centre. Journal of Hospital Infection 58 (2004): 63-77.
- Munny NN, Shamsuzzaman SM, Hossain T. Antibiotic resistance and phenotypic and genotypic detection of colistin resistance among Enterobacter species isolated from patients of a tertiary care hospital, Bangladesh. Archives of Microbiology & Immunology 5 (2021): 337-52.
- Sonia SJ, Uddin KH, Shamsuzzaman SM. Prevalence of Colistin Resistance in Klebsiella pneumoniae Isolated from a Tertiary Care Hospital in Bangladesh and Molecular Characterization of Colistin Resistance Genes among Them by Polymerase Chain Reaction and Sequencing. Mymensingh Medical Journal: MMJ 31 (2022): 733-40.
- Patel KK, Patel S. Enterobacter spp:-An emerging nosocomial infection. International Journal of Applied Research 2 (2016): 532-8.
- Bhat S, KL S, Rao AS, Rao GS. Antibacterial Susceptibility Pattern of Uropathogenic Enterobacter Species from a Tertiary Care Hospital. Journal of Krishna Institute of Medical Sciences (JKIMSU) 1 (2018): 7(4).
- Mortazavi SH, Mansouri F, Azizi M, Alvandi A, KARBASFRUSHAN A, Madadi-Goli N, et al. Prevalence of class I and II integrons among MDR Enterobacter cloacae isolates obtained from clinical samples of children in Kermanshah, Iran. Journal of Clinical & Diagnostic Research 12 (2018).
- Khajuria A, Praharaj AK, Kumar M, Grover N. Carbapenem resistance among Enterobacter species in a tertiary care hospital in central India. Chemotherapy research and practice (2014).
- Adwan G, Rabaya D, Adwan K, Al-Sheboul S. Prevalence of β-lactamases in clinical isolates of Enterobacter cloacae in the West Bank-Palestine. International Journal of Medical Research & Health Sciences 5 (2016): 49-59.
- FA, Tafneen. Detection of AMPC β-lactamases, extended spectrum β-lactamases and carbapenemases in common Enterobacter species by phenotypic and genotypic methods with their antibiotic resistance pattern (2018).
- Hong YK, Lee JY, Ko KS. Colistin resistance in Enterobacter spp. isolates in Korea. Journal of Microbiology 56 (2018): 435-40.
- Dalmolin TV, de Lima-Morales D, Barth AL. Plasmid-mediated colistin resistance: what do we know?. Journal of Infectiology and Epidemiology 21 (2018).
- Gopichand P, Agarwal G, Natarajan M, Mandal J, Deepanjali S, Parameswaran S, Dorairajan LN. In vitro effect of fosfomycin on multi-drug resistant gram-negative bacteria causing urinary tract infections. Infection and drug resistance 9 (2019): 2005-13.
- Tseng SP, Yang TY, Lu PL. Update on fosfomycin-modified genes in Enterobacteriaceae. J Microbiol Immunol Infect 52 (2017): 9-21.
- Zhang R, Huang L,Hu YY. Prevalence of fosfomycin resistance and plasmid- mediated fosfomycin-modifying enzymes among carbapenem-resistant Enterobacteriaceae in Zhejiang, China. J Med Microbiol 66 (2017): 1332-1334.
- Jiang Y, Shen P, Zhou Z, Zhang J,Yu Y, Li L Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6')-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J Antimicrob Chemother 62 (2018): 1252-1256.
- Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. Journal of medical microbiology 62 (2013): 1707-13.
- Mlynarcik P, Roderova M, Kolar M. Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur journal of microbiology 9 (2016).
- Begum N, Shamsuzzaman SM. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Tzu Chi Medical Journal 28 (2016): 94-98.
- Y, Gallah S, Hommeril B, Genel N, Decré D, Rottman M, Arlet G. Emergence of plasmidmediated fosfomycin-resistance genes among Escherichia coli isolates, France. Emerging infectious diseases 23 (2017): 1564.
- Haeili M, Javani A, Moradi J, Jafari Z, Feizabadi MM, Babaei E et al.MgrB alterations mediated colistin resisteance in Klebsiella pneumoniae isolates from iran. Front Microbiol 8 (2017): 2470.



 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
