Genetic Diversity and Quantification of Human Adenoviruses and JC Polyomaviruses in Wastewater Samples
Article Information
Lorena da Graça Pedrosa de Macena1*, Carmen Baur Vieira2, Adriana Gonçalves Maranhão1, Fernando César Ferreira1, Elba Regina Sampaio Lemos3, Marize Pereira Miagostovich1
1Laboratory of Comparative and Environmental Virology, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, RJ 21040-360, Brazil
2Department of Microbiology and Parasitology, Biomedical Institute, Fluminense Federal, University (UFF), Niterói, RJ 24210-130, Brazil
3Laboratory of Hantaviruses and Rickettsiosis, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, RJ 21040-360, Brazil
*Corresponding Author: Lorena da Graça Pedrosa de Macena, Laboratory of Comparative and Environmental Virology, Oswaldo Cruz Institute, Oswaldo, Cruz Foundation, Rio de Janeiro, RJ 21040-360, Brazil
Received: 29 September 2021; Accepted: 06 October 2021; Published: 06 November 2021
Citation:
Lorena da Graça Pedrosa de Macena, Carmen Baur Vieira, Adriana Gonçalves Maranhão, Fernando César Ferreira, Elba Regina Sampaio Lemos, Marize Pereira Miagostovich. Genetic Diversity and Quantification of Human Adenoviruses and JC Polyomaviruses in Wastewater Samples. International Journal of Plant, Animal and Environmental Sciences 11 (2021): 614-626.
View / Download Pdf Share at FacebookAbstract
Wastewater-based monitoring has been described as a non-invasive approach to assess virus distribution in a specific geographic area. In this study we assess the genetic diversity and concentration of human adenovirus (HAdV) and human polyomavirus JC (JCPyV) in wastewater samples in Rio de Janeiro, Brazil, during the Olympic Games, Rio 2016. Wastewater samples (50 mL) obtained from domestic and hospital sewages were concentrated by the skimmed milk flocculation method and processed using molecular tools. Quantitative polymerase chain reaction using the ABI PRISM 7500 Real Time TaqMan System and TaqMan Universal Master Mix II detected 18 HAdV and 17 JCPyV strains in 95% of those samples (18/19). A mean viral load of 8.6x105 genomic copies (GC)/L and 1.2x107 GC/L, was achieved for HAdV and JCPyV, respectively. Partial nucleotide sequencing using Sanger methodology revealed three HAdV species/ eight serotypes (HAdV B, D, D10, D17, D19, D22, F40 and F41) and seven genotypes/nine subtypes (JCPyV-1B, 2A, 3A, 3B, 4, 6, 7A, 8A and 8B). HAdV-D17 and -D22, as well as genotypes JCPyV-7 and -8, were detected for the first time in the country. The detection of previously undetected viruses in the region demonstrates the importance of our findings, adding data to the epidemiology of those viruses and corroborating the importance of environmental surveillance carried out from wastewater.
Keywords
Genetic diversity; HAdV; JCPyV; Wastewater-based monitoring
Genetic diversity articles; HAdV articles; JCPyV articles; Wastewater-based monitoring articles
Article Details
1. Introduction
The investigation of pathogens in untreated sewage represents an advantageous tool to determine the epidemiology and prevalence of infectious agents at the population level in a specific geographic region [1]. This approach, allows the assessment of the circulation of endemic, emerging and re-emerging pathogens shedding by infected individuals [2]. In this context, the wastewater-based monitoring has been useful for research, prevention, treatment and control of known and even unknown infectious diseases, complementing epidemiological surveillance and guiding public health action [3]. Several studies have been used this approach to monitor virus assessing virus circulation including those excreted by symptomatic and asymptomatic infected individuals [4-7]. Human adenoviruses (HAdV) and JC human polyomaviruses (JCPyV) commonly found in high concentrations of wastewater around the world [8], are both DNA viruses presenting specificity with the human host stability, persistence and wide distribution in different environmental matrices, being detected in sewage samples without seasonality [9, 10].
HAdV belong to the Adenoviridae family, genus Mastadenovirus. They are non- enveloped viruses, with a capsid of icosahedral symmetry, 90nm in diameter and double- stranded DNA of approximately 35 kb [11]. Currently, more than 49 HAdV serotypes, grouped into six species (A–F) are described based on neutralizing antibody assays and the ability to agglutinate red blood cells [12]. Adenoviruses A, F and G are related to gastrointestinal infections, B and C to respiratory infections and D to keratoconjunctivitis [13]. They can establish latent and persistent infections, with the virus being excreted for weeks after infection and regardless of the primary site of infection [14]. In addition, enteric (HAdV-F40 and 41) and non-enteric HAdV serotypes replicate in the intestine and are potentially capable of being transmitted by contaminated water [9].
JCPyV belong to the Poliomaviridae family, genus Poliomavirus, and together with the twelve virus (BKPyV, WUPyV, KIPyV, MCPyV, HPyV6, HPyV7, TSPyV, HPyV9, HPyV10, STLPyV, HPyV12 and HPyV13) constitute the polyomavirus that infects humans [15]. They are non-enveloped viruses, measuring about 40-45 nm in diameter, containing a genome composed of double-stranded circular DNA of approximately 5.1 kb [16]. Eight genotypes (JCPyV 1 to 8) and several JCPyV subtypes are described until the present study [17]. These viruses are prevalent in more than 80% of the world population, detected in environmental samples and excreted in urine, indicating the possibility of transmission from person to person and / or urine-fecal-oral in contact with contaminated surfaces, food and water [18]. Genotypic distribution seems to follow a defined pattern, determined by the excretion of a specific genotype according to the ethnic origin of each individual and not by the geographic region in which that individual is found [19, 20]. JCPyV commonly causes persistent subclinical renal lesions, but can infect secondary sites such as bone marrow, lymphoid tissue, and brain and establish latent infection [21]. Viral reactivation can occur in cases of profound impairment of cellular immunity and progressive multifocal leukoencephalopathy (PML), a rare and fatal disease, manifests itself causing multiple foci of demyelination in the brain [18]. This study aimed to assess the genetic diversity and concentration of HAdV and JCPyV in raw samples from domestic and hospital sewage in the during the Olympic Games, due to the expected large influx of people during this mass gathering event.
2. Materials and Methods
2.1 Study area and sampling site
This study was carried out in Barra da Tijuca, located on the west coast of the city of Rio de Janeiro (Brazil), from August to December 2016. Wastewater samples were collected from domestic (Residential village) and hospital (Public Health Units) sewage pipes and stored at 4°C until further processing. Residential Village consists of seven condominiums, 31 buildings and 3,604 apartments [22, 23]. General and Maternity Hospital are important Regional Emergency Centers in the West Zone of Rio de Janeiro and offer free clinical, surgical, pediatrics and obstetric care to residents and tourists [24-26].
2.2 Concentration method and virus quantification, and nucleic acids extraction and qPCR for HAdV and JCPyV
Sewage samples (42 mL) were concentrated using skim milk elution and flocculation method as previously described [27]. Briefly, before flocculation, sewage samples were previously treated with 0.25 N glycine buffer, pH 9.5 (1:2, v/v), stirred for 30min on ice, centrifuged at 8000 ×g for 30min at 4°C and added skimmed-milk solution (0.01%, w/v) to the supernatant adjusted to pH 3.5. The samples were flocculated with agitation for 8h at room temperature and the skimmed milk flakes were sedimented by centrifugation at 8000 ×g for 30min at 4°C. Finally, the pellet was resuspended with 1mL of phosphate buffer (pH 7.5).
Commercial QIAmp viral RNA Mini kit (Qiagen™, Valencia, USA) was used for nucleic acids extraction using Automatic QIAcube System according to the manufacturer’s instructions. The negative control composed of DNase/RNase-free water and positive controls were also included in each assay. Quantitative polymerase chain reaction (qPCR) was performed for quantification of HAdV and JCPyV as previously described [28, 29], using the ABI PRISM 7500 Real Time TaqMan System and TaqMan Universal Master Mix II (Applied Biosystems™, Foster City, CA).
Negative and positive (faecal suspension) controls, as well as NTC, were also included in all procedures. All samples were tested in duplicate using undiluted and 1:10 diluted nucleic acid to assess qPCR inhibitions. Samples that showed signs crossing the threshold line up to Ct ≤ 38 in both replicates and with a characteristic sigmoidal curve were considered positive. The gBlock Gene (Integrated DNA Technologies ™, Coralville, Iowa, USA) was used as standard curves for each virus analysed. Standard curves were serially diluted from 1x107 to 1x100 genomic copies per reaction and showed average values of -3.563 and -3.460 of slope, of 0.998 and 0.999 of square regression coefficient (r2) and 91% and 95% of reaction efficiencies for HAdV and JCPyV qPCRs, respectively.
2.3 Molecular characterization of HAdV and JCPyV
Conventional PCR assays were used for gene amplification of HAdV and JCPyV qPCR positive samples. HAdV positive samples were amplified in the coding region of the hexon gene using the two oligonucleotide primer pairs hex1deg/hex2deg (301pb) and hex1deg/nehex4deg (245pb) adapted from [30]. The intergenic region (VP1 and large T) was amplified in JCPyV positive samples using two oligonucleotide primer pairs EP1A/EP1A (737bp) and P1A/P2A (668bp), according to Boffil-Mas and Girones [31].
Amplicons were visualized using gel electrophoresis on a 1.5% agarose gel (UltraPure™ Agarose, Life Technologies™, Carlsbad, CA, USA) with 10mg/mL of UltraPure™ Ethidium Bromide (Life Technologies™, Carlsbad, CA, USA). Amplified DNA fragments were sequenced by the Sanger method using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit 1 (version 3.1) and the ABI Prism 3730 Genetic Analyzer (Applied Biosystems ™, Foster City, CA, USA). Nucleotide sequences were edited and aligned using the Clustal W method in Bio Edit Software 7.2.6 [32]. Sequences of HAdV and JCPyV, with size of 224bp and 624bp respectively, were compared with those available in the National Centre for Biotechnology Information (GenBank, http://www.ncbi.nlm.nih.gov/) database using the BLAST (Basic Local Alignment Search Tool).
Using the MEGA X [33], the evolutionary history was inferred using the Maximum Likelihood method to analyse sequences of both viruses. Kimura 2-parameter [34] and Tamura 3-parameter [35] were the models used for the construction of the phylogenetic HAdV and JCPyV trees, respectively. Initial trees for the heuristic search were obtained by applying the Neighbour-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. Confidence values of the internal nodes were calculated by performing bootstrap analyses with 2,000 replicates. The nucleotide sequence data reported in this study was submitted to the GenBank and received the accession numbers for HAdV (MT250569 - MT250586) and JCPyV (MT253712 - MT253728).
3. Results
HAdV and JCPyV were detected in 95% (18/19) of the samples, with minimum and maximum concentrations of 1.7x103 to 9.2x106 copies of genomics/litter (GC/L) and 2.8x104 to 1.3x108 GC/L, respectively. The results of the average concentration in all sewers showed an average load of JCPyV with 1 log more than HAdV. The logarithmic mean concentration remained stable when comparing the quantification of HAdV in domestic and hospital sewage.
In contrast, JCPyV presented 107 GC/L in hospital sewage and a concentration of approximately 106 GC/L in domestic sewage.
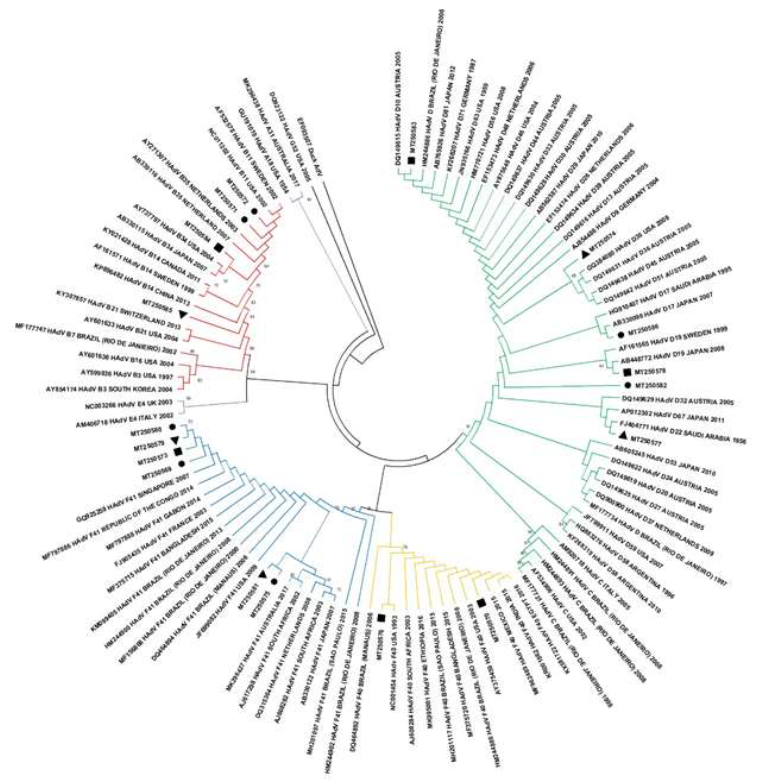
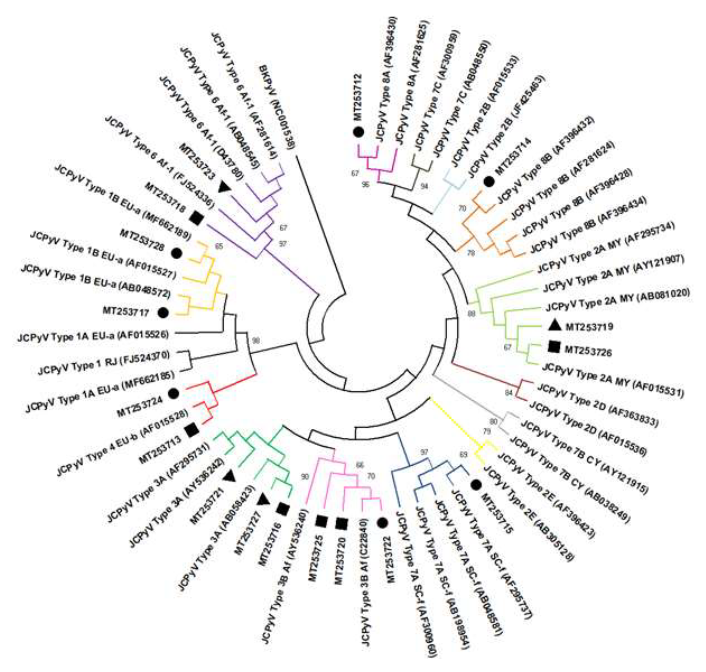
Nucleotide sequencing characterization detected three HAdV species (B, D and F), totalizing eight serotypes (HAdV B, D, D10, D17, D19, D22, F40 and F41). HAdV-F (44%) and HAdV-D (33%) were the species prevalent, being HAdV F species prevalent in hospital sewage samples while the HAdV B and D species were prevalent in domestic sewage. Regarding to JCPyV nucleotide analysis, were characterized seven genotypes and nine subtypes (JCPyV-1B, 2A, 3A, 3B, 4, 6, 7A, 8A and 8B) in 18 JCPyV positive raw sewage samples (Table 1, Figure 1 and 2).
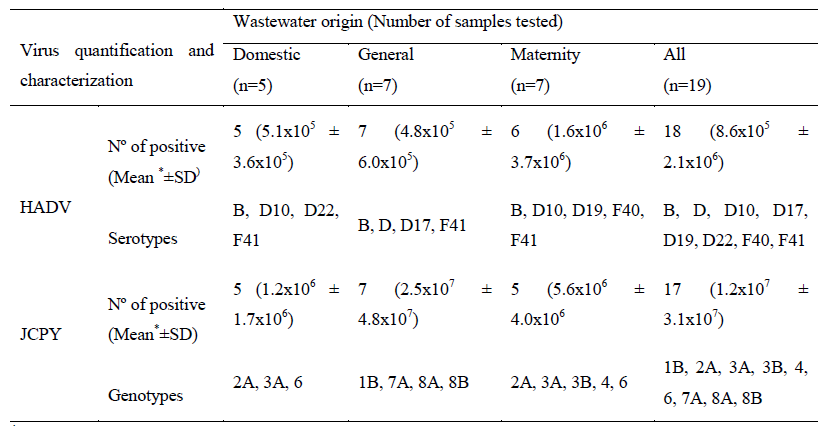
*GC/L= genome copies/Litre; SD=standard deviation
Table 1: Detection, quantification, and serotypes/genotypes of human adenovirus (HAdV) and JC polyomavirus (JCPyV) according to collection sampling sites.
Figure 1: Human adenovirus phylogenetic tree based on hexon region gene (224 base pair) showing eighteen strains found in raw sewage from Residential Village (triangle), General Hospital (circle) and Maternity Hospital (square). Maximum Likelihood method and Kimura 2-parameter model was used with 2,000 bootstrap replicates.
Figure 2: JC human polyomavirus phylogenetic tree based on intergenic region gene (624 base pair) showing seventeen strains in raw sewage found in raw sewage from Residential Village (triangle), General Hospital (circle) and Maternity Hospital (square). Maximum Likelihood method and Tamura 3-parameter model was used with 2,000 bootstrap replicates.
4. Discussion
Wastewater-based monitoring approach, even in a short period as conducted in this study, was successful for describing the occurrence of HAdV-22 and 17 and, JCPyV-7 and -8 genotypes for the first time in the country. The prevalence and quantification of both HAdV and JCPyV observed are in agreement with the findings obtained in samples of domestic and hospital sewage throughout the world [36-39]. The viral load of JCPyV, about one log higher than HAdV, was higher than that observed in previous studies carried out by our group in raw sewage (average concentration of 2.55x104 GC/L) from an urban wastewater treatment plant (WTTP) in Rio de Janeiro between 2009 and 2010 [40]. The small number of samples collected is a limiting factor in this study, not allowing a comparative assessment between sewage from different sources. According Arkhipova et al. (2018) the fact that the samples were collected during a mass event, where a large influx of individuals is expected could explain not only the possibility of an eventual introduction of a new virus in the region as well as a greater contribution of population density and transmission of infectious diseases, especially of waterborne etiological agents [41]. In this context, it is possible that the increase in the mean concentration of observed viruses, the profile of viral diversity and the pathogen-host dynamics circulating in sewage may have been influenced by the mass event [41]. Once the study area evaluated, the Barra da Tijuca neighborhood on the west coast of the city of Rio de Janeiro, housed the athletes in the Olympic Village and received 300,000 people at the Olympic Park, in addition to receiving up to 292,000 visitors at the sites of Live streaming of the Games [42].
Look upon molecular viral characterization, it was observed a similar diversity of HAdV species and serotypes between samples previously collected in the region as raw hospital sewage samples [38] and brackish from a Lagoon System samples [43] collected in 2008 and 2016, respectively. Except by HAdV-D17 and HAVdV-22, first described in the country in this study. HAdV-17 was described in sewage and mussel samples in South Africa [44], while HAdV-D22 detected in WWTP samples in Canada [45]. Both serotypes of the HAdV-D species are reported to cause outbreaks of epidemic keratoconjunctivitis (EKC) in humans on the Asian continent [46, 47].
The intraspecies co-circulation capacity of HAdV in the population and its spread in the environment was previously demonstrated [48]. Thus, depending on the number of viral infections and the immunity of the population, it is possible that one species is more detectable than another [49]. Recently, sequencing of the hypervariable regions (HVR) of the hexon gene and the long fibre gene of HAdV demonstrated the occurrence of variations in F-species circulating in India [50]. Unfortunately, the hexon region sequenced in this study did not allow us to assess the occurrence of these variants of circulating F strains. Regarding JCPyV diversity, it is remarkable the first detection of JCPyV-7 and -8 genotypes during the Olympic Games, specifically because JCPyV is used as an anthropological biomarker [51]. The detection of JCPyV-7 and -8 genotypes evidenced the presence of Asian, African, Melanesian and Polynesian ethnic and geographic ancestry foreigners. JCPyV-7A (SC strain) in Brazil was notable as it is generally identified in ethnic ancestors in East and Southeast Asia and West Africa, while JCPyV-8 (subtypes 8A and 8B) come from ancestral peoples of Melanesia (Papua New Guinea) and Polynesia [17].
Among the other genotypes detected and representatives of the Old World [52], the European (JCPyV-1 and -4), the African (JCPyV-3 and -6) and the Asian (JCPyV-2A) were characterized in the domestic and hospital sewage from different sources evaluated. These genotypes have been reported in the North, Northeast, Southeast and South of Brazil since 2000, in cerebrospinal fluid, urine from healthy kidney transplant patients and from patients with PML, as well as in wastewater from the WWTP [40, 51, 53-56]. Overall, of the eight JCPyV genotypes already described in the literature, only JCPyV-5 was not found in all the evaluated sewage samples [17]. Previously, Fumian et al (2010) demonstrated greater circulation of JCPyV-3 (Af2) genotypes, of African and Asian origin, in Rio de Janeiro [57].
5. Conclusion
The success in demonstrating populational-level infection change by detecting a new etiological agent in a given geographic region demonstrates the importance of environmental surveillance as a non-invasive, complementary approach that can support epidemiological studies.
Acknowledgements
The authors thank technical and field staff Márcia Maria Araújo Pimenta, Sérgio de Silva e Mouta Júnior (Oswaldo Cruz Foundation, RJ, Brazil), Fábio Valadão Araújo and Arnaldo Levy Lassance Cunha (Rio de Janeiro City Hall employees) for assisting in field collection and samples processing. PDTIS DNA Sequence Platform staff at FIOCRUZ for technical support in sequencing reactions.
Funding
This work was funded by Instituto Oswaldo Cruz (PAEF), Fundação de Amparo à Pesquisa do Rio de Janeiro (Faperj-E-26/202.821/2018), Conselho de Desenvolvimento Científico e Tecnológico (CNPq-Universal-406414/2016-5), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001.
This research study is under the scope of the activities of the Oswaldo Cruz Foundation (FIOCRUZ) as a Collaborating Centre of PAHO/WHO of Public and Environmental Health.
Conflict of Interest
The authors declare that they have no conflict of interest.
Authors’ Contributions
All authors critically reviewed the article for important intellectual content and approved all components of the final version. Pedrosa de Macena, L.G. contributed to the execution of the development and design of the methodology, data visualization and writing of the original sketch. Vieira, C.B. conceptualized and designed the methodology. Maranhão, A.G. and Ferreira, F.C. contributed to the realization and investigation of molecular analyses and field collection. Lemos, E.R.S. contributed resources, supervision and funding acquisition. Miagostovich, M.P. contributed to writing - review and editing, visualization, supervision, project administration and funding acquisition.
References
- Carducci A, Verani M, Battistini R, et al. Epidemiological surveillance of human enteric viruses by monitoring of different environmental matrices. Water Science and Technology 54 (2006): 239-244.
- Mao K, Zhang K, Du W, et al. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Current Opinion in Environmental Science and Health 17 (2020): 1-7.
- Magana-Arachchi DN, Wanigatunge RP. Ubiquitous waterborne pathogens in Waterborne Pathogens (Elsevier) 15-42.
- Rahman A, Kang S, Wang W, et al. Nanobiotechnology enabled approaches for wastewater based epidemiology. TrAC Trends in Analytical Chemistry 143 (2021): 116400.
- Miura F, Kitajima M, Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidem-iology: Re-analysis of patient data using a shedding dynamics model. Science of the Total Environment 769 (2021): 144549.
- Chacón L, Morales E, Valiente C, et al. Wastewater-Based Epidemiology of Enteric Viruses and Surveillance of Acute Gastro-intestinal Illness Outbreaks in a Resource-Limited Region. The American Journal of Tropical Medicine and Hygiene (2021).
- Nieuwenhuijse DF, Oude Munnink BB, Phan MVT. The Global Sewage Surveillance project consortium, Hendriksen RS, Bego A, Rees C, et al. Setting a baseline for global urban virome surveillance in sewage. Sci Rep 10 (2020): 13748.
- Rusiñol M, Girones R. Summary of Excreted and Waterborne Viruses in Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project), eds.Michigan State University, J. B. Rose, B. Jiménez Cisneros, UNESCO - International Hydrological Programme (Michigan State University).
- Allard A, Vantarakis A. Adenoviruses in Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project), eds.Michigan State University, J. B. Rose, B. Jiménez Cisneros, UNESCO - International Hydrological Programme (Michigan State University).
- Bofill-Mas S. Polyomavirus in Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project), eds.Michigan State University, J. B. Rose, B. Jiménez Cisneros, UNESCO - International Hydrological Prog-ramme (Michigan State University).
- ICTV IC on T of V. Adenoviridae. ICTV 9th Report (2011) (2021).
- Usman N, Suarez M. Adenoviruses in Stat Pearls (Treasure Island (FL): Stat Pearls Publishing).
- Heim A. Adenovirusinfektionen: Buntes Bild von Krankheiten durch eine Vielzahl von Virustypen. Monatsschr Kinderheilkd 168 (2020): 514-523.
- Desheva Y. Introductory Chapter: Human Adenoviruses in Adenoviruses, ed. Y. Desheva (IntechOpen).
- Viscidi RP, Tan CS. Polyomaviruses in Infectious Diseases (Elsevier) 1445-1448.e1.
- Moens U, Calvignac-Spencer S, Lauber C, et al. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Polyomaviridae. Journal of General Virology 98 (2017): 1159-1160.
- Torres C. Evolution and molecular epide-miology of polyomaviruses. Infection, Genetics and Evolution 79 (2020): 104150.
- Cortese I, Reich DS, Nath A. Progressive multifocal leukoencephalopathy and the spectrum of JC virus-related disease. Nat Rev Neurol 17 (2021): 37-51.
- Levican J, Levican A, Ampuero M, et al. JC polyomavirus circulation in one-year surveillance in wastewater in Santiago, Chile. Infection, Genetics and Evolution 71 (2019): 151-158.
- Stoner GL, Jobes DV, Fernandez Cobo M, et al. JC virus as a marker of human migrationto the Americas. Microbes and Infection 2 (2000): 1905-1911.
- Burrell CJ, Howard CR, Murphy FA. Polyomaviruses in Fenner and White’s Medical Virology (Elsevier) 283-288.
- Bartelt DD. The Other Side of the Medal Major sporting events in Brazil in the web of urban planning, speculation and the right to the city. Heinrich Böll Foundation (2015).
- Teixeira AL. Case study of Vila dos Atletas of the Rio 2016 Olympics: use of intangible assets to create value in megaprojects (2017).
- Couto RS. Municipal Hospital Lourenço Jorge: a study on the contribution of architecture to the therapeutic process. Public Domain Portal, Digital library developed in free software (2009).
- Lima Junior MC da G. Relatório de Gestão 2009 a 2016. Subsecretaria de Atenção Hospitalar, Urgência e Emergência. Revista Saúde em Foco (Rio de Janeiro) 1 (2016): 37.
- Prefeitura do Rio de Janeiro. BI em Internações SUS-2018-CNES 2270609. Ocupação de Leitos CNES em internações SUS (2018).
- Calgua B, Rodriguez-Manzano J, Hundesa A, et al. New methods for the concentration of viruses from urban sewage using quantitative PCR. Journal of Virological Methods 187 (2013): 215-221.
- Hernroth BE, Conden-Hansson A-C, Rehnstam-Holm A-S, et al. Environmental Factors Influencing Human Viral Pathogens and Their Potential Indicator Organisms in the Blue Mussel, Mytilus edulis: the First Scandinavian Report. Applied and Environmental Microbiology 68 (2002): 4523-4533.
- Pal A, Sirota L, Maudru T, et al. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. Journal of Virological Methods 135 (2006): 32-42.
- Allard A, Albinsson B, Wadell G. Rapid Typing of Human Adenoviruses by a General PCR Combined with Restriction Endonuclease Analysis. Journal of Clinical Microbiology 39 (2001): 498-505.
- Bofill-Mas S, Girones R. Documenting the Epidemiologic Patterns of Polyomaviruses in Human Populations by Studying Their Presence in Urban Sewage. Applied and Environmental Microbiology 66 (2000): 238-245.
- Hall T. BioEdit: an important software for molecular biology. GERF Bull Biosci 2 (2011): 60-61.
- Kumar S, Stecher G, Li M, et al. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35 (2018): 1547-1549.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16 (1980): 111-120.
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Molecular Biology and Evolution 9 (1992): 678-687.
- Bofill-Mas S, Albinana-Gimenez N, Clemente-Casares P, et al. Quantification and Stability of Human Adenoviruses and Polyomavirus JCPyV in Wastewater Matrices. Applied and Environmental Microbiology 72 (2006): 7894-7896.
- Kaas L, Ogorzaly L, Lecellier G, et al. Detection of Human Enteric Viruses in French Polynesian Wastewaters, Environmental Waters and Giant Clams. Food Environ Virol 11 (2019): 52-64.
- Prado T. Occurrence of rotavirus, adenovirus, norovirus and hepatitis A virus in sewage treatment plants in Rio de Janeiro and evaluation of viral recovery methodologies in sewage sludge (2011).
- Prado T, de Castro Bruni A, Barbosa MRF, et al. Performance of wastewater reclamation systems in enteric virus removal. Science of the Total Environment 678 (2019): 33-42.
- Fumian TM, Vieira CB, Leite JPG, et al. Assessment of burden of virus agents in an urban sewage treatment plant in Rio de Janeiro, Brazil. Journal of Water and Health 11 (2013): 110-119.
- Arkhipova K, Skvortsov T, Quinn JP, et al. Temporal dynamics of uncultured viruses: a new dimension in viral diversity. ISME J 12 (2018): 199-211.
- International Olympic Committee I. Marketing and Broadcasting Rio 2016. International Olympic Committee (2017).
- Pedrosa de Macena L da G, Castiglia Feitosa R, Vieira CB, et al. Microbiological assessment of an urban lagoon system in the coastal zone of Rio de Janeiro, Brazil. Environ Sci Pollut Res (2020).
- Vos HJ, Knox CM. The recovery and molecular identification of HAdV-D17 in raw sewage and mussel samples collected in the Eastern Cape province of South Africa. Southern African Journal of Infectious Diseases 33 (2018): 4-7.
- Vidovic S, Aly M, Flemming C, et al. First Evidence of Genotypes Ad3a16 and Ad3a18 in North America, Obtained by Genetic Analysis of Infectious Human Adenovirus from Wastewaters of Two Urban Communities in Canada. Appl Environ Microbiol 77 (2011): 4256-4259.
- Aoki K, Ishiko H, Konno T, et al. Epidemic Keratoconjunctivitis Due to the Novel Hexon-Chimeric-Intermediate 22,37/H8 Human Adenovirus. J Clin Microbiol 46 (2008): 3259-3269.
- Feng M, Smith TR, Chang RS-M, et al. Adenoviruses Isolated from Saudi Arabia: II. Pathogenicity of Certain Strains for Man. The American Journal of Tropical Medicine and Hygiene 8 (1959): 501-504.
- Demian PN, Horton KC, Kajon A, et al. Molecular identification of adenoviruses associated with respiratory infection in Egypt from 2003 to 2010. BMC Infect Dis 14 (2014): 50.
- Hage E, Espelage W, Eckmanns T, et al. Molecular phylogeny of a novel human adenovirus type 8 strain causing a prolonged, multi-state keratoconjunctivitis epidemic in Germany. Sci Rep 7 (2017): 40680.
- Chandra P, Lo M, Mitra S, et al. Genetic characterization and phylogenetic variations of human adenovirus-F strains circulating in eastern India during 2017-2020. J Med Virol 93 (2021): 6180-6190.
- Ishak R, Machado LFA, Cayres-Vallinoto I, et al. Infectious Agents as Markers of Human Migration toward the Amazon Region of Brazil. Front Microbiol 8 (2017): 1663.
- Stoner GL, Jobes DV, Fernandez Cobo M, et al. JC virus as a marker of human migrationto the Americas. Microbes and Infection 2 (2000): 1905-1911.
- Cayres-Vallinoto IMV, Vallinoto ACR, Azevedo VN, et al. Human JCV Infections as a Bio-Anthropological Marker of the Formation of Brazilian Amazonian Populations. PLoS ONE 7 (2012): e46523.
- Comerlato J, Souza-Campos F, Souza-Arantes T, et al. Distribution and genetic diversity of the human polyomaviruses JC and BK in surface water and sewage treatment plant during 2009 in Porto Alegre, Southern Brazil. Braz J Biol 77 (2016): 459-468.
- Comerlato J, Campos FS, Oliveira MT, et al. Molecular detection and characterization of BK and JC polyomaviruses in urine samples of renal transplant patients in Southern Brazil: JCPyV and BKPyV in Southern Brazil. J Med Virol 87 (2015): 52-528.
- Fink MCD, de Oliveira ACP, Romano CM, et al. Molecular characterization of human polyomavirus JC in Brazilian AIDS patients with and without progressive multifocal leukoencephalopathy. Journal of Clinical Virology 48 (2010): 6-10.
- Fumian TM, Guimarães FR, Pereira Vaz BJ, et al. Molecular detection, quantification and characterization of human polyomavirus JC from waste water in Rio De Janeiro, Brazil. Journal of Water and Health 8 (2010): 438-445.

 Impact Factor: * 3.6
Impact Factor: * 3.6 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 