Metadichol® Induced the Expression of Neuronal Transcription Factors in Human Fibroblast Dermal Cells.
Article Information
Palayakotai Raghavan R1*
1Nanorx Inc New York USA
*Corresponding author: Palayakotai Raghavan R, Nanorx Inc New York USA.
Email: raghavan@nanorxinc.com
Received: 19 September 2023; Accepted: 27 September 2023; Published: 03 October 2023
Citation:
Palayakotai Raghavan R. Metadichol® Induced the Expression of Neuronal Transcription Factors in Human Fibroblast Dermal Cells. Journal of Bioinformatics and Systems Biology. 6 (2023): 298-311.
View / Download Pdf Share at FacebookAbstract
Background: Producing neurons from fibroblast cells has the potential to treat neurodegenerative diseases, characterized by neuron loss. Neurodegenerative diseases are a growing problem in the current aging and developed world populations. Metadichol® is a nontoxic nanoemulsion of long-chain lipid alcohols currently available as an oral supplement.
Methods and Findings: In this study, Metadichol® was used to treat human fibroblasts in vitro; we subsequently evaluated changes in the expression of neuronal transcription factors by qRT-PCR and immunoblotting. We observed increased expression of critical transcription factors for neuronal development, such as ASCL1 and NGN2. ND2, NR4A2, LMX1A, LHX3. ISL1, and FOXA2.
Conclusions: These data suggest that Metadichol® is a promising putative neuronal remodeling agent. Its current availability and safety profile suggest that it could be rapidly available for in vivo testing, which has been impossible thus far.
Keywords
Machine learning, Disease, Autoimmune, Single cell RNAsequencing, Drug discovery
Machine learning articles; Disease articles; Autoimmune articles; Single cell RNA-sequencing articles; Drug discovery articles
Machine learning articles Machine learning Research articles Machine learning review articles Machine learning PubMed articles Machine learning PubMed Central articles Machine learning 2023 articles Machine learning 2024 articles Machine learning Scopus articles Machine learning impact factor journals Machine learning Scopus journals Machine learning PubMed journals Machine learning medical journals Machine learning free journals Machine learning best journals Machine learning top journals Machine learning free medical journals Machine learning famous journals Machine learning Google Scholar indexed journals Autoimmune articles Autoimmune Research articles Autoimmune review articles Autoimmune PubMed articles Autoimmune PubMed Central articles Autoimmune 2023 articles Autoimmune 2024 articles Autoimmune Scopus articles Autoimmune impact factor journals Autoimmune Scopus journals Autoimmune PubMed journals Autoimmune medical journals Autoimmune free journals Autoimmune best journals Autoimmune top journals Autoimmune free medical journals Autoimmune famous journals Autoimmune Google Scholar indexed journals Single cell RNAsequencing articles Single cell RNAsequencing Research articles Single cell RNAsequencing review articles Single cell RNAsequencing PubMed articles Single cell RNAsequencing PubMed Central articles Single cell RNAsequencing 2023 articles Single cell RNAsequencing 2024 articles Single cell RNAsequencing Scopus articles Single cell RNAsequencing impact factor journals Single cell RNAsequencing Scopus journals Single cell RNAsequencing PubMed journals Single cell RNAsequencing medical journals Single cell RNAsequencing free journals Single cell RNAsequencing best journals Single cell RNAsequencing top journals Single cell RNAsequencing free medical journals Single cell RNAsequencing famous journals Single cell RNAsequencing Google Scholar indexed journals Drug discovery articles Drug discovery Research articles Drug discovery review articles Drug discovery PubMed articles Drug discovery PubMed Central articles Drug discovery 2023 articles Drug discovery 2024 articles Drug discovery Scopus articles Drug discovery impact factor journals Drug discovery Scopus journals Drug discovery PubMed journals Drug discovery medical journals Drug discovery free journals Drug discovery best journals Drug discovery top journals Drug discovery free medical journals Drug discovery famous journals Drug discovery Google Scholar indexed journals Metadichol articles Metadichol Research articles Metadichol review articles Metadichol PubMed articles Metadichol PubMed Central articles Metadichol 2023 articles Metadichol 2024 articles Metadichol Scopus articles Metadichol impact factor journals Metadichol Scopus journals Metadichol PubMed journals Metadichol medical journals Metadichol free journals Metadichol best journals Metadichol top journals Metadichol free medical journals Metadichol famous journals Metadichol Google Scholar indexed journals neuronal transcription articles neuronal transcription Research articles neuronal transcription review articles neuronal transcription PubMed articles neuronal transcription PubMed Central articles neuronal transcription 2023 articles neuronal transcription 2024 articles neuronal transcription Scopus articles neuronal transcription impact factor journals neuronal transcription Scopus journals neuronal transcription PubMed journals neuronal transcription medical journals neuronal transcription free journals neuronal transcription best journals neuronal transcription top journals neuronal transcription free medical journals neuronal transcription famous journals neuronal transcription Google Scholar indexed journals neurogenerative articles neurogenerative Research articles neurogenerative review articles neurogenerative PubMed articles neurogenerative PubMed Central articles neurogenerative 2023 articles neurogenerative 2024 articles neurogenerative Scopus articles neurogenerative impact factor journals neurogenerative Scopus journals neurogenerative PubMed journals neurogenerative medical journals neurogenerative free journals neurogenerative best journals neurogenerative top journals neurogenerative free medical journals neurogenerative famous journals neurogenerative Google Scholar indexed journals regenerative medicine articles regenerative medicine Research articles regenerative medicine review articles regenerative medicine PubMed articles regenerative medicine PubMed Central articles regenerative medicine 2023 articles regenerative medicine 2024 articles regenerative medicine Scopus articles regenerative medicine impact factor journals regenerative medicine Scopus journals regenerative medicine PubMed journals regenerative medicine medical journals regenerative medicine free journals regenerative medicine best journals regenerative medicine top journals regenerative medicine free medical journals regenerative medicine famous journals regenerative medicine Google Scholar indexed journals fibroblasts articles fibroblasts Research articles fibroblasts review articles fibroblasts PubMed articles fibroblasts PubMed Central articles fibroblasts 2023 articles fibroblasts 2024 articles fibroblasts Scopus articles fibroblasts impact factor journals fibroblasts Scopus journals fibroblasts PubMed journals fibroblasts medical journals fibroblasts free journals fibroblasts best journals fibroblasts top journals fibroblasts free medical journals fibroblasts famous journals fibroblasts Google Scholar indexed journals
Article Details
1. Introduction
Approximately 10 million Americans suffer from neurogenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease), and Huntington’s disease. As neurodegenerative diseases begin in mid-life, in affluent societies with a low birthrate and aging populations such as the United States, Europe, and Japan, an increased incidence of neurodegenerative diseases is expected over the coming decades. Neurodegenerative disease results from neuronal damage and death in the brain or spinal cord [1]. With progressing neuron death, patients experience increasingly debilitating symptoms such as memory loss, hand tremors, and loss of the ability to walk or manage daily self-care tasks. Unfortunately, numerous neurodegenerative diseases are eventually fatal. Tens of millions of people will be impacted by these diseases in developed countries; therefore, there is an urgent need for effective treatments against their effects.
To this end, current research focuses on ways to regenerate lost neurons. Specifically, human fibroblasts have been reprogrammed into motor neurons, cholinergic neurons, dopaminergic neurons, and others by delivery of specific transcription factors (TFs) through viral vectors. In particular, the use of the BRN2, ASCL1, MYT1L, and NEUROD1 transcription factors to reprogram fibroblasts into neurons was a breakthrough in regenerative medicine [2].
Fibroblasts were chosen as target cells due to their relative abundance and ease of obtention through minimally invasive methods, allowing the generation of patient-specific cells. However, fibroblast cell cultures contain heterogeneous cell types and neural crest-derived stem cells [3] and multipotent stem cells with the ability to differentiate into neurons.
Using TFs to transform fibroblasts into neurons is similar to Yamanaka et al.’s approach, which involved direct conversion of mouse and human fibroblasts into induced pluripotent stem cells (iPSCs) using the transcription factors Oct3/4, Sox2, Klf4, and c-Myc (OSKM) [4,5]. Similarly, other groups promoted the conversion of fibroblasts harvested from patients with neurological disorders into iPSCs and, subsequently, into functional neurons [6-9].
TFs that control gene expression are critical during both embryonic development and aging. At the embryonic level, TFs control the differentiation of pluripotent stem cells into different tissue types with distinct biological functions [10]. Each neuronal TF has a role that establishes connections between neuronal cell types [11,12], as flawed neuronal development can lead to autism [13] or neurodegenerative diseases such as AD, Huntington’s disease, or schizophrenia [14-17].
In this study, summarized in Figure 1, based on a literature review, we have listed 23 genes (Table1, 2,3) that are involved in neuronal differentiation and have a critical role in neurodegenerative diseases. We then carried out analysis using the software Pathway Studio, Elsevier [18,19], to generate a gene network and list of cellular processes involved in neurogenesis (see Figure 2, Table 4). To experimentally prove this, we treated human dermal fibroblasts with Metadichol® [20], a nanoemulsion of long-chain lipid alcohols, at various doses (1 picogram, 100 picogram, 1 nanogram, and 100 nanogram) and analyzed its effect on the expression of neuronal TFs using q-RT-PCR for quantification of expressed genes and qualification with Western blot studies. Our results showed that treating fibroblasts with Metadichol® can upregulate TFs important for neuronal differentiation.
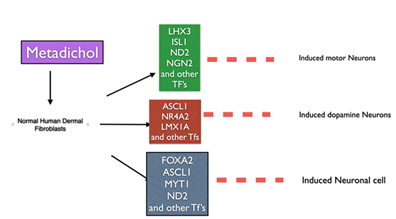
Figure 1: Expression of neuronal transcription factors
2. Materials and Methods
All work was outsourced carried out by a service provider, Skanda Life Sciences (Bangalore, India).
2.1 Chemicals and reagents
Fibroblast cells were obtained from ATCC (USA). Primary antibodies were purchased from ABclonal (Woburn, MA, USA) and Elabscience (Houston, Texas USA). Primers (Table 1) were sourced from SahaGene (Hyderabad, India). Additional reagents were obtained from Sigma-Aldrich, Bangaloore, India.
2.2 Maintenance and seeding
The cells were maintained in the appropriate medium, with or without the required supplements and 1% antibiotics, in a humidified atmosphere of 5% CO2 at 37°C. The medium was changed every other day until the cells reached confluency. The viability of the cells was assessed using a hemocytometer.
When the cells reached 70%-80% confluence, single-cell suspensions containing 106 cells/mL were prepared and seeded into 6-well plates at a density of 1 million cells per well. The cells were incubated for 24 h at 37°C in 5% CO2. After 24 h, the cell monolayer was rinsed with serum-free medium and treated with Metadichol at the concentrations described below.
2.2 Cell treatments
Different concentrations of Metadichol (1 pg/mL, 100 pg/mL, 1 ng/ml, and 100 ng/mL) were prepared in serum-free medium. Subsequently, a Metadichol-containing medium was added to predesignated wells. Control cells received medium without the drug. The cells were then incubated for 24 h. After treatment, the cells were gently rinsed with sterile PBS.
2.4 Quantitative real-time PCR (qRT-PCR)
2.4.1 RNA isolation
Total RNA was isolated from each sample using TRIzol. Approximately 1×106 cells were collected in 1.5-mL microcentrifuge tubes. The cells were thereafter centrifuged at 5000 rpm for 5 min at 4°C, and the cell supernatant was discarded. Then, 650 µL of TRIzol was added to the pellet, and the contents were mixed well and incubated for 20 min on ice. Next, 300 µL of chloroform was added, and the samples were mixed for 1-2 min by gentle inversion and incubated for 10 min on ice. Then, the samples were centrifuged at 12,000 rpm for 15 min at 4°C. The upper aqueous layer was transferred into a sterile 1.5-mL centrifuge tube, an equal amount of prechilled isopropanol was added, and the samples were incubated at −20°C for 60 min. After incubation, the mixture was centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was thereafter carefully discarded. The pellet containing RNA was washed with 1.0 mL of 100% ethanol, followed by 700 µL of 70% ethanol via centrifugation, as described above. The RNA pellet was air-dried at room temperature for 15-20 min. Then, it was resuspended in 30 µL of DEPC-treated water. The RNA concentration was quantified using a SpectraDrop (SpectraMax i3x, USA) spectrophotometer (Molecular Devices). Finally, cDNA was synthesized using reverse-transcription PCR (RT-PCR).
2.4.2 cDNA synthesis
cDNA was synthesized from 2 µg RNA using the PrimeScript cDNA synthesis kit (Takara) and oligo-dT primers according to the manufacturer’s instructions. A 20 μL reaction volume was used, and cDNA synthesis was performed on an Applied Biosystems instrument (Veriti). Then, qPCR was carried out (50°C for 30 min followed by 85°C for 5 min).
2.4.3 Primers and qPCR
The PCR mixture (at a final volume of 20 µL) contained 1 µL of cDNA, 10 µL of SYBR Green Master Mix, and 1 µM specific forward and reverse primers for the respective target genes. PCR was performed under the following conditions: an initial denaturation at 95°C for 5 min, followed by 30 cycles of secondary denaturation at 95°C for 30 s, annealing at the optimized temperature for 30 s, and extension at 72°C for 1 min. The number of cycles amplifying in the exponential range without reaching a plateau was selected as the optimal cycle number. The results were then analyzed using CFX Maestro software. Fold change was calculated using the fold change calculated using the following equation.
(ΔΔCT Method)
The comparative CT method determined the relative expression of target genes to the housekeeping gene (β-actin) and untreated control cells.
The delta CT for each treatment was calculated using the formula.
Delta Ct = Ct (target gene) - Ct (reference gene)
To compare the delta Ct of individually treated samples with the untreated control sample, the Ct was subtracted from the control to obtain the delta delta CT.
Delta delta Ct = delta Ct (treatment group) - delta Ct (control group)
The fold change in target gene expression for each treatment was calculated using the formula. Fold change = 2^ (−delta delta Ct)
2.5 Protein isolation and Western Blots
Total protein was isolated from 106 cells with RIPA buffer supplemented with the protease inhibitor PMSF (Phenuylmethyl sulfonyl fluoride). The cells were lysed for 30 min at 4°C while gently inverted. Next, the cells were centrifuged at 10,000 rpm for 15 min. The supernatant was transferred to a new tube. The Bradford method was used to determine the protein concentration, and 25 µg of protein was mixed with 1× sample loading dye containing SDS and loaded onto a gel. Proteins were then separated in Tris-glycine buffer under denaturing conditions.
The proteins were then transferred onto methanol activated PVDF membranes (Invitrogen) using the Trans-Blot Turbo system (Bio-Rad, USA). Nonspecific binding to the membranes was blocked via incubation in 5% BSA for 1 h. The membranes were then incubated overnight with the respective primary antibodies at 4°C and then with a species-specific secondary antibody for 1 h at room temperature. The blots were washed and incubated with ECL substrate (Merck) for 1 min in the dark. The images showing the results were captured at appropriate exposure settings using the ChemiDoc XRS system (Bio-Rad, USA).
Table 1: Primers used
|
Gene |
SEQUENCE |
SIZE |
Tm |
|
|
GATA-2 |
F |
GGAACCGGAAGATGTCCAACA |
169 |
60.27 |
|
R |
ATGTGTCCGGAGTGGCTGAA |
61.48 |
||
|
MYT1L |
F |
GCAGGCAGTGATGAACAACC |
201 |
59.76 |
|
R |
GGTTTGGGACTTGGGATGGT |
59.89 |
||
|
GATA-3 |
F |
TGGGCAATCAGTGTTACCGTT |
309 |
60.2 |
|
R |
CTCCGAGCACAACCACCTT |
59.93 |
||
|
PHOX2B |
F |
CCAAGGCTATTGTCGTCGCT |
159 |
60.46 |
|
R |
TGCGAAGCCAGGGAAGTTTG |
61.17 |
||
|
NR4A2 |
F |
AACACCGTCCAACATTCCTTG |
119 |
59.05 |
|
R |
GCTGCTGCATGCAAGTTTT |
58.09 |
||
|
KLF7 |
F |
CTTCTCTCGACGCCATCTCC |
247 |
59.97 |
|
R |
AGCCATCCAAAAGCCCCATT |
60.25 |
||
|
FOXA2 |
F |
ACTCGCTCTCCTTCAACGAC |
242 |
59.76 |
|
R |
CTCCCCGAGTTGAGCCTGTG |
62.51 |
||
|
NEUROG2 |
F |
CTGGGAAGAGATGATGGTGGC |
106 |
60.48 |
|
R |
GAGATTCACACGAACTGCACC |
59.54 |
||
|
LMX1A |
F |
TCTGTGTGGCTGATGGTGTT |
256 |
59.53 |
|
R |
AAGCCTTGGTGTTTCCCAGT |
59.74 |
||
|
ISL2 |
F |
TGGTCTCCTTCTCCGAGTCC |
106 |
60.32 |
|
R |
GGACTCGGCACCATACTGTT |
59.75 |
||
|
ASCL1 |
F |
GCGGCCAACAAGAAGATGAG |
308 |
59.55 |
|
R |
CCAAAGTCCATTCGCACCAG |
59.48 |
||
|
POU3F2 |
F |
TTCTCGCTTATCTCCGTGGC |
387 |
59.9 |
|
R |
GTTTCCGCCGTGATGTTCTG |
59.8 |
||
|
DLX1 |
F |
TACTTTAAGCGCACGGGGAG |
103 |
60.11 |
|
R |
CATTCGGCTCCAAACTCTCCA |
60.34 |
||
|
DLX2 |
F |
AAGTTTAGGTGCCTTTGCGG |
140 |
59.04 |
|
R |
AACTCTGTGTCCAAGTCCAGG |
59.58 |
||
|
TP53 |
F |
AGGTTGGCTCTGACTGTACC |
332 |
59.02 |
|
R |
CCCACGGATCTGAAGGGTGA |
61.26 |
||
|
LHX3 |
F |
CGAGGGGAGAGCGTTTACTG |
131 |
60 |
|
RR |
CAGTGCAGGTGGTACACGAA |
60 |
||
|
BCL11B |
F |
TTGCCAGGACTAAGCCATCC |
387 |
59.74 |
|
R |
TGCAGGGCTGAGTTACAAGG |
59.96 |
||
|
HAND2 |
F |
GACCCAGGACTCCGGAAAAG |
201 |
60.04 |
|
R |
ACGGGAGTGTCCTCTTCGTA |
59 |
||
|
NEUROD1 |
F |
TCTTCCACGTTAAGCCTCCG |
97 |
59.75 |
|
R |
CCATCAAAGGAAGGGCTGGT |
59.85 |
||
|
NEUROG1 |
F |
TCTTGGTCTGTTTCTCCGGC |
162 |
59.85 |
|
R |
GGGTCAGTTCTGAGCCAGTC |
60 |
||
|
PITX3 |
F |
GAGCACAGCGACTCAGAAAAG |
359 |
59.54 |
|
R |
CAGTTGCCGTACGAGTAGCC |
60.8 |
||
|
KL |
F |
AGGGTCCTAGGCTGGAATGT |
158 |
59.02 |
|
R |
CCTCAGGGACACAGGGTTTA |
60 |
||
|
TERT |
F |
CCCAAGTCCCTGAACTGTGT |
150 |
60.04 |
|
R |
ACATTGAAGGCCAAGGTACG |
69 |
2.6 ΔΔCT method
We determined the relative expression of the target gene in relation to a housekeeping gene (β-actin) and untreated control cells by the comparative CT method. The ΔCT for each treatment was calculated using the formula:
ΔCT = CT (target gene)-CT (reference genes).
To obtain a ΔΔCT, we subtracted individual samples in the treated and control groups as follows:
ΔΔCT = ΔCT (treatment group)-ΔCT (control group).
Similarly, we calculated the fold change in target gene expression for each treatment using the formula:
Fold change = 2^ (-ΔΔCT)
2.7 Protein isolation
We isolated total protein from 106 cells using RIPA buffer supplemented with the broad-spectrum protease inhibitor phenylmethylsulfonyl fluoride. We applied a mild inversion for 30 min at 4°C to lyse the cells and then centrifuged them at 10,000 rpm for 15 min. Finally, we transferred the supernatant to a fresh tube and determined the protein concentration using the Bradford method, where 25 µg of protein was mixed with 1x sample loading dye containing SDS and loaded on a gel. Under denaturing conditions, we separated the proteins using Tris-glycine running buffer.
2.8 Western Blotting
We transferred the proteins to methanol-activated polyvinyl difluoride membranes (Invitrogen, USA) using a Turbo transblot system (Bio-Rad, USA). We blocked the membranes with 5% BSA for 1 h and incubated them with the appropriate primary antibody for each expressed transcription study overnight at 4°C followed by a species-specific secondary antibody for 1 h at room temperature. We rinsed the blots and incubated them with enhanced chemiluminescence (ECL) substrate (Merck, USA) for 1 min in the dark and captured the images at suitable exposure settings using a ChemiDoc XRS system (Bio-Rad, USA).
4. Results
4.1 Pathway Studio analysis of neuronal TFs and gene expression
Table 2 depicts a list of TFs known to play a crucial role in neurogenesis and neuronal differentiation, and Table 3 shows a list of TFs involved in the development of brain tissue and neuronal connections [21-29].
Table 2: Fibroblast cells and reprogramming factors
|
Cell type |
Reprogramming factors |
|
Induced neurons |
PoU3F2, ASCL1, MYTL11 |
|
human induced neurons, induced dopaminergic neurons |
PoU3F2, Ascl1, MYT1L |
|
human induced neurons, induced dopaminergic neurons |
Ascll, Nurrl, LmxlA |
|
human induced neurons |
PoU3F2, Ascl1, NeuroDl, MYT1L |
|
human induced neurons, induced dopaminergic neurons |
PoU3F2, Ascl1, FoxA2, LmxlA, MYT1L |
|
Induced motor neurons |
PoU3F2, Ascl1, Lhx3, Ngn2, Isll, Ngn2, NeuroDl,MYT1L |
|
human induced neurons, induced dopaminergic neurons |
Ascll, Myt1, NeuroD2 |
|
induced neural stem cells, induced neural progenitor cells |
Oct4, Sox2, Klf4, c-Myc |
|
induced neural stem cells, induced neural progenitor cells |
Sox2, Klf4, c-Myc, |
Table 3: Transcription factors play a key role in neurogenesis.
|
Name |
Description |
|
|
KLF7 |
Kruppel like factor 7 |
control neuronal morphogenesis and promotes axon outgrowth. |
|
NR4A2 |
nuclear receptor subfamily 4 group A member 2 |
Regulates dopaminergic (DA) neuronal differentiation, survival, and maintenance |
|
DLX1 |
distal-less homeobox 1 |
Increases Interneuron GABA Synthesis, Synaptogenesis, and Dendritogenesis |
|
HAND2 |
heart and neural crest derivatives expressed 2 |
Needed for neurogenesis and subset of cell type-specific markers in the development of enteric nervous system |
|
MYT1L |
myelin transcription factor 1 like |
dysfunction results in Neuro disorders. |
|
DLX2 |
distal-less homeobox 2 |
Increases Interneuron GABA Synthesis, Synaptogenesis, and Dendritogenesis |
|
BCL11B |
BAF chromatin remodeling complex subunit BCL11B |
regulate progenitor cell proliferation differentiation, migration, and carry out integration of neural cells. |
|
ASCL1 |
achaete-scute family bHLH transcription factor 1 |
transcription factor, accessing closed chromatin to allow other factors to bind and activate neural pathways. Plays a role at early stages of development of specific neural lineages in most regions of the CNS, and of several lineages in the PNS. Essential for the generation of olfactory and autonomic neurons |
|
TP53 |
tumor protein p53 |
dysregulated p53 activity may contribute to various peripheral and brain alterations during the earliest stages of AD. |
|
NEUROG2 |
neurogenin 2 |
play an important role in neurogenesis. induces diverse neuron types from human pluripotency |
|
FOXA2 |
forkhead box A2 |
regulate both the birth of dopamine neurons and their spontaneous death, two major goals of regenerative medicine. |
|
LMX1A |
LIM homeobox transcription factor 1 alpha |
Lmx1a and Lmx1b function cooperatively to regulate proliferation, specification, and differentiation of mDA progenitors. Midbrain dopaminergic (mDA) neurons have diverse roles in regulating motor and cognitive functions |
|
NEUROD1 |
neuronal differentiation 1 |
essential for the survival and maturation of adult-born neurons |
|
ISL2 |
ISL LIM homeobox 2 |
multiple aspects of motor neuron development, including motor neuron cell body localization, motor column formation, and axon growth. and is required for survival of cranial ganglia neurons. |
|
NEUROG1 |
neurogenin 1 |
Neurog1 and Neurog2 coordinately regulate development of the olfactory system |
|
PITX3 |
paired like homeodomain 3 |
is important for the differentiation and maintenance of midbrain DA neurons during development |
|
GATA2 |
GATA binding protein 2 |
necessary and sufficient to activate the transcription factors Lmx1b and Pet1, and to induce 5-HT neurons |
|
GATA3 |
GATA binding protein 3 |
associated with early motor neuron and interneuron precursors |
|
POU3F2 |
POU class 3 homeobox 2 |
Sufficient to Convert Astrocytes into Neural Progenitors and Neurons |
|
LHX3 |
LIM homeobox 3 |
generation of locomotion, motor neurons and V2 interneurons. |
|
PHOX2B |
paired like homeobox 2B |
regulates neuronal maturation in the brain stem nuclei associated with cardiorespiratory function and in the autonomic sympathetic and enteric nervous system |
|
CAMK2D |
Calcium/calmodulin-dependent protein kinase type II delta chain |
Calcium signaling is crucial for several aspects of plasticity at glutamatergic synapses. |
|
KL |
Klotho |
tole in human brain aging. |
|
TERT |
telomerase |
preserves neuron survival and cognition |
To further understand the direct interactions among neuronal TFs, we analyzed gene networks using Pathway Studio software. Figure 2 includes protein-protein interaction maps and shows a closed feedback loop network. A complete list of cellular processes regulated by expressed genes is available in the Supplementary Material. The top five gene processes regulated by the neuronal gene set (p < e−15) were neuron development, neuron differentiation, nervous system development, stem cell differentiation, and nerve cell differentiation (Table 4).
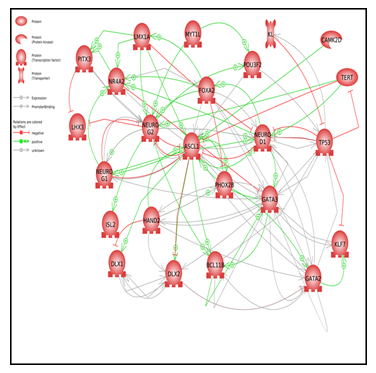
Figure 2: Pathway Studio analysis network of neuronal TFs
Table 5: q-RT-PCR raw data and TF fold changes
3.2 qRT-PCR analysis of TF expression in fibroblasts treated with Metadichol®
We performed qRT-PCR of all genes listed in Table 3 in fibroblasts treated with Metadichol®. All TFs in Table 3 were detectable, with varying changes in expression. Most TFs showed multi fold increases after treatment with Metadichol®. Down regulation was seen for protein 53 (TP53), neuronal differentiation 1 (NEUROD1), LIM homeobox 3 (LHX3), and myelin transcription factor 1 (MYT1L). Only marginal increases were observed for nuclear receptor subfamily 4, group A (NR4A2), LIM homeobox transcription factor 1 alpha (LMXA1), GATA binding protein 2 (GATA2), and distal-less homeobox 1 (DXL1).
4. Discussion
Downregulation of TP53 expression has been shown to increase the efficiency of iPSC production [30]; inactivation of the p53 pathway is a prerequisite for successful reprogramming of somatic cells into neuronal or other cell types [31]. Klotho gene (KL) expression increased 10-fold after treatment with 1 ng Metadichol®. Klotho, a type 1 transmembrane protein, has been shown to inhibit TP53 and regulate cellular senescence by repressing the p53/p21 pathway [32]. Thus, klotho prevents aging in primary human fibroblasts [33] and is neuroprotective. Maintaining Klotho levels by stimulating endogenous Klotho production may [34] be a potential therapeutic target to slow the incidence of age-related diseases.
Some TFs whose expression increased or decreased after treatment with Metadichol® play critical roles in neuronal processes and disease. For instance, NEUROD1 is expressed in aggressive neuroendocrine tumors, malignant melanoma, and prostate cancer cell lines [35]. Moreover, it is a therapeutic target for aggressive neuroendocrine cancers expressing NEUROD1 [36]. Moreover, paired-like homeobox 2b (PHOX2b) expression increased over 10-fold after treatment with 100 pg (pico gram). Metadichol®; PHOX2b has been shown to suppress neuroblastoma progression [37,38,39]. Kinesin family member 7 (KIF7) exhibited a 14-fold increase in expression after treatment with 100 ng Metadichol®. KIF7 maintains normal brain function; recent work [40] suggests that it plays a crucial role in the pathogenesis of stroke, providing vascular and neuronal protection in cases with an ischemic brain. BHLH transcription factor 1 (ASCL1) exhibited a 24-fold increase after 1 ng of treatment. ASCL1 levels are low in glioblastoma (GBM) tumors [41]; thus, increasing ASCL1 expression plays a role in controlling GBM. [42] ASCL1 induces the expression of TFs responsible for the differentiation of dopaminergic neurons, such as POU class 3 homeobox 2 (POU3F2) and forkhead box A2 (FOXA2), both of which exhibited a sixfold increase in expression after treatment with 1 ng Metadichol®. Moreover, FOXA2 plays a crucial role in the birth and death of dopaminergic neurons and in the development of PD [43], as the gradual loss of dopaminergic neurons is a hallmark of this disease. The pituitary homeobox 3 (PITX3) gene showed a sixfold increase in expression with 1 ng of Metadichol® treatment and has a similar role to FOXA2 in the differentiation and maintenance of midbrain neurons [44]. Calcium/calmodulin-dependent protein kinase 2 (CAMK2D) exhibited a nearly eightfold increase in expression after 1 ng of Metadichol® treatment. As CAMK2D can phosphorylate itself, its activity is independent of Ca2+/calmodulin and thus helpful for the formation of long-term memory [45]. GATA2 (twofold increase at 100 ng) increases the yield of serotonergic neurons [46]. POU3F2 expression increased sixfold after 1 ng of Metadichol® treatment; POU3F2 has been shown to regulate the gene coexpression network involved in schizophrenia and bipolar disorders [47]. FOXA2 [48] activates serotonergic neural differentiation pathways, leading to a 10-fold increase in serotonin production. GATA3 exhibited a sixfold increase with 100 ng of Metadichol® treatment; it enhances the neurogenic potential [49] of primary human astrocytes but cannot induce neurogenesis on its own. The expression of neurogenin 2 (NEUROG2) increased nearly threefold after treatment with 100 ng Metadichol®. Its absence halts neurogenesis [50]. BAF chromatin remodeling complex subunit (BCL11b) expression increased fivefold after treatment with Metadichol®. BCL11b is a TF with a crucial role in fetal development, as it is necessary for the differentiation and development of neuronal subtypes in the central nervous system [51]. Heart and neural crest derivative-expressed protein 2 (HAND2) exhibited a twofold increase in expression with 1 ng of Metadichol® treatment. HAND2 is involved in neurogenesis as well as in the expression of cell-specific markers in the enteric nervous system [52]. Neurogenin 1 (NEUROG1) is a TF important for sensory neuron development; it affects hair cells, having a minor impact on cochlear nuclei development [53]. Its expression increased 16-fold after treatment with 1 ng Metadichol®. ISL LIM Homeobox 2 (ISL2) exhibited an 11-fold increase after treatment with 1 ng Metadichol®. ISL2 is mainly expressed in primary sensory and motor neurons and is essential for the acquisition of motor neuron identity [54]. Telomerase (TERT) expression increased sixfold after treatment with 1 ng Metadichol®. Telomerase promotes neurogenesis [55] during neural injury and repair in patients with various diseases affecting the nervous system. Telomerase and Klotho expression in the brain can ameliorate brain aging and neurodegenerative diseases such as AD and PD [56,57,58].
4.1 Vitamin C and oxidative stress in fibroblasts treated with Metadichol®
A recent review points out the importance of ascorbate (vitamin C) and its vital role in the brain [59]. Ascorbate (vitamin C) accumulates at the highest level in brain tissues, at concentrations between 2-10 µM, while blood levels are only 40-60 mM [60]. Ascorbate is a neuromodulator of glutamine, dopamine, cholinergic (61) and GABA. Ascorbate is a cofactor in catecholamine and collagen production and is involved in the regulation of hypoxia-induced factor-1α [62.63]. Ascorbate has been shown to spare/recycle α-tocopherol in lipid bilayers [64] and erythrocytes [65].
Our previous work showed that oral administration of Metadichol® increases ascorbate levels in humans [66,67]. Dehydroascorbic acid (DHA) is the oxidized form of ascorbate and can be transported via glucose transporters of the GLUT family, such as GLUT 4. Metadichol® increases GLUT4 expression [68], suggesting that Metadichol® can enhance the ability of cells to import DHA, and once inside, it is rapidly reduced to ascorbate.
Oxidative stress in the brain is a leading cause of neurodegenerative diseases [69]. Neurons are greatly affected by ascorbate deficiency, possibly due to their 10-fold higher oxidative metabolism than other cell types [70]. Ascorbate is important for neuronal maturation and function and for protecting the brain against oxidative stress [71]. AD patients have lower ascorbate levels in plasma [72] and CSF [73] despite adequate nutritional intake. Ascorbate use was found to reduce disease incidence [74,75] and disease-related oxidative stress markers [76]. Inclusion of ascorbate in cell media could efficiently enhance the generation of mouse embryonic stem cells and iPSCs from mouse or human fibroblasts [77] compared to other antioxidants. Ascorbate might act by promoting the expression of particular genes. For example, ascorbate can facilitate cell fate reprogramming by increasing histone demethylation [78]. Moreover, ascorbate has been shown to increase the embryonic stem cell population in humans, leading to DNA demethylation at genomic loci known to undergo widespread methylation loss during reprogramming of somatic cells into iPSCs [79,80].
The findings provided in this study demonstrate that the administration of Metadichol leads to the activation of several genes associated with neuronal function. It is not possible for a single small molecule to activate all genes associated with neuronal function. There is a higher probability that Metadichol activates up field genes in the pathway, which subsequently triggers the activation of a cascade of downstream genes. Our recent study has demonstrated that Metadichol exhibits the expression of all nuclear receptors in both stem cells and fibroblast cells [81]. Nuclear receptors exhibit selective expression among neuronal populations [82].
Retinoic acid receptors (RARs) and Retinoid X receptors (RXRs) are two types of nuclear receptors that play crucial roles in mediating the effects of retinoic acid, a derivative of vitamin A. The activation of these nuclear receptors occurs by the binding of retinoic acid, which is a product of vitamin A. The involvement of retinoic acid signaling in neuronal differentiation and maturation has been demonstrated to be significant [83]. The RXRA protein plays a crucial role in the signaling pathway of retinoic acid (RA), governing the production of genes and the functioning of neurons across different brain areas. The RXRA gene [84] plays a crucial role in the proper development and functioning of the nervous system, as well as in the manifestation of behavioral abnormalities. The involvement of retinoic acid in the process of neural development is somewhat facilitated by its ability to regulate neurogenic transcription factors, such as NGN3 and PAX6 these factors are fundamental in the differentiation and development of various neuronal cell types.
Orphan nuclear receptors, namely TLX (NR2E1) and Nurr1 (NR4A2), have demonstrated their involvement in the processes of neural stem cell self-renewal and differentiation [85,86]. Although the direct regulation of NGN3 and PAX6 by these receptors is not established, their involvement in wider networks of neuronal growth and differentiation has been implicated.
Thyroid B Nuclear receptor (THRB) plays a significant role in the modulation of neuronal excitability. The activation of THRB has been demonstrated to enhance the transcription of genes implicated in the synthesis of neurotransmitters [87,88].
The nuclear receptor GR is involved in the response to stress. GR activation has been shown to decrease the expression of genes that are involved in learning and memory [89,90].
Our results demonstrate that Metadichol® enhanced the expression of many TFs associated with neuronal regulation, as identified by Pathway Studio software Interactions between neuronal factors affect processes such as neuron differentiation, maintenance of neurogenesis pluripotency, cellular senescence, and stem cell development (Figure 2). The identified genes have more interactions among themselves than expected for a random set of genes of the same size and degree distribution drawn from the genome. Such enrichment indicates that these genes share a significant biological connection through their involvement in neuronal development.
Our work was triggered by post marketing field reports from customers using Metadichol® over the last 5 years. This result suggested that symptoms in neurological syndromes such as attention-deficit hyperactivity disorder (ADHD), bipolar disorder, and autism spectrum disorders improved. Our results suggest that future work should focus on directly testing the effects of Metadichol® on patients with neurological diseases, given that it is nontoxic and has no known side effects [91,92,93]. Metadichol®, as we have shown, acts on multiple targets or disease pathways by activating nuclear receptors and and lead to a tightly bound gene network that can bring about changes in neurodegenerative diseases where such a need exists.
Acknowledgments:
We thank Dr Michelle Muller Sferalp S.A Switzerland for helpful discussions
Supplementary Files Enclosed.
- Western Blots
- Raw Q-Rt-PCR data
- Biological processes regulated by the expressed gene sets.
References
- Erkkinen MG, Geschwind MD, Kim MO, et al. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb Perspect Biol 10 (2018): 20.
- Bayreuther K, Rodemann HP, Hommel R, et al. Human skin fibroblasts in vitro differentiate along a terminal cell lineage. Proc Natl Acad Sci USA 85 (1988): 5112-
- Pang ZP, Yang N, Vierbuchen T, et al. Induction of human neuronal cells by defined transcription factors. Nature 476 (2011): 220-223.
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adultfibroblast cultures by defined factors. Cell 126 (2006): 663-
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (2007): 861-872.
- Dimos JT, Rodolfa KT, Niakan KK, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Sci 321 (2008): 1218-
- Jin J, Kwon YW, Paek JS, et al. Analysis of differential proteomes of induced pluripotent stem cells by protein-based reprogramming of fibroblasts. J. Proteome Res 10 (2011): 977-
- Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell 134 (2008): 877-
- Qiang L, Fujita R, Yamashita T, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell 146 (2011): 359-
- Nord AS, Pattabiraman K, Visel A, et al. Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85 (2015): 27-147.
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev 26 (2012): 1010-1021.
- Urbán N, Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell Neurosci 8 (2014): 396.
- De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515 (2014): 209-215.
- Dard RF, Dahan L, Rampon C. Targeting hippocampal adult neurogenesis using transcription factors to reduce Alzheimer’s disease associated memory impairments. Hippocampus 29 (2019): 579-586.
- Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington’s disease. Neurobiol. Dis. 31(2018): 298-308.
- Whitton L, Apostolova G, Rieder D, et al. Genes regulated by SATB2 during neurodevelopment contribute to schizophrenia and educational attainment. PLOS Genet 14 (2018):
- Whitton L, Cosgrove D, Clarkson C, et al. Cognitive analysis of schizophrenia risk genes that function as epigenetic regulators of gene expression. Am J Med Genet B Neuropsychiatr Genet 171 (2016): 1170-1179.
- Egorov S, Daraselia N, Mazo Pathway studio--the analysis and navigation of molecular networks. Bioinformatics 19 (2003): 2155-2167.
- SivachenkoA Y, Yuryev A, Daraselia N, et al. Molecular networks in microarray analysis. J Bioinform Comput Biol 5 (2007): 429-456.
- Raghavan PR, Policosanol Nanoparticles, US patent 8,722093 B2 (2014).
- Pang ZP, Yang N, Vierbuchen T, et al. Induction of human neuronal cells by defined transcription factors. Nature 476 (2011): 220-223.
- Pfisterer U, Kirkeby A, Torper O, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 108 (2011): 10343-
- Son EY, Ichida JK, Wainger BJ, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9 (2011): 205-
- Yoo AS, Sun A X, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476 (2011): 228-
- Vierbuchen T, Giannelli S, Valente P, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463 (2010): 1035-
- Caiazzo M, Dell'Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476 (2011): 224-
- Thier M, Wörsdörfer P, Lakes YB, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10 (2012): 473-
- Han DW, Tapia N, Hermann A, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10 (2012): 465-
- Kim J, Efe JA, Zhu S, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A 108 (2011): 7838-7843.
- Yi L, Lu C, Hu W, et al. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res 72 (2012): 5635-5645.
- Zhao T, Xu Y. p53 and stem cells: new developments and new concerns. Trends Cell Biol 20 (2013): 170-175.
- Shaker MR, Aguado J, Chaggar HK, et al. Klotho inhibits neuronal senescence in human brain organoids. npj Aging Mech Dis 7 (2021): 18.
- De Oliveira Klotho RNAi induces premature senescence of human cells via a p53/p21 dependent pathway. FEBS Lett 580 (2006): 5753-5758.
- Torbus-Paluszczak M, Bartman W, Adamczyk-Sowa M. Klotho protein in neurodegenerative disorders. Neurol Sci 39 (2018): 1677-1682.
- Osborne JK, Larsen JE, Gonzales JX, et al. NeuroD1 regulation of migration accompanies the differential sensitivity of neuroendocrine carcinomas to TrkB inhibition. Oncogenesis 2 (2013): 63.
- Osborne JK, Larsen JE, Shields MD, et al. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas by signaling molecules TrkB and NCAM. Proc Natl Acad Sci U S A 110 (2013): 6524-6529.
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 3 (2003): 203-216.
- Maris JM, Matthay KK. Molecular biology of neuroblastoma. J Clin Oncol 17 (1999): 2264-2279.
- Ke-Jie Y, Hamblin M, Yanbo F, et al. Krüpple-like factors (KLF’s) in the central nervous system: novel mediators in Stroke Brain Dis 30 (2015): 401-410.
- Xu Z, Chu X, Jiang H, et al. Induced dopaminergic neurons: A new promise for Parkinson’s diseaseRedox. Redox Biol 11 (2017): 606-
- Vue TY, Kollipara R K, Borromeo MD, et al. ASCL1 regulates neuro developmental transcription factors and cell cycle genes in brain tumors of glioma mouse models. Glia 68 (2012): 2613-
- Narayanan A, Gagliardi F, Gallotti AL, et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ 9 (2019): 1813-1831.
- Kittappa R, Chang WW, Awatramani RB, et al. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLOS Biol 5 (2007): 325.
- Li J, Dani JA, Le W. The role of transcription factor Pitx3 in dopamine neuron development and Parkinson’s Curr Top Med Chem 9 (2009): 855-859.
- Zalcman G, Federman N, Romano CaMKII isoforms in learning and memory: localization and function. Front Mol Neurosci 11 (2018): 445.
- Haugas M, Tikker L, Achim K, et al. Gata2 and Gata3 regulate the differentiation of serotonergic and glutamatergic neuron subtypes of the dorsal raphe. Development 143 (2016): 4495-4508.
- Chen C, Meng Q, Xia Y, et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med 10 (2018): 8178.
- Nefzger CM, Haynes JM, Pouton CW. Directed expression of Gata2, Mash1, and Foxa2 synergize to induce the serotonergic neuron phenotype during in vitro differentiation of embryonic stem cells. Stem Cells 29 (2011): 928-939.
- Celikkaya H, Cosacak MI, Papadimitriou C, et al. GATA3 promotes the neural progenitor state but not neurogenesis in 3D traumatic injury model of primary human cortical astrocytes. Front Cell Neurosci 13 (2019): 23.
- Hufnagel RB, Le TT, Riesenberg AL, et al. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol 340 (2010): 490-
- Lennon MJ, Jones SP, Lovelace MD, et al. Bcl11b-A Critical Neurodevelopmental Transcription Factor-Roles in Health and Disease. Front Cell Neurosci 11 (2017): 89.
- Hendershot TJ, Liu H, Sarkar A A, et al. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn 236 (2007): 93-
- Elliott KL, Pavlinkova G, Chizhikov VV, et al. Neurog1, Neurod1, and Atoh1 are essential for spiral ganglia, cochlear nuclei, and cochlear hair cell development. Fac Rev 10 (2021): 47.
- Lee H, Kim M, Kim N, et al. Slit and semaphorin signaling governed by Islet transcription factors positions motor neuron somata within the neural tube. Exp Neurol 269 (2015): 17-27.
- Li J, Liu HT, Zhao J, et al. Telomerase reverse transcriptase (TERT) promotes neurogenesis after hypoxic-ischemic brain damage in neonatal rats. Neurol Res 10 (2022): 1-
- Shim HS, Horner JW, Wu CH, et al. Telomerase reverse transcriptase preserves neuron survival and cognition in Alzheimer’s disease models. Nat Aging 1 (2021): 1162-1174.
- Saretzki G, Wan T. Telomerase in brain: the new kid on the block and its role in neurodegenerative diseases. Biomedicines 9 (2021): 490.
- Torbus-Paluszczak M, Bartman W, Adamczyk-Sowa M. Klotho protein in neurodegenerative disorders. Neurol Sci 39 (2018): 1677-1682.
- Coker SJ, Smith-Díaz CC, Dyson RM, et al. The Epigenetic Role of Vitamin C in Neurodevelopment. Int J Mol Sci 23 (2022): 1208.
- .Dhariwal KR, Hartzell WO, Levine M. Ascorbic acid and dehydroascorbic acid measurements in human plasma and serum. Am J Clin Nutr 54 (1991): 712-716.
- Rebec GV, Pierce RC. A vitamin as neuromodulator: ascorbate release into the extracellular fluid of the brain regulates dopaminergic and glutamatergic transmission. Prog Neurobiol 43 (1994): 537-565.
- Kuiper C, Dachs GU, Currie MJ, et al. Intracellular ascorbate enhances hypoxia-inducible factor (HIF)-hydroxylase activity and preferentially suppresses the HIF-1 transcriptional response. Free Radic Biol Med 69 (2014): 308-317.
- Dillon PF, Root-Bernstein RS, Lieder CM. Antioxidant-independent ascorbate enhancement of catecholamine-induced contractions of vascular smooth muscle. Am J Physiol Heart Circ Physiol 286 (2004): 2353-2360.
- Niki E, Noguchi N, Tsuchihashi H, et al. Interaction among vitamin C, vitamin E, and β-carotene. Am J Clin Nutr 62 (1995): 1322-1326.
- May JM, Qu ZC, Mendiratta S. Protection and recycling of α-tocopherol in human erythrocytes by intracellular ascorbic acid. Arch Biochem Biophys 349 (1998): 281-289.
- Raghavan Metadichol® induced high levels of vitamin C: case studies. Vitam Miner 6 (2017): 169.
- Raghavan PR. Metadichol® and vitamin C increase in vivo, an open-label study. Vitam Miner 6 (2017): 163.
- Raghavan Metadichol®, vitamin C and GULO gene expression in mouse adipocytes. Biol Med (Aligarh) 10 (2018): 426.
- Halliwell B. Oxidative stress and neurodegeneration: where are we now? J. Neurochem 97 (2006): 1634-1658.
- Moretti M, Fraga DB, Rodrigues ALS. Preventive and therapeutic potential of ascorbic acid in neurodegenerative diseases. CNS Neurosci Ther 23 (2017): 921-929.
- Margaret Rice A. Ascorbate regulation and its neuroprotective role in the brain, Trend Neurosci 23 (2005): 209-216.
- Rivière S, Birlouez-Aragon I, Nourhashémi F, et al. Low plasma vitamin C in Alzheimer patients despite an adequate diet. Int J Geriatr Psych 13 (1998): 749-754.
- Quinn JF, Montine K S, Moore M, et al. Suppression of longitudinal increase in CSF F2-isoprostanes in Alzheimer’s disease. J Alzheimers Dis 6 (2004): 93-97.
- Morris MC, Beckett LA, P A Scherr, et al. Vitamin E and vitamin C supplement use and risk of incident Alzheimer disease. Alzheimer Dis Assoc Disord 12 (1998): 121-126.
- Engelhart MJ, Geerlings MI, Ruitenberg A, et al. Dietary intake of antioxidants and risk of Alzheimer disease. JAMA 287 (2002): 3223-3229.
- Harrison FE, May JM. Vitamin C function in the brain: vital role of the ascorbate transporter SVCT2. Free Radic. Biol Med 46 (2009): 719-730.
- Esteban MA, Wang T, Qin B, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6 (2010): 7179-
- Cloos PA, Christensen J, Agger K, et al. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22 (2008): 1151140-114075.
- Chung TL, Brena RM, Kolle G, et al. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 28 (2010): 1848-1855.
- Chong, Ahearn EL, Cimmino L, et al. Reprogramming the epigenome with vitamin C. Front Cell Dev Biol 16 (2019): 128.
- Raghavan PR. Metadichol® is a nano lipid emulsion that expresses all 48 nuclear receptors in stem and somatic cells T (2022).
- Gkikas D, Moreno-Ramos AL, Haider NB, et al. The role of nuclear receptors in maintaining homeostasis of brain stem/progenitor cells. Cell Mol Life Sci 74 (2017): 4097-4120
- Norbert B. Ghyselinck Gregg Duester. Retinoic acid signaling pathways. Development 146 (2019): 167502.
- Cao H, Li MY, Li G, et al. Retinoid X Receptor α Regulates DHA-Dependent Spinogenesis and Functional Synapse Formation In Vivo. Cell Rep 31 (2020): 107649.
- Kim JY, Koh HC, Lee JY, et al. The process of dopaminergic neuronal differentiation from rat embryonic neural progenitors is facilitated with the over expression of Nurr1. J Neurochemist 85 (2003): 1443-1454.
- Jakaria M. Molecular Neurobiology 56 (2019): 5799-5814.
- Bernal J. Thyroid hormone receptors in brain development and function. Nat Clin Pract Endocrinol Metab 3 (2007): 249-59.
- Lin C, Li N, Chang H, et al.Dual effects of thyroid hormone on neurons and neurogenesis in traumatic brain injury. Cell Death Dis 671 (2020).
- Wang B, Xin N, Qian X. et al. Ahi1 regulates the nuclear translocation of glucocorticoid receptor to modulate stress response. Transl Psycharity 11(2021): 188.
- Tatro ET, Everall IP, Kaul M, et al. Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res 1286 (2009): 1- 12.
- Alemán CL, Más R, Hernández C, et al. A 12-month study of policosanol oral toxicity in Sprague Dawley rats.” Toxicol Lett 70 (1994): 77-87.
- Alemán CL, Ferreiro RM, Puig MN, et al. “Carcinogenicity of policosanol in Sprague Dawley rats: a 24 month study.” Teratog. Carcinog. Mutagen 14 (1994): 239-249.
- Alemán CL, Puig MN, Elías EC, et al. “Carcinogenicity of policosanol in mice: an 18-month study.” Food Chem Toxicol 33 (1995): 573-578.


 Impact Factor: * 4.2
Impact Factor: * 4.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 