Technological Innovations in COVID-19 Diagnostics in Singapore
Article Information
Wenfa Ng
Department of Biomedical Engineering, National University of Singapore, Singapore
*Corresponding author: Wenfa Ng, Department of Biomedical Engineering, National University of Singapore, Singapore
Received: 25 July 2023; Accepted: 01 August 2023; Published: 11 August 2023
Citation: Wenfa Ng. Technological Innovations in COVID-19 Diagnostics in Singapore. Fortune Journal of Health Sciences. 6 (2023): 285-293
View / Download Pdf Share at FacebookAbstract
SARS-CoV-2 that caused the COVID-19 pandemic is one exceptional virus with its high transmissivity and delayed onset of symptoms. This article describes the efforts put forth by the biotech industry and universities in Singapore to develop diagnostic tests that aid the detection of positive cases. Direct tests such as RT-PCR and antigen rapid test profile the virus nucleic acid and viral proteins, respectively. But, of equal importance in case detection and treatment are serological tests that measure the relative abundance of IgM and IgG which is indicative of infection phase and quality of immune response in positive cases. Other tests such as isothermal amplification, CRISPR-based diagnostics and breath tests are also in development or in emergency use and would undoubtedly provide valuable usage experience important for the development of molecular assays to detect the next pathogen of global concern.
Keywords
SARS-CoV-2, clinical diagnostics, molecular assays, antigen rapid test, antibody test, polymerase chain reaction
SARS-CoV-2 articles, clinical diagnostics articles, molecular assays articles, antigen rapid test articles, antibody test articles, polymerase chain reaction articles
Article Details
biochemistry, molecular biology, biotechnology, analytical chemistry, immunology,
1. Introduction
Pathogens have always co-existed and co-evolved with us while posing a persistent threat to our survival and well-being. On occasions, dangerous pathogens can emerge that threaten a large fraction of the population. Modern medicine and medical technologies have helped to ameliorate the threat from pathogens through detection, treatment, and vaccination, all of which have been aided by biomedical research. While modern medicine has an excellent job at protecting the world’s population from dangerous pathogens, and limiting the spread of new and emerging pathogens, on rare occasions, a new, highly infectious pathogen could break through our defences in disease surveillance and containment and overwhelms our medical systems. One such pathogen is SARS-CoV-2 that causes the COVID-19 disease.
For a pathogen that has spread worldwide and broken through our surveillance and containment system, and for which curative treatments do not exist, early detection and isolation of cases remain crucial for successful containment of SARS-CoV-2 and limiting its spread. Hence, clinical diagnostics for detecting and confirming cases of SARS-CoV-2 can play crucial roles in aiding containment of the virus, as well as provide early treatment for those infected to help retard clinical progression towards serious disease and complications. Overall, the world’s population is fortunate that, in the current pandemic, key technologies for detection of SARS-CoV-2, and informing of its viral load, and whether someone is exposed to or recovering from the virus are available to aid early use of clinical testing for containing the virus.
This review article summarizes the technological tools and innovations that have aided in the fight against COVID-19 in Singapore on the clinical diagnostic front. Besides detailing the types of diagnostics used, mechanisms as well as unique modifications to standard assays will also be described. In general, Singapore followed international research trends and invested in developing clinical diagnostics which fortuitously helped the nation mount an effective response against COVID-19. These diagnostic methods fall into the general category of polymerase chain reaction (PCR) test, antigen rapid test, and antibody (serological) tests. In addition to the established assay formats are CRISPR-based test and Breathalyzer tests. In particular, breath-based analysis may be the new frontier for rapid, non-invasive screening of new pathogens.
2. Polymerase chain reaction (PCR) tests
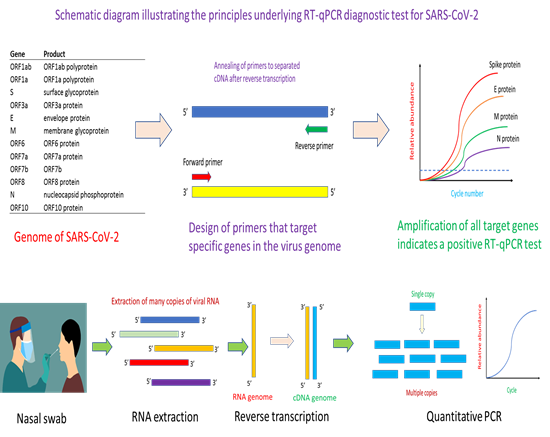
Given its sensitivity and specificity, reverse transcription-PCR (RT-PCR) test was the first tool of choice for the detection of COVID-19 infection [1, 2]. Since SARS-CoV-2 has an RNA-based genome, reverse transcription is needed to transcribe RNA to cDNA prior to PCR amplification. Up until now, RT-PCR remains the gold standard approach for the detection of COVID-19 infection. The first step in the design of a RT-PCR test kit is to obtain an annotated genome sequence of the virus (Figure 1). With automated or manual annotation of the open reading frames (or genes) in the genome, it would then form the basis for the design of primers and probe for specific targeting and amplification of genes of interest in the virus that help identifies positive cases of infection. Key to success in the RT-PCR method is the use of highly specific primers and probes that do not cross-react with similar genes from other viruses. Essentially, the primers help identify the region of a gene for PCR amplification while the probe quantifies, in a specific manner, the amount of PCR amplicons that have been generated. RT-PCR can be multiplexed for increasing its selectivity [3]. Depending on the number of genes targeted in a multiplex RT-PCR assay, correct amplification of all target genes would mean the identification of a positive case of infection [3]. In the current pandemic, many RT-PCR diagnostic kits for COVID-19 have been developed. Similar in concept, the various tests differ in the types of primers and probes used, genes targeted, as well as specificity and sensitivity of the assay [4, 5].
The key principle of the method is the use of genomic information to design primers specific to the gene of interest, whereupon a quantitative PCR assay would help confirm whether the virus is present in a patient’s sample. Essentially, the method starts with a nasal swab to obtain the viral sample. This would be subjected to an RNA extraction step to obtain the needed viral RNA for the downstream reverse transcription step. After reverse transcription, the complementary DNA (cDNA) obtained will serve as template for the binding of quantitative PCR (qPCR) primers for thermal cyclic amplification. Successful amplification of the gene of interest, and observation of fluorescence signal informs the presence of SARS-CoV-2 in the patient’s sample.
Typically, a RT-PCR test starts with obtaining the sample which could be a nasal swab (Figure 1). Other types of samples such as nasopharyngeal, oropharyngeal and throat can also be used for RT-PCR analysis for SARS-CoV-2 virus [6, 7]. The sample is dipped into a universal transfer medium (UTM) to protect the RNA nucleic acid material prior to RNA extraction [8]. Given the difficulty of obtaining reagents during the pandemic, many tests have successfully engineered a workaround that avoids the need for RNA extraction, in what is known as extraction-free RT-PCR assay [8-10]. After RNA extraction, the reverse transcriptase step helps convert the RNA into a complementary DNA (cDNA) in preparation for PCR amplification. At this step, there are two variants of RT-PCR for SARS-CoV-2 detection. One variant is a one-step approach where the reagents for reverse transcription and PCR are in the same master-mix [11, 12]. Another variant is the separation of the reverse transcription step and PCR step into a two-step approach [13]. Typically, the one-step RT-PCR approach is easier to implement and require fewer sample handling steps. The cDNA obtained after reverse transcription would be used for PCR amplification aided by binding of primers and probe to appropriate nucleic acid sequence. For quantitative PCR (qPCR), fluorescence signal from the binding of the probe to PCR amplicons increases with PCR cycle number and help generate a characteristic qPCR curve that affords identification of positive amplification of target gene. Secondly, the Ct value (which is the cycle number at which the fluorescence signal crosses a pre-defined threshold) helps provide a semi-quantitative measure of viral load in the patient’s sample. Typically, a low Ct value signifies a high viral load in the patient, while a high Ct value reflects a low viral load in the patient.
RT-PCR is a mature technology, and many biotech and diagnostic companies as well as universities and research institutes in Singapore step into the fold early in the current pandemic to develop RT-PCR based assays for detecting COVID-19. As of August 2021, a total of 12 locally designed and manufactured tests have been granted provisional authorisation by the Health Sciences Authority of Singapore. In keeping with international trends in RT-PCR diagnostics for COVID-19, extraction-free diagnostic kits were also developed by Singapore researchers, which significantly reduces sample workaround and analysis time for the diagnosis. Examples of such kits are the Resolute 1.0 and Resolute 2.0 kits. Many of the tests delivered results within 1 to 2 hours with sensitivity down to 10 to 20 copies of RNA per reaction. Also, multiplex PCR is commonly used, and has been integrated into many of RT-PCR tests developed in Singapore. Similar to other tests developed internationally, ORF1ab and N genes are the most common target genes. One example of a local test that profiles for ORF1ab and N gene is MIRXES’s Fortitude 3.0 kit. But the coronavirus’s spike (S) protein was also a target gene in a three-gene multiplex system in one of the tests. All in all, most of the tests were developed to be performed in a well-equipped laboratory with trained personnel. However, the biotech and clinical diagnostics industry in Singapore are also moving towards developing automated analytical tools and systems that complements the diagnostic kit that they developed. For example, the extraction-free Resolute 2.0 kit can be used with the RAVE automated system [14]. In another example, the VereCoV™ OneMix Detection Kit is designed for use with the VerePlex bioanalytical system [15]. Beyond automation that helps relieve sample preparation pressure, especially in mass screening, diagnostic companies in Singapore are also moving towards developing point-of-care SARS-CoV-2 diagnostics. One example of such point-of-care COVID-19 diagnostics developed in Singapore is the VitaPCR™ SARS-CoV-2 Gen 2 Assay.
3. Antigen rapid tests
While molecular nucleic acid-based tests provide sensitive and high specificity detection of the virus, the approach typically requires PCR and thermal cycling as well as specialized equipment and personnel to perform the tests. In addition, depending on the number of genes quantified and the length of the amplicons, assay time varies between 1 and 4 hours. The relatively lengthy nature of nucleic acid-based tests thus motivates the development of quicker antibody-based detection of some proteins or fragments of a virus in what is known generally as immunochromatographic antigen rapid test (ART) [16]. Such tests are specific given the highly specific nature in which antibodies bind to the target antigen.
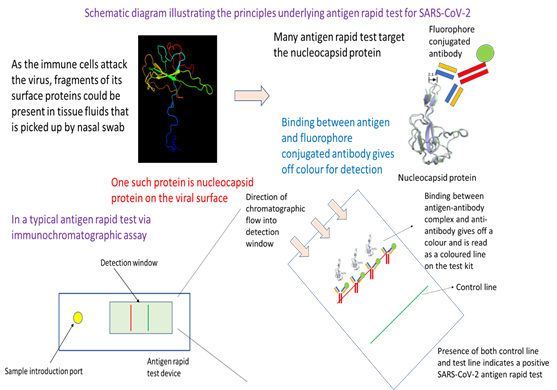
In essence, this testing approach utilizes the specificity in binding between an antibody and an antigen of SARS-CoV-2. Readout is through an antigen-antibody complex-antibody binding mediated release of colour or fluorescence that is visible or which could be detected by an instrument or plate reader. The approach starts off with the collection of a nasal swab. After dipping into a transfer medium where an antibody binds the antigen, the sample is then added to the sample introduction port of an antigen rapid test device. Capillary action from the lateral flow device moves the sample fluid down the chromatographic channel in the membrane. Binding of the antigen-antibody complex with its complement antibody at the detection line would result in colour formation that is indicative of a positive test. Emergence of the control line would indicate that the test has been successfully carried out.
The design of an ART comes from the realisation that it is possible to use specific antibody to recognise specific proteins of a virus. Such recognition can happen over a relatively short time without the aid of specialized analytical instrument or being performed in a specialized laboratory. As readout of the test can be colorimetric, which meant that such antigen rapid tests can be deployed at home or point-of-care or for pre-event screening. The key to mass adoption of such tests is in developing the technologies and methods for manufacturing these antigen rapid tests cheaply [17].
Generally, most ARTs detect the viral proteins as they are good targets for the development of specific antibodies and can help provide colorimetric readout. In the current COVID-19 pandemic, viral nucleocapsid protein (or N protein) is commonly the target antigen in tests used in Singapore for detecting the presence of intact virus or viral fragments. Antibody used to recognise the N protein is typically conjugated to colloidal gold (for colorimetric readout) or a fluorophore (for fluorescence readout), which would emit a colorimetric or fluorescence signal upon binding to capture antibodies at the test line of the immunochromatographic lateral flow assay device. To avoid the need for specialized equipment such as PCR thermocycler, ARTs are typically built on mature lateral flow assay diagnostic methodology. The principles are the same, but what differ are the antibodies used as well as the method for emitting colorimetric or fluorescence signal.
In a typical ART, the nasal swab sample is dipped into a test solution that help bring the antigen protein or fragments of the virus containing the antigen into the test solution (Figure 2). The test solution contains specific antibodies that target the viral surface antigen of interest, which in this case is the N protein. Binding of the antibody with the target antigen then create an immunocomplex that latter would be captured by the antibodies at the test line of the ART device. The next step in the test procedure involves depositing an approximate three drops of test solution with dipped nasal swab sample into the sample well of the ART device. Through rapid capillary action, the test solution containing the aforementioned immunocomplex will migrate across the nitrocellulose membrane where there are different capture antibodies at the test and control line. Capture antibodies at the control line profile for a control human antigen-antibody complex provides feedback on whether the test have been performed correctly. If a coloured band is visible at the control line after about 10-15 minutes, then the test has been performed correctly and is valid. In the case of the test line, capture antibodies there will recognise any aforementioned immunocomplex between target antigen and antibody. Presence of immunocomplex will result in a binding event at the test line, and resulting emission of a colorimetric signal which is visible as a coloured line. Together, presence of coloured bands at the test and control line means that the test result is positive, and the patient’s sample has intact SARS-CoV-2 virus or fragments of the virus. Antigen rapid test is a frontline tool in Singapore’s effort to contain the spread of COVID-19. Deployed at entry checkpoints and also available as a self-test kit for retail sale at pharmacies around the city-state, ART provides fast, reliable result to aid in pre-event screening as well as an initial screening at home to assess infection status.
4. Antibody test
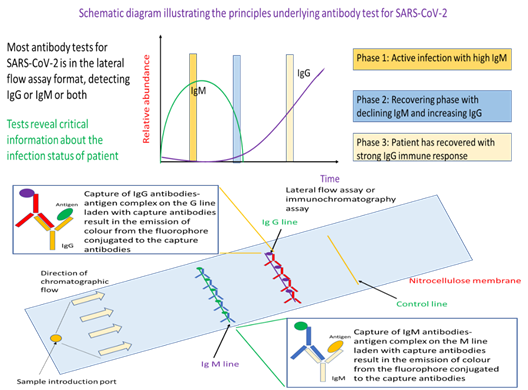
Up until now, the focus has been on technologies that perform direct detection of the virus either through testing for presence of the viral nucleic acids or detecting viral proteins. But there is another approach of testing which can, in addition, provide evidence of whether a person has infection, or to probe for the strength of the immunological reaction provoked by the virus. The latter is of particular importance for infection control and patient treatment given that different control strategies and treatment options are available for patients either in the active infection phase or someone who has partially recovered from the viral infection. In general, such antibody tests profiles for IgM, IgG, or both. Presence of IgM and no IgG represents the active phase of infection where the patient is infectious. As the patient’s immune system is activated to fight the virus, there will be production of IgG followed by a decline in IgM. Hence, detection of IgG and relatively little IgM meant that the patient is recovering from the viral infection and is less infectious.
This test has a strong immunological basis where it seeks to detect the type and concentration of antibody typical of different immune response to assess the infection status of an individual. Specifically, if IgM is detected predominantly, the individual is still in the active infection phase. On the other hand, detection of IgG with no or little IgM indicates that the person has prior exposure to the virus and is recovering. In essence, the workflow starts with a blood draw. Either whole blood, serum or plasma can be used as sample for this antibody test. The sample is added to the sample introduction port of the lateral flow assay device, where antigen for SARS-CoV-2 will be the probe for IgM or IgG. Capillary action will move the sample fluid towards the anti-antibodies that would bind IgM-antigen or IgG-antigen immunocomplex. Such binding would result in colour formation at the IgM or IgG line that would serve as readout for the test. Detection of the control line would indicate that sufficient sample fluid has reach the line, and the test is working properly. Different from RT-PCR and ARTs, antibody tests rely on whole blood, plasma, or serum as samples (Figure 3). The assay format is an established lateral flow assay, where we have the technologies to manufacture at a relatively reasonable price-point for mass deployment such as during the event of a pandemic. In a typical assay, the sample is injected into the sample introduction port of the lateral flow assay. IgM or IgG in the sample will bind probe antigens from the virus that are on the test device to form an immunocomplex. The sample will be moved via capillary action through the nitrocellulose membrane where the IgM-antigen and/or IgG-antigen immunocomplex will be separately captured by fluorophore or colloidal gold conjugated anti-IgM or anti-IgG antibodies at the respective IgM and IgG line. As the respective capture antibodies are conjugated with fluorophore or colloidal gold, binding of the target antibody-antigen immunocomplex would result in a coloured band on the test device at the respective test lines.
In contrast to antigen rapid test, serological antibody tests are not sold over the counter for at-home testing. Similar to ART, a total of four antibody tests were developed by Singapore companies. All profile for IgM and IgG antibodies for SARS-CoV-2 in a single lateral flow assay format. This is a theoretically proven technology that is modular, which means that it could be adapted to detect other pathogens of concern in future simply by changing the capture antibodies and antigens used on the nitrocellulose membrane.
5. Breathalyzer test
Current diagnostic tests still require fairly elaborate sampling and sample processing steps prior to the actual test even for the case of ART. Clearly, RT-PCR, ART and antibody tests are not suitable as diagnostic tool for mass screenings at airports or other point of entries. What is needed is a diagnostic method that is equivalent in speed to a temperature screen by an infrared camera, is low-cost, and requires little or no sample processing steps. This then motivates the development of breath analysis as a method for detecting COVID-19 given the presence of unique volatile organic compounds (VOCs) in the breath of infected patients [18, 19]. Currently, two companies, Breathonix Pte Ltd and Silver Factory Technology Pte Ltd are developing and refining the breath test for COVID-19 detection. Provisional use authorisation has been obtained for Breathonix [20] and Silver Factory Technology.
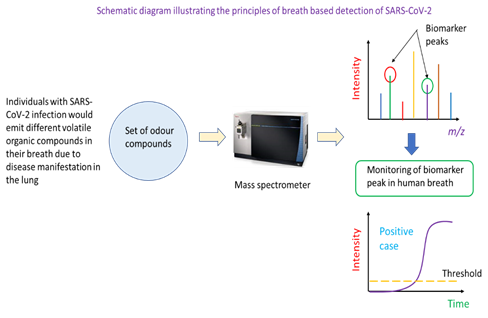
Individuals with SARS-CoV-2 infection may have a unique set of VOCs in their breath. This then constitutes the basis for the identification of infected persons using mass spectrometric analysis. In this detection method, a potential case would be asked to breathe into a breath analyser. This would be drawn into a mass spectrometer which has been programmed to monitor the ion count (or intensity) of specific mass-to-charge ratio ions. These ions are biomarkers of COVID-19 infection as they are VOCs resulting from the infection. A positive case would be called if the ion counts of specific mass ions are above a threshold for a specified analysis duration. Mass spectrometry is one of the instrumented analysis technologies that powers breath-based detection of COVID-19 infection (Figure 4). Specifically, the breath of individuals with is different in the types of VOCs present, and this can be determined through a broad-spectrum scan of compounds in the breath of healthy and infected persons. Mass ions unique to infected persons could serve as biomarkers of COVID-19 infection. In the application of the technique to real samples, the mass spectrometer would be tuned to monitor the number of ions of particular mass-to-charge ratio in the person’s breath (i.e., the ion count of a biomarker mass ion). Such monitoring can be captured and displayed as real-time ion intensity count on the instrument’s screen. Once the ion count of the mass ion biomarker peak is above a pre-determined threshold, the person could be identified as a potential COVID-19 positive case.
In another approach for breath-based detection of SARS-CoV-2 implemented in Silver Factory Technology’s TracieX technology, the phenomenon of surface-enhanced Raman spectroscopy was used to detect biomarker VOCs in the breath of patients. One important feature of the technology is decoupled sample collection and measurement, which helps reduce the risk of cross-infection. Analysis of each sample takes less than 2 minutes [21]. Despite the convenience and speed of detection and screening, any positive case will still require a RT-PCR test to be performed to confirm COVID-19 infection.
6. Other diagnostic tests
Besides the diagnostic tests described above, research is underway in Singapore to develop cheaper, faster, and more accurate diagnostics 2. Efforts in this direction include the development of CRISPR-based tests for faster, more precise, and sensitive detection as well as microfluidic devices to bring the typical lab-based RT-PCR assay to the point-of-care. In the area of CRISPR-based test, researchers are refining the technique to make the test simpler as well as able to profile for SARS-CoV-2 variants. One effort is the development of VaNGuard CRISPR-based SARS-CoV-2 test. Specifically, this test is designed to pick out variants of SARS-CoV-2 through specific guide RNA targeting each variant. Different from conventional RT-PCR, the method only requires isothermal heating and amplification at 60 to 65 oC for about 22 minutes to amplify the target nucleic acid. If target amplicons are present, these would be cleaved off by the Cas protein through recognition by the guide RNA in the subsequent incubation step. Finally, a paper test strip is dipped into the reaction mixture to provide readout where presence of two bands is indicative of a positive test [22]. Use of a paper-based test strip and lack of thermocycler meant that VaNGuard may be amenable to point-of-care application. Sensitivity of the test is about 50 copies of RNA per reaction, which is an order of magnitude less sensitive than the gold standard RT-PCR test.
Separately, Singapore researchers are also developing analytical instrumentation that could bring RT-PCR assay to the point-of-care at a cheaper price-point. Specifically, a portable microfluidic chip-based RT-PCR device known as Epidax is under development to expedite the RT-PCR process to within 1 hour. Compatible with either extraction or extraction-free assays, the method has been shown to be useful for detecting SARS-CoV-2 [23]. The system uses micro-channels to perform PCR amplification. The device uses fluorescence detection to obtain readout of qPCR test. The current sensitivity of the system is 10 RNA copies per microlitre of sample. Similarly, Cell ID in Singapore has also developed a point-of-care laptop powered real-time RT-PCR biochip that runs either the RT-PCR assay or the isothermal reverse transcriptase loop mediated isothermal amplification (RT-LAMP) assay. The test requires 10 µL of nasal swab or saliva as specimen, and can deliver a positive result in 5 minutes, and a negative result within 1 hour [24]. Available as a palm-sized device that could be powered by a laptop, the test could offer the doctor a point-of-care COVID-19 PCR test.
7. Conclusions
Overall, years of investment in development of various types of clinical diagnostics have laid down a strong intellectual foundation and have built strong capabilities in diagnostic design and development in Singapore. This is evident from how Singapore was relatively well-positioned to develop its own RT-PCR test kit for SARS-CoV-2 detection early in the pandemic. Later, research laboratories and companies in Singapore also developed ART and antibody tests based on the established lateral flow assay format. Besides these technologies, local researchers are also focusing on developing biochip-based solutions for a variety of methods ranging from RT-PCR to breathalyser test. Moving forward, with the experience gained from this COVID-19 pandemic in the development of various diagnostic technologies, Singapore is poised to be much better prepared in the event of the next Disease X.
Conflicts of interest
The authors declare no conflicts of interest.
Funding
No funding was used in this work.
References
- Udugama, Buddhisha, Pranav Kadhiresan, Hannah N. Kozlowski, Ayden Malekjahani, et al. “Diagnosing COVID-19: The Disease and Tools for Detection.” ACS Nano, March, acsnano (2020): 0c02624.
- Stang, Andreas, Johannes Robers, Birte Schonert, Karl-Heinz Jöckel, Angela Spelsberg, et al. “The Performance of the SARS-CoV-2 RT-PCR Test as a Tool for Detecting SARS-CoV-2 Infection in the Population.” The Journal of Infection 83 (2021): 237–79.
- Tombuloglu, Huseyin, Hussein Sabit, Ebtesam Al-Suhaimi, Reem Al Jindan, and Khaled R Alkharsah. “Development of Multiplex Real-Time RT-PCR Assay for the Detection of SARS-CoV-2.” PLOS ONE 16 (2021): e0250942.
- Böger, Beatriz, Mariana M Fachi, Raquel O Vilhena, Alexandre F Cobre, Fernanda S Tonin, et al. “Systematic Review with Meta-Analysis of the Accuracy of Diagnostic Tests for COVID-19.” American Journal of Infection Control 49 (2021): 21–29.
- Trevor RA, Grippon S, Chen H, Koh L, Borley D, Oladimeji P, et al. “Meta-Analysis of Rapid Direct-to-PCR Assays for the Qualitative Detection of SARS-CoV-2” (2021).
- Bwire, George M, Mtebe V Majigo, Belinda J Njiro, and Akili Mawazo. “Detection Profile of SARS-CoV-2 Using RT-PCR in Different Types of Clinical Specimens: A Systematic Review and Meta-Analysis.” Journal of Medical Virology 93 (2021): 719–25.
- Sharma, Kuldeep, Pragya Aggarwala, Deepa Gandhi, Anuniti Mathias, Priyanka Singh, et al. “Comparative Analysis of Various Clinical Specimens in Detection of SARS-CoV-2 Using RRT-PCR in New and Follow up Cases of COVID-19 Infection: Quest for the Best Choice.” PLOS ONE 16 (2021): e0249408.
- Smyrlaki, Ioanna, Martin Ekman, Antonio Lentini, Nuno Rufino de Sousa, Natali Papanicolaou, et al. “Massive and Rapid COVID-19 Testing Is Feasible by Extraction-Free SARS-CoV-2 RT-PCR.” Nature Communications 11 (2020): 4812.
- Cameron, Andrew, Nicole D Pecora, and Matthew A Pettengill. “Extraction-Free Methods for the Detection of SARS-CoV-2 by Reverse Transcription-PCR: A Comparison with the Cepheid Xpert Xpress SARS-CoV-2 Assay across Two Medical Centers.” Journal of Clinical Microbiology 59 (2021): e02643-20.
- Byrnes, Samantha A, Ryan Gallagher, Amy Steadman, Crissa Bennett, Rafael Rivera, et al. “Multiplexed and Extraction-Free Amplification for Simplified SARS-CoV-2 RT-PCR Tests.” Analytical Chemistry 93 (2021): 4160–65.
- Queiroz, Jackson Alves da Silva, Rita de Cássia Pontello Rampazzo, Edivá Basílio da Silva Filho, Gabriella Sgorlon Oliveira, Suyane da Costa Oliveira, et al. “Development of a Quantitative One-Step Multiplex RT-QPCR Assay for the Detection of SARS-CoV-2 in a Biological Matrix.” International Journal of Infectious Diseases 104 (2021): 373–78.
- Graham, Thomas GW, Claire Dugast-Darzacq, Gina M Dailey, Xavier Darzacq and Robert Tjian. “Simple, Inexpensive RNA Isolation and One-Step RT-QPCR Methods for SARS-CoV-2 Detection and General Use.” Current Protocols 1 (2021): e130.
- Bustin, Stephen A, and Tania Nolan. “RT-QPCR Testing of SARS-CoV-2: A Primer.” International Journal of Molecular Sciences 21 (2020): 3004.
- A*STAR. “A*STAR & Partners Roll out Resolute 2.0 Test and Automated Lab System RAVE.” A*STAR HQ Corporate (2020).
- NCT Magazine. “NCT Magazine April 2020 - Veredus Laboratories Supplies Coronavirus Test Kit to Singapore for Boarder (Air, Land & Sea) Surveillance and Control Against COVID-19” (2020).
- Peto, Tim, Dominic Affron, Babak Afrough, Anita Agasu, Mark Ainsworth, et al. “COVID-19: Rapid Antigen Detection for SARS-CoV-2 by Lateral Flow Assay: A National Systematic Evaluation of Sensitivity and Specificity for Mass-Testing.” EClinicalMedicine 36 (2021).
- Grant, Benjamin D, Caitlin E Anderson, John R Williford, Luis F Alonzo, Veronika A Glukhova, et al. “SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents.” Analytical Chemistry 92 (2020): 11305–9.
- Ruszkiewicz, Dorota M, Daniel Sanders, Rachel O’Brien, Frederik Hempel, Matthew J Reed, et al. “Diagnosis of COVID-19 by Analysis of Breath with Gas Chromatography-Ion Mobility Spectrometry - a Feasibility Study.” E Clinical Medicine 29 (2020).
- Giovannini, Giorgia, Hossam Haick, and Denis Garoli. “Detecting COVID-19 from Breath: A Game Changer for a Big Challenge.” ACS Sensors 6 (2021): 1408–17.
- “Singapore Provisionally Approves 60-Second COVID-19 Breathalyser Test.” Reuters, May 24, 2021, sec. Healthcare & Pharmaceuticals (2021).
- Silver Factory Technology. 2021. “TracieX.” Silver Factory Technology (2021).
- Chong, Clara. “NTU Team Develops New Covid-19 Rapid Test Kit That Can Detect Variants.” The Straits Times. March 29 (2021).
- NUS Engineering. “On-Site COVID-19 Test Results in One Hour.” NUS Engineering - National University of Singapore (blog). (2020).
- “Singapore Medtech Company Develops Portable Genetic Test for COVID-19” (2020).


 Impact Factor: * 5.814
Impact Factor: * 5.814 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 