The Cold-Active Endo-β-1,3(4)-Glucanase from a Marine Psychrophilic Yeast, Glaciozyma antarctica PI12: Heterologous Expression, Biochemical Characterisation, and Molecular Modeling
Article Information
Salimeh Mohammadia,b,*, Noor Haza Fazlin Hashima,c, Nor Muhammad Mahadid, Abdul Munir Abdul Murada
aSchool of Biosciences and Biotechnology, Faculty of Science and Technology, University Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia
bDepartment of Physics, California State University, Northridge, United State
cWater Quality Laboratory, National Hydraulic Research Institute Malaysia (NAHRIM), Ministry of Natural Resources and Environment, Lot5377, Jalan Putra Permai, 43300, Seri Kembangan, Selangor, Malaysia
dMalaysia Genome Institute, Jalan Bangi Lama, 43000, Kajang, Selangor, Malaysia
*Corresponding Author: Salimeh Mohammadi, Department of Physics, California State University, Northridge, USA
Received: 30 November 2020; Accepted: 08 December 2020; Published: 02 January 2021
Citation:
Salimeh Mohammadi, Noor Haza Fazlin Hashim, Nor Muhammad Mahadi, Abdul Munir Abdul Murad. The Cold-Active Endo-β-1,3(4)-Glucanase from a Marine Psychrophilic Yeast, Glaciozyma antarctica PI12: Heterologous Expression, Biochemical Characterisation, and Molecular Modeling. International Journal of Applied Biology and Pharmaceutical Technology 12 (2020): 279-300.
View / Download Pdf Share at FacebookAbstract
Glaciozyma antarctica is a psychrophilic yeast that was isolated from the surface of Antarctic sea ice. A key adaptation of psychrophilic microorganisms is to synthesize cold-active enzymes for survival at low temperatures. A full-length cDNA encoding β-glucanase (GaEgl) from G. antarctica PI12 was amplified by reverse-transcription polymerase chain reaction (RT-PCR). The cDNA encoded a 394-residue polypeptide with a putative signal peptide of 22 residues. Subsequently, the novel GaEgl was expressed in E. coli and purified with nickel affinity chromatography as an approximately 44 kDa protein. The biochemical characterisation of purified recombinant GaEgl (rGaEgl) revealed typical cold-active enzyme characteristics, such as maximal activity at 20 °C and pH 7.0. However, the enzyme was still active at 5-15 °C and alkaline pH values of 8-10. The activity of recombinant GaEgl was enhanced in the presence of Co2+ and Mn2+ metal ions. The Km and Vmax values of the enzyme using lichenan as the substrate were 8.87 mg mL-1 and 37.45 U mg-1, respectively. The enzymatic hydrolysis analysis of laminarin using HPLC showed that the main hydrolysis products were monosaccharides, disaccharides and trisaccharides. An analysis of the three-dimensional structure of the enzyme was carried out and compared with homologous mesophilic endo-β-1,3(4)-glucanase. The results of the comparative structural study revealed that the psychrophilic GaEgl contains longer loops, fewer hydrogen bonds and salt bridges, and a higher total solvent-accessible surface area which enhanced the protein flexibility for high catalytic efficiency at low temperatures.
Keywords
Cold active enzyme; Glaciozyma Antarctica; Endo-glucanase; 3D model; Laminarinase; Glycoside hydrolase
Cold active enzyme articles; Glaciozyma Antarctica articles; Endo-glucanase articles; 3D model articles; Laminarinase articles; Glycoside hydrolase articles
Article Details
Introduction
Psychrophiles organisms are important in global ecology because over 80% of the earth is permanently or periodically exposed to temperatures below 5 °C [1]. Psychrophiles are cold-tolerant organisms that are able to grow at very low temperatures in freezing habitats such as ice cores, deep sea waters, glaciers, snow, and mountain regions in the Antarctic [1-3]. Cold temperatures can adversely impact the architecture and growth of cells [4]. Hence, psychrophiles employ various adaptive mechanisms to withstand harsh and cold environments. The secretion of cold-adapted enzymes is a key adaptive mechanism of psychrophiles.
Cold-adapted enzymes, which are characterised by high turnover number (kcat) and catalytic efficiency (kcat/Km) at low temperatures, allow psychrophiles to compensate for low metabolic fluxes and slow reaction rates at their physiological temperatures [5]. Cold-active enzymes are more thermolabile than their thermophilic and mesophilic counterparts, which confers conformational flexibility on the active site. The structural flexibility and stability of psychrophilic enzymes are achieved through structural modifications, which include decreasing proline and arginine contents, increasing glycine content. Other modification, weakening intramolecular bonds through a reduction in the number of hydrogen bonds and salt bridges, increasing the number of hydrophobic side chains exposed to the solvent, producing long and frequent hydrophilic loops, and decreasing the compactness of the hydrophobic core [6].
Glaciozyma antarctica is an obligate psychrophilic marine yeast isolated from Antarctic sea ice [7]. It is an obligate psychrophilic yeast where the optimal growth temperature is 12 °C and not able to survive more than 20 °C [8]. It was reported G. antarctica able to produce antifreeze proteins and several cold adapted enzymes like protease and chitinase as cold adaptation mechanism [7,9,10]. In this work, we report the isolation, heterologous expression, biochemical characterisation and structural analysis of the recombinant endo-β-1,3(4)-glucanase from G. antarctica.
Beta-1,3(4)-glucanases, which belong to glycoside hydrolase (GH) family 16 and hydrolyze the β-1,3 and β-1,4 linkages in β-glucan, have received more attention in recent research. β-1,3(4)-glucanases are important biotechnological aids such as in brewing, animal feed, and food industries. As yet, a few β-1,3(4)-glucanases from mesophilic and thermophilic microorganism have been purified and characterised for industrial application; however, currently, the lack of study aboserved on cold-adapted β-1,3(4)-glucanase from a psychrophilic microorganism [11,12]. In this study, the properties of G. antarctica endo-β-1,3(4)-glucanase would facilitate the understanding of polysaccharides utilization in sea ice of Antarctica. In addition, characterisation of the cold-active β-glucanase is essential to understand the dynamics of the environments where the microorganisms live.
Material and methods
Yeast strain and cultivation
The psychrophilic yeast G. antarctica PI12 was obtained from the Culture Collection of the Universiti Kebangsaan Malaysia, Bangi, Selangor, Malaysia (strain number SKUK20041) [13]. The yeast was identified based on biochemical characteristics and the sequence of the internal transcribed spacer (ITS) as well as the LSU rRNA (accession numbers JX89896955 and JX896956) by the National Collection of Yeast Cultures, Norwich, UK [9].
Initially, G. antarctica PI12 was cultured on yeast peptone dextrose (YPD) agar (Oxoid Ltd., Hampshire, UK) for 10 days at 12 οC until a single colony was obtained. Subsequently, a single colony was inoculated in 5 mL YPD medium supplemented with 50 μg mL-1 kanamycin and 50 μg mL-1 ampicillin and grown at 12 οC for 5 days. To induce the expression of β-1,3-glucanase, a suspension of 1 × 106 cells was aseptically transferred to 100 mL yeast peptone containing 1% (w/v) lichenan as the sole carbon source, supplemented with 50 μg mL-1 ampicillin and 50 μg mL-1 kanamycin, and cultured for 8 days at 12 οC in a 250 mL Erlenmeyer flask. After centrifugation (4,629 × g, 10 min, 4 °C), the supernatant was concentrated with a Vivaspin MWCO 10 kDa concentrator (SartoriusAG, Goettingen, Germany). This concentrated supernatant was used as a crude sample for the native β-1,3-glucanase assay using 1% (w/v) laminarin and lichenan as substrates.
Full-length cDNA synthesis, construction of expression plasmid and transformation
Total RNA of G. antarctica PI12 was extracted using the TrizolTM reagent according to Bharudin et al. [14]. First-strand cDNA was synthesized using the SuperScript® III First-Strand Synthesis System (Invitrogen). Subsequently, first-strand cDNA was used as the template for full-length cDNA amplification. The specific primers to amplify the full-length sequence of β-glucanase (GaEgl) were designed based on the Genome Sequencing Survey Database of G. antarctica available at the Malaysia Genome Institute (www.genomemalaysia.gov.my/v3/). The full-length cDNA of endo-β-glucanase was amplified using primers Egl-F (5'-atgctttcctccatcacctccatca-3') and Egl-R (5'-ctacttgaggttgaagatcttgacg-3'). PCR products were analyzed by agarose-gel electrophoresis (1 % w/v) and cloned into the pGEM-T® Easy Vector (Promega, Madison, WI, USA). To express the heterologous protein in E. coli, primers were designed to amplify the GaEgl sequence encoding the mature protein (without the signal peptide) for subsequent subcloning into the expression vector. The primers used were Egl2-F (5'-cacccctctcgacgcccg-3') and Egl2-R (5'-ctacttgaggttgaagatcttgacgtagtt-3'). The PCR mixture contained 2.5 µl of first-strand cDNA, 2.5 1× PCR Buffer, 0.2 mM dNTP, 20 pmol µl-1 forward and reverse primers, 1.5 mM MgCl2, 0.5 U µl-1 Taq polymerase (Promega, Madison, WI, USA). PCRs were performed as follows: initial denaturation at 94 °C for 3 min followed by 30 cycles at 95 °C for 1 min, 68 °C for 1 min and 72 °C for 1.5 min, with a final extension at 72 °C for 5 min. The PCR product was ligated into the pET200 expression vector, which has a six-histidine tag at the 5′-region of the multiple cloning site, and the plasmid was transformed into the expression host, BL21 (DE3). To confirm the nucleotide sequence, the full-length cDNA sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Nucleotide sequence of the endo-β-1,3(4)-glucanase gene and its deduced amino acid sequence are available in GenBank with the accession number KM186795.
Sequence analysis
The molecular mass, amino acid composition, and theoretical pI were calculated using ProtParam (http://web.expasy.org/protparam/). Prediction of the signal peptide was performed using the SignalP server (http:// www.cbs.dtu.dk/services/SignalP [15]. Domain analysis was performed using the Pfam database (http://pfam.sanger.ac.uk/), whereas protein motifs were analyzed using HHrepID software (http://toolkit.tuebingen.mpg.de/hhrepid). Sequence alignments of the homologous sequences obtained from BLAST were built using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The phylogenetic tree was constructed based on a comparison of the G. antarctica endo-β-1,3-glucanase sequence with the closest endo-β-1,3-glucanase sequence that was extracted from the GenBank database (http://www.ncbi.nlm.nih.gov). A phylogenetic tree was constructed with multiple sequences using ClustalX2 and MEGA 5.2 by the neighbor-joining method (NJ) and bootstrap analysis with one thousand bootstrap replicates to assess the reliability of individual branches. The secondary structure of the protein based on the amino acid sequence was predicted using the PSIPRED program (http:// bioinf.cs.ucl.ac.uk/psipred/) and phyre2 [16].
Protein expression and purification
E. coli BL21 (DE3) cells harboring the recombinant pET-Egl plasmid were grown overnight at 37 °C in LB broth supplemented with 50 μg mL-1 The culture was inoculated (1%) into fresh LB broth containing 50 μg mL-1 kanamycin and grown at 37 °C to an optical density (OD) of 0.6 at 600 nm. The expression of the recombinant β-glucanase was induced with the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM. The culture was further grown for 18 h at 20 °C. Protein expression was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously [17]. Subsequently, to purify the rGaEgl, cells were pelleted and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM sodium chloride, 1 μg mL-1 lysozyme, 1 mM PMSF, pH 8.0). After disruption by sonication, lysates were centrifuged (18,514 × g at 4°C for 20 min) to remove cell debris. The supernatant was passed through a 0.22-μm filter and then applied to His-Trap resin (GE Healthcare, Little Chalfont , UK) equilibrated with binding buffer (20 mM sodium phosphate, 300 mM sodium chloride buffer, 10 mM imidazole, pH 8) at a flow rate of 1 mL min-1. Recombinant GaEgl was eluted with 50 mL of elution buffer (20 mM sodium phosphate, 300 mM sodium chloride buffer, 500 mM imidazole). Finally, the imidazole in the purified GaEgl solution was removed by dialysis in buffer (50 mM Tris-HCl buffer, pH 7). The collected active fractions were concentrated using Vivaspin MWCO 10 kDa concentrators (SartoriusAG) and then stored at −20 οC.
GaEgl protein identification and analysis
The eluted protein purity was determined by SDS-PAGE. Protein bands were visualized by Coomassie brilliant blue R-250 staining. The protein concentration was determined with the Bradford assay [18] using bovine serum albumin (BSA) as the protein standard. The reaction was measured at a wavelength of 595 nm. Western blotting was performed based on the detection of histidine tag proteins using a His-tagged monoclonal antibody (Sigma-Aldrich Corp, St. Louis, MO, USA. Signals were visualized using the Immun-Blot® AP Colorimetric Kit (Biorad, USA).
GaEgl was identified via liquid chromatography mass spectrometry (LC-MS/MS) using Orbitrap Fusion Tribrid Mass Spectrometer (Thermo, USA) at Malaysia Genome Institute. Briefly, targeted proteins were sliced from the SDS PAGE and equilibrated with 100 µL of 100 mM ammonium bicarbonate for 15 min, followed by the addition of an equal amount of acetonitrile. Solutions were then vortexed for 30 sec, and centrifuge for 14,000 rpm. The supernatant was removed, and the gel was then immersed in acetonitrile and dried in a vacuum centrifuge (Eppendorf, Germany). The sliced gel was suspended in 1 mM DTT for 30 min at 56°C and further rehydrated with acetonitrile for 15 min. Samples were centrifugated at 14,000 rpm; the supernatant was later discarded before resuspension with 55 mM iodoacetamide. Obtained gel was incubated in dark at room temperature for 30 min. The incubated gel treated with ammonium bicarbonate and acetonitrile followed with centrifugation at 10 000 rpm for 10 min to remove the supernatant and finally dried in vacuum. The gel particles was rehydrate in digest buffer (50 mM ammonium bicarbonate containing 12.5 ng/µL mass spectrometry grade Trypsin Gold (Promega, Madison, WI, USA) and incubated at 4°C for 30 min, then incubated the gels at 37°C for 16 h. A total of 10 µl of 50% acetonitrile/5% formic acid was added to recover the peptides and samples were agitated ultrasonically for 10 min. Digested samples were separated and analysed, with Thermo EASY-nLC separation. The aqueous mobile phase (mobile phase A) is 0.1% formic acid in the water, while the organic mobile phase (mobile phase B) contained 0.1% formic acid in acetonitrile. Peptide separations were performed with mobile phase B gradient (5-40% in 91 min, 2 min to 95% phase B, 6 min at 95% mobile phase B and back to 5% mobile phase B in 2 min). A total of 2 µL samples were loaded onto a EASY-Spray Column Acclaim PepMapTM C18 100 A0, (2µm particle size, 75µm id x 25cm) column. Mass spectrometry analysis of the separated tryptic digested peptides was performed using Orbitrap Fusion MS mass spectrometer. LCMS data were analysed using Thermo ScientificTM Proteome DiscovererTM Software Version 2.1 based on G. antarctica database. The following parameters also applied: i) Enzyme: Trypsin, ii) Fixed Modification: Carbamidomethyl (C), iii) Variable modifications: Oxidation M, Deamination N, Deamination Q, Phosphorayl STY.
Assessment of β-1,3-glucanase activity
All enzyme assays were performed in triplicate, and β-1,3-glucanase activity was measured by the dinitrosalicylic acid (DNS) method as described previously [19]. The reaction mixture (1 mL) contained 1% lichenan (Megazyme, Ireland), 50 mM Tris buffer (pH 7), and the appropriate amount of the purified enzyme. After incubation at 20 °C for 20 min, the reaction was stopped by addition of 0.5 mL of DNS. Then, the reaction mixture was boiled for 10 min in a dry bath and cooled quickly to room temperature. Enzymatic hydrolysis of lichenan was determined by measuring the absorbance at 540 nm. One unit (U) of enzyme activity was defined as the amount of the enzyme liberating, reducing sugars equivalent to 1 μmol of D-glucose in one minute under the assay conditions.
Effects of pH and temperature on enzyme activity and stability
The optimal pH for activity of the purified recombinant GaEgl was determined at 20 °C in buffers with pH values ranging from pH 4 to 10. The buffer systems were 50 mM sodium acetate (pH 4–6), 50 mM Tris-HCl (pH 6–8), and 50 mM glycine-NaOH (pH 8–9). The pH stability was estimated by measuring GaEgl activity under standard conditions (pH 7, 20 °C) after 60 min pre-incubation of the purified recombinant enzyme in buffers of pH 6.0–8.0. The optimal temperature for GaEgl activity was determined by measuring enzyme activity at temperatures ranging from 5 °C to 50 °C. The thermostability of the enzyme was determined by pre-incubating the enzyme for 60 min in 50 mM Tris-HCl (pH 7) at 15 °C, 20 °C and 25 °C without substrate for various periods and then measuring the residual enzyme activity in each case under standard conditions.
Effects of metal ions and chemical compounds on recombinant GaEgl activity
To investigate whether the enzyme required any metal ions as cofactors, the effects of various metal ions and chemical reagents on enzyme activity were tested. Each of the different chemicals and metal ions (Table 2) was added to the reaction mixture at final concentrations of 5 mM, and the rGaEgl activity was measured under standard conditions (20 °C for 20 min). The activity of rGaEgl with no addition of the metal ions or chemical reagents was set at 100%.
Substrate specificity and kinetic parameters of the GaEgl
The substrate specificity of GaEgl was evaluated with lichenan, laminarin, yeast glucan, birch wood xylan, carboxymethyl cellulose, and Avicel by measuring the enzyme activity at 20 οC in 50 mM Tris-HCl buffer at pH 7 for 20 min. The amount of reducing sugars produced was estimated using the DNS method as described above. The kinetic parameters (Km, Vmax, kcat, and kcat/Km) of the purified enzyme were determined. Different substrate (laminarin, lichenan, and barley glucan) concentrations were used, ranging from 5.0 to 30.0 mg mL-1. The reaction rate versus substrate concentration was plotted to determine whether the enzyme obeys Michaelis-Menten kinetics. The Michaelis-Menten constant (Km) and maximum velocity of substrate hydrolysis (Vmax) were determined from Lineweaver-Burk plots.
Hydrolase properties of purified rGaEgl
For analysis of laminarin hydrolysis products, all enzymatic reactions were carried out under the standard assay conditions as described above. The reaction mixtures were filtered through a 0.45-µm filter (SartoriusAG), and the products labelled with the fluorophore 2-aminobenzamide (2-AB) and analyzed via high-performance liquid chromatography (HPLC) using fluorescence detection (HPLC-FD) and glucose unit (GU) analysis [20]. A standard of glucose oligomers various degrees of polymerization/glucose units (dextran ladder) was used as a standard (gifted by Oxford Glycobiology Institute, UK). Labelling of both standards and samples was carried out as described previously [21]. Briefly, post-hydrolysis reaction samples were labeled with 20 μL of freshly prepared 0.09 M of 2-AB and 1 M sodium in acetic-acid-DMSO (3:7) mixtures followed by incubating at 65 °C for 3 hours. The 2-AB labelled samples were diluted in 200 µL acetonitrile (ACN) and extracted via HILIC (100 mg) solid-phase extraction. This involved pre-equilibration (1 mL water followed by 1 ml 85% ACN), washing (400 μL 85% ACN), and elution (3 x 100 µL water). Excess 2-AB was then removed by solvent partitioning, as described previously [22]. Extracted 2-AB labelled samples were then analyzed by HPLC-FD (fluorescence detector: Exλ 360 nm and Emλ 426 nm) using an XBridge Amide column (4.6 × 250 mm, 3.5 μm particle size), (Waters Corporation, MA, USA) [53]. The mobile phase used was solvent A (80% ACN, 60% pure water and 20% ammonium acetate pH 3.85) and solvent B (20% ACN, 60% pure water and 20% 100 mM ammonium acetate pH 3.85) run using gradient conditions. flow rates used were: t = 0, 86% solvent A (0.8 mL/min); t = 6, 86% solvent A (0.8 mL/min); t = 45, 46.6% solvent A (0.8 mL/min); t = 47, 5% solvent A (0.5 mL/min); t = 49, 5% solvent A (0.5 mL/min); t = 51, 86% solvent A (0.6 mL/min); t = 52, 86% solvent A (0.7 mL/min).
Homology modeling
Structural homology modeling of GaEgl was carried out using MODELLER9v14 [23]. The best protein template with a high sequence identity (more than 30 %) was chosen using phyre2 [24] and PSI-BLAST [25]. A total of 100 models were generated, and the model with the lowest DOPE (discrete optimized protein energy) and GA341 value (score for the reliability of a model) near or equivalent to 1 was selected as the best model and used for evaluation [23,26]. Finally, the protein structure was assessed using PROCHECK [27], Verify 3D [28], and ERRAT [29]. Model visualization was carried out using various software packages: Chimera 1.7 (http://www.cgl.ucsf.edu/chimera/), Accelrys Discovery Studio 2.5, and PyMOL graphic system.
Structure analysis
Cold-active enzymes generally have fewer hydrogen bonds and/or salt bridges, and/or a larger total accessible surface area (ASA), and/or exposed hydrophobic ASA, and/or total packing volume (TPV), and/or longer loops compared with their mesophilic and thermophilic homologues [30]. To predict the temperature-dependent activity and stability of GaEgl, these factors were compared with β-1,3(4)-glucanase from mesophilic Phanerochaete chrysosporium (2CL2) [31]. Salt bridges (distances≤4 Å) and hydrogen bonds were predicted with VMD 1.9.1 [32] and UCSF Chimera 1.2540 [33], respectively. ASA and TPV were calculated using VADAR 1.5 [34].
Results
Cloning of the full-length endo-β-1,3(4)-glucanase and sequencing analysis
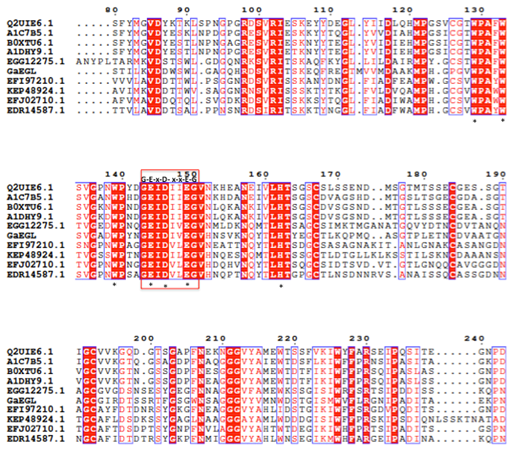
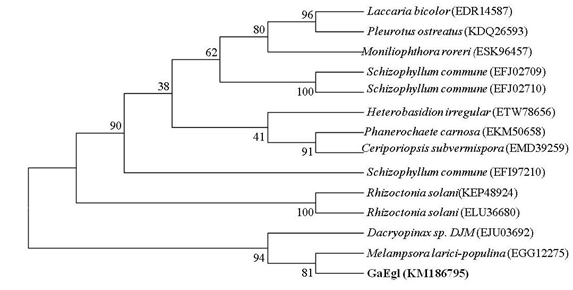
The full length cDNA fragment containing an open reading frame of 1182 bp and encoding a protein of 394 amino acids with a theoretical molecular weight of 42.8 kDa was successfully amplified and cloned into the pGEM-T easy cloning vector. A SignaIP analysis indicated the presence of an N-terminal signal peptide at residues 1–22 with a predicted cleavage site located between amino acids A22 and Q23. Prediction of the functional domains and putative active site in GaEgl with InterProScan allows the GaEgl to be classified as a new member of glycoside hydrolase (GH) family 16 (GH16). When the deduced amino acid sequence of GaEgl was compared with the other available protein sequences in GenBank, it showed a high level of similarity to fungal β-1,3-glucanases belonging to GH16. The deduced amino acid sequence of GaEgl shares the highest identity of 50% with a GH16 protein from Melampsora larici-populina 98AG31 (GenBank accession no. EGG06441), followed by Rhizoctonia solani 123E (44% identity; KEP48924) and Dacryopinax sp. DJM-731 (46% identity; EJU03692). Based on the multiple sequence alignment of GaEgl with other endo-β-1,3-glucanases, GaEgl was shown to contain the conserved amino acid motif G-E-x-D-x-x-E (Figure 1). Hence, two catalytic residues, Glu214 and Glu219 (GEIDIVE), in GaEgl G. antarctica were highly conserved among GH16 members, and these two glutamic acid residues are involved in an acid/base and nucleophile mechanism. The phylogenetic tree analyzed using bootstrapping (1000 replicates) showed that GaEgl from G. antarctica PI12 was clustered with a fungal β-1,3(4)-glucanase from M. larici-populina with a sequence identity of 50% (Figure 2).
Figure 1: Sequence alignment of GaEgl and other endoglucanases. Protein accession number (representative strain): B0XTU6 (Aspergillus fumigatus); A1DHY9 (Neosartorya fischeri); A1C7B5 (Aspergillus clavatus); Q2UIE6 (Aspergillus oryzae); EGG12275 (Melampsora larici-populina); KEP48924 (Rhizoctonia solani); EFJ02710 (Schizophyllum commune); EDR14587 (Laccaria bicolor). The conserved catalytic residues in GH16 marked with an asterisk while conserved motif represented in red box.
Figure 2: Phylogenetic tree based on the full-length amino acid sequence of G. antarctica and endo-glucanases from other fungi. The bootstrap values, indicated at the nodes, were obtained from 1000 bootstrap replicates and are reported as percentages. The numbers in parenthesis represent GenBank accession numbers.
Expression and purification of GaEgl in an E. coli expression system
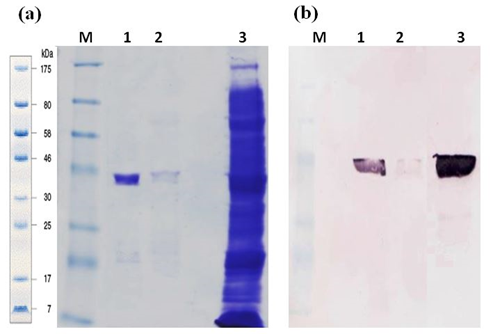
The recombinant GaEgl was produced as a soluble form in the E. coli expression system. The best induction of recombinant protein was obtained when 0.5 mM IPTG was added when the culture reached 0.5–0.6 OD600, and the bacteria were cultured at 20 °C for 18 h due to the thermolability of the enzyme. SDS PAGE analysis showed approximately ~44 kDa molecular size, corresponding to expected molecular mass and 3 kDa histidine tag. (Figure 3a). Signals were detected using the anti-His antibody, from crude and purified proteins correspond to the size (Figure 3b). LC-MS/MS analysis successfully identified three peptides matched to GaEgl sequence.
After a one-step purification procedure, the specific activity of the enzyme increased approximately 19-fold, with a recovery yield of 20.2% compared to crude lysate (Table 1). After a single pass over the Ni-Sepharose column, most of the contaminant proteins were removed. The Ni-Sepharose column had a very high affinity for the protein, and the specific activity of recombinant GaEgl increased from 1.32 U mg-1 to 23 U mg-1. The purification procedure for recombinant GaEgl is summarized in Table 1.
Table 1: Purification table of rGaEgl
|
Purification Step |
Total activity (U) |
Total protein (mg) |
Specific activity (U mg-1) |
Yield (%) |
Purification fold |
|
Crude supernatant |
86.4 |
114 |
1.32 |
100 |
1 |
|
Affinity chromatography |
9.345 |
0.41 |
23 |
20.24 |
19 |
Effects of pH and temperature on the activity of the rGaEgl
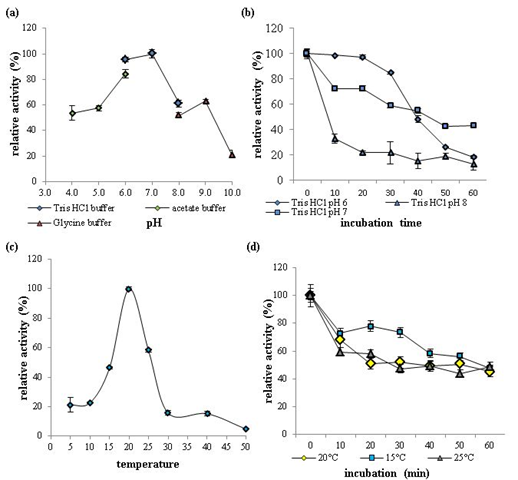
The effects of pH and temperature on recombinant GaEgl activity and stability were determined using 1% lichenan as the substrate. Over the pH range from 4 to 10, GaEgl exhibited maximum enzymatic activity at pH 7 and retained more than 50% of the maximum activity between pH 4 and 9 (Figure 4a). The pH stability assay revealed that after pre-incubation at a different pH for 1 h, GaEgl retained more than 40% of the initial activity at pH 7 (Figure 4b). These data suggested that GaEgl showed optimum activity and stability in nearly neutral pH conditions.
The optimum temperature for GaEgl was at 20 °C (Tris-HCl, pH 7.0), and GaEgl showed approximately 21% activity at 5 °C and 10 °C. The activity decreased rapidly at temperatures higher than 25 °C, and activity was almost completely lost at 50 °C (Figure 4c). The thermostability assay revealed that the half-life of the enzyme at 20 °C was just 30 min, and nearly 60% of the activity was lost after 60 min of pre-incubation at 20 °C (Figure 4d).
Effect of metal ions and chemicals on GaEgl
The activity of rGaEgl in the presence of different metal ions or chemical reagents was tested, as shown in Table 2. Enzyme activity was enhanced moderately in the presence of Co2+ and β-mercaptoethanol at a concentration of 5 mM. However, partial inhibition was observed in the presence of Cu2+, Fe2+, and Zn2+. Moreover, the addition of Li+, Mg2+, and Ni2+ metal ions strongly inhibited the recombinant enzyme activity.
Figure 4: The effects of pH and temperature on recombinant GaEgl activity and stability with 1% lichenan as the substrate. (a) Effects of pH on recombinant GaEgl activity. The assay was performed at 20 °C in buffers with pH ranging from 4.0 to 10.0. (b) pH stability of rGaEgl. The residual activity of rGaEgl was determined at 20 °C after 1 h of pre-incubation at 20°C in buffers ranging from pH 6.0 to 8.0. (c) Effects of temperature on rGaEgl activity. The assay was performed in Tris-HCl buffer (pH 7.0). (d) Thermostability of GaEgl. The enzyme was pre-incubated at 15 °C, 20 °C and 25 °C in Tris-HCl (pH 7.0), and aliquots were withdrawn at regular intervals to measure the residual activity at 20 °C.
Table 2: Effects of various metal ions and chemicals on rGaEgl activity
|
Metal ions and chemical reagents (5 mM) |
Relative activity (%) rGaEgl |
|
|
Control |
100± 1 |
|
|
Co2+ |
134± 3 |
|
|
Mg2+ |
52±1 |
|
|
Ni2+ |
51±2 |
|
|
Cu2+ |
77±0.1 |
|
|
Zn2+ |
67±7 |
|
|
Mn2+ |
105±4 |
|
|
Fe2+ |
64±20 |
|
|
PMSF |
68±1 |
|
|
Li+ |
35±0.8 |
|
|
β-mercaptoethanol |
111±0.3 |
|
Substrate specificity of GaEgl
The substrate specificity of GaEgl was examined by including polysaccharides with various linkages in reactions carried out at pH 7.0 and 20οC. The highest activity was shown towards lichenan (1.87 U mL-1), followed by yeast beta glucan (0.84 U mL-1) and laminarin (0.23 U mL-1), which are the preferred substrates (β-glucan) containing β-1,3 and mixed β-1,3/-1,4 linkages. In contrast, GaEgl was unable to hydrolyze substrates with only β-1,4 glycosidic linkages, such as CMC-Na, Avicel, and birchwood xylan. The results suggest that it is in the non-specific β-1,3(4)-glucanase (EC 3.2.1.6) category. The endo-1,3(4)-glucanases catalyze the endohydrolysis of 1,3- or 1,4-linkages in β-D-glucans when the glucose residue whose reducing group is involved in the linkage to be hydrolysed is itself replaced at C-3 [35].
Kinetic parameters of rGaEgl
The kinetic experiments were carried out by varying the substrate concentration from 0.5-30 mg mL-1. The values of the kinetic constants Km, Vmax, Kcat, and Kcat/Km of rGaEgl towards different substrates (lichenan, yeast beta glucan, and laminarin) were calculated based on the Lineweaver-Burk plot as shown in Table 3. Lichenan as a substrate utilized and a Vmax of 37.45 µmol min-1 mg-1 was recorded, followed by yeast beta glucan (33.55 µmol min-1 mg-1) and laminarin (9.25 µmol min-1 mg1) for GaEgl. The Km values of GaEgl for lichenan, yeast beta glucan and laminarin substrates were 8.87 mg mL-1, 7.71 mg mL-1, and 14.70 mg mL-1, respectively. Subsequently, the Kcat value of GaEgl was found to be the highest toward lichenan (26.83 s-1), followed by laminarin (9.25 s-1) and yeast beta glucan (2.46 s-1). The calculated values for catalytic efficiency (Kcat/Km) for lichenan, yeast beta glucan and laminarin were 3.02 mL mg−1s−1, 0.13 mL mg−1s−1 and 0.45 mL mg−1s−1, respectively. No activity detected for cellulases substrates (carboxymethylcellulose and avicel) and also xylan substrate (birch wood xylan).
Table 3: Kinetic parameters of rGaEgl with different substrates
|
Substrate |
Linkage |
Km (mg mL-1)a |
Vmax, (µmol min-1 mg-1) |
kcat s-1 |
kcat/Km mL mg-1s-1 |
|
Lichenan from Icelandic moss |
β-1,3-1,4 (1:1)b |
8.87 |
37.45 |
28.38 |
3.02 |
|
Laminarin from Laminaria digitata |
β-1,3-1,6 (7:1)b |
14.70 |
9.25 |
9.25 |
0.45 |
|
Yeast beta glucan |
β-1,3-1,4 |
7.71 |
33.55 |
2.46 |
0.13 |
|
CMC-Na |
β-1,4 |
0 |
0 |
0 |
0 |
|
Avicel |
β-1,4 |
0 |
0 |
0 |
0 |
|
Birch wood xylan |
β-1,4 |
0 |
0 |
0 |
0 |
Assays were carried out at 20 οC in 50 mM Tris buffer (pH 7.0).
a Because molecular weight of laminarins are heterogeneous, the unit, mg mL-1, is used for Km.
b The branching ratio of linkages in the substrate are shown in parentheses.
Analysis of the hydrolysis product
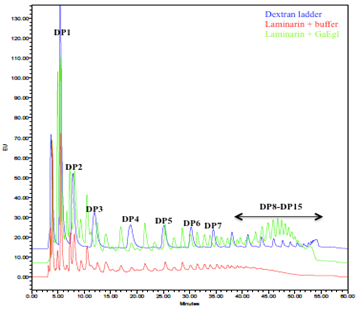
HPLC analysis in Figure 5 showed major reducing sugars observed from laminarin hydrolysis using GaEgl were monosaccharides, disaccharides and trisaccharides. The levels of other oligosaccharides from tetrasaccharides and above were increased compared to the undigested laminarin control. Besides, the unique presence of higher degree oligosaccharides from DP8-DP15 was also detected in the hydrolyzed product peaking at DP11-DP12.
Structure analysis
Based on the analysis of GaEgl with different threading methods, such as phyre2 and PSI-BLAST, the 2CL2 from Phanerochaete chrysosporium with 38% sequence similarity was selected as the best template to model the G. antarctica GaEgl. Of the 100 models built, model # 8 was selected as it showed the lowest DOPE score (31521.41797) and a GA341 value equivalent to 1. Evaluation of the modelled protein was performed using PROCHECK, Verify3D, and ERRAT. PROCHECK analyzed the conformation of the backbone by inspecting the phi/psi Ramachandran plot [27]. Based on this analysis, only 1.7 % of the residues (Pro95, Lys125, Asn203 and Thr209) were located in disallowed regions; 3.5 % of the residues were in the allowed regions, while the remaining residues (94.8 %) were located in favored regions. Using Verify3D, the compatibility of an atomic model (3D) with its own amino acid sequence (1D) was assessed and showed that 98.62 % of GaEgl residues were complementary to the 3D–1D profile (3D–1D score 0.2). A score of more than 80 % indicates that the predicted model is within the acceptable range [28]. ERRAT is another tool that is used to assess the overall quality of the model for nonbonded atomic interactions by comparing the statistics of highly refined structures. The accepted range of the ERRAT score for a good model is above 50 %, and a higher score indicates a better quality [29]. The GaEgl model had an ERRAT score of 65, which is acceptable for high quality models.
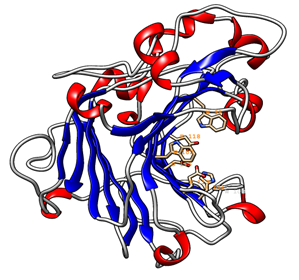
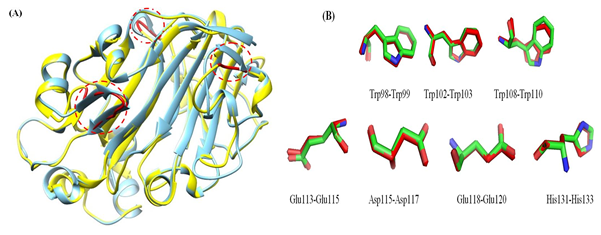
The 3D model of the GaEgl protein is shown in Figure 6. The model has similar structural features to those of other members of GH16. The GaEgl structure consists of an alternating pattern of α-helices (28.6%) and β-strands (42.8%) in its globular structure, which is considered a β-sandwich fold. The β-sandwich structure has been observed in most of the known microbial glucanase structures and is considered a common folding motif of GH16. The GaEgl 3D model was superimposed with 2CL2 as a selected template with 38% similarity and RMSD of 0.36 Ǻ and is represented in Figure 7. Moreover, the conservation of the catalytic residues between GaEgl and 2CL2 demonstrates the high accuracy of the constructed 3D model. However, a visual inspection of their superimposed structure in Figure 7 indicates two remarkable differences.
First, GaEgl has longer external loops on its surface in three different positions (THR45-LYS54, ALA106-TRP108, and LYS212-ASN214) instead of two β-sheets (THR45-SER48 and THR51-ARG55), (THR218-ASN220) and α-helix (GLY108-TRP110) on the surface of 2CL2 (as shown by red circles in the Figure 7). Second, most of the long β-sheets on the surface of the 2CL2 molecule are broken into two shorter β-sheets connected by a loop in the GaEgl molecule. Furthermore, the psychrophilicity of the GaEgl enzyme was assessed by comparative study values of total ASA, TPV, hydrogen bonds and salt bridges with a mesophilic endo-β-1,3(4)-glucanases (2CL2) (Table 4). As shown in Table 4, the total ASA and the exposed nonpolar ASA of GaEgl was greater than that of 2CL2 mesophilic β-glucanases. At the same time, the abundance of salt bridges and hydrogen bonds of GaEgl was lower than that of the mesophilic β-1,3(4)-glucanase.
Table 4: Structural characteristics of psychrophilic GaEgl with mesophilic endo-1,3(4)-glucanase
|
Parameter |
GaEgl |
2CL2 |
|
Number of salt bridges ≤4 Ǻ (formed by arginine residues) |
2 |
6 |
|
Number of hydrogen bonds |
219 |
233 |
|
Total ASA (103 Å2) |
12 |
11 |
|
Exposed nonpolar ASA (103 Å2) |
6.8 |
6.3 |
|
Total packing volume (103 Å3) |
38.2 |
37.5 |
Discussion
Glaciozyma antarctica is an obligate psychrophilic yeast that inhabits cold Antarctic ecosystems. In these ecosystems, special modifications for growth and survival are required because of the scarcity of nutrition. Glycoside hydrolases are a group of enzymes that degrade complex carbohydrate linkages to generate simple sugars. The glycoside hydrolase in fungi is required to facilitate infection, gain nutrition, and degrade organic matter in the surrounding environment. The secretion of cold adapted extracellular enzymes into the cold environment may reflect the metabolic adaptation of psychrophilic and psychrotolerant microorganisms that are speculated to be important in the biodegradation of organic matter and the cycling of essential nutrients. This idea is supported by the presence of organic carbon and nitrogen sources originating from melting glacier ice [36]. The accumulation of organic matter from the dead organism, and the secretion of organic polymers by algae and bacteria suggest that psychrophilic fungi, such as G. antarctica, would release extracellular enzyme to degrade these complex molecules into simple, consumable forms. The possibility of extracellular expression of G. antarctica PI12 as a potential β-1,3-glucanase producer was examined in an induction medium that contained 1% lichenan. The activity of the enzyme toward lichenan was the highest (2.19 UmL-1), followed by laminarin (1.1 UmL-1). This finding suggests that G. antarctica PI12 is a potential β-1,3-glucanase producer. Given the obtained results, the secreted β-glucanases may facilitate nutrient scavenging of G. antarctica in its natural habitat.
Production of extracellular β-1,3-glucanases from G. antarctica is relatively low since this yeast grows very slow. In this study, the production of recombinant 1,3-β-glucanase was conducted for a higher level of protein expression. In the present study, an endo-1,3(4)-β-glucanase gene (GaEGL) was cloned from a Glaciozyma antarctica P112 strain and successfully expressed in E. coli and biochemically characterised. The deduced amino acid sequence of GaEgl exhibited the highest identity, 50%, with a GH16 endo-1,3-1,4-β-glucanase from M. populina (98AG31). However, this new enzyme may have different properties from other family members as the sequence identity was low, which confirms the novelty of the protein.
The biochemical characterisation of recombinant GaEgl showed optimum activity at 20 °C, which is lower than the temperature reported for most fungal β-1,3(4)-glucanases, which ranges from 40 °C to 60 °C [37-39]. GaEgl still exhibited activity at low temperatures from 5 °C to 15 °C; these data show that GaEgl is a cold-adapted enzyme. The biochemical characteristic of this enzyme reflects the average temperature of G. antarctica was previously reported by Boo et al. [8]; where it can’t survive when cultured more than 20 °C.
The recombinant GaEgl showed optimum activity at pH 7.0, and the enzyme still retained its activity in alkaline conditions. This result differs from other endo-β-1,3-glucanases that have pH optimums of 4.0–6.5 and lose almost all their activity above pH 7.0 [39,41]. GaEgl has three acidic amino acid residues (E214, D216, and E219) and two neutral amino acids (I215 and I217) at the active site; conducive to acid/base catalysis.
Typical cold-active enzymes maintain activity at low temperatures with low thermostability and show unusual specificities compared to their mesophilic homologues, which is similar to the biochemical analysis and comparison of recombinant GaEgl. These biochemical features are also observed in β-1,4-mannanase (CaMan) from C. antarcticus [42] and in other cold-active enzymes, for example, β-galactosidase from Pseudoalteromonas sp. 22b [43]. However, to date, no recombinant cold-active β-1,3(4)-glucanases have been reported to show activity under low temperatures and alkaline pH conditions. Kumar et al. [44] reported that based on in silico analysis of thermophilic and psychrophilic β-galactosidase, thermophilic β-galactosidase has a marginally higher percentage of α-helix, whereas psychrophilic β-galactosidase has a higher percentage of sheet and coil region. This result suggested that more helices provide proteins with an advantage at higher temperatures. The sheet region is thermolabile and is, therefore, present in low amounts in thermophilic proteins and high amounts in psychrophilic proteins. Hence, the high percentage of beta sheets in GaEgl was in agreement with the secondary structure of other psychrophilic enzymes.
The recombinant GaEgl showed the highest activity toward lichenan (1.87 U mL-1), followed by yeast beta glucan (0.84 U mL-1) and laminarin (0.23 U mL-1). In contrast, GaEgl cannot hydrolyze CMC-Na (powdered cellulose), Avicel, and birchwood xylan containing 1,4-β-glycosidic bonds, suggesting that it is a non-specific β-1,3(4)-glucanase. This substrate specificity may be related to the different numbers of β-1,3 and β-1,4 glycosidic linkages, branching mode, and their distribution in these substrates [39]. The ratios of the β-1,3 and β-1,4 glycosidic bonds are 1:2.3-2.7 and 1:1 for β-glucan and lichenan, respectively [45], whereas the ratio of the β-1-3 and β-1-6 bonds for laminarin is 7:1. The result indicates that the activity of the recombinant enzyme was proportional to the number of β-1-4 or β-1-3 bonds in β-glucan, lichenan, and laminarin, similar to endo-1,3(4)-β-glucanase from R. miehei [39]. The recombinant GaEgl had similar substrate specificity to other endo-1,3(4)-β-glucanases from Penicillium and Paecilomyces sp. FLH30 [12,46]. All enzymes showed activity toward lichenan and laminarin but degraded neither CMC-Na nor birchwood xylan. The sequence alignment and substrate specificity revealed that GaEgl is a classical nonspecific endo-1,3(4)-β-D-glucanase, which can catalyze the hydrolysis of β-1,3 or β-1,4 glycosidic bonds. This characteristic makes it more suitable than conventional glucanases for the hydrolysis of glucans in food because these industries require the combined activities of different types of the hydrolysis products formed with laminarin via HPLC revealed that the most abundant hydrolysis products were monosaccharides, disaccharides and trisaccharides. All these products were present at elevated levels compared to the uncut laminarin. The unique presence of oligosaccharides 8-15 units (DP8-DP15) suggested that GaEgl is an endo-1,3(4)-β-glucanase, which catalyzes the endohydrolysis of 1,3- or 1,4-linkages in β-D-glucans. The hydrolysis patterns of GaEgl were similar to those enzymes previously described from Paecilomyces sp. FLH30 endo-1,3(4)-β-glucanase (Hua et al. 2011) and Aspergillus niger US368 endo-β-1,3-1,4-glucanase [47].
The kinetic parameters of GaEgl showed that lichenan achieved the highest Vmax of 37.45 µmol min−1 mg−1, with a Km value of 8.87 mg mL−1, compared with other substrates (yeast beta glucan and laminarin). A broad range of Km and Vmax values was previously reported for lichenan from different sources. β-1,3;1-4-glucanase from A. niger US368 and endo-1,3(4)-β-glucanase from R. miehei displayed Km values of 0.38 and 1.66 mg mL−1, and Vmax values of 24.02 and 7.69 U mL−1, respectively [48]. In this study, the affinity constant (Km) for lichenan was higher than those previously reported for β-1,3-glucanases from thermophile and mesophilic sources.. Interestingly, in parallel with a characterisation of cold-active GaEgl, sequence and structure analyses indicated that GaEgl had a similar sequence and structural features to the low-temperature active β-1,3(4)-glucanases but different features from mesophilic 2CL2: (1) compared with 2CL2, GaEgl had longer loops near the catalytic site and larger total ASA and TPV. These longer loops in GaEgl may increase the possible amplitude of the movement between secondary structures and result in the higher flexibility and lower thermostability of the structures [30]. A previous comparative structural analysis of ManAGN25 and PMAN, a GH 5 low-temperature active mannanase from Sphingobacterium sp. GN25, with GH 5 thermophilic and mesophilic counterparts also suggested that longer loops in ManAGN25 and PMAN could be a contributor to its adaptation to a low-temperature environment [49,50].
Structural features of psychrophilic GaEgl showed higher hydrophilic and hydrophobic accessible surface areas comparing to mesophilic counterparts. These differences revealed that the mesophilic endo-1,3(4)-β-glucanase has improved electrostatic interactions to stabilize the enzyme at higher temperatures. As reported by Tronelli et al. [6] psychrophilic enzymes tend to have more hydrophobic (nonpolar) residues in their accessible surface areas to increase the structural flexibility of the molecule at low temperatures. The existence of hydrophobic residues on the surface of a protein destabilizes its structure because of a decrease in the entropy of the water molecule. This decrease in water entropy creates cage-like structures around these residues. However, because of the decreased mobility of the released water molecules, the entropy is reduced at cold conditions [10]. In addition to the above factors, the psychrophilic GaEgl has fewer hydrogen bonds in comparison with its mesophilic counterpart. Currently, a satisfactory technique to confirm the role of the number of hydrogen bonds in the stability of the protein structure does not exist, although in mesophilic and thermophilic enzymes this type of interaction makes the protein more stable [6]. As shown in Table 4, the number of salt bridges in GaEgl was lower than in its mesophilic counterparts. Salt bridges are recognized as important factors in protein structure stabilization. The stability of a molecule is directly influenced by salt bridge disturbances [51]. Previous studies indicated that cold-adapted enzymes have a fewer salt bridges using X-ray analysis [52]. The presence of arginine in the enzyme enhances the thermostability of a molecule by providing more electrostatic interactions through their guanidine group [10]. On overall, the biochemical characterisation of GaEgl and structural prediction aided towards the understanding of how glycoside hydrolases facilitate. G. antarctica survival in the extreme conditions of the Antarctic sea ice and provide nutrition to this marine psychrophilic yeast. In addition, this study provided insights into the ecological niches of marine psychrophilic yeast. G. anatrctica and their roles in the polysaccharide decomposition of carbon cycling in marine ecosystems.
Acknowledgments
We acknowledge the support given by the Australian Antarctic Division and the Malaysian Antarctic Research Programme (MARP) of the Academy of Science, Malaysia. We thank Proteomics Core Facility, Malaysia Genome Institute, National Institutes of Biotechnology Malaysia (NIBM) for their contribution to LCMS/MS analysis.
Funding
This research was supported by a research grant from the Ministry of Science, Technology, and Innovation (MOSTI), Malaysia, under the research grants 10-05-16-MB002 and 02-05-20-SF0007.
References
- Feller G. Psychrophilic enzymes: from folding to function and biotechnology. Scientifica 2013 (2013).
- Gerday C, Aittaleb M, Bentahir M, et al. Cold-adapted enzymes: from fundamentals to biotechnology. Trends in Biotechnology 18 (2000): 103-107.
- Deming JW. Psychrophiles and polar regions. Current Opinion in Microbiology 5 (2002): 301-309.
- Foong PM, Karjiban RA, Normi YM, et al. Bioinformatics survey of the metal usage by psychrophilic yeast Glaciozyma antarctica PI12. Metallomics 7 (2015): 156-164.
- Arnórsdóttir J, Smáradóttir RB, Magnússon ÓT, et al. Characterization of a cloned subtilisin-like serine proteinase from a psychrotrophic Vibrio species. European Journal of Biochemistry 269 (2002): 5536-5546.
- Tronelli D, Maugini E, Bossa F, et al. Structural adaptation to low temperatures− analysis of the subunit interface of oligomeric psychrophilic enzymes. The FEBS Journal 274 (2007): 4595-4608.
- Hashim NH, Bharudin I, Nguong DL, et al. Characterization of Afp1, an antifreeze protein from the psychrophilic yeast Glaciozymaantarctica PI12. Extremophiles 17 (2013): 63-73.
- Boo SY, Wong CM, Rodrigues KF, et al. Thermal stress responses in Antarctic yeast, Glaciozyma antarctica PI12, characterized by real-time quantitative PCR. Polar Biology 36 (2013): 381-389.
- Hashim NH, Sulaiman S, Bakar FD, et al. Molecular cloning, expression and characterisation of Afp4, an antifreeze protein from Glaciozyma antarctica. Polar Biology 37 (2014): 1495-1505.
- Ramli AN, Mahadi NM, Shamsir MS, et al. Structural prediction of a novel chitinase from the psychrophilic Glaciozyma antarctica PI12 and an analysis of its structural properties and function. Journal of Computer-aided Molecular Design 26 (2012): 947-961.
- Bai Y, Wang J, Zhang Z, et al. A novel family 9 β-1, 3 (4)-glucanase from thermoacidophilic Alicyclobacillus sp. A4 with potential applications in the brewing industry. Applied Microbiology and Biotechnology 87 (2010): 251-259.
- Hua C, Yi H, Jiao L. Cloning and Expression of the Endo-1, 3 (4)-β-glucanase Gene from Paecilomyces sp. FLH30 and Characterization of the Recombinant Enzyme. Bioscience, Biotechnology, and Biochemistry 75 (2011): 1807-1812.
- Mohammadi S, Parvizpour S, Razmara J, et al. Structure prediction of a novel Exo-β-1, 3-Glucanase: Insights into the cold adaptation of psychrophilic yeast Glaciozyma antarctica PI12. Interdisciplinary Sciences: Computational Life Sciences 10 (2018): 157-168.
- Bharudin I, Zaki NZ, Bakar FA, et al. Comparison of RNA extraction methods for transcript analysis from the psychrophilic yeast, Glaciozyma antarctica. Malays Appl Biol 43 (2014): 71-79.
- Bendtsen JD, Nielsen H, Von Heijne G, et al. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340 (2004): 783-795.
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10 (2015): 845-858.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 (1970): 680-685.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72 (1976): 248-254.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry 31 (1959): 426-428.
- Guile GR, Rudd PM, Wing DR, et al. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Analytical Biochemistry 240 (1996): 210-226.
- Bigge JC, Patel TP, Bruce JA, et al. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Analytical Biochemistry 230 (1995): 229-238.
- Alonzi DS, Neville DC, Lachmann RH, et al. Glucosylated free oligosaccharides are biomarkers of endoplasmic-reticulum α-glucosidase inhibition. Biochemical Journal 409 (2008): 571-580.
- Eswar N, Webb B, Marti-Renom MA, et al. Comparative protein structure modeling using Modeller. Current Protocols in Bioinformatics 15 (2006): 5-6.
- Biegert A, Mayer C, Remmert M, et al. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Research 34 (2006): W335-W339.
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25 (1997): 3389-3402.
- Melo F, Sali A. Fold assessment for comparative protein structure modeling. Protein Science 16 (2007): 2412-2426.
- Laskowski RA, MacArthur MW, Moss DS, et al. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26 (1993): 283-291.
- Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253 (1991): 164-170.
- Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Science 2 (1993): 1511-1519.
- Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 75 (2006): 403-433.
- Vasur J, Kawai R, Larsson AM, et al. X-ray crystallographic native sulfur SAD structure determination of laminarinase Lam16A from Phanerochaete chrysosporium. Acta Crystallographica Section D: Biological Crystallography 62 (2006): 1422-1429.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics 14 (1996): 33-38.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry 25 (2004): 1605-1612.
- Willard L, Ranjan A, Zhang H, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Research 31 (2003): 3316-3319.
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Research 28 (2000): 304-305.
- Brizzio S, Turchetti B, De Garcia V, et al. Extracellular enzymatic activities of basidiomycetous yeasts isolated from glacial and subglacial waters of northwest Patagonia (Argentina). Canadian Journal of Microbiology 53 (2007): 519-525.
- Rapp P. Formation, separation and characterization of three β-1, 3-glucanases from Sclerotium glucanicum. Biochimica et Biophysica Acta (BBA)-General Subjects 1117 (1992): 7-14.
- McCarthy T, Hanniffy O, Savage AV, et al. Catalytic properties and mode of action of three endo-β-glucanases from Talaromyces emersonii on soluble β-1, 4-and β-1, 3; 1, 4-linked glucans. International Journal of Biological Macromolecules 33 (2003): 141-148.
- Boyce A, Walsh G. Production, purification and application-relevant characterisation of an endo-1, 3 (4)-β-glucanase from Rhizomucor miehei. Applied Microbiology and Biotechnology 76 (2007): 835-841.
- Ekinci MS, McCrae SI, Flint HJ. Isolation and overexpression of a gene encoding an extracellular beta-(1, 3-1, 4)-glucanase from Streptococcus bovis JB1. Applied and Environmental Microbiology 63 (1997): 3752-3756.
- Allardyce BJ, Linton SM. Purification and characterisation of endo-β-1, 4-glucanase and laminarinase enzymes from the gecarcinid land crab Gecarcoidea natalis and the aquatic crayfish Cherax destructor. Journal of Experimental Biology 211 (2008): 2275-2287.
- Song JM, Nam KW, Kang SG, et al. Molecular cloning and characterization of a novel cold-active β-1, 4-D-mannanase from the Antarctic springtail, Cryptopygus antarcticus. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 151 (2008): 32-40.
- Cieslinski H, Kur J, Bialkowska A, et al. Cloning, expression, and purification of a recombinant cold-adapted β-galactosidase from Antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expression and Purification 39 (2005): 27-34.
- Kumar V, Sharma N, Bhalla TC. In Silico Analysis of-Galactosidases Primary and Secondary Structure in relation to Temperature Adaptation. Journal of Amino Acids 2014 (2014).
- Teng D, Wang JH, Fan Y, et al. Cloning of β-1, 3-1, 4-glucanase gene from Bacillus licheniformis EGW039 (CGMCC 0635) and its expression in Escherichia coli BL21 (DE3). Applied Microbiology and Biotechnology 72 (2006): 705-712.
- Chen X, Meng K, Shi P, et al. High-level expression of a novel Penicillium endo-1, 3 (4)-β-D-glucanase with high specific activity in Pichia pastoris. Journal of Industrial Microbiology & Biotechnology 39 (2012): 869-876.
- Elgharbi F, Hmida-Sayari A, Sahnoun M, et al. Purification and biochemical characterization of a novel thermostable lichenase from Aspergillus niger US368. Carbohydrate Polymers 98 (2013a): 967-975.
- Elgharbi F, Hmida-Sayari A, Sahnoun M, et al. Purification and biochemical characterization of a novel thermostable lichenase from Aspergillus niger US368. Carbohydrate Polymers 98 (2013b): 967-975.
- Parvizpour S, Razmara J, Ramli AN, et al. Structural and functional analysis of a novel psychrophilic β-mannanase from Glaciozyma antarctica PI12. Journal of Computer-aided Molecular Design 28 (2014): 685-698.
- Zhang R, Zhou J, Gao Y, et al. Molecular and biochemical characterizations of a new low-temperature active mannanase. Folia Microbiologica 60 (2015): 483-492.
- Kumar S, Nussinov R. Salt bridge stability in monomeric proteins. Journal of Molecular Biology 293 (1999): 1241-1255.
- Geralt M, Alimenti C, Vallesi A, et al. Thermodynamic stability of psychrophilic and mesophilic pheromones of the protozoan ciliate Euplotes. Biology 2 (2013): 142-150.
- Neville DC, Alonzi DS, Butters TD. Hydrophilic interaction liquid chromatography of anthranilic acid-labelled oligosaccharides with a 4-aminobenzoic acid ethyl ester-labelled dextran hydrolysate internal standard. Journal of Chromatography A 1233 (2012): 66-70.


 Impact Factor: * 3.0
Impact Factor: * 3.0 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
