Expression Pattern and Clinical Significance of E2F Transcription Factors in Skin Cutaneous Melanoma
Article Information
Liuchang Tan1, Yuangang Lu1*
1Department of Plastic and Cosmetic Surgery, Army Medical Center, Daping Hospital, Army Medical University, Chongqing 400042, China
*Corresponding author: Yuangang Lu, Department of Plastic and Cosmetic Surgery, Army Medical Center, Daping Hospital, Army Medical University, Chongqing, China, No.10 Changjiang Branch Road, Chongqing 400042, China.
Received: 23 November 2022; Accepted: 30 November 2022; Published: 23 February 2023
Citation: Liuchang Tan, Yuangang Lu. Expression Pattern and Clinical Significance of E2F Transcription Factors in Skin Cutaneous Melanoma. Journal of Women’s Health and Development 6 (2023): 16-30.
View / Download Pdf Share at FacebookAbstract
Introduction: E2F is a group of genes that encodes a family of transcription factors in higher eukaryotes. Although emerging evidence indicates that E2Fs are implicated in various cancer types, the diverse expression patterns and prognostic values of E2F transcription factors in SKCM have yet to be elucidated. The purpose of this study was to enhance our knowledge concerning the role of E2Fs in SKCM patients.. Methods: GEPIA, The Human Protein Atlas, TIMER, cBioPortal, GeneMANIA, DAVID 6.8 and KEGG PATHWAY Database were utilized in this study.
Results: We observed a statistically significant increased messenger RNA (mRNA) expression in E2F1/3/5/7 compared with matched normal tissues. A significant correlation was observed between the expression of E2F7/8 and the pathological stage of SKCM patients. In addition, survival analysis revealed that SKCM patients with low transcriptional levels of E2F1/2/3/6 were associated with a significantly better prognosis. Moreover, immune infiltrations analysis showed that transcriptional levels and somatic copy number alterations (SCNA) in E2F family were significantly correlated with several immune cell recruitments, including B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells. The function of differentially expressed E2Fs and their neighboring genes were mainly linked to P53 signaling pathway, cell cycle, and oocyte meiosis.
Conclusions: Taken together, our results may provide novel strategies for the selection of prognostic biomarkers and immunotherapeutic targets in SKCM patients.
Keywords
Expression; E2F Transcription Factors; Prognosis; Skin Cutaneous Melanoma
Expression articles Expression Research articles Expression review articles Expression PubMed articles Expression PubMed Central articles Expression 2023 articles Expression 2024 articles Expression Scopus articles Expression impact factor journals Expression Scopus journals Expression PubMed journals Expression medical journals Expression free journals Expression best journals Expression top journals Expression free medical journals Expression famous journals Expression Google Scholar indexed journals E2F Transcription Factors articles E2F Transcription Factors Research articles E2F Transcription Factors review articles E2F Transcription Factors PubMed articles E2F Transcription Factors PubMed Central articles E2F Transcription Factors 2023 articles E2F Transcription Factors 2024 articles E2F Transcription Factors Scopus articles E2F Transcription Factors impact factor journals E2F Transcription Factors Scopus journals E2F Transcription Factors PubMed journals E2F Transcription Factors medical journals E2F Transcription Factors free journals E2F Transcription Factors best journals E2F Transcription Factors top journals E2F Transcription Factors free medical journals E2F Transcription Factors famous journals E2F Transcription Factors Google Scholar indexed journals Prognosis articles Prognosis Research articles Prognosis review articles Prognosis PubMed articles Prognosis PubMed Central articles Prognosis 2023 articles Prognosis 2024 articles Prognosis Scopus articles Prognosis impact factor journals Prognosis Scopus journals Prognosis PubMed journals Prognosis medical journals Prognosis free journals Prognosis best journals Prognosis top journals Prognosis free medical journals Prognosis famous journals Prognosis Google Scholar indexed journals Skin Cutaneous Melanoma articles Skin Cutaneous Melanoma Research articles Skin Cutaneous Melanoma review articles Skin Cutaneous Melanoma PubMed articles Skin Cutaneous Melanoma PubMed Central articles Skin Cutaneous Melanoma 2023 articles Skin Cutaneous Melanoma 2024 articles Skin Cutaneous Melanoma Scopus articles Skin Cutaneous Melanoma impact factor journals Skin Cutaneous Melanoma Scopus journals Skin Cutaneous Melanoma PubMed journals Skin Cutaneous Melanoma medical journals Skin Cutaneous Melanoma free journals Skin Cutaneous Melanoma best journals Skin Cutaneous Melanoma top journals Skin Cutaneous Melanoma free medical journals Skin Cutaneous Melanoma famous journals Skin Cutaneous Melanoma Google Scholar indexed journals malignant neoplasm articles malignant neoplasm Research articles malignant neoplasm review articles malignant neoplasm PubMed articles malignant neoplasm PubMed Central articles malignant neoplasm 2023 articles malignant neoplasm 2024 articles malignant neoplasm Scopus articles malignant neoplasm impact factor journals malignant neoplasm Scopus journals malignant neoplasm PubMed journals malignant neoplasm medical journals malignant neoplasm free journals malignant neoplasm best journals malignant neoplasm top journals malignant neoplasm free medical journals malignant neoplasm famous journals malignant neoplasm Google Scholar indexed journals predominantly articles predominantly Research articles predominantly review articles predominantly PubMed articles predominantly PubMed Central articles predominantly 2023 articles predominantly 2024 articles predominantly Scopus articles predominantly impact factor journals predominantly Scopus journals predominantly PubMed journals predominantly medical journals predominantly free journals predominantly best journals predominantly top journals predominantly free medical journals predominantly famous journals predominantly Google Scholar indexed journals physiological articles physiological Research articles physiological review articles physiological PubMed articles physiological PubMed Central articles physiological 2023 articles physiological 2024 articles physiological Scopus articles physiological impact factor journals physiological Scopus journals physiological PubMed journals physiological medical journals physiological free journals physiological best journals physiological top journals physiological free medical journals physiological famous journals physiological Google Scholar indexed journals Pathology articles Pathology Research articles Pathology review articles Pathology PubMed articles Pathology PubMed Central articles Pathology 2023 articles Pathology 2024 articles Pathology Scopus articles Pathology impact factor journals Pathology Scopus journals Pathology PubMed journals Pathology medical journals Pathology free journals Pathology best journals Pathology top journals Pathology free medical journals Pathology famous journals Pathology Google Scholar indexed journals diagnostic articles diagnostic Research articles diagnostic review articles diagnostic PubMed articles diagnostic PubMed Central articles diagnostic 2023 articles diagnostic 2024 articles diagnostic Scopus articles diagnostic impact factor journals diagnostic Scopus journals diagnostic PubMed journals diagnostic medical journals diagnostic free journals diagnostic best journals diagnostic top journals diagnostic free medical journals diagnostic famous journals diagnostic Google Scholar indexed journals multidimensional articles multidimensional Research articles multidimensional review articles multidimensional PubMed articles multidimensional PubMed Central articles multidimensional 2023 articles multidimensional 2024 articles multidimensional Scopus articles multidimensional impact factor journals multidimensional Scopus journals multidimensional PubMed journals multidimensional medical journals multidimensional free journals multidimensional best journals multidimensional top journals multidimensional free medical journals multidimensional famous journals multidimensional Google Scholar indexed journals
Article Details
1. Introduction
Skin Cutaneous melanoma (SKCM) is a highly aggressive malignant neoplasm that develops from uncontrolled proliferation and transformation of melanocytes. SKCM accounts for only 2% of total skin cancers while causes over 72% of deaths in skin carcinoma due to its high malignancy and invasiveness [1-5]. The incidence of cutaneous malignant melanoma has been steadily increasing over the last four decades in predominantly fair-skinned populations [6]. The 5-year and 10-year relative survival rates for patients with melanoma are 92% and 89%, respectively [7]. If an early diagnosis is made at the localized stage (stage 0/I/II), the 5-year relative survival rate for melanoma skin cancer can reach 98%, while the patient with stage IV melanoma (distant metastatic disease) has a discouraging prognosis with a 5-year survival of only 23% [8]. Despite enormous international efforts and great achievement in multidisciplinary approaches, SKCM remains a major clinical challenge worldwide and a serious public health problem, laying heavy burdens on society [9]. One of the leading causes of poor tumor prognosis is the lack of early diagnostic method and effective prognostic indicator for guiding the clinical diagnosis, prognosis, and therapy [10]. Therefore, it is imperative to explore the molecular mechanisms underlying the progression in SKCM. Early detection (screening) of melanoma can be crucial to improve prognosis and survival rates of the patients [11]. Besides, as new methods and treatment approaches for precise medicine are available, there is an urgent need for utilizing more precise and effective biomarkers to detect melanoma before distant metastasis formation and to assess the risk of relapse [12,13]. E2Fs, a set of genes that encode a family of transcription factors (TFs) in most higher eukaryotes, are generally divided into the following three subfamilies: transcriptional activator E2Fs (E2F1–E2F3), repressor E2Fs (E2F4–E2F5), and inhibitor E2Fs (E2F6–E2F8) [14,15]. Beyond simply engaging in the regulation of cell cycle, the roles of E2Fs have also been found to be involved in other numerous physiological processes, including cell proliferation [16], apoptosis [17], DNA damage repair [18], senescence [19] and autophagy [20], which are known to be crucial in tumor cell motility and tumor progression. Over the past few years, dysregulated expressions of E2F family members and their correlations with clinicopathological features and prognosis have been partly reported in human SKCM. Overexpression of E2F1 in miR-205-expressing cells has been confirmed to partially reverse the effects on melanoma cell growth and senescence [21]. E2F2 has been identified to establish the links between nicotinamide phosphoribosyltransferase (NAMPT) and p53-dependent apoptosis events in melanoma through NAMPT-SIRT1 axis [22]. Although these experimental results indicated that E2Fs may function as reliable biomarkers for SKCM, the various expression patterns, multiple biological functions, specific prognostic values and molecular pathogenesis of the majority of E2Fs members remain evasive. Meanwhile, it is of great significance to explore effective diagnostic biomarkers and therapeutic targets for SKCM. Thus, we conducted comprehensive analyses to investigate the expression pattern of every E2F family members and their relationships with clinicopathological characteristics and prognosis to further distinguish their potential roles in SKCM.
2. Methods
2.1 Ethics Statement
The study protocol was approved by the Ethics Committee for Human Study (Chongqing, China) in Daping Hospital of Army Medical University in accordance with the principles of the Declaration of Helsinki. All clinical samples were collected between 2017 and 2021 in the Department of Pathology, Daping Hospital of Army Medical University and all patients signed the informed consent form. All written informed consent was obtained as the rest data in this study were retrieved via published literature.
2.2 TCGA and GEPIA Databases
The Cancer Genome Atlas (TCGA), a public funded project that aims to improve diagnostic accuracy, treatment guideline, and ultimately prevent cancer progression, contains sequencing and pathological data for more than 30 different cancers [23]. GEPIA (Gene Expression Profiling Interactive Analysis) (http://gepia.cancer-pku.cn/about.html) is a newly generated web-based bioinformatics tool for analyzing and visualizing the RNA sequencing expression, correlation analysis, survival analysis, and dimensionality reduction analysis of 9,736 tumors and 8,587 normal samples from TCGA and Genotype-Tissue Expression (GTEx) projects using a standard processing pipeline [24]. In this study, GEPIA facilitated the analysis of relative mRNA expression of E2Fs between SKCM (n = 461) and normal tissues (n = 558). The |Log2FC| cutoff and the p value cutoff were defined as 1 and 0.01, respectively. In addition, the cumulative survival analyses and the correlations between transcriptional expression levels of E2Fs and tumor stage in SKCM were also performed using the same dataset in GEPIA.
2.3 TIMER Analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a convenient and accurate web server developed for systematically analyzing and visualizing tumor-infiltrating immune cells on the basis of TCGA database [25]. In this study, the correlations between the mRNA levels of E2Fs and the survival of SKCM patients were first performed in survival module. Then, we used TIMER to assess the correlation between E2F expression levels and abundance of immune infiltrates in SKCM dataset (n = 479). Besides, the correlations between the abundance of immune infiltrates and somatic copy number alterations (SCNA) affecting E2Fs were analyzed using a two-sided Wilcoxon Rank sum test. P-value < 0.05 was considered statically significant.
2.4 STRING
The STRING database (https://string-db.org/) focuses on collecting, scoring and integrating known and predicted protein-protein interaction (PPI) information [26]. We conducted a PPI network analysis of differentially expressed E2Fs to explore the potential interactions via STRING.
2.5 GeneMANIA
GeneMANIA (http://www.genemania.org) is a user-friendly online analysis tool that provides information for predicting the function of genes and gene sets, protein and genetic interactions, pathways, co-expression, co-localization, and protein domain similarity of submitted gene lists [27]. In this study, we applied GeneMANIA to construct a gene–gene interaction network for E2Fs.
2.6 cBioportal
cBioportal (https://www.cbioportal.org/) is an open-access web resource for interactively exploring, visualizing, and analyzing multidimensional cancer genomics data, currently providing access to data regarding the integrative analysis from 105 cancer studies in the TCGA pipeline [28]. The skin cutaneous melanoma (TCGA, Firehose Legacy) dataset, including data from 479 cases with clinical and genomic data, was selected for further analyses of E2Fs using cBioPortal. The frequency of E2F family gene alterations, mutations, putative copy number variance obtained from Genomic Identification of Significant Targets in Cancer (GISTC), mRNA expression z-scores (RNA-Seq V2 RSEM), and protein expression Z scores (reverse phase protein array [RPPA]) were performed according to the online instructions of cBioPortal. Besides, we screened Top 100 most co-related genes among SKCM patients in each family member of E2Fs using co-expression module and then generated their intersections from these 800 genes. Thereafter, we used Venn diagrams drawn by the Draw Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) to select out 38 genes that were contained in at least three subsets for further functional enrichment analysis.
2.7 Functional Enrichment Analysis
Metascape (https://metascape.org) and the Database for Annotation, Visualization and Integrated Discovery (DAVID Bioinformatics Resources 6.8; https://david.ncifcrf.gov/) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the above 38 genes most relevant to E2F family members in SKCM. The genome for homo sapiens was selected as the background parameter. P<0.05 was set as the threshold to indicate a statistically significant difference. Eventually, these results were visualized by a bioinformatics platform (http://www.bioinformatics.com.cn).
2.8 Statistical Analysis
The data were displayed with P-values, fold changes, and ranks and PMID in Table 1. Survival curves were assessed by GEPIA and TIMER with the HR and P value or Cox p-value from a log-rank test. The correlation analysis in TIMER was analyzed using the Wilcoxon test. The correlation of co-expression was adjusted by Spearman’s correlation and their strength of the correlation was determined according to the following absolute value criteria: r = 0.00–0.19 “very weak,” r = 0.20–0.39 “weak,” r = 0.40–0.59 “moderate,” r = 0.60–0.79 “strong,” r = 0.80–1.0 “very strong.” The p-value less than 0.05 were considered statistically significant.
3. Results
3.1 Transcriptional Levels of E2Fs in Patients with SKCM
So far, eight E2F family members have been identified in mammalian genome [29]. To investigate the transcriptional expression patterns of E2Fs in patients with SKCM, we first compared the transcriptional levels of E2Fs in cutaneous melanoma with those in normal tissue samples. In Talantov’s dataset, the expressions of E2F1 and E2F3 were overexpressed in cutaneous melanoma compared to normal tissue with fold changes of 2.731 and 2.648, respectively [30] (Table 1). A similar trend was observed for E2F3 in Riker’s dataset [31]: the mRNA levels of E2F3 in cutaneous melanoma (fold change = 3.146 and p–value = 6.04E-05) were significantly higher than those in normal samples (Table 1). These results indicated that the mRNA expression of E2F1/3 was distinctively upregulated in cutaneous melanoma compared with normal skin tissues, suggesting that E2Fs might play a significant role in the tumorigenesis and progression of cutaneous melanoma.
|
Gene |
Cancer type |
P-value |
Fold change |
t-test |
Sample |
Reference (PMID) |
|
|
E2F1 |
Cutaneous Melanoma |
7.67E-05 |
2.731 |
5.615 |
70 |
16243793 |
|
|
E2F3 |
Cutaneous Melanoma |
6.98E-05 |
3.146 |
5.055 |
87 |
18442402 |
|
|
Cutaneous Melanoma |
1.03E-06 |
2.648 |
10.756 |
70 |
16243793 |
Table 1: The significant changes of E2F expression in transcription level between SKCM tissues and normal skin tissues.
3.2 Relationship between the mRNA Levels of E2Fs and the Clinicopathological characteristics of Patients with SKCM
To further draw the comparisons of E2F factor in the mRNA expression each between SKCM and normal skin tissues, we first generated their expression matrix plots by utilizing multiple gene comparison module in GEPIA (Figure 1A). The density of color in each block represented the median expression value of each E2F family member, normalized by the expression value of maximum median across all blocks. Then, we applied box-plot module to quantify and visualize the transcriptional differences of E2Fs between SKCM patients and normal control. The results suggested that the transcription expression levels of E2F1/3/5/7 in SKCM patients were significantly higher than in normal samples with P < 0.01, whereas the mRNA expression levels of E2F2, E2F4, E2F6, and E2F8 were not significantly different between SKCM and normal tissues (Figure 1B, 1C). Furthermore, we also analyzed the relationship between the transcription levels of E2Fs and the tumor stage in patients with SKCM via stage-plot module. The results showed that E2F7 and E2F8 expression significantly varied among the clinical stages in SKCM (Figure 1D). Overall, the abnormal expressions of E2Fs may have a pivotal role in SKCM progression.
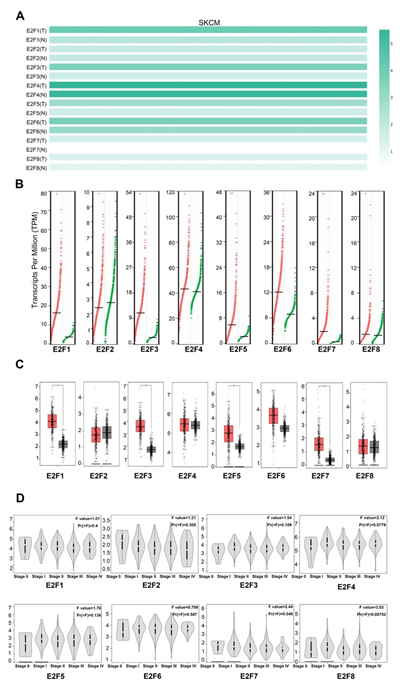
Figure 1: The expression of E2Fs in SKCM and normal skin tissues (A) The expression matrix plots (B) scatter diagram (C) box plot (D) Correlation between E2F expression and tumor stage in patients with SKCM.
3.3 Association of the increased mRNA Expression of E2F1/2/3/6 with the relatively Poor Prognosis of Patients with SKCM
To evaluate the prognostic values of differentially expressed E2Fs in the progression of SKCM, survival plots were employed to calculate the correlation between the E2Fs mRNA levels and the survival in SKCM patients (Figure 2). We first assessed the cumulative survival rates (CSR) of 462 SKCM patients with 222 dying in TIMER database and found that the E2F1/2/3/5/6 expression levels were statistically associated with their CSR (Figure 2A). To further verify the prognostic potential of E2Fs in SKCM, survival module in GEPIA dataset was applied to examine their prognostic value. The results revealed that elevated transcripts per million (TPM) levels of E2F1/2/3/6 were strongly associated with poorer overall survival (OS) rates in the high-expression group compared to the low-expression group, while lower disease-free survival (DFS) were only displayed statistical correlation with higher TPM of E2F3 and E2F6 when the cutoff was P-values < 0.05 (Figure 2B, 2C). Therefore, it is conceivable that aberrant expressions of E2F1/2/3/6 were tightly associated with the relatively deteriorative prognosis of the patients with SKCM, which was compatible with previous studies [21,22,32-35].
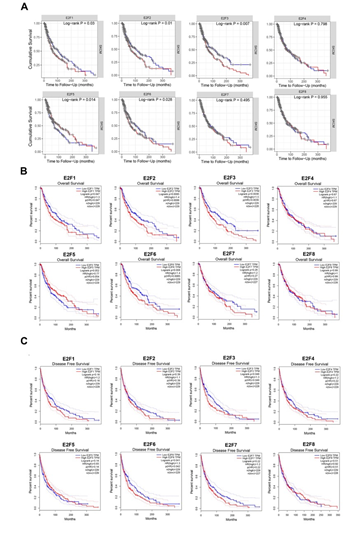
Figure 2: The prognostic value of E2F factors in SKCM (A) cumulative survival rate (TIMER) (B) overall survival (GEPIA) (C) disease free survival.
3.4 E2Fs Expression associated with the Infiltration of Immune Cell in SKCM
As tumor-infiltrating lymphocyte (TIL) grade was identified as an independent predictor of tumor stage and survival in patients with cutaneous melanoma [36], we therefore further embarked upon the correlation between E2Fs and immune cell infiltration in SKCM patients using TIMER database. E2F2 expression was positively associated with the infiltration of B cells (r = 0.202, P = 1.70e−5), CD8+ T cells (r = 0.144, P = 2.49e−3), neutrophils (r = 0.161, P = 1.26e−3) and dendritic cells (r = 0.255, P =4.89e−8), while E2F1 expression was only negatively correlated with the infiltration of macrophages (r = -0.128, P = 6.44e−3). The expression of E2F3 showed positive correlations with infiltrating levels of CD8+ T cells (r = 0.172, P = 2.98e-04) and neutrophils (r = 0.158, P = 7.59e-4). Meanwhile, there was a significant positive correlation between E2F4 expression and the infiltration of neutrophils (r = 0.29, P = 3.41e−10) as well as dendritic cells (r = 0.217, P = 3.80e−6). Moreover, the abundance of neutrophils was positively correlated with the expression of E2F5 (r = 0.308, P = 2.17e-11) and E2F6 (r = 0.312, P = 1.09e-11), respectively. E2F7 expression was positively correlated with infiltration of CD8+ T cells (r = 0.226, P = 1.82e-6) and neutrophils (r = 0.225, P = 1.39e-6), while it was negatively correlated with infiltration of CD4+ T cells (r = -0.073, P = 1.24e-1) and macrophages (r = -0.046, P = 3.28e-1). E2F8 expression exerted weak to moderate positive correlations with the immune infiltrating levels of CD8+ T cells (r = 0.307, P = 5.35e-11), neutrophils (r = 0.439, P = 9.24e-23) and dendritic cells (r =0.313, P = 1.37e-11) (Figure 3A). Somatic copy number alteration (SCNA) is a hallmark of various cancers [37], but its specific role in tumorigenesis and clinical relevance among SKCM patients remain largely unclear. Thus, we also detected the relevance between SCNA in the E2F family and immune infiltration among the patients with SKCM using the SNCA module in TIMER (Figure 3B). The data showed that high amplification in SKCM was notably correlated with the immune infiltrations of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs in both E2F1 and E2F5. Arm-level deletion was significantly associated with the infiltrations of B cells and CD4+ T cells across all the E2F transcription factors and it was prominently correlated with all the six immune cell infiltrations in E2F2. Moreover, the abundances of all six immune infiltrations in E2F3 were remarkably related to arm-level gain, whereas deep deletion was only significantly linked to the infiltration levels of CD4+ T cells in E2F2 (P < 0.05). Taken together, these somatic CNA in the E2F family suppressed the infiltrations of immune cells.
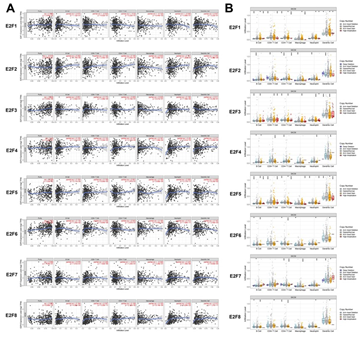
Figure 3: The correlation between E2Fs and immune infiltration in SKCM (A) The correlation between expression of E2Fs and the abundance of immune cells in SKCM (B) The role of somatic copy number alterations in E2Fs and immune cell infiltration in SKCM *P < 0.05, **P < 0.01, ***P < 0.001.
3.5 Genetic Alteration, Co-Expression, Protein-Protein Interaction (PPI) Network Analyses in Patients with SKCM
We then analyzed the E2F alterations and correlations by using the cBioPortal online tool for SKCM (The Cancer Genome Atlas, Firehose Legacy). E2Fs were altered in 263 samples of 477 patients with SKCM (55%) (Figure 4A). High mRNA was the most frequently altered type among all the alterations (33.33% across all samples), followed by multiple alterations (10.9%) and mutation (5.03%) (Table 2). According to the Oncoprint module of this dataset, the highest genetic variation rate in E2Fs was E2F3 (28%), followed by E2F1 (12%), E2F2 (9%), E2F4 (9%), E2F5 (9%), E2F6 (8%), E2F8 (6%) and E2F7 (4%) (Figure 4B). Additionally, we also calculated the correlations of E2Fs with each other by analyzing their mRNA expressions (RNA sequencing [RNA-seq] version (v.)2 RSEM) via the co-expression module for SKCM. The results indicated significant and positive correlations as follows: E2F1 with E2F2 (r = 0.57, P = 9.58e-43) and E2F7 (r = 0.39, P = 2.05e-18); E2F2 with E2F1 (r = 0.57, P = 9.58e-43) and E2F8 (r = 0.32, P = 1.37e-12); E2F3 with E2F5 (r = 0.31, P = 5.50e-12) and E2F6 (r = 0.36, P = 1.03e-15); E2F5 with E2F3 (r = 0.31, P = 5.50e-12); E2F6 with E2F3 (r = 0.36, P = 1.03e-15); E2F7 with E2F1 (r = 0.39, P = 2.05e-18); and E2F8 with E2F2 (r = 0.32, P = 1.37e-12) (Figure 4C).
|
Alteration |
Frequency |
|
Mutation |
5.03% (24 cases) |
|
Amplification |
3.35% (16 cases) |
|
Deep Deletion |
0.84% (4 cases) |
|
mRNA High |
33.33% (159 cases) |
|
mRNA Low |
1.68% (8 cases) |
|
Multiple Alterations |
10.9% (52 cases) |
Table 2: Alteration type detailed summary of E2Fs in SKCM (TCGA, Firehose Legacy).
3.6 Gene altered in 55.14% of 477 Cases
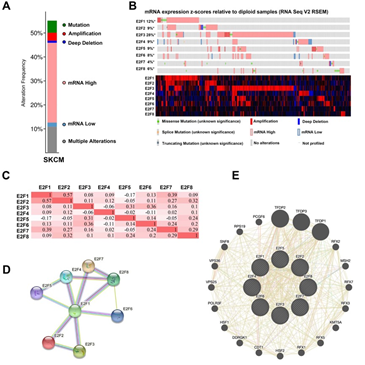
Figure 4: Genetic alteration, Oncoprint, co-expression, protein-protein interaction analyses and neighbor gene network of E2Fs in patients with SKCM. (A) Alteration type summary of different expressed E2Fs in SKCM. (B) Oncoprint visual summary of alteration on a query of E2F family members. (C) Correlation heat map of different expressed E2Fs in SKCM. (D) Protein-protein interaction analysis of E2Fs (E) Gene–gene interaction network of E2Fs.
To explore the potential interactions among E2Fs, we first conducted a PPI network analysis of differentially expressed E2Fs with STRING database. The results indicated that 8 nodes and 12 edges were obtained with PPI enrichment P-value: 1.55e-14 in their PPI network (Figure 4D). To further explore the networks and functions of eight E2Fs, we then applied GeneMANIA to construct their PPI network analysis (Figure 4E). Each node symbolized a gene, the size of the node indicated the strength of the interaction, the colors of nodes represented the possible functions of the respective genes and the color of the connection lines between nodes served as the types of gene–gene interaction. A total of 20 nodes representing genes and 457 links were associated with E2Fs in shared protein domains, co-localization, prediction, physical interactions, co-expression, pathway and genetic interactions. The top 10 genes related to E2Fs were TFDP2 (transcription factor Dp-2), TFDP1 (transcription factor Dp-1), TFDP3 (transcription factor Dp-3), COPS4 (COP9 signalosome subunit 4), PSMD12 (proteasome 26S subunit, non-ATPase 12), KMT5A (lysine methyltransferase 5A), PSMD13 (proteasome 26S subunit, non-ATPase 13), MSH2 (mutS homolog 2), CDT1 (chromatin licensing and DNA replication factor 1) and RFX2 (regulatory factor X2). More specifically, TFDP2 was associated with E2F1/2/3/4/6 regarding physical interaction and shared a pathway interaction with E2F1/4. There also existed associations between TFDP1 and E2F1/2/3/4/5/6 concerning physical interaction, and a genetic interaction was observed between TFDP1 and E2F2. Besides, MSH shared connections with E2F3/5 in terms of co-localization. Except for KMT5A and MSH2, other eight related genes had shared protein domains with E2F1/2/3/4/5/6/7/8. Further functional analysis revealed that these genes were significantly correlated with transcription initiation from RNA polymerase II promoter (FDR = 7.28E-8), followed by DNA-templated transcription, initiation (FDR = 1.02E-7), core promoter binding and G1/S transition of mitotic cell cycle (FDR = 5.23E-5).
3.7 Functional Enrichment Analysis of E2Fs in SKCM Patients
In order to elucidate the underlying mechanism of E2Fs in SKCM, enrichment analyses were performed to analyze the functions of differentially expressed E2Fs and their neighboring genes in SKCM. Utilizing the co-expression module in cBioPortal, we first screened top 100 most co-related genes in each family member of E2Fs among SKCM patients and then generated their intersections from these 800 genes. Next, we selected out 38 genes that were closely associated with E2F alterations and functions for further enrichment analysis. The functions of E2Fs and the genes related to E2F alterations in SKCM were predicted by analyzing gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in Metascape (https://metascape.org) and DAVID (https://david.ncifcrf.gov/). Figure 5 showed the results of the functional enrichment analysis obtained from Metascape. As presented in Figures 5A, B, the functions of differentially expressed E2Fs and their neighboring genes were mainly enriched in chromosome segregation, cell cycle, microtubule cytoskeleton organization and DNA repair (Table 3). To better identify densely connected network components of differentially expressed E2Fs and their neighboring genes in SKCM, the PPI network and the Molecular Complex Detection (mCODE) components were analyzed via Metascape. The MCODE networks identified for the gene lists were gathered in Figure 5C, D. Then, we extracted the most significant mCODE components from the PPI network of E2Fs in SKCM. Pathway and process enrichment analyses were also applied to each MCODE component independently, and the functional roles of target genes were mainly focused on chromosome segregation, nuclear division and cell cycle. Besides, the three best-scoring terms of the functional description in the corresponding components by P-value were retained, including mitotic anaphase, resolution of sister chromatid cohesion as well as mitotic metaphase and anaphase.
|
GO |
Category |
Description |
Count |
% |
Log10 (P) |
Log10 (q) |
|
GO: 0007059 |
GO Biological Processes |
Chromosome segregation |
19 |
50 |
-26.21 |
-21.85 |
|
R-HAS-1640170 |
Reactome Gene Sets |
Cell Cycle |
22 |
57.89 |
-25.37 |
-21.48 |
|
GO: 0010564 |
GO Biological Processes |
regulation cytoskeleton organization |
18 |
47.37 |
-17.73 |
-14.45 |
|
GO: 0000226 |
GO Biological Processes |
Microtubule cytoskeleton organization |
14 |
36.84 |
-13.59 |
-10.55 |
|
GO: 0006281 |
GO Biological Processes |
DNA repair |
12 |
31.58 |
-11.14 |
-8.26 |
|
WP2446 |
WikiPathways |
Retinoblastoma Gene in Cancer |
7 |
18.42 |
-10.58 |
-7.74 |
|
GO: 0007088 |
GO Biological Processes |
regulation of mitotic nuclear division |
7 |
18.42 |
-9.91 |
-7.15 |
|
GO: 0051321 |
GO Biological Processes |
meiotic cell cycle |
8 |
21.05 |
-8.87 |
-6.2 |
|
M176 |
Canonical Pathways |
PID FOXM1 PATHWAY |
5 |
13.16 |
-8.66 |
-6.02 |
|
GO: 0090068 |
GO Biological Processes |
positive regulation of cell cycle process |
8 |
21.05 |
-8.22 |
-5.64 |
|
GO: 0071103 |
GO Biological Processes |
DNA conformation change |
8 |
21.05 |
-7.71 |
-5.17 |
|
M242 |
Canonical Pathways |
PID AURORAA PATHWAY |
4 |
10.53 |
-7.06 |
-4.57 |
|
GO: 0000910 |
GO Biological Processes |
cytokinesis |
6 |
15.79 |
-6.97 |
-4.5 |
|
GO: 0071539 |
GO Biological Processes |
protein localization centrosome |
3 |
7.89 |
-4.92 |
-2.68 |
|
GO: 0031109 |
GO Biological Processes |
microtubule polymerization or depolymerization |
4 |
10.53 |
-4.72 |
-2.54 |
|
GO: 0006513 |
GO Biological Processes |
protein monoubiquitination |
3 |
7.89 |
-4.01 |
-1.02 |
|
GO: 1902275 |
GO Biological Processes |
regulation of chromatin organization |
3 |
7.89 |
-2.68 |
-0.69 |
Table 3: Top 17 clusters with the representative enriched terms of E2Fs and their neighboring genes in SKCM (Metascape).
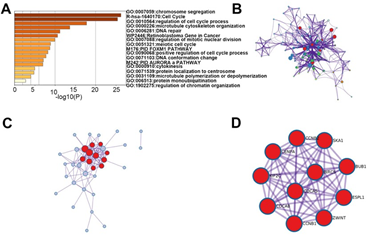
Figure 5: The enrichment analysis of different expressed E2Fs and 38 most frequently altered neighbor genes in SKCM (Metascape). (A) Bar graph of top 17 enriched terms across different expressed E2Fs and 38 most frequently altered neighboring genes, colored by p values. (B) Network of enriched terms, colored by cluster ID. (C-D) PPI and MCODE components identified in different expressed E2Fs and 38 most frequently altered neighboring genes. Pathway and process enrichment analysis has been applied to each MCODE component independently, and the three best-scoring terms by p-value have been retained as the functional description of the corresponding components, shown in the tables underneath corresponding network plots.
We additionally validated our results using DAVID 6.8. GO enrichment analysis. GO analysis in DAVID database predicted the functional roles of target genes based on three aspects: processes (BP), cellular components (CC), and molecular functions (MF). Among the 10 most highly enriched functions in the BP category, cell division, sister chromatid cohesion and mitotic sister chromatid segregation were remarkably associated with the tumorigenesis and progression of SKCM (Figure 6A). Nucleus, nucleoplasm, spindle microtubule, chromosome, centromeric region, cytosol, cytoplasm, chromosome kinetochore, microtubule, midbody and chromosome passenger complex were the 10 most highly enriched items in the CC category (Figure 6B). As for the MF category, protein binding, microtubule binding, chromatin binding, ATP binding and DNA binding were notably regulated by E2Fs alterations and their neighboring genes in SKCM (Figure 6C). Finally, we utilized KEGG analysis to define the possible pathways linked to the functions of E2F alterations and the frequently altered neighbor genes. The top 5 KEGG pathways for the E2F family members and their neighboring genes were shown in Figure 6D. Among these pathways, cell cycle and P53 signaling pathway were involved in the tumorigenesis and pathogenesis of melanoma according to the KEGG pathway database (Figure 7 A-B).
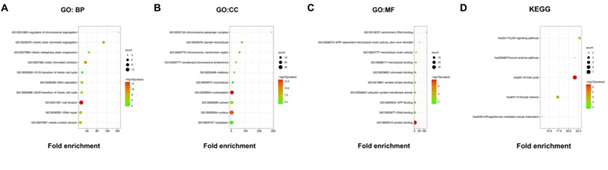
Figure 6: The Functions of E2Fs and Genes Significantly Associated with E2F Alterations in SKCM patients. The functions of E2Fs and genes significantly associated with E2F alterations were predicted by the analysis of gene ontology (GO) by DAVID (Database for Annotation, Visualization and Integrated Discovery) tools GO enrichment analysis predicted the functional roles of target host genes based on three aspects, including (A) biological processes (BP) (B) cellular components (CC) and (C) molecular functions (MF) (D) Kyoto Encyclopedia of Genes and Genomes (KEGG).
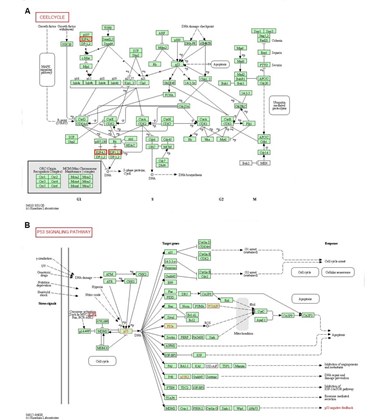
Figure 7: Cell cycle and P53 signaling pathway regulated by the E2F alteration in melanoma. The cell cycle (A) and P53 signaling pathway (B) are shown.
4. Discussion
The E2F family, a group of genes encoding a set of transcription factors in higher eukaryotes, has been involved not only in cell cycle regulation but also in DNA synthesis. Recently, accumulative evidences have suggested that the dysregulation of E2Fs is involved in the occurrence and development of several malignancies [38-40]. Although some E2F family members have been confirmed to play promising roles in SKCM by experimental data [21,22,41,42], further systematic bioinformatics analysis of SKCM remains to be elucidated. The aim of our study was to detect the possible connections among the expression patterns, clinicopathological characteristics, prognostic values, immune cell infiltration, genetic alteration, co-expression, protein-protein interaction, and functional enrichment analysis of different E2F factors in SKCM, thereby providing strategies for the development of clinical medicine, optimizing treatment methods and extending the lifetimes for the patients with SKCM. Among the E2F transcription factors, E2F1 is most extensively studied in melanoma studies [21,32,43-45]. E2F1, a key regulator of cell cycle and survival, is often overexpressed or hyperactivated in a variety of tumors, including melanoma [44]. E2F1 may contribute to the cooperation with alterations in promoting cell proliferation through the MAPK or Rb pathways among most types of melanoma [43]. Either inhibition or block of E2F1 activity has been found to induce cell death of metastatic melanoma [32]. Besides, E2F1 has been identified as a crucial mediator of Hedgehog/GLI- iASPP axis to regulate melanoma cell growth and survival [46]. Our results showed that E2F1 mRNA was highly up-regulated in SKCM compared with that in normal control based on data from Oncomine, TCGA datasets and GEPIA. Although E2F1 expression was not lined with the clinical stages, elevated expression of E2F1 was significantly associated with a worse CSR and OS in SKCM, consistent with a previous finding showing that overexpression of E2F1 was common in high-grade tumors and was associated with poor patient survival [47]. E2F2 and E2F3 are also closely linked to the occurrence and development of melanoma. For instance, E2F2 has been shown to involve in the NAMPT/SIRT1 axis to govern melanoma cells proliferation and apoptosis resistance [22]. Moreover, E2F3 has been found to promote cancer growth and is overexpressed through copy number variation in human melanoma [48]. In our study, the transcription expression levels of E2F3 in SKCM patients were significantly higher than in normal samples, whereas the mRNA expression levels of E2F2 were not significantly different between SKCM and normal tissues. Further survival analysis indicated that transcriptional expression levels of E2F2 and E2F3 were associated with CSR and OS in patients with SKCM, while DFS rate in high E2F3 expression group was lower than in low E2F3 expression group.
E2F4 and E2F5, the transcriptional suppressors of E2F transcription factors, have been reported to play an essential role in pocket protein-mediated G1 control of cycling cells [49]. Increased levels of E2F4-RB repressive complexes is associated with replicative senescence in normal melanocytes [50]. E2F5 has been reported to have an inverse correlation with the expression of miR-205 that is a tumor suppressor in melanoma [21]. The data in this report found that the mRNA expression of E2F5 in SKCM was higher than that in normal tissues and its aberrant high expression level was only significantly correlated with CSR. No significance was observed between the mRNA expression of E2F4 and prognostic value (CRS, OS and DFS) in the patients with SKCM. E2F6, one of the unique E2F family members, is considered as a transcriptional repressor, working independently of the pRB family [51,52]. The absence of E2F6 has been reported to be advantageous to melanoma cells, as it can cause unusual proliferation arrest when overexpressed [53,54]. Although there is no significance of E2F6 in mRNA level between SKCM and normal skin tissues, its mRNA level is related to all the prognostic value (CRS, OS and DFS) evaluated in this study, which suggests that E2F6 could be proposed as an effective diagnostic biomarker in patients with SKCM.Synergistic function of E2F7 and E2F8 is thought to be essential for cell survival and embryonic development [55]. Although their synergistic effects are crucial to suppress stress-induced skin cancer [56], less is known about their significance in patients with SKCM. In the presented study, the mRNA levels of E2F7 in SKCM were significantly overexpressed whereas the E2F8 mRNA levels in SKCM did not differ from normal tissues. Meanwhile, the transcriptional expressions of E2F7/8 varied as the clinical stages progressed in SKCM, but had no association with CSR, OS and DFS. Therefore, future studies are required to explore possible functions of E2F7 and E2F8 in patients with SKCM.
Tumor-infiltrating lymphocyte (TIL) grade is an independent predictor of survival in patients with melanoma and patients with a pronounced TIL infiltrate have an excellent prognosis [36]. Since multiple E2Fs expressions were significantly correlated with prognostic values, we then explored their immune cell infiltration in SKCM. The transcription level of E2F1 was negatively correlated with infiltration of macrophages while E2F4/5/6/7/8 expressions were weak to moderate positively correlated with the infiltration of neutrophils. Meanwhile, CD8+ T cells was positively correlated with the expression of E2F6/7/8 and there also existed a positive correlation between dendritic cells and E2F2/4/8. Notably, B cells were found to only share a positive correlation with E2F2. Taken together, the correlation between the expression of E2Fs and the abundance of immune cell indicates that E2Fs may be involved in the regulation of tumor immunity in SKCM. Moreover, the relevance between SCNA in the E2F family and immune infiltration among the patients with SKCM were detected using the SNCA module in TIMER. High amplification, arm-level deletion and arm-level gain were the most prevalent SCNAs in SKCM and we found that the infiltrations of immune cells were inhibited by these somatic CNA in the E2F family. To further clarify the genetic alteration, potential function, and potential biological interaction and reaction networks of the E2F family members in SKCM, we explored the molecular characteristics of E2Fs. We first calculated the percentages of genetic alterations in E2F family members for SKCM and the result showed that they varied from 4 to 28% for individual E2F family members. Among all the alterations, elevated mRNA was the most frequently altered type, followed by multiple alterations and mutation. Besides, the co-expression module among the differentially expressed E2Fs indicates that E2Fs may play a synergistic role in the tumorigenesis and progression of SKCM. In addition to the above, the alterations in E2F genes among SKCM patients were associated with several common cancer-related pathways, such as P53 signaling pathways and cell cycle. These finding are consistent with previous studies, where E2Fs are highly regulated throughout cell cycle and P53 signaling pathway [17,22,41,42,57]. Moreover, the functions of E2F alterations and the frequently altered neighboring genes in SKCM were also found to share connections with Fanconi anemia pathway, oocyte meiosis and progesterone mediated oocyte maturation, which were responsible for tumor proliferation and invasion [58-60]. Therefore, E2F transcription factors may regulate the tumorigenesis and progression of SKCM via these target pathways. In the near future, we propose to explore the extent and clinical implications, further decipher the E2F transcriptional signatures between patients and tumor regions and discuss how profiling the E2F expression pattern could help clinicians for precision medicine, new prognostic signatures, and methodical therapy regimen for patients with SKCM.
Author Contributions
Conceptualization, methodology, investigation, formal analysis, software and writing—original draft, Liuchang Tan; writing—review, editing and supervision, Yuangang Lu. All authors reviewed the results and finally approved this version of the manuscript.
Declaration of Competing Interest
The authors declared no conflicts of interest in this study.
Acknowledgements
The results presented here were based on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
This work was supported by National Natural Science Foundation of China (82272763).
Reporting Checklist
The authors have completed the TRIPOD reporting checklist.
Conflicts of Interest
All authors have completed the ICMJE uniform disclosure form. The authors have no conflicts of interest to declare.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
References
- Liu Y, Sheikh MS. Melanoma: Molecular Pathogenesis and Therapeutic Management. Mol Cell Pharmacol 6 (2014): 228.
- Tang J, Fewings E, Chang D, et al. The genomic landscapes of individual melanocytes from human skin. Nature 586 (2020): 600-605.
- van der Weyden L, Brenn T, Patton EE, et al. Spontaneously occurring melanoma in animals and their relevance to human melanoma. J Pathol 252 (2020): 4-21.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 70 (2020): 7-30.
- Singh BP, Salama AK. Updates in Therapy for Advanced Melanoma. Cancers (Basel) 8 (2016).
- Memon A, Bannister P, Rogers I, et al. Changing epidemiology and age-specific incidence of cutaneous malignant melanoma in England: An analysis of the national cancer registration data by age, gender and anatomical site, 1981–2018. The Lancet Regional Health – Europe 2 (2021).
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66 (2016): 271-289.
- Rebecca VW, Somasundaram R, Herlyn M. Pre-clinical modeling of cutaneous melanoma. Nat Commun 11 (2020): 2858.
- Tripp MK, Watson M, Balk SJ, et al. State of the science on prevention and screening to reduce melanoma incidence and mortality: The time is now. CA Cancer J Clin 66 (2016): 460-480.
- Dong D, Mu Z, Zhao C, et al. ZFAS1: a novel tumor-related long non-coding RNA. Cancer Cell Int 18 (2018): 125.
- Voss RK, Woods TN, Cromwell KD, et al. Improving outcomes in patients with melanoma: strategies to ensure an early diagnosis. Patient Relat Outcome Meas 6 (2015): 229-242.
- Broggi MAS, Maillat L, Clement CC, et al. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J Exp Med 216 (2019): 1091-1107.
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 372 (2015): 793-795.
- Hazar-Rethinam M, Endo-Munoz L, Gannon O, et al. The role of the E2F transcription factor family in UV-induced apoptosis. Int J Mol Sci 12 (2011): 8947-8960.
- Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. EMBO J 23 (2004): 4709-4716.
- Miles WO, Dyson NJ. RB-loss puts focus on Myc. Nat Cell Biol 17 (2015): 968-969.
- Irwin M, Marin MC, Phillips AC, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407 (2000): 645-648.
- Blanchet E, Annicotte JS, Lagarrigue S, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 13 (2011): 1146-1152.
- Rajarajacholan UK, Thalappilly S, Riabowol K. The ING1a tumor suppressor regulates endocytosis to induce cellular senescence via the Rb-E2F pathway. PLoS Biol 11 (2013): e1001502.
- Haim Y, Bluher M, Slutsky N, et al. Elevated autophagy gene expression in adipose tissue of obese humans: A potential non-cell-cycle-dependent function of E2F1. Autophagy 11 (2015): 2074-2088.
- Dar AA, Majid S, de Semir D, et al. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 286 (2011): 16606-16614.
- Zhao H, Tang W, Chen X, et al. The NAMPT/E2F2/SIRT1 axis promotes proliferation and inhibits p53-dependent apoptosis in human melanoma cells. Biochem Biophys Res Commun 493 (2017): 77-84.
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 490 (2012): 61-70.
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45 (2017): W98-W102.
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 77 (2017): e108-e10.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47 (2019): D607-D13.
- Mostafavi S, Ray D, Warde-Farley D, et al. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 9 (2008): S4.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2 (2012): 401-404.
- Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell 127 (2006): 871-874.
- Talantov D, Mazumder A, Yu JX, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res 11 (2005): 7234-7242.
- Riker AI, Enkemann SA, Fodstad O, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 1 (2008): 13.
- Rouaud F, Hamouda-Tekaya N, Cerezo M, et al. E2F1 inhibition mediates cell death of metastatic melanoma. Cell Death Dis 9 (2018): 527.
- Ghosh R, Nadiminty N, Fitzpatrick JE, et al. Eugenol causes melanoma growth suppression through inhibition of E2F1 transcriptional activity. J Biol Chem 280 (2005): 5812-5819.
- Xia Y, Zhou Y, Han H, et al. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol 234 (2019): 19592-19601.
- Deng T, Kuang Y, Wang L, et al. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochem Biophys Res Commun 387 (2009): 611-616.
- Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 30 (2012): 2678-2683.
- Franch-Exposito S, Bassaganyas L, Vila-Casadesus M, et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. Elife 9 (2020).
- Zhou Q, Zhang F, He Z, et al. E2F2/5/8 Serve as Potential Prognostic Biomarkers and Targets for Human Ovarian Cancer. Front Oncol 9 (2019): 161.
- Liu XS, Gao Y, Liu C, et al. Comprehensive Analysis of Prognostic and Immune Infiltrates for E2F Transcription Factors in Human Pancreatic Adenocarcinoma. Front Oncol 10 (2020): 606735.
- Huang YL, Ning G, Chen LB, et al. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag Res 11 (2019): 1725-1740.
- Haddad MM, Xu W, Schwahn DJ, et al. Activation of a cAMP pathway and induction of melanogenesis correlate with association of p16(INK4) and p27(KIP1) to CDKs, loss of E2F-binding activity, and premature senescence of human melanocytes. Exp Cell Res 253 (1999): 561-572.
- Bandyopadhyay D, Medrano EE. Melanin accumulation accelerates melanocyte senescence by a mechanism involving p16INK4a/CDK4/pRB and E2F1. Ann N Y Acad Sci 908 (2000): 71-84.
- Roberts JD. E2F1 amplication and genetic heterogeneity in melanoma. Cancer Biol Ther 5 (2006): 691-692.
- Nelson MA, Reynolds SH, Rao UN, et al. Increased gene copy number of the transcription factor E2F1 in malignant melanoma. Cancer Biol Ther 5 (2006): 407-412.
- Rocca MS, Benna C, Mocellin S, et al. E2F1 germline copy number variations and melanoma susceptibility. J Transl Med 17 (2019): 181.
- Pandolfi S, Montagnani V, Lapucci A, et al. HEDGEHOG/GLI-E2F1 axis modulates iASPP expression and function and regulates melanoma cell growth. Cell Death Differ 22 (2015): 2006-2019.
- Alla V, Engelmann D, Niemetz A, et al. E2F1 in melanoma progression and metastasis. J Natl Cancer Inst 102 (2010): 127-133.
- Feng Z, Peng C, Li D, et al. E2F3 promotes cancer growth and is overexpressed through copy number variation in human melanoma. Onco Targets Ther 11 (2018): 5303-5313.
- Gaubatz S, Lindeman GJ, Ishida S, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell 6 (2000): 729-735.
- Bandyopadhyay D, Timchenko N, Suwa T, et al. The human melanocyte: a model system to study the complexity of cellular aging and transformation in non-fibroblastic cells. Exp Gerontol 36 (2001): 1265-1275.
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3 (2002): 11-20.
- Pei Y, Banerjee S, Sun Z, et al. EBV Nuclear Antigen 3C Mediates Regulation of E2F6 to Inhibit E2F1 Transcription and Promote Cell Proliferation. PLoS Pathog 12 (2016): e1005844.
- Halaban R, Cheng E, Smicun Y, et al. Deregulated E2F transcriptional activity in autonomously growing melanoma cells. J Exp Med 191 (2000): 1005-1016.
- Gaubatz S, Wood JG, Livingston DM. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci U S A 95 (1998): 9190-9195.
- Li J, Ran C, Li E, et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell 14 (2008): 62-75.
- Thurlings I, Martinez-Lopez LM, Westendorp B, et al. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene 36 (2017): 829-839.
- Xu W, McArthur G. Cell Cycle Regulation and Melanoma. Curr Oncol Rep 18 (2016): 34.
- Nepal M, Che R, Zhang J, et al. Fanconi Anemia Signaling and Cancer. Trends Cancer 3 (2017): 840-856.
- Bourseguin J, Bonet C, Renaud E, et al. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells. Sci Rep 6 (2016): 36539.
- Majerus MA. The relationship between the cancer cell and the oocyte. Med Hypotheses 58 (2002): 544-551.


 Impact Factor: * 1.1
Impact Factor: * 1.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 