Comparing two Different RNA Production Databases show Similar Patterns of Age-Related Changes and Demonstrates how Ontogenesis Defines Aging
Article Information
Lev Salnikov*1, Saveli Goldberg2, Eugene Pinsky3, Heena Rijhwani3, Michael Golitsyn4
1AntiCA Biomed, San Diego, CA 92111
2Department of Radiation Oncology, Mass General Hospital, Boston, MA 02115
3Department of Computer Science, Met College, Boston University, Boston, MA 02215
4Department of Computer Science, Brandeis University, MA 02453
*Corresponding author: Lev Salnikov, AntiCA Biomed, 7917 Ostrow St, San Diego, CA 92111, United States.
Received: 15 March 2024; Accepted: 20 March 2024; Published: 29 March 2024.
Citation: Lev Salnikov, Saveli Goldberg, Eugene Pinsky, Heena Rijhwani, Michael Golitsyn. Comparing two Different RNA Production Databases show Similar Patterns of Age-Related Changes and Demonstrates how Ontogenesis Defines Aging. Journal of Bioinformatics and Systems Biology. 7 (2024): 101-107.
View / Download Pdf Share at FacebookAbstract
Most authors studying the causes of aging link them to the ontogenesis program, but the question of the mechanisms linking these phenomena remains open. In this paper we continue the statistical analysis of RNASeq results of the whole genome of Mus musculus during their lifetime by comparing two different databases, GSE132040 and GSE34378 submitted to NCBI. We hypothesize that the implementation of a developmental program by cells and their transition to active function is a major mechanism of aging. We focus our attention on the dynamics of two functional groups of the cell genome that perform the main tasks of the organism: its selfsupply, based on the function of the group of housekeeping genes (HG), and functional specialization, provided by the group of integrative genes (IntG). We used the value of the HG/IntG production ratio in both databases to compare the results in terms of absolute values of RNA production. In both databases, this ratio had a strong tendency to decrease with age. We also used the data obtained by calculating the deviation from the mean value for each of the genes in both databases. This approach allowed us to compare the age-related dynamics of RNA production in relative values independent of absolute values. The results confirm the presence of a strong tendency to increase the production of all genes of the IntG group with a simultaneous decrease in the production of all HG genes in both databases with increasing age. The obtained results were also confirmed by the data on cluster analysis of gene production dynamics.
Keywords
Aging; RNA-Seq data analysis; Ontogenesis program; Housekeeping genes.
Aging articles; RNA-Seq data analysis articles; Ontogenesis program articles; Housekeeping genes articles.
Article Details
1. Introduction
To the present day researchers studying aging, have not reached a consensus on its causes. It remains unclear which processes occurring during aging are related to its causes and which are their consequences [1]. Although most authors dealing with the problems of aging connect them with the ontogenesis program [2,3,4,5,6,7], the question of the mechanisms linking these phenomena remains open. From our point of view, aging is embedded in the very principle of multicellular organization. For the purpose of developing specialized functions and tissues of the organism, the ontogenesis program uses not the whole cellular genome, but only a part of it. Thus, it is possible to distinguish two functionally different parts of the organism genome: the part responsible for functional differentiation provided by the integrative group of genes (IntG) and the part responsible for infrastructural support of cells, or housekeeping genes (HG) [8].
Proponents of the stochastic theory of aging base their views on the data showing both an increase in the amount of DNA damage in the cells and an increase in the amount of entropy in the organism with age [9,10,11]. However, the known fact of practical "immortality" of unicellular organisms leads to the conclusion that aging itself as a biological process appeared together with multicellular organisms and is the result of the way of its organization. An indispensable condition for the development of any multicellular organism is the presence of a developmental program in its genome, which determines the entire course of ontogenesis. We understand the program of ontogenesis as a strictly defined sequence of IntG gene expression with the assignment of advantage in the expenditure of cellular resources for the formation of the organism and its achievement of maximum development by the beginning of reproductive age. In our ideas, we proceed from two main provisions: the resource capabilities of any cell are always limited; the main goal of the ontogenesis program is to achieve the maximum competitive advantage by the time the organism reaches reproductive age, since natural selection is aimed precisely at this stage of development [12,13]. This advantage, obtained at a critical moment for the survival of the species, is paid for by the suppression of autonomous and regenerative potential in the future. This situation is essentially pleiotropic [14,15], but it refers not to individual genes, but to the HG and IntG groups that we have identified. Here it is very important to note that the ontogenesis program exists and controls not the entire cellular genome, but only its IntG part. We proceed from the statement that insufficient level of repair is the main cause of aging. In the case of a multicellular organism, its aging is also caused by a decrease in resources required by its cells for their repair and tissue regeneration and starts at the moment when organism recovery begins to be incomplete [16]. Understanding exactly how this deficit arises is our primary concern.
One of the main indicators of genome function is RNA production [17,18,19,20,21,22]. In our previous works, we analyzed this parameter, where we showed a relative decrease in the level of absolute values of RNA production in the infrastructural part of the genome (HG), especially pronounced after the end of the fertility period [23]. Given that we analyzed only one database, we decided to perform a comparative analysis of two databases on age-related dynamics of RNA production to more reliably validate our aging model. When analyzing the data presented in our previous work, their irregularity was noticeable. Thus, the number of genes that produce more than 80% of all RNA production is only 4,850, or 13.6% of the total number of genes. Such a scatter in the activity of genes leads to the fact that studying the age dynamics of RNA production in absolute values we obtain a distorted result, where genes with high productivity have a great influence on it. Taking into account that the main objective of the study is to assess the age dynamics of RNA production rather than its magnitude, for its assessment and comparison in this work we use a relative value, namely, the percentage of deviation from the average value of RNA production for each individual gene. This allows us not only to level out the non uniformity in the level of production of individual genes, but also to get an idea of their total age dynamics in the functional groups HG and IntG. The main goal of this work is to compare the data we obtained in our previous work with a similar database to confirm our main results and to analyze in parallel the dynamics of RNA production in the functional parts of the HG and IntG genome. The main parameter supporting our views is the confirmation of the switch of cell resources from RNA production of HG genes to IntG genes with age, which we verify in this paper. The results of the comparative analysis presented in this paper provide a broader foundation based on experimental data to confirm our views on the causes and mechanisms of aging.
2. Materials and Methods
To analyze the results of RNA sequencing of the Mus musculus genome production, we used the same material as in our previous article. In this study we used the data and results presented in thearticles by the authors [24,25]. The RNA-Seq data on the Mus musculus mouse genome transcriptome is available in the GEO repository, under the accession number GSE132040 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132040).The analyzed database contains the the RNA-Seq data from 17 tissue types: Brain, BAT, Bone, GAT, Heart, Kidney, Lung, Marrow, MAT, Pancreas, SCAT, Skin, Small, Spleen, WBC, Limb, Liver. For the age dynamics a new group of experimental mice was used each time they reached the age of 1, 3, 6, 9, 12, 15, 18, 21, 24 and 27 months.
As a material for comparison, we used the data and results presented in the articles by the authors [26]. The RNA-Seq data on the C57BL/6J female mice genome transcriptome is available in the GEO repository, under the accession number GSE34378 (https://www.ncbi.nlm.nih.gov/bioproject/?term=GSE34378) . For the age dynamics a new group of experimental mice was used each time they reached the age of 3, 6, 12, 18, 24, 30 months (we converted the original age in weeks into months). Database contains the RNA-Seq data from 5 tissue types: Brain, Kidney, Liver, Lung, Spleen. For both databases the Mus musculus RNA-seq was converted into FASTQ format and quantified the expression for each gene using Salmon, where GRCm39(Genome Reference Consortium Mouse Reference 39) was used as a reference transcript for all samples. The tissue types and their labeling are also taken from the database we mentioned earlier. Quality control and initial data normalization were used in the process of extracting them from the database. The quantification output was processed and computed in order to obtain the mean and standard deviation for the gene expression level. Total number of genes from the database GSE132040 was 35,630, for the database GSE34378 was 35 283 genes total. Selection into the HG gene part for both databases was performed according to the Housekeeping and Reference Transcript Atlas (HRT Atlas v1.0, (www.housekeeping.unicamp.br) [27]. When the HG list was compiled, all variants of genes from those already listed were additionally included. Total number of the Housekeeping genes in database GSE132040was 5,101. All remaining genes (30,529) were assigned to the IntG part. Total number of the Housekeeping genes in database GSE34378was 6,433. All remaining genes (28,850) were assigned to the IntG part. Genes with the same name in both groups were removed from the total list of the whole genome. Comparisons were made only for tissues present in both databases: Brain, Kidney, Liver, Lung, Spleen. When comparing the results, we used the age points present in both databases, from 3 to 24 months. Given the difference between databases in the methods used to obtain absolute values of RNA production, we used the percentage deviation of each gene's production from its mean value to compare results, thus moving away from absolute values of production.
We conducted clustering analysis, using his generally accepted methodology [28]on percentage variation data denoting the normalized deviation of a gene from its mean at each time point. In the first round of clustering, K*Means algorithm with euclidean distance was used to get gene trajectories. A comparison of the number of clusters (K) VS sum, or squared errors (SSE) using an elbow plot gave the optimal number of clusters to be 4. Each gene trajectory depicts the cluster id a gene from a particular tissue belongs to at each age. We then clustered these gene trajectories using K*Means with Hamming distance. Optimal number of clusters was found to be 3 for the second round of clustering, where we compared patterns of HG and IntG genes in these groups of gene trajectories.
3. Results
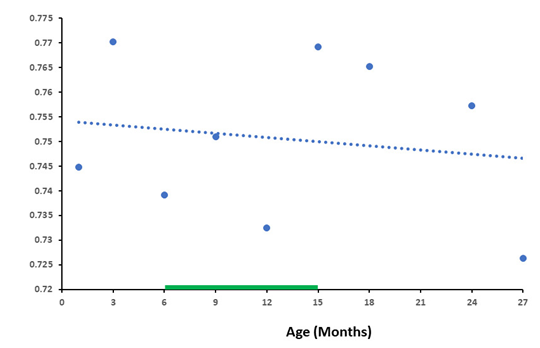
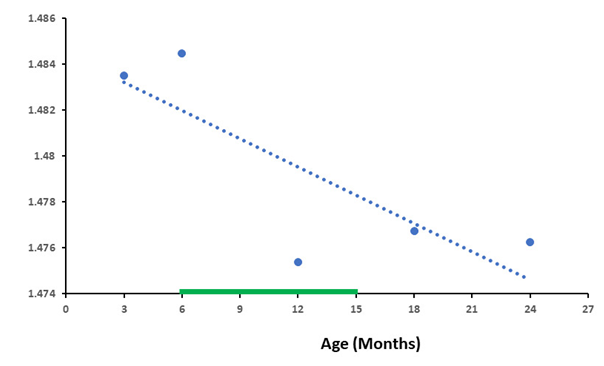
As the results show, the compared databases were close in their baselines in both databases, both the number of genes and the differences in RNA production between HG and IntG were quite similar. Total number of genes from the database GSE132040 was 35,630, for the database GSE34378 was 35,283 genes total. Housekeeping genes in database GSE132040 were 5,101 and 6,433 in database GSE34378. The results allow us to conclude that the differences in the level of RNA synthesis between the HG and IntG parts of the genome were statistically highly significant at all age points presented (P-value < 0.0001) throughout the entire observation period, from 3 to 27 months, and their total values showed a close trajectory. We did not use age greater than 27 months, which is not available in the GSE132040 database. In addition, the probability of death increases dramatically in Mus musculus after reaching 24 months of age [29,30], distorting the results obtained. As shown in one of our previous work [31], the data obtained at the extremely old age mainly characterize the genetic features of the group of long-livers who managed to undergo age selection. Absolute values of RNA production by genes in the analyzed databases are presented in different forms. For the GSE132040 database, a direct count value was used, while for GSE34378 the results were presented as the logarithm of the value obtained. Given these differences, we used the ratio of RNA production between the HG/IntG gene groups for each database, rather than their absolute values, to compare results between bases in terms of averaged RNA production. Moreover, this ratio directly demonstrates the degree of RNA production switching with age. We used the linear approximation method [32] to graphically display the results of the age-related dynamics of RNA production and analyze it. The results of the ratio of absolute values of RNA production between HG/IntG gene groups for each database are presented in Figure 1 and Figure 2.
As can be seen from the graphs in Figure 1 and Figure 2, the ratio of RNA production between the HG/IntG gene groups decreased with age in both databases, although this decrease was more pronounced in the GSE34378 database.
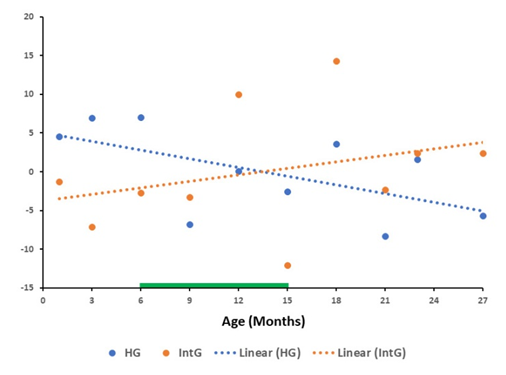
The graph in Figure 3 shows the results of the overall trend of changes relative to the mean value in the GSE132040 database for each gene of all genes of the HG and IntG functional group in Mus musculus. The approximating lines show a pronounced tendency for RNA production to decrease relative to the mean value with age in the group of all HG genes and an opposite tendency for the production of all genes in the IntG group. In this group of genes, there is a pronounced tendency for an increase in RNA production by all genes of this group with increasing age.
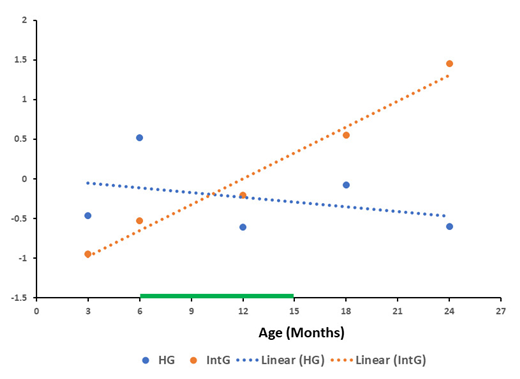
The results obtained for the overall dynamics of changes relative to the mean value in the GSE34378 database for each gene of the functional groups HG and IntG shown in Figure 4 were almost similar to the data obtained for the GSE132040 database. In both plots, there was an increase in RNA production of all IntG group genes with a simultaneous decrease in the production of all HG genes.
The results of cluster analysis of RNA production dynamics are presented in Table 1. For the cluster analysis, we used the results of the percentage deviation from the mean value for each of the genes. The table contains the distribution of the three obtained clusters for each database.
|
Database GSE132040 Cluster number |
Percentage (%) of HG genes |
Percentage (%) of genes IntG |
|
0 |
3.23 |
96.77. |
|
1 |
20.21 |
79.79. |
|
2 |
97.94. |
2.06. |
|
Database GSE34378 Cluster number |
Percentage (%) of HG genes |
Percentage (%) of genes IntG |
|
0 |
0.00 |
100.0 |
|
1 |
100.0 |
0.00 |
|
2 |
31.61. |
68.39.00 |
Table 1: The table shows the results of cluster analysis of the GSE132040 and GSE34378 databases as measured by the percent deviation of RNA production from the mean for each gene in the HG and IntG groups. The result represents how many percent of the genes in these groups fall into each cluster.
The resulting cluster distributions demonstrate that the functional groups of HG and IntG genes change along significantly different trajectories and are almost equally distributed across clusters in both databases. It should be noted that the genes in Database GSE34378 are better distributed across clusters, although the difference is insignificant, but the results for this database are stronger when compared.
4. Discussion
We set out to compare the results on RNA production between two databases (GSE132040 and GSE34378) performed by different groups of authors. Given the different methods of estimating absolute values used in these databases, it was necessary to use indicators that do not depend on them. For this purpose, we used two indicators: the results of the ratio of absolute values of RNA or HG/IntG production, which simply show the relative value of their ratio changing with time and the percentage deviation from the mean value of production for each gene at each age point. The main conclusion from our results comparing two different RNA production databases is the presence of the same patterns of age-related changes in the functional groups of HG and IntG genes, confirming the obvious link between organism aging and ontogenesis. Based on our theoretical model of aging, one of the main criteria for its verification is the matter of redistribution of the level of RNA synthesis and related resources between the functional parts of the HG and IntG genome that we have identified during ontogenesis, leading to a deficit of reparative capabilities of cells. In our previous work, we obtained a result indicating a statistically significantdifference in the dynamics of decreased activity in a part of HG genes compared to IntG by the age of 18 months. These results are consistent with the data on changes in RNA production obtained by other authors [33,34]. The significance of such a decrease in the part of the genome responsible for cellular infrastructure is most clearly seen when comparing the results obtained with the data on Mus musculus mortality rate. Comparing the data on the Mus musculus mortality rate with the results obtained, we note that their mortality rate begins to increase significantly from 15 months of age, exactly when the production of HG genes reaches its minimum value. Starting from the age of 18 months the actual risk of mortality begins to rise, increasing more than twofold to the age of 24 months [35,36]. Figure 3 and Figure 4 clearly show that the intersection of the HG and IntG gene groups approximating the age dynamics of activity falls practically in the middle of the fertility period, where the optimal ratio of their activity is located. One of the specific features of the GSE34378 database is that it represented by only five types of tissues: Brain, Kidney, Liver, Lung, Spleen, forcing us to limit our analysis to them only. It is noteworthy that all these tissues are highly differentiated tissues with low mitotic index (MI).
In the paperson aging, very much attention is paid to cells and tissues with high mitotic index. These properties allow such cells to retain a high regenerative potential and make them a target for the rejuvenation experiments [37,38,39]. From our point of view, only affecting the epigenetic mechanisms of cells with high MI is clearly insufficient to achieve true rejuvenation of the organism. On the other hand, the majority of the organism consists of organs and tissues composed of post mitotic cells and cells with low MI, which supply senescent cells in the course of aging. There are many works devoted to the aging processes in such cells, their influence on the surrounding tissues and the immune system response to them. The contribution to the aging processes and negative effect of senescent cells they have, been shown in a number of articles [40,41,42,43]. Thus, making a comparison only on tissues with low MI we deal with tissues that essentially determine the aging processes in the organism. The presented results show the main trend, demonstrating a decrease in RNA production in all genes of the HG functional group with a simultaneous increase in all IntG genes, both in absolute values (Figure 1 and Figure 2) and by the results of deviation from the mean values (Figure 3 and Figure 4) for both databases. It should be noted that the results obtained for the GSE34378 database show significantly more pronounced trends for all indicators. This difference is caused by the fact, that genetically homogeneous mice of the C57BL/6J line were used in this database, which allowed significantly reduce the scatter of the obtained data.
The obtained changes in gene activity confirm our assumptions about the implementation of the ontogenesis program based on unilateral stimulation of the activity of genes of the IntG functional group. It is the constant increase in the consumption of cellular resources at integrative functional costs that determines the main mechanism of aging, which is embedded in the very nature of genome functioning in the conditions of highly integrated multicellularity. Both for individual cells and for the organism as a whole there is a zone of functional optimum. This is the state at which the programmed level of development is accompanied by the lowest metabolic costs for its maintenance. Both for individual cells and for the organism as a whole this occurs at different times. The organism as a whole system of specialized functions reaches its functional optimum always later than its individual cells. Such mismatch naturally leads to "pushing" of the organism cells out of the optimum zone, which leads to a similar situation for the organism as a whole. Let us repeat that here we are talking about the system-forming principle of highly specialized multicellularity, on which it is built, and which determines the course of development of a multicellular organism [44].
The obtained data confirm the validity of the theoretical model of aging proposed earlier, allowing us to transfer it to the rank of a hypothesis. In addition, the shown significance of changes in infrastructural genes allows us to use their dynamics as a criterion for the effectiveness of influences on the aging processes of the organism. It is possible to try to find certain medicinal substances stimulating the production of infrastructural genes, although it is unlikely that this will help to achieve a long-term effect. It is possible to radically change the aging processes only by interfering in the organism's development program, purposefully changing its genetic code. Other approaches to solving this problem are also possible, changing the point of attachment of the impact, but it will still be aimed at changing the development program recorded in cellular DNA, or more precisely in its IntG part of the genome.
Conflict of Interest
Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
Salnikov has proposed the theoretical model of aging based on functional genome partition and the role of ratio of the genome two functional parts activity in ontogenesis and aging. He also wrote the bulk of the article. S. Goldberg and H. Rijhwani did the statistical analysis of the data. M. Golitsyn performed data extraction and participated in data analysis. E. Pinsky organized the work and participated in the data analysis. All authors have made a contribution to prepare the article for publication. All authors reviewed, revised and approved the final version of the manuscript.
Authors Approvals
All authors have seen and approved the manuscript. The manuscript hasn't been accepted or published elsewhere.
Funding
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilized in the production of this manuscript.
Conflicts of interest
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- De Magalhães JP. Distinguishing between driver and passenger mechanisms of aging. Nat Genet (2024).
- Walker RF. A Mechanistic Theory of Development-Aging Continuity in Humans and Other Mammals. Cells 11 (2022): 917.
- Larocca D, Lee J, West MD, et al. No Time to Age: Uncoupling Aging from Chronological Time. Genes 12 (2021): 611.
- De Magalhães JP, Church GM. Genomes optimize reproduction: aging as a consequence of the developmental program. Physiol (Bethesda) 20 (2005): 252-259.
- Bilinski T, Bylak A, Kukula K, et al. Senescence as a trade-off between successful land colonisation and longevity: critical review and analysis of a hypothesis. PeerJ 9 (2021): e12286.
- West MD, Sternberg H, Labat I, et al. Toward a unified theory of aging and regeneration. Regen Med 14 (2019): 867-886.
- Gems D, Singh Virk R, De Magalhães JP. Epigenetic Clocks and Programmatic Aging (2023).
- Salnikov L, Baramiya MG. The Ratio of the Genome Two Functional Parts Activity as the Prime Cause of Aging. Front Aging 1 (2020).
- Soto-Palma C, Niedernhofer LJ, Faulk CD, et al. Epigenetics, DNA damage, and aging. J Clin Invest 132 (2022): e158446.
- Perevoshchikova K, Fedichev PO. Differential Responses of Dynamic and Entropic Aging Factors to Longevity Interventions. BioRxiv (2024).
- Kinzina ED, Podolskiy DI, Dmitriev SE, et al. Patterns of Aging Biomarkers, Mortality, and Damaging Mutations Illuminate the Beginning of Aging and Causes of Early-Life Mortality. Cell Rep 29 (2019): 4276-4284.
- Gems D, Kern CC, Nour J, et al. Reproductive Suicide: Similar Mechanisms of Aging in C. elegans and Pacific Salmon. Front Cell Dev Biol 9 (2021): 688788.
- Kirkwood TBL, Holliday R. The evolution of ageing and longevity. Proc. R. Soc. London Ser. B Biol Sci. 205 (1979): 531–546.
- Williams GC. Pleiotropy, natural selection and the evolution of senescence. Evol 11 (1957): 398-411.
- Salnikov L, Baramiya MG. From Autonomy to Integration From Integration to Dynamically Balanced Integrated Co-existence: Non-aging as the Third Stage of Development. Front Aging 2 (2021).
- Santra M, Dill KA, De Graff AMR. Proteostasis collapse is a driver of cell aging and death. Proc Natl Acad Sci U S A 116 (2019): 22173-22178.
- Salnikov L, Goldberg S, Pinsky E. Perspectives for Using RNA Sequencing Analysis of the Genome to Understand the Mechanisms of Aging. J Gerontol Geriatr Med 9 (2023): 185.
- Meyer DH & Schumacher B. BiT age: A transcriptome-based aging clock near the theoretical limit of accuracy. Aging Cell 20 (2021).
- Stoeger T, Grant RA, McQuattie-Pimentel AC, et al. Aging is associated with a systemic length-associated transcriptome imbalance. Nat Aging (2022).
- Stegeman R, Weake VM. Transcriptional Signatures of Aging. J Mol Biol 429 (2017): 2427-2437.
- Lu JY, Simon M, Zhao Y, et al. Comparative transcriptomics reveals circadian and pluripotency networks as two pillars of longevity regulation. Cell Metab 34 (2022): 836-856.
- Ferreira M, Francisco S, Soares AR, et al. Integration of segmented regression analysis with weighted gene correlation network analysis identifies genes whose expression is remodeled throughout physiological aging in mouse tissues. Aging 13 (2021): 18150-18190.
- Salnikov L, Goldberg S, Rijhwani H, et al. The RNA-Seq data analysis shows how the ontogenesis defines aging. Front Aging 4 (2023): 1143334.
- Tabula Muris Consortium, Overall coordination, Logistical coordination, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nat 562 (2018): 367-372.
- Schaum N, Lehallier B, Hahn O, et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nat 583 (2020): 596-602.
- Jonker MJ, Melis JP, Kuiper RV, et al, Life spanning murine gene expression profiles in relation to chronological and pathological aging in multiple organs. Aging Cell 12 (2013): 901-909.
- Hounkpe BW, Chenou F, De Lima F, et al. HRT Atlas v1.0 database: redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res 49 (2021): D947-D955.
- Jaeger A, Banks D. Cluster analysis: A modern statistical review. WIREs Computational Statistics 15 (2023): e1597.
- Gavrilova NS, Gavrilov LA. Biodemography of old-age mortality in humans and rodents. J Gerontol A Biol Sci Med Sci 70 (2015): 1-9.
- Hughes BG, Hekimi S. Different Mechanisms of Longevity in Long-Lived Mouse and Caenorhabditis elegans Mutants Revealed by Statistical Analysis of Mortality Rates. Genet 204 (2016): 905-920.
- Salnikov L, Goldberg S, Pinsky E. The RNA Genome Sequencing Demonstrates Increased Production of Metabolic Genes in Late Ages in Highly Differentiated Tissues. Med Res Arch 12 (2024).
- Fatima SS, Wooldridge M, Jennings NR. A linear approximation method for the Shapley value. Artificial Intelligence 172 (2008): 1673-1699.
- Anisimova AS, Meerson MB, Gerashchenko MV, et al. Multifaceted deregulation of gene expression and protein synthesis with age. Proc Natl Acad Sci U S A 117 (2020): 15581-15590.
- Debès C, Papadakis A, Grönke S, et al. Ageing-associated changes in transcriptional elongation influence longevity. Nat 616 (2023): 814–821.
- Gavrilova NS, Gavrilov LA. Biodemography of old-age mortality in humans and rodents. J. Gerontol. A Biol Sci Med Sci 70 (2015): 1–9.
- Brust V, Schindler PM, Lewejohann L. Lifetime development of behavioural phenotype in the house mouse (Mus musculus). Front Zool 12 (2015): S17.
- Zhang W, Qu J, Liu GH, et al. The ageing epigenome and its rejuvenation. Nature Reviews Molecular Cell Biology. Nat Res (2020).
- Olova N, Simpson DJ, Marioni RE, et al. Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell 18 (2019).
- Lapasset L, Milhavet O, Prieur A, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev 25 (2011): 2248-2253.
- Van Deursen JM. The role of senescent cells in ageing. Nat 509 (2014): 439-46.
- Childs BG, Gluscevic M, Baker DJ, et al Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov 10 (2017): 718-735.
- Mylonas A, O'Loghlen A. Cellular Senescence and Ageing: Mechanisms and Interventions. Front Aging 3 (2022): 866718.
- Huang W, Hickson LJ, Eirin A, et al. Cellular senescence: the good, the bad and the unknown. Nat Rev Nephrol 18 (2022): 611–627.
- Salnikov L. Aging is a Side Effect of the Ontogenesis Program of Multicellular Organisms. Biochemist (Mosc) 87 (2022): 1498-1503.


 Impact Factor: * 4.2
Impact Factor: * 4.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 