Natural Selection Footprint in Novel Coronavirus: A Genomic Perspective of SARS-COV2 Pandemic and Hypothesis for Peptide-Based Vaccine
Article Information
Mojtaba Shekarkar Azgomi1≠, Leila Mohammadnezhad1≠, Marco Pio La Manna1*, Francesco Dieli1&, Nadia Caccamo1&
1Central Laboratory for Advanced Diagnosis and Biomedical Research (CLADIBIOR) and Department of Biomedicine, Neurosciences and Advanced Diagnostic (Bi.N.D.); University of Palermo, Palermo 90127, Italy
≠M.S. A. and L.M. contributed equally to this work.
&F.D. and N.C. share last authorship for this work.
*Corresponding Author: Marco Pio La Manna, PhD, Central Laboratory of Advanced Diagnosis and Biomedical Research, University of Palermo, Via del Vespro 129, Palermo 90127, Italy.
Received: 31 May 2021; Accepted: 08 June 2021; Published: 13 July 2021
Citation:
Mojtaba Shekarkar Azgomi, Leila Mohammadnezhad, Marco Pio La Manna, Francesco Dieli, Nadia Caccamo, Natural Selection Footprint in Novel Coronavirus: A Genomic Perspective of SARS-COV2 Pandemic and Hypothesis for Peptide-Based Vaccine. Journal of Biotechnology and Biomedicine 4 (2021): 108-123.
View / Download Pdf Share at FacebookAbstract
We retrospective analyzed in silico the binding affinity of SARS-CoV-2 peptides to MHC class I HLA-A, -B, and –C molecules in different countries with high and low morbidity and mortality rates. We used the bioinformatics approach to screen 18260 SARS-CoV-2 epitopes that have significant affinity for different MHC class I alleles and found approximately five thousand predicted nonamers to bind different alleles. Those predicted epitopes show a different significant affinity for occurring MHC I alleles. regarding HLA frequencies within different populations that can vary due to differences in their evolutionary histories, we showed that those alleles have different correlations with SARS-CoV-2 pandemic in 22 countries based on different mortality and morbidity rate. There was a strong negative correlation between morbidity and mortality rates and the frequency of HLA-A*24, HLA-C*06, and HLA-B*5, while a strong positive correlation is detected between HLA-A*02, HLA-B*38, HLA-C*04, and HLA-C*08. We speculate that HLA class I polymorphism, by governing the set of viral peptides presented to CD8+ T cells, influences the outcome of SARS-Cov-2 infection. Finally, we were able to draw a footprint of natural selection on MHC I alleles based on the significantly different affinity of the predicted peptides for known alleles. Our data showed that the HLA class I genetic background and the study epitope prediction should be taken into account for the generation of epitope-based vaccine or diagnostic tools.
Keywords
SARS-CoV-2; CD8+ T cells; MHC class I; In silico analysis; Peptides
SARS-CoV-2 articles SARS-CoV-2 Research articles SARS-CoV-2 review articles SARS-CoV-2 PubMed articles SARS-CoV-2 PubMed Central articles SARS-CoV-2 2023 articles SARS-CoV-2 2024 articles SARS-CoV-2 Scopus articles SARS-CoV-2 impact factor journals SARS-CoV-2 Scopus journals SARS-CoV-2 PubMed journals SARS-CoV-2 medical journals SARS-CoV-2 free journals SARS-CoV-2 best journals SARS-CoV-2 top journals SARS-CoV-2 free medical journals SARS-CoV-2 famous journals SARS-CoV-2 Google Scholar indexed journals CD8+ T cells articles CD8+ T cells Research articles CD8+ T cells review articles CD8+ T cells PubMed articles CD8+ T cells PubMed Central articles CD8+ T cells 2023 articles CD8+ T cells 2024 articles CD8+ T cells Scopus articles CD8+ T cells impact factor journals CD8+ T cells Scopus journals CD8+ T cells PubMed journals CD8+ T cells medical journals CD8+ T cells free journals CD8+ T cells best journals CD8+ T cells top journals CD8+ T cells free medical journals CD8+ T cells famous journals CD8+ T cells Google Scholar indexed journals MHC class I articles MHC class I Research articles MHC class I review articles MHC class I PubMed articles MHC class I PubMed Central articles MHC class I 2023 articles MHC class I 2024 articles MHC class I Scopus articles MHC class I impact factor journals MHC class I Scopus journals MHC class I PubMed journals MHC class I medical journals MHC class I free journals MHC class I best journals MHC class I top journals MHC class I free medical journals MHC class I famous journals MHC class I Google Scholar indexed journals In silico analysis articles In silico analysis Research articles In silico analysis review articles In silico analysis PubMed articles In silico analysis PubMed Central articles In silico analysis 2023 articles In silico analysis 2024 articles In silico analysis Scopus articles In silico analysis impact factor journals In silico analysis Scopus journals In silico analysis PubMed journals In silico analysis medical journals In silico analysis free journals In silico analysis best journals In silico analysis top journals In silico analysis free medical journals In silico analysis famous journals In silico analysis Google Scholar indexed journals Peptides articles Peptides Research articles Peptides review articles Peptides PubMed articles Peptides PubMed Central articles Peptides 2023 articles Peptides 2024 articles Peptides Scopus articles Peptides impact factor journals Peptides Scopus journals Peptides PubMed journals Peptides medical journals Peptides free journals Peptides best journals Peptides top journals Peptides free medical journals Peptides famous journals Peptides Google Scholar indexed journals virus articles virus Research articles virus review articles virus PubMed articles virus PubMed Central articles virus 2023 articles virus 2024 articles virus Scopus articles virus impact factor journals virus Scopus journals virus PubMed journals virus medical journals virus free journals virus best journals virus top journals virus free medical journals virus famous journals virus Google Scholar indexed journals homozygosity articles homozygosity Research articles homozygosity review articles homozygosity PubMed articles homozygosity PubMed Central articles homozygosity 2023 articles homozygosity 2024 articles homozygosity Scopus articles homozygosity impact factor journals homozygosity Scopus journals homozygosity PubMed journals homozygosity medical journals homozygosity free journals homozygosity best journals homozygosity top journals homozygosity free medical journals homozygosity famous journals homozygosity Google Scholar indexed journals infection articles infection Research articles infection review articles infection PubMed articles infection PubMed Central articles infection 2023 articles infection 2024 articles infection Scopus articles infection impact factor journals infection Scopus journals infection PubMed journals infection medical journals infection free journals infection best journals infection top journals infection free medical journals infection famous journals infection Google Scholar indexed journals mortality articles mortality Research articles mortality review articles mortality PubMed articles mortality PubMed Central articles mortality 2023 articles mortality 2024 articles mortality Scopus articles mortality impact factor journals mortality Scopus journals mortality PubMed journals mortality medical journals mortality free journals mortality best journals mortality top journals mortality free medical journals mortality famous journals mortality Google Scholar indexed journals Bioinformatic articles Bioinformatic Research articles Bioinformatic review articles Bioinformatic PubMed articles Bioinformatic PubMed Central articles Bioinformatic 2023 articles Bioinformatic 2024 articles Bioinformatic Scopus articles Bioinformatic impact factor journals Bioinformatic Scopus journals Bioinformatic PubMed journals Bioinformatic medical journals Bioinformatic free journals Bioinformatic best journals Bioinformatic top journals Bioinformatic free medical journals Bioinformatic famous journals Bioinformatic Google Scholar indexed journals
Article Details
1. Introduction
In December 2019, the world experienced the outbreak of a novel coronavirus, when the first case was reported with respiratory-related symptoms in Wuhan, Hubei, China, and then has spread to other countries [1]. The viral genome was then fully sequenced [2] and showed similarity, but distinct composition to the genomes of two other SARS-CoV and MERS-CoV coronaviruses, that have been pandemic in 2002 and 2011, respectively. The new virus was officially termed "2019 novel coronavirus” while the disease that it causes was termed “the Corona Virus Disease 2019” (COVID-19) by World Health Organization (WHO) [3], but then the International Committee on Taxonomy renamed the virus as “Severe Acute Respiratory Syndrome Coronavirus-2” (SARS-CoV-2) [4]. Based on the genome sequence, SARS-CoV-2 is a member of genus Betacoronavirus subgenus (Sarbecovirus) [5] and shares approximately 79% homology with SARS-CoV at the nucleotides level, with ∼72% nucleotide sequence similarity in the spike (S) protein [6]. The pathogenesis of COVID-19 is still under investigation. SARS-CoV-2 and SARS-CoV enter host cells through ACE2 receptors (1) while MERS-CoV uses dipeptidyl peptidase (DPP)-4 [7].
Previous studies in chronic infections have highlighted the role of CD8+ T cells, as a powerful effector mechanism to eliminate the virus, but also because they differentiate to long-lasting memory, that provide protective responses against the subsequent infection [8]. Major Histocompatibility Complex (MHC) class I genes play critical roles in determining the outcome (i.e., susceptibility or resistance to the infection). Association between human MHC (HLA) alleles and the outcome of viral infections have been documented. HIV-infected patients who are heterozygous for certain HLA class alleles, progress more slowly to AIDS and have a lower mortality rate [9]. Similarly, other studies have shown a direct relationship between susceptibility to infection and increased HLA homozygosity in a genetically isolated population [10]. Moreover, Ying, M. et al. [11] suggested that patients expressing the HLA-B*46 allele had a more severe course of hemorrhagic fever with renal syndrome (HFRS) upon Hantan virus (HTNV) infection with respect to the control group. Another study demonstrated a reduced risk of developing Dengue hemorrhagic fever (DHF) associated with HLA-A*68 and HLA-DRB1*08 alleles in a Sri Lankan Population [12]. Finally, cerebral malaria was significantly more frequent in patients expressing HLA-A*30 and HLA-A*33 alleles [13].
Based on this evidence we aimed to speculate on the impact of HLA class I allele polymorphism on the severity, mortality, and morbidity of COVID-19. We have analyzed the distribution of HLA class I alleles in countries with the diverse extent of the COVID-19 pandemic, their potential role in SARS-CoV-2 CD8+ T cell epitope recognition in silico, and speculate on how this knowledge may impact future epitope-based vaccine or diagnostic tools development [14-20].
2. Material and method
2.1 In silico Peptide prediction
The amino acid sequence of the 29882 bp, SARS-CoV-2 complete genome (2019-nCoV/USA-WA1-A12/2020), was received in FASTA format from NCBI Protein Database (MT020880). This large genome codes ten essential proteins (Table 1). Because of the novel nature of 2019nCoV, we used machine learning methods and constructed models to predict peptide-HLA interactions with different HLA class I alleles, which have a higher frequency in the chosen population (Table 2). We used the artificial neural networks (ANNs) method for predicting the binding affinity of peptides [21].
NetMHC 4.0 Server (http://www.cbs.dtu.dk/services/NetMHC-4.0/) was used to identify CTL epitopes within the ten different essential protein sequences [22]. The threshold for strong binders was set as % Rank of <0.5 and the threshold for weak binders was set as % Rank of >2. The highest scoring epitopes (SB) for each HLA supertype were selected for analysis of binding affinity. Based on this method a list of potential nonamer-peptides has been created which consists of a total of 18219 peptides (Table 3).
2.2 Subjects
We used a retrospective study of a cohort of SARS-CoV-2 countries reported by WHO [23]. To find a correlation between alleles that have high affinity for predicted SARS-CoV-2 nonamer-peptides and covid-19 pandemic. We selected two different cohort groups (Table 2 and Table 4), the first cohort group consists of HLA allele selection of infected people belonging to different countries, and this group was further divided into a subgroup with high and low mortality and morbidity rate (Table2). The second cohort group consists of patients that were used to test the correlation between allele frequency and mortality morbidity rates (Table 4). This study method was tested in three different periods: from the beginning of the Covid-19 pandemic until March 2020, until April 2020, and until July 2020. The latest update of data is presented here which included reports until July 12th, 2020. Allele frequencies of candidate place (first and second group) have been collected using Allele Frequency Net Database (AFND) [24].
2.3 Allele frequency of HLA I genes in different study subjects
According to the hypothesis that different frequencies of HLA class I alleles that can vary due to differences in their evolutionary histories in the different populations; can be associated with mortality and morbidity rates, the most frequent alleles in the two different cohort groups with high and low mortality and morbidity rates have been chosen (Table 5). Mortality and morbidity rates are calculated by the following formula:
We used the calculated rate for further analysis based on the prevalence of the disease (Table 4). The population with a higher rate of mortality and morbidity included Lombardy (Italy), Wuhan (China) and Tehran (Iran), and population with a low rate of mortality and morbidity included Saudi Arabia, Germany, and Sweden (Table 2).
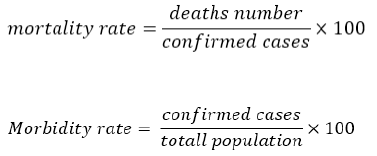
We have chosen the most frequent HLA class I alleles in group 1 and based on allele frequency (Table 2).
A total of 30 alleles were selected as candidates for nonamers epitope prediction (HLA-A*01, HLA-A*02, HLA-A*03, HLA-A*11, HLA-A*23, HLA-A*24, HLA-A*68, HLA-B*07, HLA-B*08, HLA-B*14, HLA-B*15, HLA-B18, HLA-B35, HLA-B38, HLA-B40, HLA-B44, HLA-B46, HLA-B50, HLA-B51, HLA-B*52, HLA-C*0303, HLA-C*0401, HLA-C*0501, HLA-C*0602, HLA-C*0701, HLA-C*0702, HLA-C*0802, HLA-C*1203, HLA-C*1402, HLA-C*1502).
Two separate analysis were used to test our hypothesis. binding affinity in nano-Molar units and frequency of predicted nonamers that can be recognized by different HLA class I alleles. The affinity of nonamers was analyzed using Kruskal–Wallis one-way ANOVA test. p-values < 0.05 were considered significant. The second analysis regarded the correlation between HLA class I alleles (selected from first analysis) with mortality and morbidity rates has been run on the second group: Pearson correlation coefficient with a one-tailed p-value and 90% of the confidence interval was used for allele correlation.
|
Accession Id |
Protein name |
Length Bp |
Percentage of total genome |
HLA-A |
HLA-B |
HLA-C |
|
Envelope protein |
75 |
0.25% |
0.86% |
1.00% |
0.99% |
|
|
Membrane Glycoprotein |
222 |
0.74% |
0.79% |
1.00% |
0.99% |
|
|
Nucleocapsid phosphoprotein |
419 |
1.40% |
1.93% |
2.72% |
2.79% |
|
|
orf1ab polyprotein |
7976 |
26.69% |
77.44% |
77.38% |
75.38% |
|
|
ORf3a protein |
275 |
0.92% |
5.16% |
3.39% |
4.07% |
|
|
ORF6 protein |
61 |
0.20% |
0.36% |
0.50% |
0.33% |
|
|
ORF8a protein |
121 |
0.40% |
1.07% |
0.67% |
0.76% |
|
|
Orf10 protein |
38 |
0.13% |
0.57% |
0.94% |
0.52% |
|
|
surface glycoprotein and spike |
1273 |
4.26% |
11.82% |
12.40% |
14.16% |
|
|
total seq |
10460 |
35.00% |
||||
|
total seq |
29882 |
Table. 1: SARS-Cov-2 protein and total of predicted nonamers and different MHC I genes.
|
HLA-A |
HLA-B |
HLA-C |
|||||
|
HLA |
Allele Freq |
HLA |
Allele Freq |
HLA |
Allele Freq |
||
|
Group A |
Lombardi |
A*02 |
26.80% |
B*35 |
13.90% |
C*07 |
18.80% |
|
A*03 |
11.80% |
B*18 |
9.90% |
C*04 |
15.90% |
||
|
A*01 |
11.70% |
B*44 |
9.10% |
C*06:02 |
10.40% |
||
|
A*24 |
10.90% |
B*51 |
8.70% |
||||
|
Wuhan |
A*02 |
31.00% |
B*40 |
15.70% |
C*01 |
24.50% |
|
|
A*11 |
29.30% |
B*15 |
15.60% |
C*03:04 |
14.90% |
||
|
A*24 |
17.80% |
B*46 |
13.50% |
C*07:02 |
11.60% |
||
|
A*30 |
5.40% |
B*14 |
10.80% |
C*04 |
7.50% |
||
|
Tehran |
A*02 |
18.30% |
B*35 |
19.10% |
C*07 |
19.20% |
|
|
A*24 |
12.00% |
B*51 |
13.70% |
C*12 |
16.70% |
||
|
A*03 |
11.80% |
B*52 |
5.60% |
C*15 |
14.80% |
||
|
A*01 |
10.90% |
B*38 |
5.30% |
C*04 |
13.90% |
||
|
Group B |
Saudi Arabia |
A*02 |
28.90% |
B*51 |
19.30% |
C*07 |
24.90% |
|
A*68 |
10.00% |
B*50 |
16.30% |
C*06 |
20.10% |
||
|
A*24 |
8.00% |
B*08 |
10.00% |
C*15 |
12.60% |
||
|
A*26 |
7.40% |
B*07 |
8.10% |
C*04 |
10.60% |
||
|
Germany |
A*02 |
28.20% |
B*07 |
13.50% |
C*07:01 |
20.90% |
|
|
A*03 |
15.40% |
B*08 |
13.50% |
C*03:04 |
10.50% |
||
|
A*01 |
14.40% |
B*44 |
9.20% |
C*02:02 |
10.30% |
||
|
A*24 |
10.10% |
B*40 |
8.60% |
||||
|
Sweden |
A*02 |
32.90% |
B*07 |
14.10% |
C*07 |
31.90% |
|
|
A*03 |
16.80% |
B*44 |
12.70% |
C*03 |
18.90% |
||
|
A*01 |
13.90% |
B*08 |
12.10% |
C*04 |
10.10% |
||
|
A*24 |
9.60% |
B*15 |
10.60% |
||||
Table 2: First cohorts’ group for allele Selection: the population was divided into two groups on the basis of HLA alleles frequency. Group A: population with a high rate of mortality and morbidity based on the WHO report; Group B: population with a low rate of mortality and morbidity based on the WHO report.
|
T Cell predicted peptide |
High affinity for HLA I (N) |
Strong binder HLA I (N) |
|
HLA-A |
4376 |
1437 |
|
HLA-B |
6523 |
1832 |
|
HLA-C |
7320 |
2163 |
|
Total number |
18219 |
5432 |
Table 3: Number of nonamers for SARS-Cov2 proteins with binding affinity for HLA Class I molecules (High affinity= the peptides with significant affinity, Strong binder= the peptides that have strong affinity for epitope and MHC binding).
|
|
population |
confirmed cases |
deaths |
mortality rate |
Morbidity rate |
|
India |
1,364,562,908 |
820,916 |
22,123 |
2.69% |
0.06% |
|
Saudi Arabia |
34,218,169 |
226,486 |
2,151 |
0.95% |
0.66% |
|
Jordan |
10,721,796 |
1,173 |
10 |
0.85% |
0.01% |
|
China |
1,403,482,160 |
85,487 |
4,648 |
5.44% |
0.01% |
|
Albania |
2,845,955 |
3,371 |
89 |
2.64% |
0.12% |
|
Croatia |
4,076,246 |
3,532 |
117 |
3.31% |
0.09% |
|
Iran |
8,36,03,884 |
252,720 |
12,447 |
4.93% |
0.30% |
|
Germany |
83,166,711 |
198,556 |
9,060 |
4.56% |
0.24% |
|
Belgium |
11,528,375 |
62,469 |
9,782 |
15.66% |
0.54% |
|
Italy |
60,238,522 |
242,639 |
34,938 |
14.40% |
0.40% |
|
Spain |
47,329,981 |
253,908 |
28,403 |
11.19% |
0.54% |
|
France |
67,081,000 |
161,275 |
29,907 |
18.54% |
0.24% |
|
Finland |
5,498,027 |
7,279 |
329 |
4.52% |
0.13% |
|
UK |
66,796,807 |
288,137 |
44,650 |
15.50% |
0.43% |
|
Argentina |
45,376,763 |
94,060 |
1,787 |
1.90% |
0.21% |
|
Brazil |
211,778,013 |
1,800,827 |
70,398 |
3.91% |
0.85% |
|
Japan |
125,930,000 |
21,502 |
982 |
4.57% |
0.02% |
|
Ecuador |
17,524,324 |
67,209 |
5,031 |
7.49% |
0.38% |
|
peru |
32,824,358 |
319,646 |
11,500 |
3.60% |
0.97% |
|
Sweden |
10,348,730 |
74,898 |
5,526 |
7.38% |
0.72% |
|
Republic of Korea |
51,780,579 |
13,417 |
289 |
2.15% |
0.03% |
Table 4: Second cohorts’ group for allele correlation analysis consisting of eleven countries based on their mortality and morbidity rate. The confirmed cases and deaths number is based on WHO report until 11th of April. Number of populations is based on united nation last report Nations WPP-PD-U. 2019 Revision of World Population Prospects 9 November 2019; Available from: https://population.un.org/wpp/.
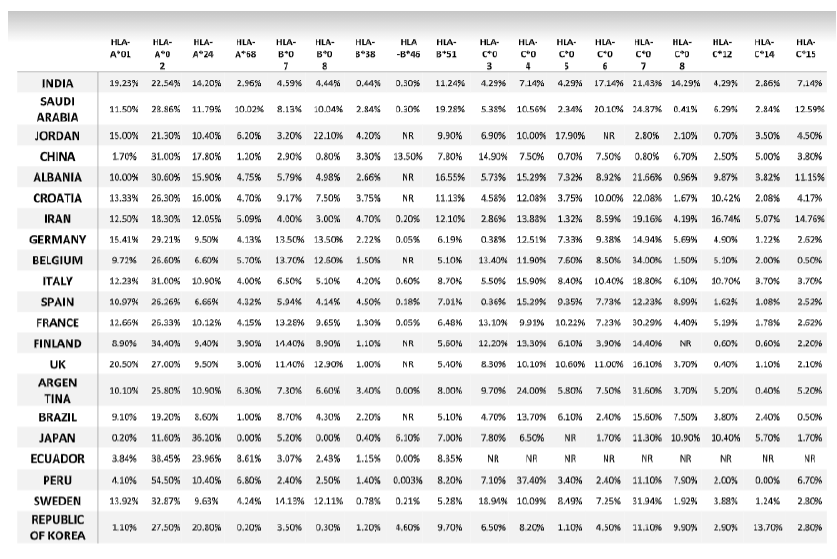
Table. 5: Allele frequency of second cohort of SARS-CoV-2 infected countries reported by WHO.
*NR: allele frequency was Not reported in Allele Frequency Net Database.
|
Pearson r correlation |
|||
|
allele |
Correlation factor |
Vs Mortality |
Vs Morbidity |
|
HLA-A*01 |
r |
0.2273 |
0.05488 |
|
90% confidence interval |
-0.1551 to 0.5504 |
-0.3210 to 0.4158 |
|
|
R squared |
0.05164 |
0.003012 |
|
|
HLA-A*02 |
r |
0.02707 |
0.4413 |
|
90% confidence interval |
-0.3458 to 0.3925 |
0.08588 to 0.6970 |
|
|
R squared |
0.0007331 |
0.1947 |
|
|
HLA-A*24 |
r |
-0.2829 |
-0.4685 |
|
90% confidence interval |
-0.5906 to 0.09656 |
-0.7143 to -0.1199 |
|
|
R squared |
0.08004 |
0.2195 |
|
|
HLA-A*68 |
r |
-0.06578 |
0.349 |
|
90% confidence interval |
-0.4248 to 0.3112 |
-0.02335 to 0.6364 |
|
|
R squared |
0.004326 |
0.1218 |
|
|
HLA-B*07 |
r |
0.4478 |
0.1823 |
|
90% confidence interval |
0.09394 to 0.7012 |
-0.2005 to 0.5169 |
|
|
R squared |
0.2005 |
0.03325 |
|
|
HLA-B*08 |
r |
0.1413 |
0.02803 |
|
90% confidence interval |
-0.2406 to 0.4853 |
-0.3449 to 0.3933 |
|
|
R squared |
0.01996 |
0.0007855 |
|
|
HLA-B*38 |
r |
-0.1121 |
-0.0747 |
|
90% confidence interval |
-0.4623 to 0.2684 |
-0.4321 to 0.3030 |
|
|
R squared |
0.01256 |
0.005579 |
|
|
HLA-B*46 |
r |
-0.16 |
-0.5295 |
|
90% confidence interval |
-0.5766 to 0.3226 |
-0.7952 to -0.09330 |
|
|
R squared |
0.02559 |
0.2804 |
|
|
HLA-B*51 |
r |
-0.4901 |
-0.1505 |
|
90% confidence interval |
-0.7277 to -0.1474 |
-0.4925 to 0.2317 |
|
|
R squared |
0.2402 |
0.02266 |
|
|
HLA-C*03 |
r |
0.2666 |
0.09891 |
|
90% confidence interval |
-0.1372 to 0.5944 |
-0.3022 to 0.4703 |
|
|
R squared |
0.07107 |
0.009782 |
|
|
HLA-C04 |
r |
-0.1104 |
0.5719 |
|
90% confidence interval |
-0.4793 to 0.2916 |
0.2347 to 0.7863 |
|
|
R squared |
0.01219 |
0.3271 |
|
|
HLA-C05 |
r |
0.3474 |
-0.0176 |
|
90% confidence interval |
-0.06215 to 0.6568 |
-0.4155 to 0.3860 |
|
|
R squared |
0.1207 |
0.0003097 |
|
|
HLA-C*06 |
r |
-0.05918 |
0.07537 |
|
90% confidence interval |
-0.4494 to 0.3500 |
-0.3357 to 0.4623 |
|
|
R squared |
0.003502 |
0.00568 |
|
|
HLA-C*07 |
r |
0.3229 |
0.3315 |
|
90% confidence interval |
-0.07622 to 0.6328 |
-0.06665 to 0.6385 |
|
|
R squared |
0.1042 |
0.1099 |
|
|
HLA-C*08 |
r |
-0.08358 |
-0.2325 |
|
90% confidence interval |
-0.4688 to 0.3283 |
-0.5794 to 0.1857 |
|
|
R squared |
0.006985 |
0.05404 |
|
|
HLA-C*12 |
r |
-0.06214 |
-0.1335 |
|
90% confidence interval |
-0.4410 to 0.3355 |
-0.4972 to 0.2700 |
|
|
R squared |
0.003861 |
0.01783 |
|
|
HLA-C*14 |
r |
-0.2459 |
-0.466 |
|
90% confidence interval |
-0.5799 to 0.1588 |
-0.7241 to -0.09344 |
|
|
R squared |
0.06048 |
0.2171 |
|
|
HLA-C*15 |
r |
-0.4622 |
0.1069 |
|
90% confidence interval |
-0.7218 to -0.08864 |
-0.2949 to 0.4766 |
|
|
R squared |
0.2136 |
0.01143 |
|
Table 6: Correlation between different alleles and mortality/morbidity rate.
3. Results
3.1 Bioinformatic-based prediction of SARS-CoV-2 epitopes binding to HLA-A alleles
A total of 4417 SARS-CoV-2 peptides showed a significant affinity to selected HLA-A chosen alleles. Among these nonamer epitopes, 1451 (32%) had a strong binding affinity based on a 2% Rank threshold for weak binders and 0.5% Rank threshold for strong binders. The most frequent HLA-A allele in our database was HLA-A*02, but the affinity of the nonamer epitopes for HLA-A*02 was significantly lower (even extremely lower) than for all other selected alleles (p-value=0.0001) (Figure 1A). Approximately 31% of nonamer epitopes have a strong binding affinity to HLA-A*01 and HLA-A*68. Interestingly, the countries with a high frequency of these two alleles showed a low mortality and morbidity rate. Pearson correlation analysis showed a strong positive correlation between HLA-A*02 and morbidity rate (Figure 2A and Table 6), where the correlation between HLA-A*24 and morbidity rate were significantly negative (p-value = 0.0322), whereas HLA-A*02 showed a significantly positive correlation with morbidity rate (p-value = 0.0452).
3.2 Bioinformatic-based prediction of SARS-CoV-2 epitopes binding to HLA-B alleles
A total of 6523 SARS-CoV-2 peptides showed a significant affinity to selected HLA-B alleles and 1840 (28%) of these had a strong binding affinity. Our in-silico analysis showed that similarly to HLA-A, the most frequent HLA-B alleles, such as HLA-B*35 or HLA-B*40 had the lowest binding affinity for nonamer epitopes (Figure1B) and only 13% epitopes were recognized by HLA-B*35 and B*40. The countries with a lower prevalence of the disease showed different allele frequency patterns, which comprised HLA-B*07, *08 or B*51. As shown in Figure 1B, a small number of peptides (4%) had a strong binding affinity to HLA-B*51, while 26.20% of the predicted nonamer epitopes display a strong affinity binding to HLA-B*08. The correlation between selected alleles with higher affinity for nonamers HLA-B epitopes (HLA-B*07, HLA-B*08, HLA-B*38, HLA-B*51) and mortality and morbidity rates were estimated. HLA-B*07 showed a significantly positive correlation versus mortality rate (p-value = 0.0418) and HLA-B*38 alleles showed a negative correlation with morbidity rates, while HLA-B*51 showed a statistically significant negative correlation with mortality (p-value =0.0241) and strong negative correlation versus morbidity rates (Figure 2B and table 6).
3.3 Bioinformatic-based prediction of SARS-CoV-2 epitopes binding to HLA-C alleles
There was no significant difference in the number of SARS-CoV2 nonamers that have strong binding affinity to HLA-C alleles (Figure 1C), but 3 allele clusters could be observed, as reported in Table 2 and Figure 1C; the most frequent allele among 22 countries is HLA-C*07, but our data showed an affinity that was lower than other alleles and interestingly the correlation between this allele and mortality rate was strongly positive (Figure 2C). On the other hand, HLA-C*04 which had a strong affinity for predicted SARS-CoV2 nonamers showed the significantly strongest positive correlation versus morbidity rate. According to our results, these two alleles may contribute to the spread and mortal function of this new virus. Contrastingly, the third cluster showed a different result. The HLA-C*14 and -C*15 showed a low affinity for predicted SARS-CoV2 nonamers but HLA-C*14 showed a significantly negative correlation with morbidity (p-value =0.0443) and HLA-C*15 significant negative correlation with mortality (p-value =0.0463), which again confirm our hypothesis that high affinity of SARS-CoV2 epitopes with HLA-C alleles may contribute to spread and fatal disease progression of this new virus.
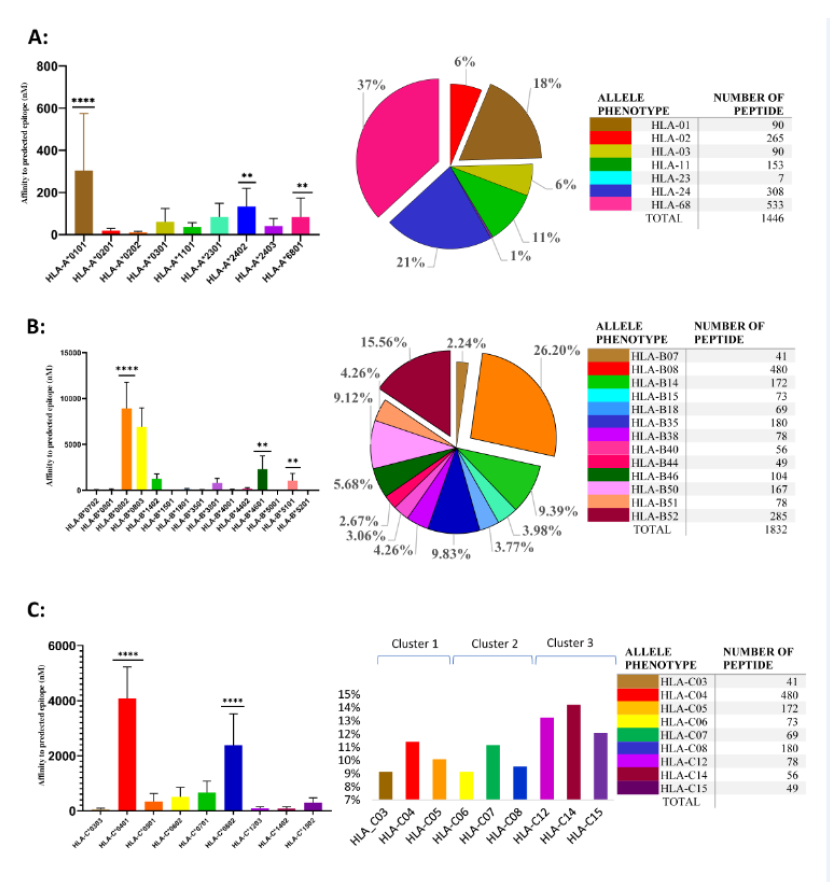
Figure. 1: SARS-CoV-2 epitopes predicted to bind to HLA class I alleles.
On the left side is shown the affinity of peptides that bind to different HLA alleles. X axis represents allele type and Y axis represents affinity of peptides that bind to different alleles type(nM). Shown is median affinity and bars indicate Standard Deviation. On the middle, are shown the SARS-CoV-2 nonamers that can be recognized by different HLA-A alleles (% of total strong binder to different HLA alleles phenotype). On the right side, are shown numbers of SARS-CoV-2 nonamers that can be recognized by different HLA-A (A), HLA-B (B), and HLA-C (C).**p<0.01 and ****p<0.0001.
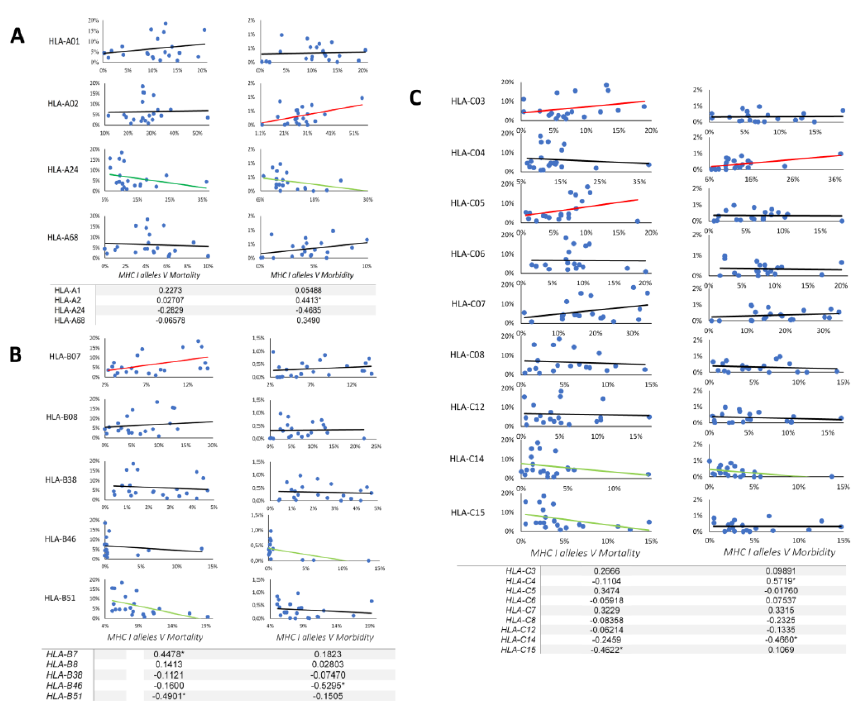
Figure. 2: Correlation between HLA class I alleles frequency and morbidity/mortality rate.
Correlation has been calculated based on Pearson coefficient with one-tailed p-value and 90% of confidence interval. Each graph represents the correlation between HLA-A (A), HLA-B (B) and HLA-C (C) alleles and morbidity/mortality rate. X axes indicate allele frequency and Y-axes indicate the morbidity/mortality rates. The red line indicates positive correlation, the green line indicates negative correlation and the black one indicates no correlation. The table represents each allele correlation with a 90% confidence interval and * indicate the significant p value <0.05 where significant for alpha = 0.1. The complete correlation data can be seen in table 6.
4. Discussion
During evolution, living species have adapted to environmental constraints such as different pathogens according to the mechanism of natural selection. Here we showed that different frequencies of HLA class I alleles that can vary due to the different evolutionary histories in the different populations, may contribute to the spread of this new coronavirus because the different alleles can have different affinity for pathogen epitope and consequently elicit different adaptive immune responses. Understanding the pathogenesis of SARS-CoV-2 infection, including the role of immunogenetics, is essential not only for the development of new strategies to treat and prevent this novel infection, but also for vaccine development [25, 26]. Here, we have used bioinformatics and in silico approaches to evaluate associations between HLA alleles and SARS-CoV-2 epitopes in different populations with different HLA Class I alleles frequency.
The vast majority of HLA-A and -B alleles fall into one of 9 supertypes [28], for example, HLA-A*6802 and HLA-A*0201 have exact matches in the B and F pockets and A*0301, A*1101 and A*6801 belong to the same HLA A supertype [27]. But, they are structurally far from HLA-A*0201 allele. Based on the AFND database, the HLA-A*02 is the most frequent allele almost in all candidate populations, but we show here that HLA-A*02 has a low affinity for SARS-CoV-2 nonamer epitopes. In contrast, HLA-A*01, A*24, and A*68 alleles, despite having a lower frequency with respect to HLA-A*02 allele, have the highest binding affinity for the mentioned viral nonamers. Take into consideration that HLA-A*24 and HLA-A*68 have more affinity in comparison to HLA-A*02, it can be considered that this polymorphism might favor SARS-CoV-2 peptide binding to B and F pockets and consequently promote activation of CTLs CD8+ T virus-specific cells. In our study, we show a strong negative correlation between morbidity rate and HLA-A*24 allele at partial support of this possibility. Notably, several HLA-A*24-restricted epitopes derived from the influenza virus have been also identified in human studies [28, 29], and another study suggested the role of HLA-A*24-restricted CD8+T cell responses against 2009 pH1N1 [30].
In different populations, alleles of the HLA-B*07 superfamily, including HLA-B*0702 HLA-B*35, HLA-B*51, and HLA-B*53, preferentially select peptides with a proline residue in P2 [31]. In some instances, this small allele polymorphism can provide an advantage for a given population. Here, we show that HLA-B*07 has a low affinity for the predicted SARS-Cov-2 epitopes but, another member of the same supertype family, HLA-B*5101, displays a high binding affinity. Kawashima et al. [32] reported that HLA-B*51 is associated with slow disease progression to AIDS; accordingly, we show here that there is a strong negative correlation between HLA-B*51 and SARS-CoV-2 mortality and morbidity rate. We speculate that individuals that express HLA-B*51 and HLA-B*08 alleles are not affected as much as other people expressing other HLA alleles. Concerning HLA-C alleles, HLA-C*0801 correlates with the susceptibility to SARS-CoV-2 [33], while HLA-C*06 had a strong negative correlation with mortality rate. Interestingly, HLA-A*24-B*51-C*06 are the most frequent extended HLA class I haplotype in Albania [33], where 475 confirmed COVID-19 infection cases and 24 deaths were reported by WHO [23].
In conclusion, our in-silico study, although very preliminary, suggests the possibility that HLA class I polymorphism, by selecting potential SARS-Cov-2 epitopes capable to induce protective CD8+ T cell responses, may account for the diverse mortality and morbidity rates documented in different countries that screening during almost 10 months of this tragic pandemic. Due to the important role of CD8+ T cells in SARS-CoV-2 immunity, a specific antiviral cytotoxic immune response induced by viral peptides should be considered. We suggest that the mentioned peptides could be used for peptide-based vaccine development and could be evaluated for the development of diagnostic tools with high sensitivity and specificity.
Funding
This work was supported by grants from the European Commission within the Horizon2020 Programmed TBVAC2020 [Horizon 2020 cod 643381].
Conflict of Interest
All the authors declare that no conflict of interests exist.
Acknowledgments
This work was supported by grants from the European Commission within the Horizon2020 Programmed TBVAC2020. The text represents the authors’ views and does not necessarily represents position of the European Commission, which will not be liable for the use made of such information.
Author’s contribution
Conceived and wrote the paper: MSA and LM, revised the paper FD and NC and MPLM.
References
- Prompetchara E, Ketloy C, Palaga TJAPJAI. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific Journal of Allergy and Immunology 38 (2020): 1-9.
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 579 (2020): 265-269.
- Lai C-C, Shih T-P, Ko W-C, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents 55 (2020): 105924.
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5 (2020): 536-544.
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 395 (2020): 565-574.
- Zhang Y-Z, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 181 (2020): 223-227.
- Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (2020): 270-273.
- McMaster SR, Wilson JJ, Wang H, et al. Airway-Resident Memory CD8 T Cells Provide Antigen-Specific Protection against Respiratory Virus Challenge through Rapid IFN-γ Production. J Immunol 195 (2015): 203-219.
- Carrington M. HLA and HIV-1: Heterozygote Advantage and B*35-Cw*04 Disadvantage. Science 283(1999): 1748-1752.
- Black FL, Schiffman G, Pandey JP. HLA, Gm, and Km Polymorphisms and Immunity to Infectious Diseases in South Amerinds. Immunogenetic Risk Assessment In Human Disease: S. Karger AG. p. 206-16.
- Ma Y, Yuan B, Yi J, Zhuang R, Wang J, Zhang Y, et al. The Genetic Polymorphisms of HLA Are Strongly Correlated with the Disease Severity after Hantaan Virus Infection in the Chinese Han Population. Clinical and Developmental Immunology 2012 (2012): 1-9.
- Malavige GN, Rostron T, Rohanachandra LT, et al. HLA Class I and Class II Associations in Dengue Viral Infections in a Sri Lankan Population. PLoS ONE 6 (2011): e20581.
- Lyke KE, Fernández-Vina MA, Cao K, et al. Association of HLA alleles with Plasmodium falciparum severity in Malian children. Tissue Antigens 77 (2011): 562-571.
- Taylor PM, Askonas BA. Influenza nucleoprotein-specific cytotoxic T-cell clones are protective in vivo. Immunology 58 (1986): 417-420.
- Melendi GA, Zavala F, Buchholz UJ, et al. Mapping and Characterization of the Primary and Anamnestic H-2d-Restricted Cytotoxic T-Lymphocyte Response in Mice against Human Metapneumovirus. Journal of Virology 81 (2007): 11461-11467.
- Slütter B, Pewe Lecia L, Kaech Susan M, et al. Lung Airway-Surveilling CXCR3hi Memory CD8+ T Cells Are Critical for Protection against Influenza A Virus. Immunity 39 (2013): 939-948.
- Schmidt ME, Knudson CJ, Hartwig SM, et al. Memory CD8 T cells mediate severe immunopathology following respiratory syncytial virus infection. PLOS Pathogens 14 (2018): e1006810.
- Graham BS, Bunton LA, Wright PF, et al. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. The Journal of Clinical Investigation 88 (1991): 1026-1033.
- Channappanavar R, Fett C, Zhao J, et al. Virus-Specific Memory CD8 T Cells Provide Substantial Protection from Lethal Severe Acute Respiratory Syndrome Coronavirus Infection. Journal of Virology 88 (2014): 11034-11044.
- Chen WH, Cross AS, Edelman R, et al. Antibody and Th1-type cell-mediated immune responses in elderly and young adults immunized with the standard or a high dose influenza vaccine. Vaccine 29 (2011): 2865-2873.
- Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Science 12 (2003): 1007-1017.
- Larsen MV, Lundegaard C, Lamberth K, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8 (2007): 424.
- Organization WH. Coronavirus disease 2019 (COVID-19): situation report, 72 (2020).
- González-Galarza Faviel F, Takeshita Louise YC, Santos Eduardo JM, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Research 43 (2014): D784-D788.
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? Journal of Evolutionary Biology 16 (2003): 363-377.
- He Y, Xiang Z. Databases and In Silico Tools for Vaccine Design. Kortagere S, editor. Totowa, NJ: Humana Press (2013): 115-127.
- Sidney J, Peters B, Frahm N, et al. HLA class I supertypes: a revised and updated classification. BMC Immunology 9 (2008): 1.
- Alexander J, Bilsel P, del Guercio M-F, et al. Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Human Immunology71 (2010): 468-474.
- Assarsson E, Bui H-H, Sidney J, et al. Immunomic Analysis of the Repertoire of T-Cell Specificities for Influenza A Virus in Humans. Journal of Virology 82 (2008): 12241-12251.
- Liu J, Zhang S, Tan S, et al. Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals. Journal of Virology 86 (2012): 13281-13294.
- Sidney J, del Guercio MF, Southwood S, et al. Several HLA alleles share overlapping peptide specificities. The Journal of Immunology 154 (1995): 247.
- Chen Y-MA, Liang S-Y, Shih Y-P, et al. Epidemiological and Genetic Correlates of Severe Acute Respiratory Syndrome Coronavirus Infection in the Hospital with the Highest Nosocomial Infection Rate in Taiwan in 2003. Journal of Clinical Microbiology 44 (2006): 359.
- Sulcebe G, Cuenod M, Sanchez-Mazas A, et al. Human leukocyte antigen-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in an Albanian population from Kosovo. International Journal of Immunogenetics. 40 (2013): 104-117.


 Impact Factor: * 5.3
Impact Factor: * 5.3 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 