Captive Snakes from Brazil as Carriers of Multidrug-Resistant Enterococci
Article Information
Juliana Moraes da Silva Heck¹, Janira Prichula2, Rosana Huff1, Roberto Baptista de Oliveira4, Thiago Silva-Soares5, Jeverson Frazzon3, Ana Paula Guedes Frazzon¹
1Post-Graduation Program in Agricultural and Environmental Microbiology, Microbiology, Immunology, and Parasitology Department, Institute of Basic Health Sciences, Federal University of Rio Grande do Sul, Porto Alegre, RS, Brazil
2Gram-positive Coccus Laboratory, Federal University of Health Sciences of Porto Alegre, Porto Alegre, Brazil
3Laboratory of Biochemistry and Molecular Biology of Microorganisms, Federal University of Rio Grande do Sul, Porto Alegre
4Museum of Natural Sciences, Rio Grande do Sul State Department of Environment and Infrastructure, Porto Alegre, Brazil
5Museum of Natural Sciences of the South of Espírito Santo, Federal University of Espírito Santo, Jerônimo Monteiro, Brazil
*Corresponding Author: Ana Paula Guedes Frazzon, Post-Graduation Program in Agricultural and Environ-mental Microbiology, Microbiology, Immunology, and Parasitology Department, Institute of Basic Health Sciences, Federal University of Rio Grande do Sul, Porto Alegre, RS, Brazil
Received: 28 July 2021; Accepted: 16 August 2021; Published: 26 August 2021
Citation: Juliana Moraes da Silva Heck, Janira Prichula, Rosana Huff, Roberto Baptista de Oliveira, Thiago Silva-Soares, Jeverson Frazzon, Ana Paula Guedes Frazzon. Captive Snakes from Brazil as Carriers of Multidrug-Resis-tant Enterococci. International Journal of Plant, Animal and Environmental Sciences 11 (2021): 503-523.
View / Download Pdf Share at FacebookAbstract
Brazil has one of the most diverse herpetofauna and snakebites are an important health issue. The oral cavity of snakes harbored a wide range of bacteria. Enterococci have been isolated from animals, however, few studies have taken in snakes. In this sense, the present study aimed to evaluate Entero-coccus spp. and their virulence attributes including antimicrobial resistance in oral cavities of healthy snake species in Brazil. Oral swabs from wild and captive snakes were screened for enterococci distri-bution, antimicrobial susceptibility, resistance and virulence genes, and CRISPRs elements by PCR. Overall, 116 enterococci were detected and Enterococcus faecalis was dominant in all snake species, followed by E. faecium, E. avium, and E. hirae. Interestingly, no resistant enterococci were detected in wild snakes. In contrast, captive snakes were found to be carriers of resistant strains, including resistance to erythromycin, rifampicin, norfloxacin, ciprofloxacin, and tetracycline. Enterococcus faecium (50%) and E. faecalis (15.78%) isolates were multi-drug-resistant. Erythromycin resistance genes, the msrC and ermB, were detected in 13.33% and 6.67% of the isolates, respectively. The tetM (70%), tetL (30%) and tetS (10%) genes were detected in the tetracycline-resistant strains. Among the virulence genes, gelE was the most frequent in all strains. CRISPR1-cas, orphan CRISPR2, and CRISPR3-cas elements were present in 16.03%, 15.79%, and 18.31% of the isolates, respectively. No antibiotic resistance was associated with CRISPRs. In conclusion, resistant enterococci in captive snakes are the result of confinement, antibiotic therapy and human contact. Resistant bacteria in captive snakes provide crucial information about public health safety.
Keywords
Enterococci; Maldi-TOF; Antimicrobial resistance; Virulence genes; CRISPRs; Snakes
Enterococci articles; Maldi-TOF articles; Antimicrobial resistance articles; Virulence genes articles; CRISPRs articles; Snakes articles
Article Details
1. Introduction
Snakes play an important role in maintaining balance in the ecosystem. The snakes diet ranges from inver-tebrates to vertebrates; in wildlife they eat a wide variety of animals including snails, insects, fish, frogs, lizards, snakes, amphibians, birds, rodents, bats, primates, and eggs of lizards and birds [1, 2]. Snakes are reptiles belonging to the order Squamata and sub-order Serpente. There are more than 3,900 species of snakes found in the world [3]. In Brazil, the diversity of ophidians is approximately 405 species, distributed into ten families: Anomalepididae, Leptotyphlopidae, Typhlopidae, Aniliidae, Tropidophiidae, Boidae, Viperidae, Elapidae, Colubridae e Dipsadidae [2, 4, 5]. These species are found in all Brazilian biomes, and some are kept in captive conditions, like zoos and serpent scientific breeders for poison extraction and subsequent production of antivenom [2, 4-6].
Not all snakes are venomous, in fact, 600 species are venomous and only 200 can kill or significantly wound a human. Snakebite envenoming is a major public health issue in the developing world; clinical reports have revealed that snakebites are a neglected public in many countries, with major impacts in Africa, Asia and Latin America [7]. According to data from the Brazilian Ministry of Health, during the period of 2009-2013, 144,060 snakebites were recor-ded in Brazil (an average of 28,812 cases per year), with an average mortality of 119 per year [8]. The deaths are caused by poisoning, as well as the snake mouth is colonized by bacteria that can be transmitted to the bitten patient through the skin injury associated with the bite, and may cause secondary infection along with envenomation [9]. Interesting, clinically relevant bacterial species have been found in the oral microbiota and bite wounds from snakes worldwide [10-12]. Diverse studies have revealed a mixture of both aerobic and anaerobic bacterial species in the oral cavity of snakes [13-16]. Panda et al. [17] identified Gram-negative and Gram-positive bacteria, including clinical pathogens such as Bacillus spp., Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus epidermidis in Indian cobra (Naja naja).
Enterococcus spp. are facultative anaerobic bacteria, belonging to the Phylum Firmicutes. Currently, the genus is composed of more than 50 species [18], with E. faecalis predominant in the gastrointestinal tract of humans and other mammals, followed by E. faecium, E. hirae, E. durans, E. casseliflavus, E. gallinarum, and E. mundtii [19]. These genera are also found in oral cavity and urinary tract of humans and other animals. They can also be found in different environments such as soil, water, sewage and plants [18]. However, enterococci are also important opportunistic pathogens for humans due to virulence factors and antibiotic resistance [20]. They represent the second most common cause of hospital-acquired infections, particularly affecting the urinary tract, wounds, and soft tissues. Researches have shown that enterococci species were isolated from human wound infections caused by dogs, cats, bears, and snake bites [15-21]. Enterococcus spp. were the most common pathogens isolated in infected bite wounds and oral microbiota of Naja atra in Taiwan [12]. Huang et al. [11], investigating bacterial infection associated with snakebites in central Taiwan, identified Enterococcus spp. as one of the most common pathogens. Chen et al. [10], analyzing snakebite from Northern Taiwan medical center, identified the Enterococcus spp. as the most frequently pathogens in the wound. In Brazil, group D streptococci (enterococci) were isolated in the abscesses at the site of Bothrops spp. bite [22].
Due to their remarkable ability to adapt to environ-mental conditions and ubiquity, enterococci have been used as sentinel organisms for tracking trends in resistance to antimicrobials [23]. Resistant enterococci have been isolated from captive and wild animals worldwide [24-30] and rare studies regarding snakes [15-21]. This could be justified by the difficulty to manipulate these animals, and also observing them in the wild environment since they make unseen movements in fields and forests [31]. Despite Brazil having one of the most diverse herpetofauna, studies evaluating bacteria in snakes' oral cavities are scarce, and most of them are associated with abscesses caused by bites of snakes [32-34]. This is the first study to report enterococci in the oral cavity of captive and wild snakes of several species in Brazil. We evaluated the antimicrobial susceptibility and virulence determinants of enterococci isolated from oral cavities of snake species in Brazil. The study intends to address if the snakes can be a reservoir of antibiotic-resistant enterococci that can spread through people and animals, contributing with information for public health safety.
2. Materials and Methods
2.1 Oral snakes samples collection
Fourteen oral swab samples were collected from wild and captive snake species (Table 1). Seven wild snakes were captured in the Pacotuba National Forest (FLONA- Pacotuba; 20º45’9.71”S, 41º17’21.27”W) – Espírito Santo state, and Caparaó National Park (20° 25'10"S, 41°48'54") – Serra do Caparaó, in the border between the states of Espírito Santo and Minas Gerais, southeastern Brazil. Sampling technique the active search (visual encounter survey protocol), between March and May 2019, were used. Six different wild snakes species were captured: Thamno-dynastes strigatus, Leptophis ahaetulla, Pseudablabes patagoniesis, Oxyrhopus petolarius, Erythrolamprus poecilogyrus, and Bothrops jararaca. After collec-tion, the wild snakes were returned to nature.
Captive snakes (n = 7), belong to serpent scientific breeder of the Museum of Natural Sciences of the Rio Grande do Sul State Department of Environment and Infrastructure (MCN), Porto Alegre, Brazil, were handled using a snake hook, and the sampling were collected in January and May 2019 (Figure 1). To avoid adding a source of stress for the healthy snakes, the samples were collected during the routine proce-dures of the breeding facility, which follows all the international standards of animal welfare and biosecurity. Six different captive snake species were selected: Philodryas olfersii, E. poecilogyrus, Oxyr-hopus rhombifer, T. strigatus, Bothrops diporus and B. jararaca.
Oral swabs were stored in Stuart transport medium (Oxoid™) and transported to the laboratory for microbiological analyses. The sampling was perfor-med following regulations established by the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio), System Authorization and Information on Biodiversity (SISBIO) n° 300675 and n° 52838.
Table 1: Description of wild (FLONA de Pacotuba and Caparaó) and captive (MCN) snakes that oral samples were collected.
|
Habitat |
Species (common name) |
Family |
N1 |
Collection |
Diet |
|
Wildlife/ FLONA |
Bothrops jararaca (jararaca)
|
Viperidae |
01 |
05/03/2019 |
F Frogs, rodents [35] |
|
Erythrolamprus poecilogyrus (Goldbauch-Buntnatter) |
Dipsadidae |
01 |
05/03/2019 |
Frogs, fish, lizards and rodents [36] |
|
|
Leptophis ahaetulla (parrot snake)
|
Colubridae |
01 |
04/24/2019 |
Frogs and lizards [37] |
|
|
Oxyrhopus petolarius (false-coral) |
Dipsadidae |
01 |
03/20/2019 |
Lizards, rodents and bird eggs [38] |
|
|
Pseudablabes patagoniensis (Patagonia green racer)
|
Colubridae |
02 |
05/03/2019 |
Amphibians, frogs, birds, lizards, mammals, fish and snakes [35] |
|
|
Wildlife/ Caparaó |
Thamnodynastes strigatus (coastal house snake)
|
Dipsadidae |
01 |
04/24/2019 |
Frogs, lizards and mammals [39] |
|
Captive/ MCN |
Bothrops diporus (jararaca-pintada)
|
Viperidae |
02 |
01/13/2019 |
Wistar rats [40] |
|
Bothrops jararaca (jararaca)
|
Viperidae |
01 |
01/13/2019 |
Wistar rats [40] |
|
|
Erythrolamprus poecilogyrus (Goldbauch-Buntnatter) |
Dipsadidae |
01 |
05/24/2019 |
Fish [40] |
|
|
Oxyrhopus rhombifer (Amazon false coral snake)
|
Dipsadidae |
01 |
05/19/2019 |
Wistar rats [6] |
|
|
Philodryas olfersii (South American green racer)
|
Colubridae |
01 |
01/13/2019 |
Wistar rats [40] |
|
|
Thamnodynastes strigatus (coastal house snake)
|
Dipsadidae |
01 |
01/20/2019 |
Wistar rats [40] |
- N: number of animals
2.2 Isolation and identification of enterococci from the oral cavities of captive and wild snakes
Oral swabs were pre-processed according to Prichula et al. [27]. Twenty colony-forming units were randomly selected from each sample. Phenotypic criteria, such as size/volume, shape, color, Gram staining, catalase production, capacity to growth at 45 °C and bile aesculine reaction, were used to separate the enterococci group and the non-enterococcal strains [41].
Selected pure colonies were stored in a stock solution of skin milk 10% (Difco, Sparks, MD, USA) and 10% glycerol (Neon Comercial Ltda, São Paulo, SP, BR) at -20 °C. Collected bacteria were identified by matrix-assisted laser ionization and desorption technique (MALDI-TOF) applied to Enterococcus, according to Sauget et al. [42]. MALDI-TOF analysis was per-formed using a LT Bruker microflex mass spec-trometer (Bruker Daltonik GmbH) and spectra were automatically identified using BrukerBioTyper ™ 1.1 software.
Strains not identified by MALDI-TOF were submitted to species-specific PCR assay. Total DNA extraction was carried out by a physical-chemical method [43], with a total volume of 25 µL, containing: 100 ng of DNA template, 1X PCR buffer (10 mM Tris–HCl [pH 9.0], (Invitrogen, Carlsbad, CA, USA), 1.5 mM of MgCl2 (Invitrogen, Carlsbad, CA, USA), 200 μM of dNTPs (Ludwig Biotecnologia), 0.4 μM of each primer (Invitrogen, Carlsbad, CA, USA), 1.0 U of Taq polymerase (Invitrogen®). PCR conditions for all amplification reactions were as follows: initial denaturation at 94 °C for 5 min.; followed by 35 cycles of denaturation at 94 °C for 1 min.; the appropriate annealing temperature for each species (as listed in Supplementary Table 1) for 1 min.; extension at 72 °C for 1 min.; and final extension at 72 °C for 5 min.
2.3 Antibiotic resistance profiles of enterococci strain isolated from oral samples of snakes
All strains were screened for antibiotic susceptibility by Kirby-Bauer disk diffusion method according to Clinical and Laboratory Standards Institute [44]. Eleven antibiotics commonly used in clinical and veterinary medicine were evaluated: ampicillin 10 μg (AMP), ciprofloxacin 5 μg (CIP), chloramphenicol 30 μg (CHL), erythromycin 15 μg (ERI), gentamicin 120 μg (GEN), nitrofurantoin 300 μg (NIT), norfloxacin 10 μg (NOR), rifampicin 5 μg (RIF), streptomycin 300 μg (EST), tetracycline 30 μg (TET) and vancomycin 30 μg (VAN). Minimum inhibitory concentration (MIC) of vancomycin was determined by broth microdilution and interpretation of the results was performed following CLSI guidelines [45]. Staphylococcus aureus ATCC 25923 and E. faecalis ATCC 29212 strains were used as quality control of disks. Isolates that showed a resistance profile to one, two, and three or more classes of antimicrobials were classified as: single-resistant (SR), double-resistant (DR), and multidrug-resistant (MDR), respectively [46]. Intermediate-resistant strains were grouped in the resistant strains.
2.4 Detection of virulence, resistance-associated genes and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) in enterococci by PCR
The presence of virulence genes, such as ace (adhesin to collagen of E. faecalis), cylA (cytolysin) and gelE (gelatinase) was determined in all enterococcal isolates. On the other hand, only erythromycin- and tetracycline- resistance phenotypes were examined for the presence of macrolide (ermB and msrC) and tetracycline (tetL, tetM and tetS) resistance genes, respectively. PCR reactions followed the protocol described by Santestevan et al. [28]. Primers are described in Supplementary Table 1, with the appropriate annealing temperatures.
The presence of Type II CRISPRs elements (CRISPR1-cas, CRISPR2-orfan, and CRISPR3-cas) were investigated by PCR in all enterococcal samples. Primers for CRISPRs genes reported by Palmer and Gilmore [45] were used in PCR reactions. The primers and annealing temperatures used are listed in Supplementary Table 1. The PCR was performed as described by Huescas et al. [47].
3. Results
3.1 Enterococci species in the oral cavities of captive and wild snakes species from Brazil
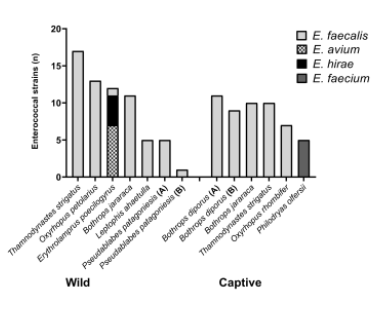
A total of 116 enterococci (64 from wild and 52 from captive snakes) were recovered from 13 oral samples of snakes belonging to the species including T. strigatus, L. ahaetulla, P. patagoniesis, O. rhombifer, O. petolarius, P. olfersii, B. diporus and B. jararaca. Only in one sample of captive snake belonging to E. poecilogyrus species was not detected enterococci.
As result, among the 116 Enterococcus spp. reco-vered, the most frequently isolated species were E. faecalis (78.45%), followed by E. faecium (12.07%), E. avium (6.03%), and E. hirae (3.45%).
Differences in the distribution of enterococci species were detected amongst the two groups of snakes, as shown in Figure 2. Among the 64 enterococci isolates from wild snakes, the species E. faecalis (82.81%; n = 53), E. avium (10.93%; n = 7), and E. hirae (6.25%; n = 4) were identified. On the other hand, 52 enterococci were isolated from captive snakes belonging to E. faecalis (73.07%; n = 38) and E. faecium (26.92%; n = 14).
3.2 Resistance profile in enterococci from wild and captive snakes
The enterococci isolated from wild snakes were susceptible to all antimicrobial agents tested. In contrast, of the 52 strains isolated from captive snakes, 45 (86.53%) were resistant to at least one antimicrobial agent tested. Strains showed resistance to erythromycin (57.69%), rifampicin (50%), ciprofloxacin/norfloxacin (30.77%), tetracycline (19.23%), nitrofurantoin (13.46%), and chloramphe-nicol (5.77%).
The percentages of DR and MDR strains isolated were 32.69% and 25%, respectively (Table 2). Of the 13 MDR strains, six (15.78%) were E. faecalis and seven (50%) were E. faecium. Interesting, one E. faecalis isolated from captive B. diporus showed resistance to six different antimicrobials tested (norfloxacin; chloramphenicol; erythromycin; nitro-furantoin; rifampicin; tetracycline) (Table 3).
Table 2: Antimicrobial resistance profiles among enterococci isolated from oral samples of captivity snakes.
¹Antimicrobials: CIP/NOR, ciprofloxacin/norfloxacin; CHL, chloramphenicol; ERY, erythromycin, NIT, nitrofurantoin; RIF, rifampicin, TET, tetracycline.
²Profiles: SR, single-resistant; DR, double-resistant; MDR, multidrug-resistant.
Table 3: Antimicrobial resistance phenotypic profile of Enterococcus spp. isolated from oral samples of captive snakes.
|
Number of resistant enterococci by snake species |
|||||||
|
Profile1 |
Antimicrobials2 |
Species |
B.d3 |
B.j4 |
O.r.5 |
P.o6 |
T.s7 |
|
SR |
RIF |
E. faecalis |
3 |
1 |
|||
|
E. faecium |
1 |
||||||
|
TET |
E. faecalis |
||||||
|
ERY |
E. faecalis |
1 |
3 |
||||
|
E. faecium |
1 |
||||||
|
NIT |
E. faecium |
1 |
|||||
|
NOR-CIP |
E. faecalis |
1 |
2 |
||||
|
DR |
ERY/RIF |
E. faecalis |
7 |
||||
|
CLO/NOR |
E. faecalis |
1 |
|||||
|
ERI/NIT |
E. faecalis |
1 |
1 |
||||
|
CLO/ERY |
E. faecalis |
1 |
|||||
|
ERY/NOR |
E. faecalis |
1 |
|||||
|
RIF/NIT |
E. faecalis |
1 |
|||||
|
RIF/NOR |
E. faecalis |
1 |
1 |
||||
|
RIF/TET |
E. faecalis |
1 |
|||||
|
TET/ERY |
E. faecium |
1 |
|||||
|
MDR |
TET/RIF/ERI |
E. faecium |
2 |
||||
|
RIF/ERY/CIP-NOR |
E. faecium |
4 |
|||||
|
E. faecalis |
2 |
1 |
1 |
||||
|
TET/RIF/ERY/NOR |
E. faecium |
1 |
|||||
|
TET/RIF/ERI/NIT |
E. faecalis |
1 |
|||||
|
TET/RIF/CLO/ERY/NOR/NIT |
E. faecalis |
1 |
|||||
- SR: single-resistant; DR: double-resistant; MDR: multidrug-resistant. 2. Antimicrobials: ERY, erythromycin; CIP, ciprofloxacin; NOR, norfloxacin; RIF, rifampicin; NIT, nitrofurantoin; CHL, chloramphenicol; TET, tetracycline. 3. B.d: Bothrops diporus (jararaca-pintada); 4. B.j: Bothrops jararaca (jararaca); 5. O.r: Oxyrhopus rhombifer (Amazon false coral snake); 6. P.o: Philodryas olfersii (South American green racer) and 7: T.s.: Thamnodynastes strigatus (coastal house snake).
3.3 Occurrence of resistance and virulence-associated genes and Clustered Regularly Inter-spaced Short Palindromic Repeats (CRISPRs) in enterococci
The frequency of erythromycin-resistant strains (n = 30) positive for the ermB and msrC genes were 6.67% (n = 2) and 13.33% (n = 4), respectively (Supple-mentary Table 2). Among the 10 tetracycline-resistant enterococci, seven (70%) were positive to tetM gene, three (30%) to tetL gene, and one (10%) to tetS gene (Supplementary Table 2).
Virulence genes were detected among all enterococci species. The gelE was the most frequent (59.48%; n = 69), followed by ace (57.76%; n = 67), and cylA (1.72%; n = 2). The gelE gene presented a higher percentage in wild snakes, while ace and cylA genes showed a similar frequency between the snakes Supplementary Table 3).
CRISPR1-cas, orphan-CRISPR2, and CRISPR3-cas elements were positive in 16.03%, 15.79%, and 18.31% of the strains, respectively (Table 4). The orphan-CRISPR2 was detected at a low frequency in enterococci strains collected from captive snakes and CRISPR3-cas in wild snakes. CRISPR1-cas was fou-nd in similar frequency among the strains. No antibiotic resistance was associated with CRISPRs elements.
Table 4: Number (%) of CRISPRs elements identified in enterococci isolated from oral samples of wild and captive snakes.
|
Number (%) CRISPRs elements |
||||
|
Habitat |
Species (n) |
I |
II |
III |
|
Captive |
E. faecalis (38) |
2 (5.26) |
2 (5.26) |
11 (2.94) |
|
E. faecium (14) |
8 (57.14) |
0 |
3 (21.42) |
|
|
Subtotal (52) |
10 (19.23) |
2 (3.84) |
14 (26.92) |
|
|
Wildlife |
E. avium (7) |
0 |
0 |
0 |
|
E. hirae (4) |
1 (25) |
0 |
0 |
|
|
E. faecalis (53) |
6 (11.32) |
16 (30.18) |
5 (9.43) |
|
|
Subtotal (64) |
7(10.93) |
16 (25) |
5 (7.81) |
|
|
Total (116) |
17(14.65) |
18 (15.51) |
19 (16.37) |
|
4. Discussion
4.1 Enterococci species occurrence and dis-tribution in oral cavities of captive and wild Bra-zilian snake species
In this study, we detected the enterococci genus, bacteria of clinical relevance known as multidrug-resistant nosocomial pathogens, in snake species from Brazil. A few studies have previously examined the oral microorganisms from captive and wild Brazilian snake species [33, 34, 48]. Fonseca et al. [33] detected the presence of diverse bacterial, including clinical pathogens such as coagulase-negative staphylococci, Bulkolderia sp., Moraxella sp., Proteus sp., S. aureus, and Yersinia enterocolitica in oral samples of several captive snakes species. Jorge et al. [34] detected the presence of group D streptococci (Enterococcus spp.) in oral samples of B. jararaca. Currently, in relation to wild snakes, there is only one study that isolated Pseudomonas aeruginosa and Proteus vulgaris from oral samples of Crotalus durissus terrificus snakes in Brazil [48].
Enterococcus faecalis was the most common enterococcal species detected in oral samples of captive and wild snakes in this study. The results observed here are in agreement with the literature, Padhi et al. [13] identified E. faecalis as the most frequent enterococci species in the oral cavity of free-living vipers (Echis carinatus) in Orissa, India. Plentz et al. [49] collected 46 samples from boid snake species and also identified E. faecalis as one of the most frequent species in oral and traqueal samples of Python bivittatus. A microbiological study carried out by Gatti et al. [50] in Argentina analyzed the oral cavity of free-living B. alternatus, B. neuwiedi, B. ammodytoides, B. jararaca and B. jararacussu and found 37 bacterial strains; among them, six were E. faecalis and one Enterococcus sp. The other enterococci species isolated here have already been found in samples of amphibians, reptiles, mammals, and birds [18, 27, 28, 30].
The diet of snakes ranges from invertebrates to vertebrates, and varies widely among species, some being generalist and preying on a wide variety of prey categories, while others are highly specialized [1, 2, 36]. There is a distinct difference between the snakes diet of captive and wild snakes. One of the greatest differences is the availability of food variety or lack of it. Whereas in the wild they have high dietary diversity, in captivity they are fed with a low dietary diversity composed of small rodents (Wistar rats) or fish. These differences in the diet may have contri-buted to the distribution of enterococci species among the snakes evaluated in this study.
4.2 Multidrug-resistant enterococci in captive snakes and absence of resistant strains in wild snakes
The antimicrobial susceptibility profile showed that only captive snakes revealed resistant enterococci colonizing the oral cavity. The absence of resistant enterococci in samples from wild snakes may be associated with two factors in the wildlife: (i) the snakes can go without eating for about six months, thus reducing exposure to microorganisms; and (ii) the snakes try to avoid human contacts, being less exposed to impacts of anthropogenic activities. Our findings were consistent with other studies that evaluated the antimicrobial susceptibility of bacteria isolated from the oral cavity of wild snakes [51-53]. Shaikh et al. [51] also observed that Gram-positive and Gram-negative bacteria isolated from venomous snakes, in India, were susceptible to antimicrobials. Artavia-León et al. [52] found that the vast majority of wild snake isolates in Costa Rica showed antibiotic susceptible microorganisms. A recent study with presumed Naja spp. bites in Vietnam found large amounts of susceptible E. faecalis strains isolated from local wounds [53].
However, as evidenced in this work, captive snakes revealed multidrug-resistant enterococci colonizing the oral cavity. The occurrence of MDR strains has been associated with the proximity of animals to human activities, since enterococci are sentinel species [24, 54]. In the captive environment, feeding, use of antibiotics in a therapeutic manner, and human contact may have a major impact on the resistance of enterococci from captivity snakes. Other studies have associated resistant-enterococci isolated from animals with the proximity of human activities and/or to the environmental resistance [25-28, 55-57]. Previous studies examining the oral microbiota of captive snakes found high incidences of antibiotic resistance traits [17, 58, 59]. In India, N. naja captured from various localities (households) of Odisha were found to be harbouring antibiotic-resistant bacteria [17].
As shown by Hejnar et al. [58], resistant Steno-trophomonas maltophilia strains were isolated from captive snakes. Besides, Salmonella enteritidis isolated from edible snakes showed resistance to most drugs, but susceptibility to tetracycline and amikacin [59].
The emergence of MDR clinical pathogens such as enterococci are well-recognized to be one of the most important current public health issues [60]. Broad spectrum antibiotics are usually prescribed following snakebite and wound infection after cobra bites worldwide. Prophylactic antibiotic administration in snake bitten patients is recommended to prevent secondary infections from animal bites, and according to international guidelines amoxicillin-clavulanate is recommended [61].
However, to avoid the selection of pathogenic bacteria resistant to drugs, studies have been showing that antibiotic administration in snake bitten patients should be considered only in those with severe local signs of envenomation, or empiric use in those having local or general signs of infection, regardless of the degree of envenoming [61].
4.3 Determinants of virulence and antibiotic resistance genes in enterococci isolated from wild and captive snakes from Brazil
Tetracycline and erythromycin are prescribed in veterinary medicine [62, 63]. The isolation of tetracycline and erythromycin-resistant enterococci in captive snakes can be related to the administration of antibiotics in these animals, as well as in rodents. In the present study, tetL, tetM and tetS genes were detected in tetracycline-resistant and ermB and msrC genes were present in erythromycin-resistant enterococci strains. The frequency of these genes detected in the present study is congruent with the results obtained in previous studies conducted on Enterococcus strains isolated from wild and captive animals [24, 27, 28, 54].
Genes likely important for colonization in many contexts, but also studied for coding virulence traits were revealed in this study. The gelE gene was detected in enterococci from samples of snakes of the both groups, although it was more prevalent in wild snakes while ace and cylA genes had a similar prevalence in both groups. Our data corroborate other studies that recovered E. faecalis isolated from diverse origins over the past 100 years and showed a prevalence of the gelE and ace genes in genomes of clinical and environmental strains [26]. The presence of ace genes may be associated with the permanence of strains in the oral cavity of snakes, as it encodes an adhesion to collagen, aiding in the colonization and permanence of host cells. In contrast, the low fre-quency of the cylA gene in the analyzed samples corroborates with recent studies that recovered enterococci for animals, such as mammals [30], reptiles [27], birds [26-27] and insects [25]. The virulence genes in the snake strains analyzed in this study may demonstrate a symbiotic characteristic between strains and the host.
In clinical, MDR E. faecium and E. faecalis are asso-ciated with CRISPR defects [18, 45]. In this study, we observed that there was not a direct association between the absence of CRISPR–Cas and the presence of resistance in enterococci isolated from captive snakes. Therefore, further studies involving the analysis of the whole genome sequencing of these isolates might elucidate the genetic aspects of CRISPRs in enterococci strains isolated from captive and oral snake species in Brazil.
5. Conclusion
In conclusion, this work advances our understanding of the nature and ecology of enterococci in wild and captive snake species in Brazil. Our data showed that enterococci seem to be a natural member of the oral microbiota of these animals, although the presence of resistance traits in captive animals indicate that human contact and confinement may be important factors in the spread of resistant enterococci. There-fore, further studying monitoring the resistant strains on the oral cavity of these animals constitutes important for snakebite management to determine public health safety plans.
Acknowledgments
The authors are grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico do Brasil (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundações de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES-/VALE) and from Rio de Janeiro (FAPERJ) and Instituto Chico Mendes de Conservação da Bio-diversidade (ICMBio).
This work was performed within the framework of CAPES (a foundation linked to the Brazilian Ministry of Education that operates in the expansion and consolidation of stricto sensu graduate programs in all Brazilian states), sponsored by CNPq (# 407886/2018-4, and # 302574/2017-4), and FAPES/VALE and FAPERJ (#01/2015 and # 527/2016).
Declaration of Competing Interest
None to be declared
References
- Bernarde PS, Abe AS. Hábitos alimentares de serpentes em Espigão do Oeste, Ron-dônia, Brasil. Biota Neotropica 10 (2010): 167-173.
- Martins M, Oliveira ME. Natural history of snakes in forests of the Manaus regions. Central Amazônia, Brazil. Herpetological Natural History 6 (1998): 78-150.
- Uetz P, Freed P, Aguilar R. et al. (eds.) The Reptile Database (2021).
- Sampaio ILR, Santos CP, França RC, et al. Ecological diversity of a snake assemblage from the Atlantic Forest at the south coast of Paraíba, northeast Brazil. Zookeys 2 (2018):107-125.
- Costa HC, Bérnils RS. Répteis do Brasil e suas Unidades Federativas: lista de espécies. Herpetologia Brasileira 1 (2018): 11-57.
- Grego KF, Vieira SEM, Vidueiros JP, et al. Maintenance of venomous snakes in cap-tivity for venom production at Butantan Institute from 1908 to the present: a scoping history. Journal of Venomous Animals and Toxins including Tropical Diseases 27 (2021): 1-7.
- World Health Organization [Internet]. Sna-kebite envenoming (2021).
- da Silva AM, Bernarde PS, de Abreu LC. Accidents with poisonous animals in Brazil by age and sex. Journal of Human Growth and Development 25 (2015): 54-62.
- Brenes-Chacón H, Ulloa-Gutierrez R, Soriano-Fallas A, et al. Bacterial infections associated with Viperidae snakebites in children: a 14-year experience at the Hospital Nacional de Niños de Costa Rica†. American Journal of Tropical Medicine and Hygiene 5 (2019): 1227-1229.
- Chen C, Wu K, Chen C, et al. Bacterial infection in association with snakebite: a 10-year experience in a northern Taiwan medical center. Journal of Microbiology, Immunology and Infection 44 (2011): 456-460.
- Huang L, Wang J, Huang J, et al. Wound infections secondary to snakebite in central Taiwan. Journal of Venomous Animals and Toxins including Tropical Diseases 18 (2011): 272-276.
- Mao Y, Chuang H, Shih C, et al. An inves-tigation of conventional microbial culture for the Naja atra bite wound and the comparison between culture-based 16S Sanger sequencing and 16S metagenomics of the snake oropharyngeal bacterial micro-biota. PLOS Neglected Tropical Diseases 15 (2021): 1-8.
- Padhi L, Panda SK, Mohaprata PP, et al. Antibiotic susceptibility of cultivable aerobic microbiota from the oral cavity of Echis carinatus from Odisha (India). Microbial Pathogenesis 143 (2020): 104121
- Résière D, Olive C, Kallel H, et al. Oral microbiota of the snake Bothrops lanceolatus in Martinique. International Journal of Environmental Research and Public Health 15 (2018): 2122.
- Abrahamian FM, Goldstein EJC. Micro-biology of animal bite wound infections. Clinical Microbiology Reviews 24 (2011): 231-246.
- Jho YS, Park DH, Lee JH, et al. Iden-tification of bacteria from the oral cavity and cloaca of snakes imported from Vietnam. Laboratory Animal Research 27 (2011): 213-217.
- Panda SK, Padhi L, Sahoo G. Oral bacterial flora of Indian cobra (Naja naja) and their antibiotic susceptibilities. Heliyon 4 (2018): 1008.
- Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts: Eye and Ear Infirmary (2014): 3-25.
- List of prokaryotic names with standing in nomenclature [Internet]. Genus Enteroco-ccus.
- Selleck EM, van Tyne D, Gilmore MS. Pathogenicity of Enterococci. Microbiology Spectrum 7 (2018): GPP3-0053-2018.
- Wagener M, Naidoo M, Aldous C. Wound infection secondary to snakebite. South African Medical Journal 29 (2017): 315-319.
- Jorge MT, Ribeiro LA, da Silva MLR, et al. Microbiological studies of abscess com-plicating Bothrops snakebite in humans: a prospective study. Toxins 32 (1994): 743-748.
- Cho S, Barrett JB, Frye JG, et al. Anti-microbial resistance gene detection and plasmid typing among multidrug resistant enterococci isolated from freshwater environments. Microorganisms 8 (2020): 1338.
- de Araujo GO, Huff R, Favarini MO, et al. Multidrug resistance in enterococci isolated from wild Pampas Foxes (Lycalopex gymnocercus) and Geoffroy's Cats (Leo-pardus geoffroyi) in the Brazilian Pampa Biome. Frontiers in Veterinary Sci-ence 4 (2020): 1-11.
- Huff R, Pereira RI, Pissetti C, et al. Anti-microbial resistance and genetic relationships of enterococci from siblings and non-siblings Heliconius erato phyllis caterpillars. PeerJ 27 (2020): 1-20.
- Prichula J, van Tyne D, Schwartzman J, et al. Enterococci from wild Magellanic penguins (Spheniscus magellanicus) as an indicator of marine ecosystem health and human impact. Applied and Environmental Microbiology 17 (2020): 1-19.
- Prichula J, Pereira RI, Wachholz GR, et al. Resistance to antimicrobial agents among enterococci isolated from fecal samples of wild marine species in the southern coast of Brazil. Marine Pollution Bulletin 105 (2016): 51-57.
- Santestevan NA, Zvoboda DA, Prichula J, et al. Antimicrobial resistance and virulence factor gene profiles of Enterococcus spp. isolates from wild Arctocephalus australis (South American fur seal) and Arctocephalus tropicalis (Subantarctic fur seal). World Journal of Microbiology and Biotechnology 21 (2015): 1935-1946.
- Mannu L, Paba A, Daga E, et al. Comparison of the incidence of virulence determinants and antibiotic resistance between Entero-coccus faecium strains of dairy, animal and clinical origin. International Journal of Food Microbiology 88 (2003): 291-304.
- Poeta P, Costa D, Sáenz Y, et al. Char-acterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal. Journal of Veterinary Medicine. B, Infectious Diseases and Veterinary Public Health 52 (2005): 396-402.
- Fraga R, Lima AP, Prudente ALC, et al. Guia de cobras da região de Manaus – Amazônia Central. Manaus: INPA (2013): 303.
- Andrade JG, Pinto RNL, Andrade AL, et al. Estudo bacteriológico de abscessos causados por serpentes do gênero Bothrops. Revista do Instituto de Medicina Tropical de São Paulo 31 (1989): 363-367.
- Fonseca MG, Moreira WMQ, Cunha KC, et al. Oral microbiota of brazilian captive snakes. Journal of Venomous Animals and Toxins including Tropical Diseases 28 (2009): 54-60.
- Jorge MT, Mendonça JS, Ribeiro LA, et al. Flora bacteriana da cavidade oral, presas e veneno de Bothrops jararaca: possível fonte de infecção no local da picada. Revista do Instituto de Medicina Tropical de São Paulo 32 (1990): 6-10
- Sazima I. Natural history of the jararaca pitviper, Bothrops jararaca, in southeastern Brazil. In: Campbell JA, Brodie ED, editors. Biology of the pitvipers. Texas: Selva (1992): 199-216.
- Marques OAV, Eterovic A, Sazima I. Serpentes da Mata Atlântica - guia ilustrado para serra do mar. 1nd ed. Ribeirão Preto (SP): Holos (2001).
- Oliver JA. The relationships and zoogeo-graphy of the genus Thalerophis Oliver. Bulletin of the American Museum of Natural History 92 (1948): 157-280.
- Vitt LJ. Communities. In: Seigel RA, Collins JT, Novak SS, editors. Snakes: ecology and evolutionary biology. New York: Macmillan Publishers (1987): 335-365.
- Ruffato R, Di-Bernardo M, Maschio GF. Dieta de Thamnodynastes strigatus (Serp-entes, Colubridae) no sul do Brasil. Phyllo-medusa 2 (2003): 27-34.
- Melgarejo-Gimenez AR. Criação e manejo de serpentes. In: Andrade A, Pinto SC, Oliveira RS, editors. Animais de laboratório. Rio de Janeiro: Fiocruz (2002): 175-199.
- Teixeira LM, Carvalho MG, Facklam RR, et al. In: Versalovic J, Carroll KC, Funke G, editors. Manual of clinical microbiology. Washington, DC: ASM Press (2011): 350-364.
- Sauget M, Valot B, Bertrand X, et al. Can Maldi-tof mass spectrometry reasonably type bacteria? Trends in Microbiology 25 (2017): 447-455.
- Depardieu F, Perichon B, Courvalin P. Detection of the van alphabet and iden-tification of enterococci and staphylococci at the species level by multiplex PCR. Journal of Clinical Microbiology 42 (2004): 5857-5860.
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for anti-microbial susceptibility testing – twelve-six edition. Wayne: CLSI document 100S (2019): 251.
- Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio 12 (2010): e00227-10.
- European Food Safety Authority and Euro-pean Centre for Disease Prevention and Control. The European Union Summary Report on antimicrobial resistance in zoo-notic and indicator bacteria from humans, animals and food in 2011. European Food Safety Journal 11 (2013): 3196.
- Huescas CGY, Pereira RI, Prichula J, et al. Frequency of clustered regularly interspaced short palindromic repeats (CRISPRs) in non-clinical Enterococcus faecalis and Entero-coccus faecium strains. Brazilian Journal of Biology 79 (2019): 460-465.
- Junior RSF, Siqueira AK, Campagner MV, et al. Comparison of wildlife and captivity rattlesnakes (Crotalus durissus terrificus) microbiota. Pesquisa Veterinária Brasileira 29 (2009): 999-1003.
- Plentz B, Schmidt V, Grosse-Herrenthey A, et al. Characterisation of the aerobic bacterial flora of boid snakes: application of MALDI-TOF mass spectrometry. Veterinary Record 176 (2015): 285.
- Gatti EA, Stanchi N, Arias DR, et al. Estudio de la actividad antibiótica del veneno de serpiente Bothrops (Ophidia: Viperidae: Cro-talinae). Avances en Ciencias Veterinarias 13 (2010): 25-29.
- Shaikh IK, Dixit PP, Pawade BS, et al. Assessment of cultivable oral bacterial flora from important venomous snakes of India and their antibiotic susceptibilities. Current Microbiology 74 (2017): 1278-1286.
- Artavia-León A, Romero-Guerrero A, Sancho-Blanco C, et al. Diversity of aerobic bacteria isolated from oral and cloacal cavities from free-living snakes species in Costa Rica Rainforest. International Scholarly Research Notices (2017): 1-9.
- Ngo ND, Le QX, Anh PQ, et al. Clinical features, bacteriology, and antibiotic treat-ment among patients with presumed Naja bites in Vietnam. Wilderness & Environ-mental Medicine 31 (2020): 151-156.
- Grassotti TT, Zvoboda DA, Costa LFX, et al. Antimicrobial resistance profiles in Entero-coccus spp. isolates from fecal samples of wild and captive black capuchin monkeys (Sapajus nigritus) in South Brazil. Frontiers in Microbiology 9 (2018): 2366.
- Oravcova V, Janecko N, Ansorge A, et al. First record of vancomycin-resistant Entero-coccus faecium in Canadian wildlife. Environmental Microbiology 6 (2014): 210-211.
- Lozano C, Gonzalez-Barrio D, Camacho MC, et al. Characterization of fecal vanco-mycin resistant enterococci with acquired and intrinsic resistance mechanisms in wild animals, Spain. Microbial Ecology 72 (2016): 813-820.
- Yahia HB, Chairat S, Hamdi N, et al. Antimicrobial resistance and genetic lineages of faecal enterococci of wild birds: emergence of vanA and vanB2 harbouring Enterococcus faecalis. International Journal of Antimicrobial Agents 52 (2018): 936-941.
- Hejnar P, Kolár M, Sauer P. Antibiotic resistance of Stenotrophomonas maltophilia strains isolated from captive snakes. Folia Microbiol (Praha) 55 (2010): 83-87.
- Xia Y, Li H, Shen Y. Antimicrobial drug resistance in Salmonella enteritidis isolated from edible snakes with pneumonia and its pathogenicity in chickens. Frontiers in Veterinary Science 7 (2020): 463.
- Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature 543 (2017) : 15.
- Résière D, Gutiérrez JM, Névière R, et al. Antibiotic therapy for snakebite envenoming. Journal of Venomous Animals and Toxins including Tropical Diseases 26 (2020): 1-2.
- Schafhauser BH, Kristofco LA, de Oliveira CMR, et al. Global review and analysis of erythromycin in the environment: Occurrence, bioaccumulation and antibiotic resistance hazards. Environmental Pollution 238 (2018): 440-451.
- Rudra P, Hurst-Hess K, Lappierre P, et al. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrobial Agents and Chemotherapy 62 (2018): e00119-e00118.
- Sedgley C, Nagel A, Shelburne CE, et al. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Archives of Oral Biology 50 (2005): 575-583.
- Medeiros AW, Pereira RI, Oliveira DV, et al. Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil. Brazilian Journal of Microbiology 45 (2014): 327-332.
- Sutcliffe J, Tait-Kamradt AE, Wondrack L. Streptococcus pneumoniae and Strepto-coccus pyogenes resistant to macrolides but sensitive to clindamycin: A common resis-tance pattern mediated by an efflux system. Antimicrobial Agents and Chemotherapy 40 (1996): 1817-1824.
- Werner G, Hildebrandt B, Witte W. The newly described msrC gene is not equally distributed among all isolates of Entero-coccus faecium. Antimicrobial Agents and Chemotherapy 45 (2001): 3672-3673.
- Frazzon APG, Gama BA, Hermes V, et al. Prevalence of antimicrobial resistance and molecular characterization of tetracycline resistance mediated by tet(M) and tet(L) genes in Enterococcus spp. isolated from food in Southern Brazil. World Journal of Microbiology and Biotechnology 26 (2010): 365-370.
- Aarestrup FM, Agerso Y, Gerner-Smidt P, et al. Comparison of antimicrobial resistance phenotypes and resistance genes in Entero-coccus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagnostic Microbiology and Infectious Disease 37 (2000): 127-137.
- Shankar V, Baghdayan AS, Huycke MM, et al. Infection derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infection and Immunity Journal 67 (1999): 193-200.
- Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Applied and Environ-mental Microbiology 67 (2001): 1628-1635.
Supplementary Material
Supplementary Table 1: Primers used in the PCR reactions carried out for detection of enterococci species (E. faecalis and E. faecium), resistance (ermB, msrC, tetL, tetM, tetS), virulence (ace, cylA, and gelE), and CRISPRs genes (CRISPR1, CRISPR2, and CRISPR3).
|
Gene |
Nucleotide sequence (5′-3′) |
AT¹ (°C) |
Size (bp²) |
Reference |
|
|
E. faecalis |
E16s-F |
CCGAGTGCTTGCACTCAATTGG |
66 |
136 |
[64] |
|
E16s-R |
CTCTTATGCCATGCGGCATAAAC |
||||
|
E. faecium |
EM1A-F |
TTGAGGCAGACCAGATTGACG |
62 |
172 |
[65] |
|
EM1B-R |
CGGAAGTGATGCTTCCTACTG |
||||
|
Erythromycin |
ermB_F |
GAAAAGGTACTCAACCAAATA |
52 |
547 |
[66] |
|
ermB_R |
AGTAACGGTACTTAAATTGTTTAC |
||||
|
msrC 3 |
AAGGAATCCTTCTCTCTCCG |
52 |
343 |
[67] |
|
|
msrC 4 |
GTAAACAAAATCGTTCCCG |
||||
|
Tetracycline |
tetL_F |
ACTCGTAATGGTGTAGTTGC |
58 |
625 |
[68] |
|
tetL_R |
TGTAACTCCGATGTTTAACACG |
||||
|
tetM_F |
GTTAAATAGTGTTCTTGGAG |
52 |
657 |
[69] |
|
|
tetM_R |
CTAAGATATGGCTCTAACAA |
||||
|
tetS_F |
TGGAACGCCAGAGAGGTATT |
58 |
720 |
[69] |
|
|
tetS_R |
ACATAGACAAGCCGTTGACC |
||||
|
Adhesion |
ace1_F |
AAAGTAGAATTAGATCACAC |
57 |
320 |
[29] |
|
ace2_R |
TCTATCACATTCGGTTGCG |
||||
|
Cytolysine |
cylA_TE17 |
TGGATGATAGTGATAGGAAGT |
54 |
517 |
[70] |
|
cylA_TE18 |
TCTACAGTAAATCTTTCGTCA |
||||
|
Gelatinase |
gelE_TE9 |
ACCCCGTATCATTGGTTT |
50 |
402 |
[71] |
|
gelE_TE10 |
ACGCATTGCTTTTCCATC |
||||
|
CRISPRs |
crispr1_F |
CAGAAGACTATCAGTTGGTG |
55 |
783 |
[52] |
|
crispr1_R |
CCTTCTAAATCTTCTTCATAG |
||||
|
crispr2_F |
CTGGCTCGCTGTTACAGCT |
55 |
variable |
[52] |
|
|
crispr2_R |
CCAATGTTACAATATCAACCA |
||||
|
crispr3_F |
GCTGAATCTGTGAAGTTACTC |
50 |
258 |
[52] |
|
|
crispr3_R |
CTGTTTTGTTCACCGTTGGAT |
¹AT: annealing temperatures; ²bp: base pair.
Supplementary Table 2:
Distribution of erythromycin- and tetracycline-resistance genes in the enterococci isolated from oral samples of captivity snakes.
|
Specie |
Number (%) of strains positive for resistance genes |
||||||||||
|
Erythromycin |
Tetracycline |
||||||||||
|
R* |
msrC |
ermB |
R* |
tetL |
tetM |
tetS |
|||||
|
E. faecalis |
21 |
3 (14.29) |
2 (9.52) |
6 |
1 (16.67) |
3 (50) |
1 (16.67) |
||||
|
E. faecium |
9 |
1 (11.11) |
0 |
4 |
2 (50) |
4(100) |
0 |
||||
|
Total |
30 |
4 (13.33) |
2 (6.67) |
10 |
3 (30) |
7 (70) |
1 (10) |
||||
*R, number of resistant strains.
Supplementary Table 3:
Number (%) of virulence genes among enterococci isolated from oral samples of wild and captive snakes.
|
Habitat |
Strains (n) |
Number (%) of positive enterococci |
||
|
ace |
cylA |
gelE |
||
|
Wildlife |
E. avium (7) |
4 (57.14) |
0 |
6 (85.71) |
|
|
E. hirae (4) |
0 |
0 |
4 (100) |
|
|
E. faecalis (53) |
32 (60.38) |
1 (1.89) |
42 (79.25) |
|
|
Subtotal (64) |
36 (56.25) |
1 (1.56) |
52 (81.25) |
|
Captive |
E. faecalis (38) |
21 (55.26) |
1 (2.63) |
13 (34.21) |
|
|
E. faecium (14) |
10 (71.43) |
0 |
4 (28.57) |
|
|
Subtotal (52) |
31 (59.62) |
1 (1.92) |
17 (32.69) |
|
Total (116) |
67 (57.76) |
2 (1.72) |
69 (59.48) |
|

 Impact Factor: * 3.6
Impact Factor: * 3.6 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 