Corona Outbreak: Is it Nature’s wake-up Call?
Article Information
Kapinder1, Kriti Bhardwaj1, Anita Kamra Verma2*
1Department of Zoology, Deen Dayal Upadhyaya Gorakhpur University, Gorakhpur, 273009, Uttar Pradesh, India
2Nanobiotech lab, Kirori Mal College, University of Delhi, Delhi-110007, India
*Corresponding Author: Anita Kamra Verma, 2Nanobiotech lab, Kirori Mal College, University of Delhi, Delhi-110007, India
Received: 13 December 2020; Accepted: 24 December 2020; Published: 04 January 2021
Citation: Kapinder, Kriti Bhardwaj, Anita Kamra Verma. Corona Outbreak: Is it Nature’s wake-up Call?. International Journal of Applied Biology and Pharmaceutical Technology 12 (2020): 301-321.
View / Download Pdf Share at FacebookAbstract
Abstract
Corona pandemic triggered by SARS-CoV-2, the Severe Acute Respiratory Syndrome Corona Virus-2 has swept the world and created unique health-emergencies. Highly unrelated issues may have cumulated for a crisis of this magnitude to have occurred and can be aptly correlated to climatic and biodiversity changes that are deeply interlinked. It has emerged due to failing human health caused by climatic changes, over urbanization, and civilisations endangering the natural habitats on which they depend. As the pandemic progresses, a plethora of scientific and technical news keeps getting updated, causing a lot of anxieties on the therapeutics of corona virus and prior knowledge makes correlations among demography, climate and regional availabilities to manage the spread of the infection. This epidemic further altered the life style; causing huge losses and endangered the sustenance and livelihood of people. The review brings forth a consolidated story of the zoonotic origin of SARS-COV-2, its molecular structure, and discusses the ecological factors responsible for pandemic outbreak of Covid-19 and the strategies applied for the treatment. The list of “zoonotic diseases” includes bird flu, Ebola, HIV, Hendra, Zika, Hendra, SARS, and MERS. Having bats as its initial host, Corona then got communicated to humans directly or via a intermediate biological host. For any disease to become pandemic, ecological factors such as high population growth rate, malnutrition and unhygienic conditions, poor health sectors, environmental changes are responsible. Several environmental factors such as decrease in air-particulate matter, lowered emissions of CO2 and NO2 primarily because of lockdown leading to negligible human activities have caused reduction in air and water pollution in various countries. Undeniably, the worldwide air pollution is linked to industrialization, urbanization, consumption of excess fossil fuels etc. The unprecedente
Keywords
SARS-CoV-2; Zoonotic diseases; Ecological factors; Nurture; Therapeutic strategies
SARS-CoV-2 articles; Zoonotic diseases articles; Ecological factors articles; Nurture articles; Therapeutic strategies articles
Article Details
Introduction
The Corona pandemic that has swept the world is definitely a man-made crisis. It has emerged due to failing human health, over urbanization, civilisations endangering the natural habitats on which they depend. Highly unrelated issues may have cumulated for a crisis of this magnitude to have occurred and can be aptly correlated to climatic and biodiversity changes that are deeply interlinked. This scenario may have erupted due to our ignorance or gross negligence whatever be the reason, it is failure due to our reluctance to respect inter-dependence between humans, between humans and animal species and the bio-resources in general. In the past years SARS appears as an outbreak in 2003, which originated in China in Guangdong [1], and was transmitted to humans from bats using civets as intermediary hosts. Middle East Respiratory Syndrome (MERS) surfaced in 2012 causing fresh epidemic, where camels were the intermediary hosts and originated in Saudi Arabia [2]. In year 2019 Novel Corona virus is originated from Wuhan, Hubei province of China, wherein it originated in bats, using pangolin as hosts transmitted to humans. But now-a-days because of absence of host and it has fast mutation rate leads to the direct human to human transfer without intermediary host.
ICTV (International Committee on Taxonomy of viruses) termed Corona virus as Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV2) based on the previous outbreak by SARS in 2003. WHO provided the name as COVID-19 (corona virus outbreak in 2019) on 11 February 2020, following the guidelines from OIE and FAO. Corona is named as such because of his appearance of spherical virions with an inner and surface projections shows similarity with solar corona which means crown [3]. This virus belongs to Coronavirinae family in the Nidoviriales order having four different genus are discovered- α(alpha), β(beta) which originated from the bats origin while γ (gamma), δ (delta) are obtained from the pigs, and bird origin [3]. Human corona virus had a group includes hCoV-229E, OC-43, NL-63, and HKU-1, SARS-CoV and the Middle East Respiratory Syndrome Corona-virus (MERS-CoV). SARS-CoV has been detected in gastrointestinal tract, saliva, sputum and urine so generally the route of infection includes mouth, eyes, and nose. The symptoms shown by the host includes pneumonia symptoms like fatigue and nasal congestion, high body temperature upper respiratory tract infections, with a gestation period of 5-7 days. In the infected peoples the inflammatory markers are enhanced such as proinflammatory cytokines and C-reactive protein. Out of different genus only some are fatal for the human beings such as beta strain are the most virulent affecting the respiratory and digestive system adversely.
As China reported that the Wuhan outbreak was epidemiologically connected to a large animal market that reveals the zoontic origin of the covid19 [4], has currently affected over 200 nations worldwide and has been acknowledged as a pandemic. Present scenario in mid-June 2020 specifies ~8.2 million people being infected people worldwide [5]. Over 50% of the corona positive people have completely recovered, ~40% continue as active cases, and additional 55,000 people may have reached the critical stage, necessitating hospitalization and ventilators as respiratory support. Developing countries including Brazil, Russia and India were affected, not sparing the developed nations like USA, Spain, Italy, UK which were severely affected. USA recorded their maximum number of affected patients and reported ~10% mortality rate. Although, China, is now indicating downslide in infected cases with reports of single-digit mortality records. The world has been arrested owing to lockdown being imposed to arrest the virus transmission from human to human to control the infection from spreading. Seeing the meteoric rise in the COVID-19 infected cases, the World Health Organization(WHO) has correctly stated Corona as a pandemic and a global-health emergency [6].
Coronaviruses (CoVs) represents a collection of viruses enveloped by a lipid -protein coat, encapsulating a positively charged single-stranded RNA [7]. SARS-CoV-2 causes COVID-19, the infective form responsible for its pathogenesis when compared to its earlier reported form SARS-CoV (2002), as well as when compared to Middle East respiratory syndrome coronavirus (MERS-CoV, 2013). A holistic approach is necessary to appreciate the structure, virulence, molecular mechanism of pathogenesis, to achieve therapeutic strategies to ameliorate the symptoms in terminally ill patients [8].
CoVs belongs to Family Coronaviridae, and order Nidovirales. Taxonomically, divided in four genera namely α-, β-, γ-, and δ-coronaviruses [9]. While, mammals get infected by α- and β- CoVs, the γ-coronaviruses mostly infect the avians, and both aves and mammals are susceptible to δ-coronaviruses. MERS-CoV, SARS-CoV, bovine coronavirus (BCoV), mouse hepatitis coronavirus (MHV), human coronavirus OC43, bat coronavirus HKU4, and importantly SARS-CoV-2, represent the β-coronaviruses [10]. Altogether, the three strains of coronaviruses namely MERS-, SARS-and SARS-CoV-2 are spread via zoonotic transmission and are transmitted to humans by proximity or close contact. There is an exponential growth of the infected as the primary reproduction number (R0) representing individual-to-individual transmission of SARS-CoV-2 is approximately 2.6 [11].
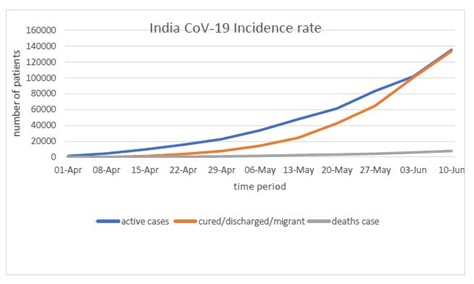
That corona which originates in Wuhan had been mutated at the faster rate because of its positive sense strand which directly interferes with the host protein machinery. The data provided by the WHO situation report by 2:32 PM CET June 10, 2020 total cases 7127753 are confirmed out of which 87835 are cases added in last 24 hours. Total mortality had 407159 out of which 2763 in last 24 hours (Figure 1). The scenario in India as presented by MyGov Corona updates as on 10 June at 8 Am, Total active cases are 135206, cured/Discharged/migrated cases 133632 and death cases are 7745 (Figure 2).
Data obtained from research shows that the enveloped diameter is of 60 to 140nm showing some cases of polymorphism [12]. As genomic studies had been made on it which unpacked the homology of bat corona virus isolated RaTG13 strain MN996532 was 96% but the other isolates show not more than 80%. Recent data which obtained from the pangolin and bat is of 99% which indicate towards the transfer of virus from bat to humans is via the intermediary host pangolin still more research in this field is pending to give the more appropriate answer [13].
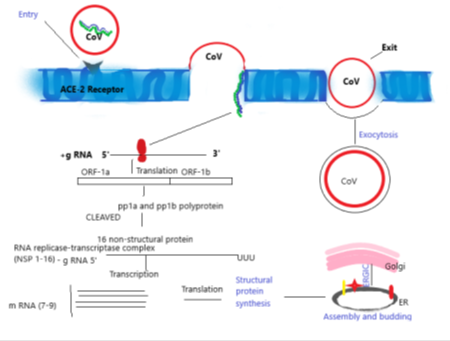
The transmission includes infectious droplets via respiratory mode mainly through cough and sneezes [14,15]. The semen or body fluids are major reservoirs of virus including urine, sweat and respiratory secretions [16]. As the virus enters the host, the initial target is enterocytes and pneumocytes and starts their division of genetic material [14,15]. Other target organs include nephric tubules, tubular epithelial cells of excretory system, cells of defense system, and brain neuronal cells because of the presence of ACE2 receptors [14]. The route used to infect humans was supposedly receptor-mediated endocytosis through the angiotensin-converting enzymes II [17].
Origin of nCoV-19
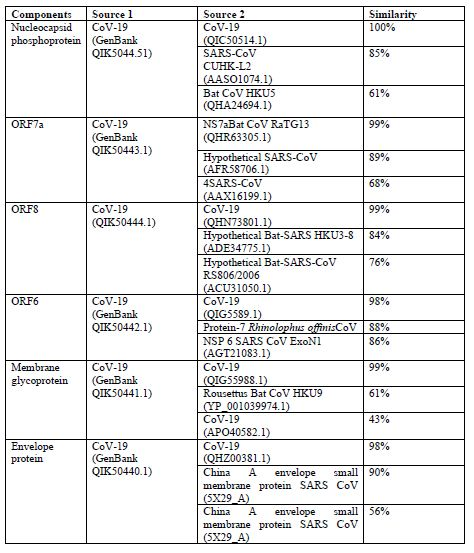
The origin of nCoV-19 analysis done by using different CoV-19 genes from GenBank and following outcomes are occurred which provides significant data for the result [18] (table1):
From above data we can predict that CoV-19 is originated from mutations within the corona virus family.
Molecular structure of corona virus
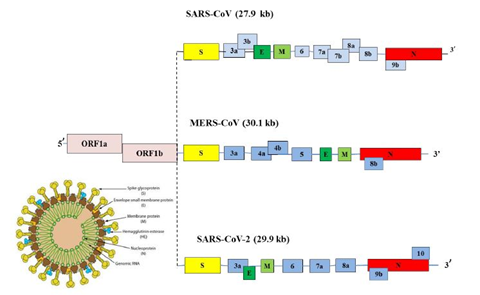
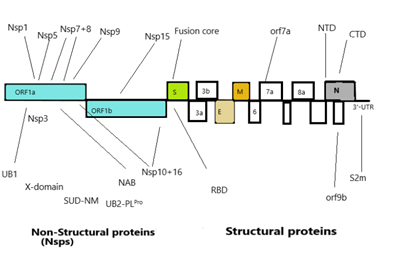
Corona virus genomic size is of about 26 to 32 Kb [17]. Ultrastructure of the virus indicates presence of four structural genes which encode four different proteins (Figure3) [19,20]. The ORF1a, ORF1b, S, OEF3, E, M, N are the genes in 5’ to 3’ direction as written order in the RNA genome of CoV (Figure 4) [20]. The ORFa/b gene segment cover up the 2/3rd part of the RNA genome, which function to perform the two viral replicase polyproteins (PP1a and PP1ab) [21]. These PPs forms sixteen mature non-structural proteins (NSPs) which take part in different viral functions [22] involving replicase transcriptase complex formation. The other genes than PPs gene participate in mRNA formation which further produces the structural proteins (Figure 3) [20,22]. The structural proteins and their functions are given below:
Nucleocapsid protein (N): The conserved structure found in the strains of corona virus family for N-protein [20]. N-arm, Central linker and the C-tail are the three intricately dis-ordered regions (IDRs) of N-protein [23]. The structural and functional domains of N-protein are the NTD (N-Terminal Domain) and CTD (C-Terminal Dimerization Domain). Functionally, CTD is responsible for dimerization, whereas NTD performs RNA binding. Arginine and serine residues are found in large quantity in CL region and their phosphorylation sites also have large number. Nucleocapsid protein oligomerization and N- M protein interactions are done with the help of C- terminal IDRs [23].
Functionally the N protein maintain the RNP complex and its formation [23]. They regulate the transcription and replication of viral RNA. With the help of EF1α-mediated action, it inhibits protein translation in host body. Alteration of host cell metabolism including host cell cycle (N proteins inhibits CDK4), and apoptosis [23]. By inhibition of cytokinesis it can inhibit the proliferation in cells [20]. Interaction of 5-monophosphate and NTD RNA-binding site maybe further used for designing RNA-binding inhibitors. Oligomerization process completes the CTD N-protein oligomeric site which is having tail peptide N377-389 and nearly at 330μM concentration of viral titer, inhibition is found [24]. The 3-dimensional structure including all complex has been seen from PBD is 4LMC, 4LM9, 4LM7, 4LI4 respectively. N220, a type of nucleocapsid protein peptide, have binding affinity to human MHC-1 in T2 cells, and leads to the activation of T-cells (cytotoxic). Some other important peptides are NP111, NP331 and NP351 [24].
Spike glycoprotein
The spike glycoprotein has a detrimental role in pathogenesis as its main target is to bind the host cell membrane via RBD [25]. Infection gets initiated through the sticking the virion S protein to the cell of host. S protein has three subunits -S1, S2, and S2 that have different function for adherence and consists of 1273amino acid residues While, S1 subunit facilitates attachment of virions to the cell membrane by binding to human ACE2 that is responsible for the onset of infection [26]. Simultaneously, a conformational change of the S protein is initiated after the virion’s endosomal entry [27]. For the purpose of vaccine development, it is essential to appreciate the dynamics of the conformational change as this fluidity of structure of the target alters the immune response significantly [28]. Probably the S protein mutations facilitate conformational changes, that alters antigenicity. Numerous mutations have been reported in the binding region of S1 receptor of SARS-CoV-2, its interface with the ACE2 is highly conserved in civet, swine, humans, as well as bats, with the exception of mouse ACE2 [26,29,30]. S2, the other sub-unit acts as a fusion protein enabling the fusion of mammalian cell-membrane with the virion. The assumption is that during fusion, there are three states in which the S2 protein exists: i) Native state/pre-fusion state, leading to ii) an intermediate pre-hairpin state, and lastly iii) resulting in the post-fusion hairpin state. Interestingly, these dynamic states of conformation fine-tune the virion-entry mechanism into the host, this gives us the opportunity window to target and develop effective therapy [28]. The cleaved portion of S2’ subunit of S protein acts as the fusion peptide [31]. Intense similarity ~99%, has been observed in the sequence of stalk spike of S2 in SARS-CoV-2 to bat SARS-like CoVs with human SARS-CoV, signifying that broad-spectrum antivirals could be developed for use against the viral S2 subunit, that may effective against COVID-19 treatment [7,32]. The greatest variability has been observed in the RBD of the spike protein of SARS-CoV-2. Again, the MSA profile suggested that the three CoVs i.e., SARS-CoV, BAT-CoV HKU3 and SARS-CoV-2, share a characteristic pattern of evolution, although, MERS-CoV exhibits a diverse composition of RBD amino acid sequence from the earlier cited virus strains. The stretched receptor binding motif of RBD enables virus binding to the receptor of host indicating divergence and involvement of several pathogenic mechanisms. In previous outbreaks of SARS and MERS have suggested that the HR1 of HCoV spike (S) protein can also act as an inhibitory site.
The spike protein (S): It appears in the form of trimer, type I-TM protein. S-protein is divided into three parts such as ectodomain (ED) region, the TM region, and the intracellular domain, that is an integral part of the intracellular short tail [33]. The ED comprises of a receptor binding S1 domain (three S1 heads) and a subunit S2 (trimeric stalk), with membrane fusion on C-terminal. The exterior of the virion shows a geometric arrangement of the spike protein, making it appear like a corona on the exterior [33]. The spike proteins have glycans which attribute DC/L-SIGN virus infection. These glycosylation asp- linked sites are 109, 118, 158, 227, 589 and 699 [34]. S-protein is multifunctional in nature, plays a definite part in internalization of the virus in the host cell [35]. During the initial phase, the S1 domain recruitsthe respective host receptor namely ACE2 in SARS-CoV and PP4 in MERS-CoV, and the S2 domain is responsible for initiating the merging of the host and viral membranes, thereby, facilitating the entry of virus genome into the host. Because of this property it is used as target in research view for producing viable medicines [35]. Spike proteins also provide signals to the host cells for entry of corona virus. The main components of S1 Domainare NTD and CTD. It functions as antigen on the viral surface having receptor binding domain (RBD) [36,37]. R453 residue of RBD of spike protein interacts with K341 of ACE-2 receptor of host involving 14 amino acid of SARS-CoV and 18 residues of ACE-2 [37]. The S2 domain consists of seven repeat regions- HR1 & 2, and the lyophilic fusion peptide [37].
An envelope protein (E): The membrane Envelope (E) proteins represent a small group of proteins, comprising ~75aa, that assist in viral morphogenesis, assembly formation and triggers release of virions [38,39] in CoV having two domains- the hydrophobic domain and the charged cytoplasmic domain. But their structure is polymorphic in different strains [40]. This E protein plays specific role in CoV morphogenesis, to be more precise during association and dissociation to the host surface. This protein found around the endoplasmic reticulum and golgi bodies [41]. Oligomerization is the main function of E protein that leads to formation of ion channels. Works in tandem with other proteins and modifies their functions. It acts as the virulence factor, and is essential for gathering and budding processes [40].
Membrane (M) protein: M proteins represent conserved structural proteins having a length of ~222 amino acid that function in consensus with N, E, and the S proteins, and have a key role in packaging of viral RNA [42]. These proteins exhibit a stretch of conserved amino acids that proposes a shared architecture. Since the M proteins provide distinct shape and stable structure to virus, this protein is in abundance in the virus. The M protein MSA data indicates conserved sequence across the four strains, SARS-CoV, BAT-CoV and SARS-CoV-2. Nevertheless, there is substantial deviation in the M protein sequence of MERS-CoV. M proteins characteristically exhibit three trans-membrane proteins TM domains [43] with C-terminal long, inside and N-terminal short, outside [44]. They help in viral assembly through multiple interactions such as M-M, M-S, M-N proteins [40]. Responsible for maintaining the shape of the viral envelope [40] and preserves viral homeostasis. It takes part in assembly and disassembly of virus-like particles [40] and activates kappa pathway and INF-beta pathway [45].
3C-like protease: This 3CLpro having two identical sub-units join together and forms homodimer having cysteine-histidine dyad on active site having protease activity. It cleaves 11 sites in p1 position of PP1a and PP1ab and gives a mature protein for the further division of viral genome [46]. The major protease of both the SARS-CoV, and SARS-CoV-2, exhibit higher similarity (~96%) in the genome sequence signifying a shared evolution. The key protease of MERS-CoV lacks sequence similarity with the other strains of virus, indicating a distinct evolutionary divergence. Replicase polyprotein 1ab likewise comprises of macro, papain-like, and chief protease domains with an additional catalytic domain-RdRp that serves as an RNA-binding poly-protein. Also, the MSA of replicase poly-protein 1ab signifies a higher level of conserved sequence across all four strains of CoVs.
Once the virus has gained entry into the human cells, the S- protein of CoV binds to the ACE-2 receptors present in the outer surface which causes the viral RNA entering the host cell to transcribe and translate into structural and non-structural protein. The translation products are ORF-1a and ORF-1b, which further synthesises pp1a and pp1b polyproteins which cleaved to finalised into 16 non-structural protein. This step is further proceeded in the direction of assembly, finally budding into ER’s lumen, or intermediate compartment of Golgi (ERGIC) and the newly assembled viruses go out via exocytosis (Figure 5).
Probable receptors facilitating the entry of Corona virus’s into host cell:
The sequential steps for internalization of corona virus maybe as follows:
❖ Adherence to cell membrane of host: ACE-2 receptor is the main gateway where the viral spike protein binding occurs for the entry [30].
❖ Fusion to penetrate the cell endosome to the nucleus: Transmembrane protein serine protease 2 help in the fusion of S-protein by priming inside host cell endosome for induction of life cycle [26].
Furin receptor secretes furin enzyme which cleaves S-protein leads to activation of this protein which leads to more virulent behaviour and host cell becomes more susceptible [26]. As same as CD-26 (DPP4) of host body interacts with S1 domain of CoV S-protein which causes weak immune response and virulence increases.
Non-structural proteins(non-SPs): Apart from the envelope forming structural protein, the genome further encodes several non-structural proteins that perform various functions including replication and assembly formation [47]. Non-SPs contribute in pathogenesis of virus by modifying the initial regulation of transcription regulation, immunomodulation, helicase activity, trans-activation of gene and repudiating the anti-viral response [48,49,50]. Few key functions of non-SPs in SARS-CoV-2are enumerated in Table 1. Analysis of reported data suggests that the SARS-CoV-2 has a cluster of inherently dis-ordered regions that under native conditions are deficientin an organized tertiary structure. The Nsp3 (920–1020) of the N-terminal region indicates enhanced propensity to be dis-ordered. However, the reported analysis offers an insight into the proteome of non-SPs along with the un-structured region of the SARS-CoV-2 poly-protein, that inadeptly helps appreciate the source of viral infection, ie. the structure-based drug design and discovery to establish interaction of proteins of SARS-CoV-2 with the host membrane inthe diverse biological milieu.
Probable targets for drugs Human CoVs indicate insignificant protein machinery but it plays an important role in pathogenesis as it facilitates the entry of virus in host. As discussed earlier, an envelope protein (E), a spike glycoprotein(S), nucleoprotein (N), a membrane protein (M), and the two iso-forms of the replicase poly-protein, specifically, 1a and 1ab are probable targets for drugs. The foremost protease-domain along with 1ab- RNA-binding belonging to RdRp domain that is present in the replicase poly-protein is critical for establishing pathogenesis. The sequences analysis of the possible drug targets by multiple sequence alignment (MSA) indicates the associations amongst viruses, with regard to conservation of sequence and divergence. Similarity index as predicted by BLASTp search specified that length similarity was present in the coding regions of the bat-SL-CoVZXC21and bat-SL-CoVZC45, with minor deletions/insertions excluding the longer spike protein. There were twelve coding regions namely 1ab, S, 3, E, M, 7, 8, 9, 10b, N, 13 and 14. Majority of the proteins exhibited higher sequence similarity amid CoV of bat-origin and SARS-CoV-2 with approximately 90% sequence homology for1a, around 86% identity to 1ab, none the less minor sequence homology, ~ 80% for spike protein, including ~73.2% for protein 13. High similarity of Protein1ab was observed amid SARS-CoV and SARS-CoV-2, although protein 3a and 8b-, the additional SARS-CoV-2genes are contiguous to SARS-CoVs. Amongst all, RdRp shows dissimilarity [51]. A methodical assessment indicated substantial variations in the human-infectious CoV proteins: SARS-CoV expresses protein 8a, but is lacking in SARS-CoV-2; interestingly, protein 8b is ~121 amino acid, i.e., lengthier in SARS-CoV-2).Substitutions of about 380 amino acidhave been recognized between SARS-CoV-2,along with the contiguous arrangements of SARS and SARS-like CoVs, that shows deviation in the pathogenic and functional characteristics of SARS-CoV-2. Substitutions were identified primarily in the nsp3, nsp2, and protein S; whereas, nsp7, nsp13, envelope, matrix, or accessory protein sp6 and 8b were conserved. Based on protein sequence similarity, it canbe inferred that three viral species, namely, SARS-CoV, bat-SL-CoVZC45, and bat-SL-CoVZXC21 are very similar to SARS-CoV-2.
Therapeutic Strategies
Data collected from different sources regarding treatment on the basis of different structural components and their interaction with certain chemicals provide a way for effective treatment of CoV and helps identify potential targets.
Inhibitory action of different chemicals against N protein
N proteins maybe used as possible drug targets, as they bind to the viral RNA via the core RNA-binding domain that is ~140 amino acid in length in a “beaded string” style [10]. N Proteins have a key role in packaging of RNA of the virus in the ribo-nucleocapsid [42]. SARS-CoV-2 possesses a very conserved N protein across CoVs, it shares ~90% sequence homology with SARS-CoV.The interaction of M protein with viral genome facilitates the formation of viral assembly that further augments the transcription and replication of the virus [52]. Analysis of N protein from the SARS-CoV, BAT-CoV, and SARS-CoV-2 indicate regions that are conserved and similar identities. Therefore, antibodies against SARS-CoV may possibly recognize and bind to SARS-CoV-2. Similar homology was observed for MERS-CoV, where the regions of minor variations in sequence signifies its evolutionary divergence. In a humanized CoV-OC43 model, the three-dimensional structure of HCoV-OC43 N-NTD complex with ribonucleoside-5’-monophosphate is generally used to identify the distinct ribonucleotide-binding groove. N-protein’s RNA-binding affinity was reduced that hindered the replication process of virus. On the basis of the above structure two tyrosine residues (tyr124 and tyr126) interacted with RNA bases through hydrogen-bonding and stacking interactions [24]. PJ34 at 10µM inhibits replication of CoV, effectively interfering with RNA-binding capacity of HCoV-OC43 N protein, aiming the N-NTD ribonucleotide-binding pocket [24].
Replicase
Replicase poly-protein is an important enzyme that promotes cleavage of the host RNA, facilitating viral replication [53]. As mentioned afore, in the viral genome, the non-structural ORFs namely 1a and 1ab, overlap most of the nucleotide sequences. Multi-fumctional replicase poly-proteins accomplish several functions attributed to viral pathogenesis [54]. But, the key function is to regulate viral transcription and replication. The three domains of replicase poly-protein are papain-like-, macro-, and the main protease domains. Again, MSA data interprets the conserved region of 1a, when compared to other domains, excluding the protease, where the sequence shows least identity and maximum divergence.
Inhibitory activity against protease have been observed by drugs like decahydro-isoquinolin fused-ring, that functions as an inhibitory scaffold. Substitution at the 4-postion of the phenyl group in the N-biphenyl acyl derivative gave a slightly more potent inhibitor that may have some interactions with R188I SARS 3CLpro (the structural protease in CoV) [55]. 4-Nitrile-based peptidomimetic compound is yet another example where it shows the N-terminal protective groups of various peptide length (eg: Cbz-AVLQ-CN with an N-terminal carbo-bezyloxy group) that inactivates the 3CLpro autocleavage of SARS-CoV at IC50 value is 4.6-49μM. This action is taken place by forming a covalent bond with the catalytic cys145 residue and AVQL peptide forms a no. of stable interactions with the S1-S4 substrate binding pockets [56].
In a quest to identify inhibitors, a broad-spectrum peptide designed as spectrum of OC43-HR2P was successfully used. This inhibitor was derived from the HR2 domain of HCoV-OC43, and exhibits broad spectrum fusion-inhibitory activity against the corona family [32]. For example: EK1 was optimized as a form of OC43-HR2P, having the property to form a six-helix bundle structure with both a short alpha-CoV and a long beta-CoV, thereby successfully acting as pan-CoV fusion inhibitors [32]. Further, a recent study identified an antibody blocking domain A-mediated hemogglutination of erythrocytes with an in-vitro neutralizing activity of MERS-CoV S pseudo-virion; and a partial in-vivo prophylactic protection of mice, challenged with a lethal dose of MERS-CoV was observed. Also, Sialosides bind to MERS-CoV S using a different site, that suggests that coronavirus S glycoprotein may have independently evolved the ability to recognize the sialosides using domain A, similar to their ability to engage different proteinaceous receptors via domification of domain B. This clearly indicates that the identified sialoside-binding groove represents a specific origin of viability of inhibitors of MERS-CoV [57].
Envelope protein
E protein is an attractive drug target from the array of SARS-CoV-2 structural proteins [58]. Viroporins are formed by the E proteins, that form lipid-protein pores in the host cell-membrane and maybe useful for ion transport. E protein sequences possess highly conserved regions across all four strains namely SARS-CoV, BAT-CoV and SARS-CoV-2, exhibiting minor deviations in the sequence of the envelope proteins of MERS-CoV.
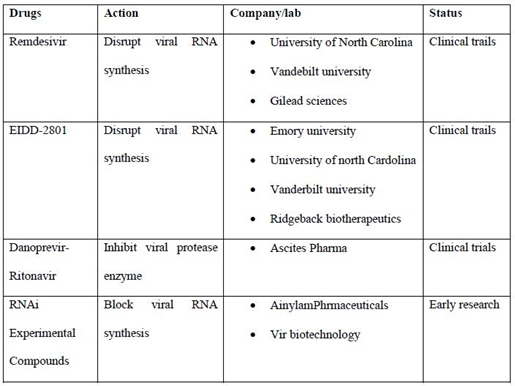
List of some drugs which are widely used in the treatment and their status (table 2):
Universal impact of COVID-19 with respect to ecology; nature’s wake-up call
The overexploitation of natural resources and nature by humans has been witnessed by the world, leading to several intra and inter-national meetings and conferences without significant consequences or plan to save the environment. Nevertheless, the forced lockdown due to COVID-19, has thrust more than half of the human population indoors, with practically nothing to do in the name of saving the environment, but trying desperately to get clear the environment from the pandemic causing virus. Probably, this is Mother Nature wake up call, as well as a chance to rebound, when the human are is idle, and that maybe the highest contribution of the human race towards reviving nature. Although, every organism forms an integral part of the world ecosystem, it is definitely dominated by human race, who inadvertently takes advantage of their absence. Excess urbanization has led to wild animals moving across cities, roads, and intruding human habitation. Humans are witnessing spontaneous variations in nature observing nature’s revival. The Air Quality Index (AQI) indicates a change in favour of Mother Nature. Is a naturally mutated strain of Covid-19 that was introduced by nature from bat to humans [59] a tactical plan of Mother nature or a wakeup call for humanity!.
Wildlife and biodiversity
The imposed quarantine has confined all humans into their homes; allowing the wild-life, a life time opportunity to enjoy the never seen freedom. It has been observed that in human-populated zones, a ‘rewildling’ of urban areas leading to free movements of wildlife occurred. Real time data was issued by Wildlife Institute of India, with an app named “Lockdown Wildlife Tracker” to share the free wildlife movement within the zones where humans were restricted. This free app conveniently helps to track on the comfortable movements of wild animals owing to the lockdown. The data is freely available, can be stored and used for education, scientific research, and to develop strategies for wildlife conservation [60]. Some examples relating to human dominated zones being used by wildlife indicate spotting of coyotes on the Golden Gate Bridge in San Francisco, USA (coyotes are timid by nature and wary of traffic), homes in Washington have deer grazing near their homes, in Bergamo and Barcelona, wild boars are treading boldly, peacocks are seen dancing in the rain, have been observed parading through Bangor, goats through Llandudno and sheep in Wales [61]. The number of vehicles on roads is far less than previous years leading to fewer kill of wildlife by accidents. Reportedly, last year the annual casualty as road kill cases in UK alone ~30,000 deer, ~100,000 hedgehogs, ~50,000 badgers and ~100,000 foxes, will definitely expected to be much lower offering protection to them [62].
Nature’s bounce back
The nature’s flora and fauna seems to have benefitted from the lockdown. This clearly puts the problem of exploitation of natural resources on the fore front, [63]. This should alert the humans to reduce the burden on natural habitats and allow the rejuvenation of natural resources. Variety of colourful insects have emerged and found hovering over plants especially flowers. Tiny moles have been observed on ground lining the footpaths which is perpetually dominated by walking humans. Plenty of wild flowers are seen blooming across the countryside probably due to less pollution [64]. It specifies that given a chance, nature can regenerate much faster without human intrusion. A good take home lesson would be to impose stringent rules post-COVID-19 era to ensure lockdown at regular intervals to nurture the nature.
Ecological factors responsible for pandemic outbreaks
The major problem which we all face in some last outbreaks is the host shifting of viral infections. Due to unavailability of their primary host, they are forced to adapt themselves or change their genome through evolutionary process involving mutations to shift their host which can be easily available such as human having large population size. There are some factors which contribute in host shifting:
Population size: The microparasite reproduction rate is directly proportional to the available primary host reproductive rate. The large population size of human beings is serving a large door for new microparasites to enter into them and becomes the temporarily primary host. The reproductive rate of human population is very high and easily available for the survival of microparasites in the absence of their own primary host [65].
Malnutrition and unhygienic conditions: The major problem world faces during any pandemic outbreaks is the poverty and famishment [66]. During an outbreak disease, the human body should be strong in physical fitness, mental and psychological conditions. To break the chain of transmission the various country governments emphasize on the complete lockdown which is a necessary step taken towards pandemic situation. This lockdown give time to scientist to develop vaccine against CoV. Because of this lockdown various jobs and income sources are disturb which leads to low economic people to suffer from lack of money causes irregular food habits. The side effects of lockdown are malnutrition and these poor people to become susceptible for infection of CoV as well as other infections. These people even not in condition to maintain hygiene for a longer duration.
Less developed health sector: Those countries which are in the criteria of developing countries are still on the way to develop improved health sectors such as hospitals, testing kits, personal protection equipment. For various outbreaks these countries are facing too much of economic loss, mortality of their frontline warriors, and due to less research facilities more time require for development of vaccine or medicine against microparasite infections.
Environmental changes: The population is growing with much high rates and continuously interfering with the environment in the form of industrialization, converting forests into crop lands and project lands for their own benefits. This leads to deleterious effect on primary host favorable survivability so the primary host number decreases and the microparasites are tends to find a new host which are easily available in the surrounding environment.
Overall view of past to present outbreaks and their present situation
Influenza outbreak: Influenza virus have different subtypes which are H1N1, H2N2, H3N3, H5N1 and H7N9. All subtypes have different incubation period and virulence. H5N1 and H7N9 have high death rates in comparison to other whereas H3N3 shows high virulence behavior. The primary hosts of these viruses are wild aquatic birds and virus was travelling in human blood by about 12 years and mutated itself to adopt the human body as new primary host and causes a lot of death troll in different phases of world influenza pandemic outbreaks. Still exist in population with decreased virulence and less morbidity [67].
HIV/AIDS outbreak: This virus comes from simian immunodeficiency virus (SIV). Total 26 different species of apes are exposes similar patterns of virus genomes. Chimpanzees show SIVcdz and sooty mangabeys shows SIVsm which are main causes of AIDS in humans. This virus also comes in zoontic origin. Still exist in human population but the transmission routes are known so occurrence is under controlled [68]. A lot of medicines are now available which control the immunity loss done by this infection.
SARS Outbreak: The primary host of SARS are bats and somehow it transfers to humans via intermediate host as civets. 81th deleterious mutation in virus weakens it and causes less virulence characteristic. In a research it was found that part of genome which was deleted is a major factor used against host antiviral response.
MERS Outbreak: The primary host is also bat and it transfer to humans via camel as intermediary host. This virus also belongs to betacoronavirus family and causes respiratory illness and ultimately death.
Nipah Outbreak: The primary host was also bat and it transmitted via pigs in humans of Malaysia. Their patients are asymptomatic to acute respiratory syndrome and even fatal encephalitis also. The spread of virus is mainly due to un-hygienic conditions. Still there is no vaccine available for it [69].
Ebola outbreak: Also originated from bat via intermediate hosts, such as monkeys, non-human anthropoids or pigs. When humans consumed milk, raw or improper cooked food materials that virus also invade into them. Symptoms shows headache, muscular pain, nausea and sore throat, etc [70]. No specific vaccine is available till date.
Zika virus outbreak: This virus was first isolated from the cage rhesus monkey of Uganda. Later on the primary host becomes Ades mosquito which bites in daytime. This virus in previous time is specifies to only certain areas of Uganda. Symptoms included the pregnant women which face Zika virus their new born babies shown microcephaly few months after the birth [71].
COVID-19 outbreak: Also originated from bat and transmitted via pangolin or some unknown sources. Continues till date. A lot of research started to find the way to reduces it effects and virulence.
During last outbreaks which we all seen is that the host shifting is opportunistic in nature. The human population is very large so now it easy to become the primary host for a variety of microparasites. In this journey the microparasite will face a lot of problems to accomplish within the human body. The human body has a network of immune system which can easily identify the parasite and mechanism used by them to spread their colonies. Due to the immune response the human body starts preparing itself against invaders. To solve this problem, the microparasites have to mutate fast to continue their progeny and during this phase the virulent genes also mutated and the virulence parameter decrease with every cycle. In the whole scenario, the microparasite loses their genomic composition and tried to go back to their original primary host. If the tendency of growth of human population is same as such then in upcoming future the human will faces a lot of microparasites with decreasing interepidemic period.
Conclusion
The list of “zoonotic diseases” includes bird flu, Ebola, HIV, Hendra, Zika, Hendra, SARS, and MERS. COVID-19 has supposedly originated in bats, transmitted to humans using another animal host, probably at the market in Wuhan, where they trade live animals. It is already established that Ebola virus originated in central Africa due to climatic changes, when excessive use of land compelled chimpanzees and bats to co-exist in concerted food areas. Again, urbanization of fruit bats owing to habitat loss is directly responsible for spread of Hendra virus. There is loss of habitat, shift in climatic zones that causes excess migration of wildlife that enhances their possibility of interaction with other species that they may have never encountered otherwise. These man-caused climatic changes are happening globally leading to emergence of novel diseases and spread causing pandemics.To reduce the chances of such outbreaks the human population, there is urgent need to shift towards the sustainable development in spite of urbanization. In this sustainable development, humans need to provide place for other organisms which may play the role of primary host for these parasites. Population control is another problem that requires urgent control measures as they utilize more and more land to fulfil their primary needs because of which a lot of biodiversity (flora and fauna) is becoming extinct due to overuse. The air and water pollution have definitely reduced and perhaps even saved lives in this process, but this epidemic that has killed innumerable lives definitely should not be perceived as a positive impact on environmental change. Essentially, it may be noted that it is not definite whether the dip in emission will be long lasting or transient?. What if the carbon and nitrogen levels revert back when the epidemic finally wanes, and other pollutants will again mar the skies? To protect ourselves from such pandemics, we need to develop our health sectors, control our population growth rate, and move towards the sustainable development and maintain our biodiversity. Do we consider Covid-19 just a chance occurrence of a neo-infectious disease or is it the result of our continued exploitation of nature. Nonetheless, it is definitely nature’s alarm bells ringing to wake-up humanity before it is too late.
Acknowledgement
We are thankful to Deen Dayal Upadhyaya Gorakhpur University and Kirori Mal College, University of Delhi for providing facilities to complete this work.
References
- Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. The Lancet 362 (2003): 1353-1358.
- Memish ZA, Cotten M, Meyer B, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerging Infectious Diseases 20 (2014): 1012.
- Velavan TP, Meyer CG. The COVID-19 epidemic. Tropical Medicine & International Health 25 (2020): 278.
- Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research 7 (2020): 1-10.
- https://www.worldometers.info/coronavirus/
- Malik YS, Sircar S, Bhat S, et al. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Veterinary Quarterly 40 (2020): 68-76..
- Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 12 (2020): 244.
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. Journal of Medical Virology 92 (2020): 418-423.
- Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host & Microbe (2020).
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. InCoronaviruses (2015): pp. 1-23..
- Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 8 (2020): 488-496.
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). InStatpearls [internet] (2020).
- Jin Y, Yang H, Ji W, et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 12 (2020): 372.
- Guo Y, Korteweg C, McNutt MA, et al. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Research 133 (2008): 4-12.
- Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. The American Journal of Pathology 170 (2007): 1136-1147.
- Ding Y, He LI, Zhang Q, et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 203 (2004): 622-630.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (2020): 270-273.
- Dawood AA. Mutated COVID-19, may foretells mankind in a great risk in the future. New Microbes and New Infections (2020): 100673.
- Rottier PJ. The coronavirus membrane glycoprotein. InThe Coronaviridae (1995): 115-139.
- Prajapat M, Sarma P, Shekhar N, et al. Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology 52 (2020): 56.
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology 17 (2019): 181-192.
- McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses 4 (2012): 2902-2923.
- Chang CK, Lo SC, Wang YS, et al. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discovery Today 21 (2016): 562-572.
- Lin SY, Liu CL, Chang YM, et al. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. Journal of Medicinal Chemistry 57 (2014): 2247-2257.
- Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581 (2020): 215-220.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2020): 271-280.
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (2020): 1260-1263.
- Walls AC, Park YJ, Tortorici MA, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell (2020): 281-292.
- Chana JF, Yip CC, To KK. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific 2 COVID-19-RdRp/Hel realtime reverse transcription-polymerase chain reaction assay validated 3 in vitro and with clinical specimens. J Clin Microbiol (2020): 00310-00320.
- Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367 (2020): 1444-1448.
- Qing E, Gallagher T. SARS Coronavirus Redux. Trends in Immunology 41 (2020): 271-273.
- Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Science Advances 5 (2019): eaav4580.
- Belouzard S, Millet JK, Licitra BN, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4 (2012): 1011-1033.
- Han DP, Lohani M, Cho MW. Specific asparagine-linked glycosylation sites are critical for DC-SIGN-and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. Journal of Virology 81 (2007): 12029-12039.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology 3 (2016): 237-261.
- Yuan Y, Cao D, Zhang Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8 (2017): 15092.
- Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nature Reviews Microbiology 7 (2009): 226-236.
- Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. Journal of Virology 81 (2007): 2249-2262.
- Li S, Yuan L, Dai G, et al. Regulation of the ER Stress Response by the Ion Channel Activity of the Infectious Bronchitis Coronavirus Envelope Protein Modulates Virion Release, Apoptosis, Viral Fitness, and Pathogenesis. Frontiers in Microbiology 10 (2020): 3022.
- Venkatagopalan P, Daskalova SM, Lopez LA, et al. Coronavirus envelope (E) protein remains at the site of assembly. Virology 478 (2015): 75-85.
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virology Journal 16 (2019): 1-22.
- Tang T, Bidon M, Jaimes JA, et al. Coronavirus membrane fusion mechanism offers as a potential target for antiviral development. Antiviral Research (2020): 104792.
- Suzuki T, Otake Y, Uchimoto S, et al. Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis. Viruses 12 (2020): 183.
- Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. Journal of Virology 84 (2010): 11418-11428.
- Wang Y, Liu L. The Membrane Protein of Severe Acute Respiratory Syndrome Coronavirus Functions as a Novel Cytosolic Pathogen-Associated Molecular Pattern To Promote Beta Interferon Induction via a Toll-Like-Receptor-Related TRAF3-Independent Mechanism. Mbio 7 (2016): e01872-15.
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal 281 (2014): 4085-4096.
- Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Research 176 (2020): 104742.
- Xue B, Blocquel D, Habchi J, et al. Structural disorder in viral proteins. Chemical Reviews 114 (2014): 6880-6911.
- J Alsaadi EA, Jones IM. Membrane binding proteins of coronaviruses. Future Virology 14 (2019): 275-286.
- Tang C, Deng Z, Li X, et al. Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure. Viruses 12 (2020): 215.
- Müller C, Schulte FW, Lange-Grünweller K, et al. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses. Antiviral Research 150 (2018): 123-129.
- Minakshi R, Padhan K, Rehman S, et al. The SARS Coronavirus 3a protein binds calcium in its cytoplasmic domain. Virus Research 191 (2014): 180-183.
- Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. Journal of Virology 94 (2020).
- Graham RL, Sparks JS, Eckerle LD, et al. SARS coronavirus replicase proteins in pathogenesis. Virus Research 133 (2008): 88-100.
- Ruan Z, Liu C, Guo Y, et al. Potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) of SARS-CoV-2 (2020).
- Shimamoto Y, Hattori Y, Kobayashi K, et al. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorganic & Medicinal Chemistry 23 (2015): 876-890.
- Chuck CP, Chen C, Ke Z, et al. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. European Journal of Medicinal Chemistry 59 (2013): 1-6.
- Park YJ, Walls AC, Wang Z, et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nature Structural & Molecular Biology 26 (2019): 1151-1157.
- Anderson RM, Heesterbeek H, Klinkenberg D, et al. How will country-based mitigation measures influence the course of the COVID-19 epidemic?. The Lancet 395 (2020): 931-934.
- Wild Life Institute of India. Lockdown Wildlife Tracker. https://wii.gov.in/ (retrieved on 23.04.2020).
- Loring K. In San Francisco, coyotes are your wildest neighbours (2020).
- Newburger E, Jeffery A. As coronavirus restrictions empty streets around the world, wildlife roam further into cities. CNBC Environment, Londres (2020).
- Swain SS, Sahoo A, Paital B, et al. Vitamin-C and anti-HIV drug darunavir as combinatorial drug against COVID-19. Front. Biosci. (2020) (accepted).
- Child D. The positive impacts on the environment since the coronavirus lockdown began. https://www.standard.co.uk/news/world/positive-impact-environment-coronavirus-lockdowna4404751.html (2020).
- May RM. Ecological aspects of disease and human populations. American Zoologist 25 (1985): 441-450.
- Pimentel D, Cooperstein S, Randell H, et al. Ecology of increasing diseases: population growth and environmental degradation. Human Ecology 35 (2007): 653-668.
- Park JE, Ryu Y. Transmissibility and severity of influenza virus by subtype. Infection, Genetics and Evolution 65 (2018): 288-292.
- Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends in molecular medicine 18 (2012): 182-192.
- Field HE. Bats and emerging zoonoses: henipaviruses and SARS. Zoonoses and Public Health 56 (2009): 278-284.
- Holmes EC, Dudas G, Rambaut A, et al. The evolution of Ebola virus: Insights from the 2013–2016 epidemic. Nature 538 (2016): 193-200.
- Kindhauser MK, Allen T, Frank V, et al. Zika: the origin and spread of a mosquito-borne virus. Bulletin of the World Health Organization 94 (2016): 675.


 Impact Factor: * 3.0
Impact Factor: * 3.0 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
