Kaleidoscope: A Bioinformatics Web Application for In Silico Exploration of Omics Data
Article Information
Khaled Alganem1#, Ali S. Imami2#, Smita Sahay2#, Hunter Eby2, Taylen O. Arvay2, Mackenzie Abel2, Xiaolu Zhang2, William Brett McIntyre3,4, Jiwon Lee3, Christy Au- Yeung3, Roshanak Asgariroozbehani3,4, Roshni Panda3, Rammohan Shukla2, Sinead M. O'Donovan2, Margaret Hahn3-7, Jarek Meller8-12, Robert McCullumsmith2,13*
1Almar Biosciences Incorporated, Fremont, California, USA.
2Department of Neurosciences, University of Toledo College of Medicine, Toledo, Ohio, USA.
3Centre for Addiction and Mental Health, Toronto, Ontario, Canada.
4Institute of Medical Sciences, University of Toronto, Toronto, Ontario, Canada.
5Department of Psychiatry, University of Toronto, Toronto, Ontario, Canada.
6Banting and Best Diabetes Centre, Toronto, Ontario, Canada.
7Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, Canada.
8Division of Biomedical Informatics, Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio, USA. 9Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH, USA 10Department of Environmental Health, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA.
11Department of Electrical Engineering and Computing Systems, University of Cincinnati College of Medicine,
Cincinnati, OH, USA.
12Department of Informatics, Nicolaus Copernicus University, Torun, Poland.
13Neurosciences institute, ProMedica, Toledo, Ohio, USA.
#Authors contributed equally.
Corresponding Author: Robert McCullumsmith, Deptartment of Neurosciences, University of Toledo College of Medicine and Life Sciences, 3000 Arlington Ave. Mail Stop 1007, Block Health Science Building, Toledo, OH 43614
Received: 12 September 2023; Accepted: 19 September 2023; Published: 28 November 2023;
Citation: Khaled Alganem, Ali S. Imami, Smita Sahay, Hunter Eby, Taylen O. Arvay, Mackenzie Abel, Xiaolu Zhang, William Brett McIntyre, Jiwon Lee, Christy Au-Yeung, Roshanak Asgariroozbehani, Roshni Panda, Rammohan Shukla, Sinead M. O'Donovan, Margaret Hahn, Jarek Meller, Robert McCullumsmith. Kaleidoscope: A Bioinformatics Web Application for In Silico Exploration of Omics Data. 6 (2023): 327-338.
View / Download Pdf Share at FacebookAbstract
Background: Exploring research questions through in silico analysis of genomic, transcriptomic, and/or proteomics (omics) data is an essential initial phase of understanding the pathophysiology of medical disorders and requires the use of bioinformatics. Currently, numerous publicly available bioinformatics tools present appealing features to facilitate this endeavor; however, applications are constrained by outdated user interfaces that are difficult to use. Thus, a streamlined, user-friendly, data exploration platform is pertinent in effectively investigating medical disorders and treatment options in silico.
Results: We developed an R Shiny web application called Kaleidoscope to address this challenge. The application offers access to several omics databases and bioinformatics tools to analyze these data, allowing users the ease and flexibility to explore research questions in silico. The application is straightforward to use with a unified interface and offers the ability to upload user-defined datasets. We demonstrate the application features such as building protein-protein interaction networks, generating gene expression and enrichment data across cell-subtypes in the brain, and curating the top differentially expressed genes across schizophrenia versus control transcriptomic datasets with a starting query of the DISC1 (Disrupted in schizophrenia 1) gene.
Conclusion: Kaleidoscope provides easy access to several bioinformatics databases under a unified user interface to explore biomedical research questions in silico. Currently, the application focuses on neuropsychiatric disorders; however, with the flexibility of uploading user-defined datasets, the capability of answering biomedical questions surrounding any medical disorder exists. The web application is opensource and freely available at https://cdrl.shinyapps.io/Kaleidoscope/. In conclusion, Kaleidoscope streamlines the process of in silico data exploration and expands the efficient use of omics tools to stakeholders without specific bioinformatics expertise.
Keywords
Kaleidoscope, Bioinformatics, In Silico, Omics, Exploration, Database, Integration, R Shiny. Application.
Kaleidoscope articles; Bioinformatics articles; In Silico articles; Omics articles; Exploration articles; Database articles; Integration articles; R Shiny. Application articles.
Article Details
1. Introduction
Large biological omics datasets are continuing to be deposited into publicly available repositories [1]. In conjunction, an ever-increasing number of bioinformatics tools are being developed to process, analyze, and view a spectrum of biological datasets [2]. Although meant to be helpful, the rapidly growing availability of bioinformatics databases is an impediment for scientists, often hindering discovery, especially for users who are not well versed in the bioinformatics field [3, 4]. We have observed that many bioinformatics tools and databases relevant to investigating biomedical disorders remain underutilized. While a few of these tools are widely recognized such as Bioconducter [5], BioJava [6], and SAMtools [7], some researchers avoid utilizing them due to their extensive scope or intricate user interfaces, as they may be intimidating to novice users.
To address this gap in the field, we developed Kaleidoscope: an interactive R Shiny web application. Kaleidoscope is an easy-access, open source, user-friendly platform designed for in silico data exploration of biological datasets through the BrainRNA-Seq, STRING, iLINCS, BrainCloud, GTEx, BrainAtlas, GWAS, and IDG databases (Fig 1). Additionally, our platform contains over 200 disease-related differential gene expression datasets readily available for analysis under the "Lookup" feature. Integrating multiple datasets and databases in a single platform facilitates the user's interactive exploration of diverse molecular biology and biomedical research questions. This data exploration tool has already yielded intriguing observations that supplement existing hypotheses, generate new ones, and direct future studies [8-11].
Our exploratory, interactive, omics data analysis platform is thoughtfully designed with a clean, intuitive interface to cater to a wider range of investigators who may not be well-acquainted with these specialized tools. We leverage application programing interfaces (APIs) to seamlessly access and harmonize omics data and present it via meaningful visualizations. Additionally, the application offers the capability of uploading user- defined datasets, accommodating the specific research interests of each user. This feature sets our tool apart from others, as the process of discovering, curating, formatting, and analyzing datasets can be both time- consuming and require specialized expertise. By streamlining our application to simplify the data acquisition and formatting stages, we enable investigators to focus on their research questions, sparing them unnecessary delays in preceding data processing steps.
As the community of end-users continues to expand, we anticipate a sizable expansion in the number and type of datasets hosted on our platform, ultimately increasing the breadth of data available for exploration as well as a collaborative environment where researchers from different disciplines may contribute to and benefit from the collective knowledge pool. Researchers may expect an ever-growing catalog of datasets encompassing an array of diseases, tissues, and experimental conditions, enhancing the platform′s utility for hypothesis-driven investigations. In essence, Kaleidoscope embodies the intersection of cutting-edge bioinformatics and user-centric design, aimed at empowering scientists, regardless of their background. As we look ahead, we are committed to continually enhancing the platform's capabilities, refining its user experience and creating a community of diverse researchers dedicated to data-driven discovery. In the subsequent sections of this paper, we provide a comprehensive overview of Kaleidoscope's features, functionalities, and practical applications to illustrate its potential to catalyze innovative medical research.
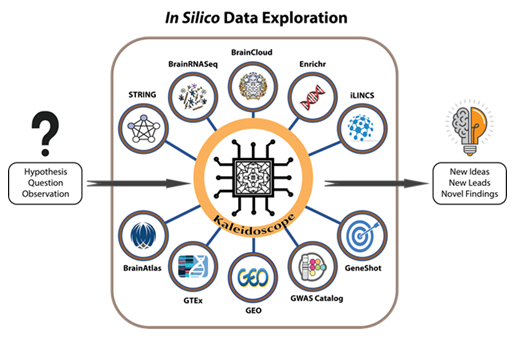
Figure 1: A workflow model for using Kaleidoscope to perform in silico exploratory data analyses. Kaleidoscope integrates multiple bioinformatic tools to streamline the process of in silico data exploration. Currently, end-users have the flexibility of selecting from any combination of the following databases to answer a specific research question or to generate a hypothesis based on output from Kaleidoscope:
1) BrainRNA-Seq generates cell-specific gene expression data in human and mouse brain tissues.
2) The Search Tool for the Retrieval of Interacting Genes (STRING) provides protein-protein interaction networks. 3) The Integrative Library of Integrated Network-Based Cellular Signatures (iLINCS) provides gene knockdown transcriptional signatures and generates L1000 signatures as input for further exploration within the iLINCS database. 4) The "Lookup" feature contains disease and treatment-specific databases in specified modules and generates heatmaps containing differentially expressed genes within selected datasets. 5) BrainCloud extracts gene expression patterns in the brain across the lifespan of healthy humans. 6) The Genotype-Tissue Expression (GTEx) tool explores tissue-specific gene expression patterns. 7) BrainAtlas assesses gene expression levels in different brain cell types. 8) The Genome-Wide Association Study (GWAS) Catalog is utilized to search for previously published genome-wide association studies. 9) The Illuminating the Druggable Genome (IDG) tool enhances the user's understanding of the druggable genome: the subset of genes and proteins in an organism's genome that has the potential to be targeted by drugs. 10) Finally, the "Report" feature in Kaleidoscope allows users to create one curated and concise report of their exploratory findings.
1.1 Brain RNA-Seq
The Brain RNA-Seq database provides insight into the gene expression profiles within isolated and purified cells from both human and rodent cortical tissue [12, 13]. Our application accesses this database, providing users a comprehensive repository of cell-specific mRNA data. While the original Brain RNA-Seq database offers its own web interface, its functionality is limited to individual gene searches. In contrast, our platform permits the inquiry of multiple genes simultaneously, with the added benefit of visually displaying gene expression ratios across all cortical cell types. The BrainRNA-Seq tool within our platform allows for the exploration of target genes across distinct brain cell types, ultimately providing insight on the role of cell-specific functions. Investigators also have the capability to compare expression profiles between humans and rodent brain tissues, further enhancing the versatility and utility of our platform for neuroscience research.
1.2 Search Tool for the Retrieval of Interacting Genes (STRING)
STRING is a comprehensive database and widely used resource for the exploration and analysis of known and predicted protein-protein interactions (PPIs) and functional associations among proteins [14]. This tool is designed to help researchers understand how proteins interact within biological systems and the roles they play in cellular processes, pathways, and diseases. On the back end, the PPI networks are built based on computation prediction models, knowledge transfer between organisms, and aggregated interactions from other databases. This tool within our application allows users to input one or a few genes of interest and retrieve entire PPI networks, ultimately providing deeper knowledge regarding the functional properties of and relationships among a set of genes. Furthermore, our tool utilizes the STRING Representational State Transfer Application Programming Interface (REST API) on the back end allowing users to seamlessly access and manipulate data such as specifying the stringency of the predicated associations by selecting the following: an appropriate score cutoff, the desired number of connected nodes (proteins), and the desired organism. The score refers to confidence scores assigned to PPI networks generated by STRING, helping users assess the reliability of each interaction. Our application efficiently communicates the complex results from STRING by displaying the PPI network, a figure legend providing context for the different edges in the output network, a table listing all proteins in the network with a brief description of each, and the confidence score for the network [15].
1.3 The Library of Integrated Network-Based Cellular Signatures (iLINCS)
The iLINCS resource and multi-omics profiling database focuses on integrating and providing access to cellular signature data. Cellular signatures refer to the profiles of gene expression, protein abundance, and other cellular responses to various perturbations or treatments [16]. For transcriptional datasets, iLINCS utilizes a high-throughput gene expression profiling technology known as the LINCS 1000 (L1000). The L1000 was developed to efficiently measure the expression levels, displayed as relative log2-fold change (LFC) values, of a core set of 978 landmark genes. These landmark genes are a reduced representation of the full transcriptome, and the LFC values reflect the expression of the genes under a specified treatment conditions: gene knockdown, gene over-expression, and drug treatments [17]. This reduced representation of the transcriptome allows for cost-effective, scalable gene expression profiling across a range of experimental conditions.
Kaleidoscope allows users to generate L1000 signatures across any user-specified datasets for any treatment condition. The L1000 signature is available from Kaleidoscope as a tab delimited file and may be uploaded to the integrative LINCS (iLINCS) portal to perform perturbagen connectivity analysis. Additionally, for a specific set of user-defined genes, the module will return a table with each knockdown and overexpression signature available per gene from the iLINCS database, specified by the signature ID. Cell lines and direct links to the signature web page on the iLINCS web portal are also available in this table. Additionally, the user may query genes within the module to generate figures displaying the number of gene knockdown and/or overexpression signatures available in iLINCS as well as the number of signatures per organ type (e.g., lung, liver, kidney, etc.).
1.4 Lookup
The "Lookup" tool within our web application hosts various modules, including many neuropsychiatric disorders, each containing numerous datasets that have been meticulously curated from the Gene Expression Omnibus (GEO): an open-source functional genomics data repository [1], allowing end users the capability of understanding the differential expression of genes queried within datasets of interest. The current modules in Lookup include schizophrenia, major depressive disorder, asthma, dopamine signaling, antipsychotics, antidepressants, post-traumatic stress disorder, Alzheimer's disease, insulin signaling inhibition, bipolar disorder, ketosis, aging, microcystin, renal associated databases, and myositis. The curated datasets cover several substrates, including stem cells, postmortem brain, and animal models. At present, the application has over 200 datasets. Prior to being placed in their respective modules, datasets were processed using well-established differential expression analysis R packages. Specifically, limma was used to process microarray datasets, and edgeR or DESeq2 were used to process RNASeq datasets [18-20]. Datasets were automatically harmonized by calculating the empirical cumulative probabilities of the LFC values of genes within each dataset [21]. This feature of the application is expandable, as user curated datasets may be uploaded and modules may be created to reflect a group of datasets in a specific domain, thus increasing the diversity of disease domains hosted on the platform.
Kaleidoscope displays the results from the "Lookup" tool across three tabs: Summary, Heatmap, and Correlation. The Summary tab displays a table with LFC values and p-values, where each row represents a gene from the input list of genes and each column represents a dataset from the selected module(s). The Heatmap tab visually represents the results from this table, where the x-axis of the figure represents genes, the y-axis represents the dataset in which the gene was differentially expressed, and the colors represent the degree to which a gene is differentially expressed, where red represents upregulation, and blue represents downregulation, based on a cutoff LFC threshold that can be adjusted by the user. These heatmaps utilize unsupervised hierarchical clustering for both genes and datasets. The clustering allows patterns of similar changes of expression across the list of genes and the datasets that were selected to be identified. Finally, the Correlation tab provides a concordance scores matrix, calculated using Spearman correlation analysis based on the LFC values. The user may calculate the concordance scores based on the input list of genes or the full list of genes in the datasets. For each correlation analysis between two datasets, the test is applied using only the observations of genes found in both datasets. The colors in the figure denote the direction of the concordance scores, where red represents negative correlation (high discordance), and blue represents positive correlation (high concordance).
This feature of the software allows the user to lookup expression patterns from publicly available datasets to explore and/or validate interesting patterns of expression changes. For instance, an analysis that returns above average gene expression changes (indicated by increased LFC values) across multiple datasets would strongly indicate that a gene, or a panel of genes, may be driving the disease process. Additionally, the "Lookup" feature may be used to complement a user's own transcriptome and/or wet lab-based study, as they may conduct a quick and easy in silico analysis utilizing the datasets in Lookup that match their disease of interest to assess whether they can replicate or confirm their expression or lab findings [8].
1.5 BrainCloud
The BrainCloud database was developed through a collaboration between the Lieber Institute and The National Institute of Mental Health to give a global perspective on the role of the human transcriptome in cortical development and aging [22]. Specifically, the database explores the temporal dynamics and genetic control of transcription and DNA methylation in the human dorsolateral prefrontal cortex in postmortem tissue. The tissue was collected from 269 subjects without neuropathological or neuropsychiatric disorders. Kaleidoscope accesses this data generated by BrainCloud, allowing users to analyze patterns of expression in the brain for target genes in healthy humans. For each gene of interest, the BrainCloud feature in Kaleidoscope generates a graph showing the normalized expression across the human lifespan, with the x-axis representing age and the y-axis representing expression levels. Four additional graphs are generated allowing users to analyze expression patterns based on the following age ranges: fetal: 14-20 gestational weeks, infant: 0-6 months, child: 1-18 years, and adults: 18-80 years.
1.6 Genotype-Tissue Expression (GTEx)
The GTEx database is a comprehensive atlas of tissue-specific gene expression and regulation data. The database is constructed based on samples that were collected from 54 non-diseased tissues from almost 1,000 individuals, primarily for molecular assays including whole genome and transcriptome sequencing [23]. Its purpose is to allow investigators to understand how genetic variations contribute to gene expression differences across various tissues and how these variations may be linked to disease. The GTEx tool within our application allows the user to query a list of genes to understand their expression levels across all tissues that were analyzed in the GTEx database. Kaleidoscope displays the results in a bar graph format as well as a heatmap, where the median TPM (Transcripts Per Kilobase Million) of each gene in the list is shown based on tissue-type. Users may select to view their data based on the unsupervised hierarchical clustering or log transformation methods. Ultimately, this module allows researchers to easily investigate and visualize tissue-specific expression patterns, identify candidate genes for disease studies, and explore the role of genetic variants in human health and disease.
1.7 BrainAtlas
The Allen Brain Atlas is a collection of online databases that provides detailed information about the gene expression patterns in the mouse and human brain. The human collection contains RNA-Seq data derived from intact nuclei in frozen brain specimens that may be utilized to survey cell type diversity in the human medial temporal gyrus. In total, 15,928 nuclei from 8 human tissue donors ranging in age from 24-66 years were analyzed. Analysis of these transcriptional profiles reveals approximately 75 transcriptionally distinct cell types, subdivided into 45 inhibitory neuron types, 24 excitatory neuron types, and 6 non-neuronal types [24].
Using Kaleidoscope, a list of target genes may be queried across all these clusters of cell-subtypes to examine their enrichment and/or expression levels. The BrainAtlas tool within the application displays a bar graph with expression levels separated by cell-subtype. It also displays a heatmap utilizing unsupervised hierarchical clustering to show the differential expression of the target genes across the cell-subtype clusters. Lastly, the BrainAtlas tool also has a feature to display the log transformed data. Ultimately, users may use this tool to gain an understanding of gene expression patterns and/or enrichment in specific cell-subtypes as well as overall brain development and function.
1.8 Genome-Wide Association Study (GWAS)
The GWAS Catalog is supported by the National Human Genome Research Institute. This is the largest curated collection of all published genome-wide association studies. It currently contains 3,841 publications and 126,603 genetic variant-phenotype associations, and these numbers are constantly increasing [25]. Through our GWAS tool within our application, researchers interested in genetic variants associated with complex traits and diseases in humans may query a gene or a list of genes. Kaleidoscope will display the number of associated SNPs, associated traits, the type of SNP (e.g., intron variant, intergenic variant, 3' UTR variant, regulatory region variant, other variant), and a table with all significant single-nucleotide polymorphisms (SNPs) associated with the input gene(s). The table contains the gene name, risk allele (rsID), chromosome position, studied disease or phenotype, SNP type, p-value, and a direct link to the study. Two distinct figures will also be displayed: the first shows the top overlapping disease/phenotype traits and the second is an interactive Sankey graph that represents the flow rate between the list of genes, SNP type, and disease/phenotype traits.
1.9 Illuminating the Druggable Genome (IDG)
The IDG database, supported by the National Institute of Health, aims to characterize the understanding of the druggable genome: the subset of genes and proteins in an organism's genome that have the potential to be targeted by small molecules, such as drugs, for therapeutic purposes [26]. The database contains targets about which virtually nothing is known. The overall goal of the IDG initiative is to catalyze research in the areas of biology that are currently understudied but have a very high potential to impact human health. For each gene or gene set queried within the IDG module, Kaleidoscope will display its protein structure, development level, publication statistics, disease associations, pathways the gene is involved in, associations between the target and diseases, as well as any associated ligands. Links are also provided so the user may further explore proteins encoded by the gene, possible upstream protein functions, and details regarding cellular pathways the gene may be involved in, ultimately either answering/supplementing the researcher's initial research question, or allowing them to utilize this collective information to form a novel hypothesis.
1.10 Back End Tools: Enrichr and GeneShot
Enrichr and GeneShot are two tools that were developed by The Ma'ayan Laboratory, Icahn School of Medicine at Mount Sinai (27, 28). Enrichr is a tool for performing gene set enrichment analysis across many databases [27]. GeneShot is a search engine for search terms and gene mentions based on arbitrary text queries of published literature [28]. Kaleidoscope integrates the APIs of these tools for gene set enrichment analyses.
1.11 Report
Finally, the "Report" feature of Kaleidoscope allows users to curate one document containing all relevant output for genes of interest. Users may select which data they would like to include in their report by selecting the genes and bioinformatic tools of interest. This report may be downloaded for further interpretation and exploration.
2. Discussion
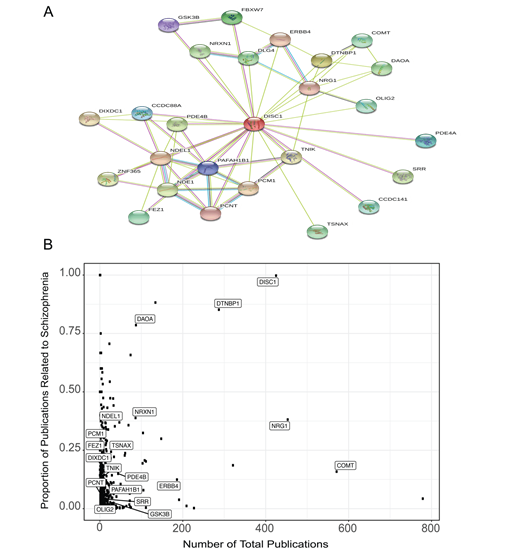
To highlight some of the key capabilities of the features in our application, we conducted an example session commencing with a single gene of interest. This demonstration displays the abundance of information that may be harnessed through Kaleidoscope. Our selection for this exercise was the Disrupted in schizophrenia 1 (DISC1) gene, a prominent candidate gene associated with the risk of developing major mental disorders [29, 30]. Expanding from this single gene, we constructed an intricate PPI network using the STRING tool, specifying "human" as the target species, establishing a score cutoff of 500, and limiting the number of desired nodes to 25 (Fig 2A). This action generated a table presenting succinct gene descriptions, along with association scores under the different criteria (e.g., experimental database, co-expression, text mining etc.) (Supplementary Table 1). Leveraging the gene list extracted from the PPI network, we delved into other tools within Kaleidoscope. Employing GeneShot, we probed the relevance of the disorder "schizophrenia" within the context of our PPI network across publications. Fig 2B elegantly displays both the number of publications referencing each gene and the proportion of publications that also mention "schizophrenia." As expected, the DISC1 query garnered a substantial proportion of publications linked to schizophrenia. Notably, some genes within the network emerged as relatively understudied in terms of their association with schizophrenia, including serine racemase (SSR), oligodendrocyte transcription factor 2 (OLIG2), and glycogen synthase kinase-3 beta (GSK3B).
Figure 2: DISC1-associated protein-protein interaction (PPI) network genes and mentions in the schizophrenia literature. A. The PPI network generated from the Search Tool for the Retrieval of Interacting Genes (STRING) database for DISC1 and its 25 closest associated proteins. B. A scatterplot generated from the GeneShot tool representing the associations between the DISC1-associated PPI network of genes and schizophrenia. The proportion was calculated as the number of papers that mention "DISC1" and "schizophrenia" divided by the number of papers that mention "DISC1."
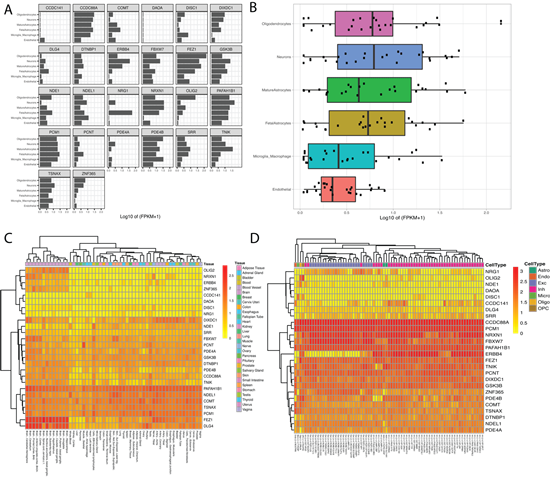
Next, the BrainRNA-Seq tool was employed to explore DISC1 cell-subtype specific gene expression. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) for DISC1 in both human and mice are shown for multiple neuronal cell types (e.g., neurons, astrocytes, endothelial cells, oligodendrocyte progenitor cells) (Figure S1). We additionally queried our gene set extracted from the DISC1-associated PPI network in BrainRNA-Seq to assess cell-subtype specific expression for multiple targets, using the multiple targets input. Fig 3A displays the expression for each gene by cell-subtype. Figure 3B displays the boxplot figure generated when using the multiple target option. We observed higher levels of expression for this network in neurons and oligodendrocytes compared to astrocytes, macrophages, and endothelial cells. Given that oligodendrocytes have been linked to myelin dysfunction and neurocircuitry abnormalities in schizophrenia [31], our results were not surprising.
We continued our demonstration with the BrainAtlas feature to explore the expression profiles of our target genes within various cell types. This feature yielded an informative table and gene expression heatmap for the queried target genes. The table presented the range of count per million (CPM) values for each gene across the various cell types, incorporating all sub-clusters within each cell type. The associated heatmap, generated with unsupervised hierarchical clustering for genes and cell-type sub clusters, revealed the expression levels for each gene by tissue-type (Figure 3C).
In parallel, we queried our genes of interest using the GTEx feature to investigate tissue-specific gene expression levels. Unsurprisingly, samples from brain tissues demonstrated a tendency to cluster together, suggesting a shared gene expression profile among brain tissues compared to other tissue types for our genes of interest. Additionally, many genes of interest in our network including OLIG2, NRXN1 and DLG4 showed enrichment in brain tissues (Figure 3D).
Figure 3: Display of transcription enrichment utilizing Kaleidoscope’s BrainRNA-Seq, GTex, and BrainAtlas tools. A.BrainRNA-Seq results showing expression levels (log2(FPKM+1)) across five different cell types in human brain tissue for each gene of interest. B. BrainRNA- Seq generated boxplot showing the distribution of gene expression levels for the DISC1-associated genes of interest. C. BrainAtlas heatmap highlighting differential gene expression levels in different tissues. D. GTEx heatmap showing gene expression levels across various neuronal cell-subtypes.
Next, we demonstrated the capabilities of the Lookup feature in Kaleidoscope. We assessed the relative expression changes of the same set of DISC-1 associated genes across schizophrenia transcriptional datasets [32-45]. Several figures and tables are displayed, most importantly a heatmap of LFC values and an associated table of gene expression changes across the different datasets (Figure 4A, Supplementary Table 2). The results indicate a clear dysregulation in the expression of our target genes, consistent with previous findings demonstrating their dysfunction in schizophrenia [46-48]. A correlation matrix is also presented that
shows the association between the DISC-1 associated PPI network of genes and select transcriptomic datasets within the Lookup schizophrenia module (Figure 4B). Finally, we demonstrated the ability of the iLINCS tool in Kaleidoscope to generate a table of signatureIDs, cell lines, and a direct link to the iLINCS webpage based on an initial query for knockdown signatures of the DISC-1 associated target genes (Supplementary Table 3).
Figure 4: Figure 4: Differential gene expression of the DISC1-associated target genes across 34 schizophrenia datasets. A. Heatmap with LFC values between cases and controls across curated schizophrenia datasets. The datasets are grouped by their sample level (Region: samples taken from tissues, Cell: samples taken from pools of isolated cells). B. Spearman correlation analysis showing the association between the LFC for the DISC1-associated genes and the schizophrenia datasets. Blue represents high concordance and red represents high discordance.
3. Conclusions
As demonstrated, Kaleidoscope is a useful platform for conducting in silico exploration of omics data. Through this application, users have the capacity to generate a PPI network, analyze the number of publications that are associated with this network and disease of interest, generate differential expression data by cell-type, explore the distribution of expression based on cell-type, produce heatmaps to better visualize gene expression and/or enrichment data, perform correlation analyses between the gene network and disease datasets of interest, and much more, for any user-defined gene or gene set of interest. The seamless integration of multiple bioinformatic databases in one web application makes Kaleidoscope versatile and user friendly. Notably, Kaleidoscope has recently been utilized to perform in silico replication analyses of publicly available datasets, highlighting its efficacy [8-11, 49-54]. This platform has been used to augment research findings, including investigations of bioenergetic gene expression dysregulation and adenosine system dysregulation in schizophrenia [8, 9]. It has also been instrumental in understanding the abnormal regulation of protein prenylation in schizophrenia [10]. Most recently, Kaleidoscope facilitated an investigation into the alteration of glutamate transporter interacting proteins in schizophrenia, major depressive disorder, and amyotrophic lateral sclerosis [11]. Collectively, these examples demonstrate the advantages of having a platform that streamlines the complex process of in silico data exploration. Kaleidoscope makes bioinformatics tools accessible to a broader range of users, empowering them to rigorously test and investigate scientific inquiries and discoveries in silico.
Availability and requirements
Project name: Kaleidoscope.
Webpage: https://cdrl.shinyapps.io/Kaleidoscope/
Project home page: https://github.com/CogDisResLab/kaleidoscope
Operating system(s): Platform independent.
Programming language: R.
Other requirements: e.g., Dependent R packages.
List of abbreviations
API: Application programing interface
PPI: Protein-protein interaction
STRING: Search Tool for the Retrieval of Interacting Genes
MTG: middle temporal gyrus
RPKM: Reads Per Kilobase Million
TPM: Transcripts Per Kilobase Million
eQTL: Expression quantitative trait loci
SNP: Single nucleotide polymorphism
LINCS: The Library of Integrated Network-Based Cellular Signatures
iLINCS: Integrative LINCS GWAS: genome-wide association study
UTR: untranslated region
FPKM: Fragments Per Kilobase of transcript per Million mapped reads
CPM: count per million
Declarations
Ethical approval and consent to participate
Not applicable
Consent for publication
Not Applicable
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GitHub repository, http://github.com/CogDisResLab/Kaleidoscope/
The application is available at https://cdrl.shinyapps.io/Kaleidoscope/
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by NIMH R01 MH107487, and MH121102
Authors’ contribution
KA, ASI, and SS contributed equally to the work presented. KA and ASI developed and designed the software. KA, SS, and RM wrote the manuscript. SS revised and edited the manuscript. HE revised and edited the figures. RS, SO, AF, MH, JM. TA and RM provided feedback on the application. KA, ASI, SS, HE, MA, XZ, WM, JL, CA, RA, and RP curated the datasets. All authors read and approved the final manuscript.
Acknowledgements
Not applicable
References
- Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol 1418 (2016): 93-110.
- Duck G, Nenadic G, Filannino M, et al. A Survey of Bioinformatics Database and Software Usage through Mining the Literature. PloS one 11 (2016): 0157989.
- Cannata N, Merelli E, Altman RB. Time to organize the bioinformatics resourceome. PLoS Comput Biol 1 (2005): 76.
- Wren JD, Bateman A. Databases, data tombs and dust in the wind. Bioinfo 24 (2008): 2127-2128.
- Sepulveda JL. Using R and Bioconductor in Clinical Genomics and Transcriptomics. J Mol Diagn 22 (2022): 3-20.
- Lafita A, Bliven S, Prli? A, et al. BioJava 5: A community driven open-source bioinformatics library. PLoS Comput Biol 15 (2019): e1006791.
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map formatand SAMtools. Bioinformatics 25 (2009): 2078-2089.
- Sullivan CR, Mielnik CA, O'Donovan SM, et al. Connectivity Analyses of Bioenergetic Changes in Schizophrenia: Identification of Novel Treatments. Mol Neurobiol 56 (2019): 4492-4517.
- Moody CL, Funk AJ, Devine E, et al. Adenosine Kinase Expression in the Frontal Cortex in Schizophrenia. Schizophrenia bulletin (2019).
- Pinner AL, Mueller TM, Alganem K, et al. Protein expression of prenyltransferase subunits in postmortem schizophrenia dorsolateral prefrontal cortex. Translational psych 10 (2020): 3.
- Asah S, Alganem K, McCullumsmith RE, et al. A bioinformatic inquiry of the EAAT2 interactome in postmortem and neuropsychiatric datasets. Schizophrenia Res (2020).
- Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 34 (2014): 11929-11947.
- Zhang Y, Sloan SA, Clarke LE, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89 (2016): 37-53.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47 (2019): 607-613.
- Von Mering C, Jensen LJ, Snel B, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 33 (2005): 433-437.
- Pilarczyk M, Najafabadi MF, Kouril M, et al. Connecting omicssignatures of diseases, drugs, and mechanisms of actions with iLINCS. bioRxiv (2019).
- Keenan AB, Jenkins SL, Jagodnik KM, et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 6 (2018): 13-24.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43 (2015): e47.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (2010): 139-140.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15 (2014): 550.
- Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016 (2016).
- Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 478 (2011): 519-523.
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45 (2013): 580-585.
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nat 489 (2012): 391-399.
- Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47 (2019): 1005-1012.
- Kropiwnicki E, Binder JL, Yang JJ, et al. Getting Started with the IDG KMC Datasets and Tools. Curr Protoc 2 (2022): e355.
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14 (2014): 128.
- Lachmann A, Schilder BM, Wojciechowicz ML, et al. Geneshot: search engine for ranking genes from arbitrary text queries. Nucleic Acids Res 47 (2019): 571-577.
- Blackwood DH, Fordyce A, Walker MT, et al. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet 69 (2001): 428-433.
- Bentea E, Depasquale EAK, O'Donovan SM, et al. Kinase network dysregulation in a human induced pluripotent stem cell model of DISC1 schizophrenia. Mol Omics 15 (2019): 173-188.
- Takahashi N, Sakurai T, Davis KL, et al. Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog Neurobiol 93 (2011): 13-24.
- Xu Y, Yao Shugart Y, Wang G, et al. Altered expression of mRNA profiles in blood of early-onset schizophrenia. Sci Rep 6 (2016): 16767.
- Wen Z, Nguyen HN, Guo Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515 (2014): 414-418.
- Gandal MJ, Haney JR, Parikshak NN, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Sci 359 (2018): 693-697.
- Hoffman GE, Hartley BJ, Flaherty E, et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun 8 (2017): 2225.
- Roussos P, Katsel P, Davis KL, et al. A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen psychiatry 69 (2012): 1205-1213.
- Torrey EF, Webster M, Knable M, et al. The stanley foundation brain collection and neuropathology consortium. Schizophrenia Res 44 (2004): 151-155.
- Arion D, Corradi JP, Tang S, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry 20 (2015): 1397-1405.
- Pietersen CY, Mauney SA, Kim SS, et al. Molecular profiles of pyramidal neurons in the superior temporal cortex in schizophrenia. J Neurogenet 28 (2014): 53-69.
- Pietersen CY, Mauney SA, Kim SS, et al. Molecular profiles of parvalbumin-immunoreactive neurons in the superior temporal cortex in schizophrenia. J Neurogenet 28 (2014): 70-85.
- Barnes MR, Huxley-Jones J, Maycox PR, et al. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J Neuroscience Res 89 (2011): 1218-1227.
- Maycox PR, Kelly F, Taylor A, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry 14 (2009): 1083-94.
- Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Human Mol Genet 14 (2005): 241-253.
- Iwamoto K, Kakiuchi C, Bundo M, et al. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry 9 (2004): 406-416.
- Wu X, Shukla R, Alganem K, et al. Transcriptional profile of pyramidal neurons in chronic schizophrenia reveals lamina-specific dysfunction of neuronal immunity. bioRxiv (2020).
- Labrie V, Fukumura R, Rastogi A, et al. Serine racemase is associated with schizophrenia susceptibility in humans and in a mouse model. Human Mol Genet 18 (2009): 3227-3243.
- Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biological psychiatry 60 (2006): 132-140.
- Funk AJ, Mielnik CA, Koene R, Newburn E, Ramsey AJ, Lipska BK, et al. Postsynaptic Density-95 IsoformAbnormalities in Schizophrenia. Schizophrenia bulletin. 2017;43(4):891-9.
- Schoonover KE, Farmer CB, Morgan CJ, et al. Abnormalities in the coppertransporter CTR1 in postmortem hippocampus in schizophrenia: A subregion and laminar analysis. Schizophrenia Res 228 (2021): 60-73.
- Schoonover KE, Roberts RC. Markers of copper transport in the cingulum bundle in schizophrenia. Schizophrenia Res 228 (2021): 124-133.
- Su RC, Breidenbach JD, Alganem K, et al. Microcystin-LR (MC-LR)Triggers Inflammatory Responses in Macrophages. International Journal of Molecular Sci 22 (2021): 9939.
- Sullivan CR, Mielnik CA, O’Donovan SM, et al. Connectivity analysesof bioenergetic changes in schizophrenia: identification of novel treatments. Mol Neurobiol 56 (2019): 4492-4517.
- Wu X, Shukla R, Alganem K, et al. Transcriptional profile of pyramidalneurons in chronic schizophrenia reveals lamina-specific dysfunction of neuronal immunity. Molecular psychiatry 26 (2021): 7699-7708.
- Zhang X, Wolfinger A, Wu X, et al. Gene Enrichment Analysis ofAstrocyte Subtypes in Psychiatric Disorders and Psychotropic Medication Datasets. Cells 11 (2022): 3315.



 Impact Factor: * 4.2
Impact Factor: * 4.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 