Molecular Mechanisms of Resistance to Osimertinib from Liquid Biopsies of Plasma and Pleura Effusion in EGFR Mutant NSCLC Patients
Article Information
Ning Li1*, Shuhang Wang1, Huiyao Huang1, Minghui Zhang2, Zicheng Yu3, Feng Xie4, Shidong Jia4, Yixuan Ren5, Danyi Wen5
1Clinical Trial Center, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
2Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin, China
3GenePlus-Shenzhen, Shenzhen, China
4Huidu Shanghai Medical Sciences, Shanghai, China
5Shanghai LIDE Biotech, Shanghai, China
*Corresponding Author: Ning Li, Clinical Trial Center, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, 100021, Beijing, China
Received: 07 April 2020; Accepted: 03 May 2021; Published: 21 May 2021
Supplementary Files
Citation: Shuhang Wang, Huiyao Huang, Minghui Zhang, Zicheng Yu, Fend Xie, Shidong Jia, Yixuan Ren, Danyi Wen, Ning Li. Molecular Mechanisms of Resistance to Osimertinib from Liquid Biopsies of Plasma and Pleura Effusion in EGFR Mutant NSCLC Patients. Journal of Cancer Science and Clinical Therapeutics 5 (2021): 235-249.
View / Download Pdf Share at FacebookAbstract
In the past decade, tyrosine kinase inhibitors (TKI) have been developed and widely-approved by countries to treat advanced lung cancer patients with sensitizing epidermal growth factor (EGFR) mutations, and substantially improved patients’ prognosis compared to traditional chemotherapies. However, a large proportion of patients who initially experienced ostensibly remarkable therapeutic outcomes still developed inevitable drug resistance eventually, including those received newly developed third-generation TKIs. Previous studies have demonstrated the value of plasma cell-free DNA (cfDNA) in monitoring patients’ response to therapies in lung cancer. However due to several physiological reasons and DNA degradation, DNA released from tumor cell apoptosis in lung was hard to maintain at a high concentration and good quality before being collected from venous blood, leading to the loss of pivotal mutational information. Pleural effusion is a potential substitute to overcome these problems.
However, limited studies have been reported to characterize the consistency of genetic alterations found in these two sample types, and due to the modest sample sizes, the mechanisms underlying third-generation TKI resistance remains elusive. Herein we drew 8ml venous blood and collected 20ml pleural effusion by pleural effusion drainage from 8 lung cancer patients, and performed hybrid-capture based next generation sequencing assays on 18 matched plasma cfDNA, pleural effusion cells genomic DNA and pleural effusion supernatant cfDNA samples, we have shown that only sequencing either plasma or pleural effusion sample resulted in the missing of mutations in key cancer drivers. Sequencing both pleural effusion and plasma yielded a more concrete genetic landscape for monitoring tumor response. We have also exhibited that somatic mutations and copy number changes in PI3K/AKT pathway, NOTCH signaling pathway, he
Keywords
EGFR; TKI; Osimertinib; Liquid Biopsie
EGFR articles; TKI articles; Osimertinib articles; Liquid Biopsie articles
EGFR articles EGFR Research articles EGFR review articles EGFR PubMed articles EGFR PubMed Central articles EGFR 2023 articles EGFR 2024 articles EGFR Scopus articles EGFR impact factor journals EGFR Scopus journals EGFR PubMed journals EGFR medical journals EGFR free journals EGFR best journals EGFR top journals EGFR free medical journals EGFR famous journals EGFR Google Scholar indexed journals TKI articles TKI Research articles TKI review articles TKI PubMed articles TKI PubMed Central articles TKI 2023 articles TKI 2024 articles TKI Scopus articles TKI impact factor journals TKI Scopus journals TKI PubMed journals TKI medical journals TKI free journals TKI best journals TKI top journals TKI free medical journals TKI famous journals TKI Google Scholar indexed journals Osimertinib articles Osimertinib Research articles Osimertinib review articles Osimertinib PubMed articles Osimertinib PubMed Central articles Osimertinib 2023 articles Osimertinib 2024 articles Osimertinib Scopus articles Osimertinib impact factor journals Osimertinib Scopus journals Osimertinib PubMed journals Osimertinib medical journals Osimertinib free journals Osimertinib best journals Osimertinib top journals Osimertinib free medical journals Osimertinib famous journals Osimertinib Google Scholar indexed journals Liquid Biopsie articles Liquid Biopsie Research articles Liquid Biopsie review articles Liquid Biopsie PubMed articles Liquid Biopsie PubMed Central articles Liquid Biopsie 2023 articles Liquid Biopsie 2024 articles Liquid Biopsie Scopus articles Liquid Biopsie impact factor journals Liquid Biopsie Scopus journals Liquid Biopsie PubMed journals Liquid Biopsie medical journals Liquid Biopsie free journals Liquid Biopsie best journals Liquid Biopsie top journals Liquid Biopsie free medical journals Liquid Biopsie famous journals Liquid Biopsie Google Scholar indexed journals lung cancer articles lung cancer Research articles lung cancer review articles lung cancer PubMed articles lung cancer PubMed Central articles lung cancer 2023 articles lung cancer 2024 articles lung cancer Scopus articles lung cancer impact factor journals lung cancer Scopus journals lung cancer PubMed journals lung cancer medical journals lung cancer free journals lung cancer best journals lung cancer top journals lung cancer free medical journals lung cancer famous journals lung cancer Google Scholar indexed journals DNA degradation articles DNA degradation Research articles DNA degradation review articles DNA degradation PubMed articles DNA degradation PubMed Central articles DNA degradation 2023 articles DNA degradation 2024 articles DNA degradation Scopus articles DNA degradation impact factor journals DNA degradation Scopus journals DNA degradation PubMed journals DNA degradation medical journals DNA degradation free journals DNA degradation best journals DNA degradation top journals DNA degradation free medical journals DNA degradation famous journals DNA degradation Google Scholar indexed journals pleural effusion articles pleural effusion Research articles pleural effusion review articles pleural effusion PubMed articles pleural effusion PubMed Central articles pleural effusion 2023 articles pleural effusion 2024 articles pleural effusion Scopus articles pleural effusion impact factor journals pleural effusion Scopus journals pleural effusion PubMed journals pleural effusion medical journals pleural effusion free journals pleural effusion best journals pleural effusion top journals pleural effusion free medical journals pleural effusion famous journals pleural effusion Google Scholar indexed journals dacomitinib articles dacomitinib Research articles dacomitinib review articles dacomitinib PubMed articles dacomitinib PubMed Central articles dacomitinib 2023 articles dacomitinib 2024 articles dacomitinib Scopus articles dacomitinib impact factor journals dacomitinib Scopus journals dacomitinib PubMed journals dacomitinib medical journals dacomitinib free journals dacomitinib best journals dacomitinib top journals dacomitinib free medical journals dacomitinib famous journals dacomitinib Google Scholar indexed journals afatinib articles afatinib Research articles afatinib review articles afatinib PubMed articles afatinib PubMed Central articles afatinib 2023 articles afatinib 2024 articles afatinib Scopus articles afatinib impact factor journals afatinib Scopus journals afatinib PubMed journals afatinib medical journals afatinib free journals afatinib best journals afatinib top journals afatinib free medical journals afatinib famous journals afatinib Google Scholar indexed journals olmutinib articles olmutinib Research articles olmutinib review articles olmutinib PubMed articles olmutinib PubMed Central articles olmutinib 2023 articles olmutinib 2024 articles olmutinib Scopus articles olmutinib impact factor journals olmutinib Scopus journals olmutinib PubMed journals olmutinib medical journals olmutinib free journals olmutinib best journals olmutinib top journals olmutinib free medical journals olmutinib famous journals olmutinib Google Scholar indexed journals
Article Details
1. Introduction
In the past decade, tyrosine kinase inhibitors (TKI) have been developed and widely-approved by countries to treat advanced lung cancer patients with sensitizing epidermal growth factor (EGFR) mutations, and substantially improved patients’ prognosis compared to traditional chemotherapies [1-4]. In clinical trials, first-generation TKIs including gefitinib, carboplatin-paclitaxel and second-generation TKIs including afatinib, dacomitinib both yielded remarkable response rates and therapeutic outcomes initially, nonetheless acquired resistance was observed in a large proportion of patients [5-8]. The commonest mechanism of acquired resistance was indicated to be EGFR T790M mutation, and based on this, third-generation TKIs were developed [1].
Third-generation TKIs include osimertinib, nazartinib, olmutinib, PF-06747775, YH5448, avitinib and rociletinib [1, 9], and in China, only osimertinib has been approved to date [10]. Objective response rates and progression-free survival of EGFR T790M positive patients were observed significantly improved with osimertinib [11, 12]. However, acquired resistance was again experienced eventually by many patients. Studies suggested potential molecular mechanisms underlying resistance encompassing BRAF V600E, EGFR G796D, EGFR C797S, EGFR L718Q mutations and MET amplification etc as reported [13-17]. Recent studies revealed that a wide range of other genetic changes might also contribute to the resistance to third-generation TKIs, such as HER2 amplifications, PIK3CA mutations and several gene fusions [18, 19]. Given the highly heterogenous mechanisms behind the resistance to third-generation TKIs, a few studies published recently suggested that sequencing both plasma cfDNA and tissue biopsies might result in an optimal sensitivity in predicting the resistance [20, 21]. Another recent case study reported primary resistance was observed in a patient who was detected with EGFR T790M mutation from sequencing traditional plasma liquid biopsy, suggesting plasma cfDNA might not be sufficient to detect these possible resistant mechanisms [22]. In addition, though studies have demonstrated multiple possible mechanisms underlying third-generation TKI resistance and even proposed potential fourth-generation inhibitor EAI045 to circumvent this issue [13-17, 23], our understandings were still largely limited by the modest sample sizes in these studies, and a whole picture still awaits exploration given that most of these studies were based on individual cases. Moreover, there is an urgent need to develop a novel tool to predict patients’ response to third-generation TKIs and monitor the therapeutic outcomes in a real-time manner, considering tissue biopsies were believed to be Janus-faced due to their invasive nature and high risk of inducing implantation metastasis [24].
In this regard, non-invasive liquid biopsies have been developed. Cell-free tumor DNA (ctDNA) in plasma has gradually become a commonly used tool in the therapeutic arsenal for real-time clinical surveillance [25-30]. However, tumor-derived ctDNA in several specific cancer types might be restrained from entering the venous blood, for instance ctDNA from brain tumors is mostly unable to enter the blood because of the blood-brain-barrier, and ctDNA released from tumor cell apoptosis in lung is hard to maintain at a high concentration and good quality before being collected in venous blood [31]. Based on these, cell-free DNA in tissue-specific bodily fluids were also utilized, such as cerebrospinal fluid and pleura effusion [32, 33]. Previous reports have demonstrated the value of plasma ctDNA in monitoring patients’ response to therapies in lung cancer [34]. Nevertheless, whether plasma or pleura effusion could be solutions to monitor patients’ resistance to third-generation TKIs still needs to be addressed. Herein by using targeted-capture next-generation sequencing (NGS), we profiled the genetic alterations in 15 matched plasma cfDNA, pleura effusion cells genomic DNA, pleura effusion supernatant cfDNA samples from 8 lung cancer patients, trying to answer whether comparable mutational landscapes could be obtained from plasma and pleural effusion biopsies, and reason the possible mechanisms underlying the acquired drug resistance, rendering it promising to guide personalized treatment decisions, screen suitable candidates for third-generation TKIs and develop novel drugs to overcome the drug resistance.
2. Methods and Materials
2.1 Patient enrollment and ethics
8 patients diagnosed with lung cancer were enrolled from Cancer Hospital Chinese Academy of Medical Sciences from 2014 to 2020. Detailed clinicopathological characteristics of all patients with the timing of progression and sample collections in relation to third-generation TKIs administration were shown in Table S1. All patients harbored EGFR T290M mutations determined by fluorescent PCR and underwent third-generation TKI therapies as indicated in Table S1 before developing resistance afterwards. Then matched peripheral plasma and pleura effusion samples were collected in EDTA Vacutainer tubes (BD Diagnostics, NJ, USA) for cfDNA extraction and purification within 3h. Written informed consent was collected and this study was approved by the Institutional Review Board (IRB) of Cancer Hospital Chinese Academy of Medical Sciences and conducted in accordance with the Declaration of Helsinki.
2.2 Plasma cfDNA NGS testing
Circulating cell-free DNA (cfDNA) testing was performed using the 600-gene PredicineaATLAS assay in a College of American Pathologist (CAP)-accredited laboratory in China (Huidu Shanghai Medical Sciences Ltd.) in 2019-2020.
2.3 Plasma cfDNA extraction
cfDNA was extracted using QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany) from plasma samples. Quantity and quality of the purified cfDNA were checked using Qubit fluorimeter and Bioanalyzer 2100 (Thermo Fisher Scientific, MA, USA).
2.4 Library preparation, capture and sequencing
5 to 20ng of extracted cfDNA was prepared for library construction including end-repair, dA-tailing, adapter ligation, and PCR amplification. The amplified DNA libraries with sufficient yields proceeded to hybrid capture. In brief, the library was hybridized overnight with the panel probes. Unbound fragments were then washed away. The purified libraries were QCed with Bioanalyzer 2100 and then paired-end 2x150bp sequenced using the Illumina sequencing platform.
2.5 Variant calling
Variants were called using a Predicine in-house developed analysis pipeline, which starts from the raw sequencing data and outputs the final mutation calls. Briefly, the pipeline first performed adapter trimming, barcode checking, and correction. Cleaned paired FASTQ files were outputted by the in-house pipeline and further aligned to the human reference genome build hg19 using BWA alignment tool. Consensus bam files were then derived by merging paired-end reads originated from the same molecules (based on mapping location and unique molecular identifiers) as single strand fragments. Single strand fragments from the same double strand DNA molecules were further merged as double stranded. Both sequencing and PCR errors were deeply suppressed during this process.
Candidate variants, consisting of point mutations, small insertions and deletions, were identified across the targeted regions covered in the panel. A variant identified in cfDNA was considered a candidate somatic mutation only when (i) the number of distinct fragments containing a particular mutation was at least 0.25% of the total distinct fragments. For hotspot variants, the number of distinct fragments containing a particular mutation was at least 0.1% of the total distinct fragments; and (ii) the variant was not present in public databases of common germline variants including 1000 genomes, ExAC, gnomAD with population allele frequency >0.5%. Candidate somatic mutations were further filtered on the basis of gene annotation to identify those occurring in protein-coding regions. Intronic and silent changes were excluded, while mutations resulting in missense mutations, nonsense mutations, frameshifts, or splice site alterations were retained. Mutations annotated as benign or likely benign in the ClinVar database were also filtered out.
Copy number variations were estimated at the gene level. The pipeline calculated the on-target unique fragment coverage, which was first corrected for GC bias, and was then adjusted to the probe level bias (estimated from a pooled reference). Each adjusted coverage profile was self-normalized (assuming diploid of each sample) first and then compared against correspondingly adjusted coverages from a group of normal reference samples to estimate the significance of the copy number variant. Amplification or deletion of gene copy number with an absolute z-score > 3 were called.
3. Result
3.1 Somatic point mutation identified in liquid biopsies
Targeted-capture NGS yielded an average sequencing depth of 20,000x from all samples. A total of 161 somatic single nucleotide variations (sSNV) and small insertions and deletions (Indels) were identified. Within expectation, EGFR was detected with the highest mutation frequencies (15/18, 83.3%) among all samples, including all plasma, pleural effusion cells and pleural effusion supernatant samples. Other recurrent mutations with high prevalence include mutations in canonical cancer-associated genes, encompassing BCR (6/18, 33.3%), SUFU (6/18, 33.3%), DNMT3A (5/18, 27.8%), TP53 (5/18, 27.8%), CTNNB1 (4/18, 22.2%), FAT1(4/18, 22.2%), CTSLTR2 (4/18, 22.2%) etc (Figure 1). As for mutation frequencies among all patients, a specific gene was considered mutated in a patient if this mutation was detected in any of the above three sample types from the patient. In this regard, EGFR was detected with mutations in 7/8 (87.5%) patients. Other recurrently mutated genes included TP53 (4/8, 50.0%), BCR (3/8, 37.5%), SUFU (3/8, 37.5%), DNMT3A (3/8, 37.5%), CTNNB1 (2/8, 25.0%), FAT1 (2/8, 25.0%), CYSLTR2 (2/8, 25.0%). Mapped into Reactome signaling pathway database (accessed in Oct. 2020), mutations were observed enriched in pathways of growth factor receptor (EGFR, CBL, CTNNB1, AKT1, CTNNA1, PGR, EP300, PIK3CA, PDGFRA, NTRK1, NTRK2) (7/8, 87.5%), DNA repair pathways (ERCC6, FANCC, BRCA1, APEX1, ERCC1, TP53) (5/8, 62.5%), hedgehog signaling pathway (SUFU, GLI1) (4/8, 50.0%), NOTCH signaling pathway (NOTCH1, NOTCH3) (2/8, 25.0%) and etc.
We then compared the consistency of mutation detection among three matched samples from individual patients. In result, a total of 16 mutations were shared by all three matched samples from the same patients. 7 mutations were found only shared between plasma and pleura effusion supernatant, while 4 mutations were shared between pleura effusion cells and supernatant. A total of 22, 9, and 21 mutations were private to plasma, pleura effusion cells and pleura effusion supernatant respectively. Specifically, in P1, four mutations were shared by all the three samples (BCR, SUFU, FAT1, CYSLTR2). DNMT3A mutation was detected in both plasma and pleura effusion supernatant, while DOT1L, C7orf76 and EGFR mutation was only detected in pleura effusion supernatant. 5 mutations were shared by all the samples in P2, including mutations in EGFR, TGFBR2, FLCN, ERBB3 and BRCA1. Mutations in APEX1, NTRK2 and TBX3 were observed in both plasma and pleura effusion supernatant. 10 mutations were private to the plasma, while 2 and 9 mutations were private to pleura effusion cells and supernatant respectively. In P3, 4 mutations were shared by all three samples. Mutations in DNMT3A and TET2 were identified in both plasma and pleura effusion supernatant, whereas NKX3-1 mutation was shared by pleura effusion cells and supernatant. In addition, 2 and 6 mutations were found private to the plasma and pleura effusion supernatant samples. P4 harbored mutations in EGFR and GLI1 in both of the plasma and pleura effusion supernatant samples, while 1, 2 and 3 mutations were detected privately in the plasma, pleura effusion cells and supernatant accordingly. In P6, only EGFR mutation was shared across all the three samples. BCR, SUFU and ALK mutations were shared between two pleura effusion samples whereas TP53 mutation was shared between plasma and pleura effusion supernatant. 10 and 5 mutation were observed private in plasma and pleura effusion cells respectively. Among all patients excluding two with only plasma samples collected, 22/45 (number of private mutations in plasma/number of all mutations in plasma; 48.9%), 9/29 (number of private mutations in pleural effusion cells/number of all mutations in pleural effusion cells; 31.0%) and 21/48 (number of private mutations in pleural effusion supernatant/number of all mutations in pleural effusion supernatant; 43.8%) mutations were found private to plasma, pleural effusion cells and pleural effusion supernatant accordingly.
3.2 Somatic copy number variations identified in liquid biopsies
A total of 54 copy number variations were identified from all samples (Figure 2). EGFR was observed with the highest frequencies of variations (8/18, 44.4%), where all 8 changes were amplifications. Other recurrent amplifications were loss of PPP2R2A (4/18, 22.2%), gain of AR (3/18, 16.7%), loss of BRCA2 (3/18, 16.7%), gain of CCND1 (3/18, 16.7%), loss of CDKN2A (3/18, 16.7%), loss of PTEN (3/18, 16.7%), gain of CD274 (3/18, 16.7%), loss of RB1 (3/18, 16.7%) and etc (Figure 2). Among all patients, amplifications of EGFR were identified in 4/8 (50.0%) patients. Loss of PPP2R2A, BRCA2, CDKN2A, RB1 were all found in 3/8 (37.5%), while gain of AR, CCND1, AKT3 were identified in 2/8 (25.0%) patients. Pathway analysis revealed the enrichment of EGFR-dependent growth factor receptor pathway (BCL2, CCND1, AKT3, EGFR) (5/8, 62.5%) and PI3K/AKT signaling pathway (PIK3CB, FGFR4, PTEN, PIK3CA, AKT3, ERBB2, MET, EGFR) (5/8, 62.5%).
Surprisingly, relatively less consistent mutational patterns of somatic CNVs were observed among plasma, pleural effusion cells and pleural effusion supernatant. In P1, somatic CNVs were only identified in pleural effusion supernatant, including gain of EGFR and loss of BRCA2, CDKN2A, PTEN, RB1 and ATM. P2 harbored amplifications of EGFR and CD274 in all of the three samples and loss of PPP2R2A, PTEN, gain of AR, FGFR4 and BCL2 in only plasma and pleural effusion supernatant. Loss of RB1, BAP1 were observed private in pleural effusion supernatant while loss of NF2 was specific to the plasma sample. In P3, only EGFR amplification was found shared between two pleural effusion samples while other CNVs including loss of PPP2R2A, BRCA2, CDKN2A and gain of MET were observed private to pleural effusion supernatant. Similarly, P4 harbored most of the somatic CNVs only in pleural effusion supernatant, including gain of AR, CCND1, AKT3, CCND2, PIK3CA, MYCN and loss of PPP2R2A, BRCA2, CDKN2A, RB1, BAP1, IKZF1, while gain of EGFR and CRKL were both identified in pleural effusion cells and pleural effusion supernatant. In P6, gain of CCND1 and ERBB2 were found in both of the two pleural effusion samples, while gain of AKT3 and PIK3CB were found specific to pleural effusion supernatant and plasma samples respectively. In summary, 26/38 (68.4%; number of private CNVs in pleural effusion supernatant/number of all CNVs in pleural effusion supernatant) somatic CNVs detected in pleural effusion supernatant were not detected in the other matched samples, and only 2/9 (22.2%; number of private CNVs in plasma/number of all CNVs in plasma) CNVs in plasma were found tissue-specific. While all 7 CNVs detected in pleural effusion cells could also be observed in either matched plasma or pleural effusion supernatant sample. In regard of CNVs, pleural effusion supernatant and plasma might be more suitable biopsies yielding more comprehensive mutational information.
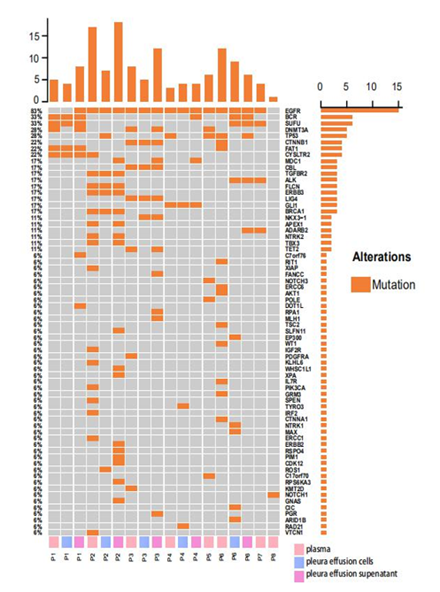
Figure 1: Somatic SNVs and Indels detected from plasma, pleural effusion cells and pleural effusion supernatant from 8 EGFR-mutated lung cancer patients; panel on the top demonstrates the number of mutations detected in each sample; panel at the right indicates the numbers of mutations identified in respective genes; panel at the bottom shows the sample ID where P represents patient, and plasma, pleura effusion cells and pleura effusion supernatant were labeled in pink, blue and violet accordingly.
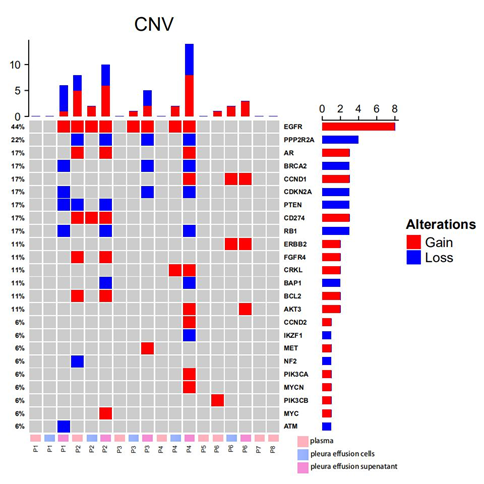
Figure 2: Somatic CNVs detected from plasma, pleural effusion cells and pleural effusion supernatant from 8 EGFR-mutated lung cancer patients; the panel on the top illustrates the number of CNV detected in each sample; the panel at the right indicates the number of CNV identified in each gene; the panel at the bottom exhibits the sample ID, where P stands for patient, and plasma, pleura effusion cells and pleura effusion supernatant were labeled in pink, blue and violet accordingly.
4. Discussion
Osimertinib showed efficacy superior to that of standard EGFR-TKIs in the first-line treatment of EGFR mutation-positive advanced NSCLC [35]. However, all patients eventually developed resistance to osimertinib. Several mechanisms of resistance to osimertinib have been described, such as acquired KRAS mutations, EGFR T790M loss and targetable gene fusions [36]. Basically, the resistance mechanisms are divided into EGFR-dependent and EGFR-independent. EGFR-dependent mechanisms include EGFR rare mutation [37] whereas examples of EGFR-independent mechanisms include alternative kinase activation [38, 39] and histological transformation [40]. Other reported mechanisms of acquired mechanisms include mutations like BRAF V600E, EGFR G796D, EGFR C797S, EGFR L718Q, MET amplification etc [13-17]. Previous studies have characterized the consistency between pleural effusion and plasma samples in lung cancer patients. A real-world research published recently have demonstrated that pleural effusion supernatant detected actionable mutations with higher sensitivity than plasma samples in M1a disease but not in M1b/c disease, showing different mutational profiles in these two samples [41]. Besides, the study also showed that pleural supernatant outperformed plasma in identifying mutations that confer resistance to first and second generation TKIs [41]. These results were relatively consistent with the present studies, and our results further substantiated that pleural effusion might detect mutations with an improved sensitivity in some patients while plasma might be preferred in the others, indicating that sequencing both plasma and pleural effusion might yield optimal sensitivity. Also, we enriched Jin et al’s findings that pleural effusion could be used in detecting resistance to first and second generation TKIs, by showing that possible mechanisms to third-generation TKI were also observed. Besides, studies exhibited only a low amount of circulating tumor cells could be detected in plasma of lung cancer patients compared to other cancer types [31], indicating that pleural effusion samples were indeed better choice for liquid biopsies in lung cancer patients. Recent researches also demonstrated the use of various types of liquid biopsies to characterize genetic changes in lung cancer patients. Song et al showed that comparable mutational landscapes were observed in pleural effusion-derived exosomal DNA and pleural effusion cfDNA, suggesting that pleural effusion exosomal DNA could be a potential alternative source for genetic testing [42]. Lee et al reported that extracellular vesicle-derived DNA (EV-DNA) from pleural effusion detected EGFR mutations with higher sensitivity than pleural effusion cfDNA and tissue biopsies, indicating that pleural effusion EV-DNA might suitable in predicting resistance to TKIs [43]. In our study, we have showed in addition to plasma cfDNA and pleural effusion supernatant, pleural effusion cells could also be used in genetic testing, providing with an alternative source of biopsy for lung cancer.
In general, somatic SNVs detected across three sample types were considered consistent. Mutations in canonical cancer driver genes were mostly shared by matched samples, including EGFR, BCR, SUFU, FAT1, ERBB3 and BRCA1, indicating that these mutations might be clonal mutations that occurred at an early stage of tumorigenesis, so that they existed in most of the tumor cells making it easy to detect from either plasma or pleural effusion, and they might play pivotal roles in driving tumor progression. However, TP53, another essential cancer driver gene, was only identified in pleural effusion cells in P2 and plasma in P4, suggesting that some key drivers might be missed when sequencing only one of the sample types. Therefore, testing pleural effusion in addition to plasma might be able to provide a more comprehensive and concrete mutational landscape of tumors. Notably, we also noticed that in two patients, EGFR mutation was not observed in some of the samples, namely plasma and pleural effusion cells of P1 and the plasma sample of P8, while fluorescent PCR result indicated they harbored EGFR T290M mutation. This discrepancy was possibly due to the difference in the sequencing techniques utilized, and again indicated that sequencing only one liquid biopsy might not be sufficient to reveal the whole picture of genetic alterations in patients.
We also noted that the two components of pleural effusion samples harbored relatively different mutations in some patients, such as P2 and P6, and this was conceivably reasonable, considering about the different source of DNA contained in these two components. DNA from the pleural effusion supernatant were mainly cell-free DNA that released from cell apoptosis, necrosis, exosome secretion, and other cellular activities [43, 44]. In comparison, DNA from pleural effusion cells were mainly genomic DNA from a variety of cells in pleural effusion [45], including immune cells, tumor cells etc. Sequencing both pleural effusion cells and supernatant might be able to avoid missing any key mutations that potentially contribute to drug resistance and compromise therapeutic outcomes, and also helps get a better knowledge of the detailed tumor micro-environment. Intriguingly, more mutations were found shared between matched plasma and pleural effusion supernatant than between plasma and pleural effusion cells, indicating that plasma and pleural effusion supernatant might shared a substantially similar contents, possibly released by tumor cell apoptosis. Also, more private mutations were found in these two sample types compared to pleural effusion cells, suggesting that compared to cellular gDNA, cfDNA could better reveal the heterogeneity of the tumor samples and yield a systemic overview of the genetic alterations in patients. Few CNVs were detected in plasma and pleural effusion cells samples. A possible explanation is the tumor DNA fraction in plasma and pleural effusion was considerably low, and targeted-capture sequencing could only cover limited chromosomal segments, therefore the detection of somatic CNVs might not be sensitive as the whole-exome sequencing [34] of tissue biopsies. Another limitation of this study was the modest sample size. Whether deep WES sequencing of a larger size of liquid biopsy cohort can accurately detect somatic CNVs awaits further investigations.
Looking at the somatic SNVs and CNVs, we proposed several potential mechanisms underlying patients’ resistance to third-generation TKIs. First of all, mutations in PI3K-AKT signaling pathway was observed recurrently mutated, and copy number changes in related genes AKT3, ERBB2, PTEN etc were identified in several patients. In previous studies, PI3K/AKT pathway was indicated to be the down-stream signaling pathway orchestrated by EGFR. PI3K encodes proteins to function with RAS and ANG1/TIE2 and further regulate PTEN functions to induce ADK and mTOR signaling transduction, finally facilitating cellular survival and prevent apoptosis [46-50]. It was also suggested that PI3K-AKT pathway mutations were stringently associated with patients’ resistance to targeted therapies in breast cancer [47]. PI3CA mutations were also an indicator of chemoresistance [51]. NOTCH signaling pathway is another pathway frequently mutated in our cohort, and was indicated to play pivotal roles in various cancer types. Essentially, NOTCH reacts to signaling from cyclin D and Sox-9 on the plasma membrane of the tumor cell, and triggering down-stream beta-catenin/Tcf-4 signaling and eventually enticing tumor cell proliferation [52-56]. Mutations in hedgehog pathway, regulated by interaction of Hh, Ptc and Smo proteins and functioning to trigger Kif/sufu/Gli signaling were also related to cell cycle, survival and proliferation in cancer [57-59]. Of particular interest, we also observed high prevalence of somatic SNVs and CNVs in genes involved in DNA damage repair, including ERCC6, FANCC, BRCA1, APEX1, ERCC1 etc, which was indicated to be a powerful indicator of patients’ response to PARP inhibitors in ovarian and breast cancers [60-62] as well as therapies targeting DNA damage checkpoint in gastric cancer [63]. Based on these findings, we concluded that these signaling pathways might be potential mechanisms underlying patients’ acquired resistance to third-generation TKIs, and they might also be the candidate targets to develop novel therapies in third-generation TKI resistant patients.
In summary, mutations identified by pleural effusion supernatant was quite consistent to plasma samples. The small sample size is the major limitation of this study, more investigations with larger sample sizes are needed to gain a deeper understanding in the mechanisms of third-generation TKI resistance and the clinical practice of liquid biopsies of lung cancer. Private mutations detected in plasma, pleural effusion cells or pleural effusion supernatant indicated that sequencing single liquid biopsies might not be sufficient to obtain a concrete and comprehensive picture of the genetic alterations in lung cancer patients. Sequencing pleural effusion in addition to plasma can help avoid missing important alterations related to drug resistance. Somatic mutations and copy number changes in PI3K/AKT pathway, NOTCH signaling pathway, hedgehog signaling pathway and genes involved in DNA damage repair might be potential mechanisms underlying third-generation TKI resistance, serving as the potential therapeutic targets to develop novel drugs for TKI resistant patients.
References
- Chee-Seng Tan, Nesaretnam Barr Kumarakulasinghe, Yi-Qing Huang, et al. Third generation EGFR TKIs: current data and future directions. Mol Cancer 17 (2018): 29
- Jianlin Xu, Bo Jin, Tianqing Chu, et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: a real-world study in China. Lung Cancer 96 (2016): 87-92.
- Becker A, Crombag L, Heideman D A M, et al. Retreatment with erlotinib: regain of TKI sensitivity following a drug holiday for patients with NSCLC who initially responded to EGFR-TKI treatment Eur J Cancer 47 (2011): 2603-2606.
- Tony S Mok, Yi-Long Wu, Sumitra Thongprasert, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361 (2009): 947-957.
- Youngwook Kim, Jeonghun Ko, ZhengYun Cui, et The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Ther 11 (2012): 784-791.
- Wu S-G, Shih J-Y. Management of acquired resistance to EGFR TKI–targeted therapy in advanced non-small cell lung cancer. Molecular Cancer 38 (2018).
- Sullivan I, Planchard D J. Next-generation EGFR tyrosine kinase inhibitors for treating EGFR-mutant lung cancer beyond first line. Front Med (Lausanne) 3 (2017): 76.
- Ma C, Wei S, Song Y J J. T790M and acquired resistance of EGFR TKI: a literature review of clinical reports. J Thorac Dis 3 (2011): 10-18.
- Darren AE Cross, Susan E Ashton, Serban Ghiorghiu, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer 4 (2014): 1046-1061
- Wu B, Gu X, Zhang Q J J. Cost-effectiveness of osimertinib for EGFR mutation–positive non–small cell lung cancer after progression following first-line EGFR TKI therapy 13 (2018): 184-193.
- Tony S Mok, Yi-Long Wu, Myung-Ju Ahn, et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med 376 (2017): 629-640.
- Mok T S, Wu Y-L, Papadimitrakopoulou V A J T N E. Osimertinib in EGFR T790M-positive lung cancer. N Engl J Med 376 (2017): 1993-1994.
- Huang Y, Gan J, Guo K, et al. Acquired BRAF V600E mutation mediated resistance to osimertinib and responded to osimertinib, dabrafenib, and trametinib combination therapy 14 (2019): e236-e237.
- Di Zheng, Min Hu, Yu Bai, et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget 8 (2017): 49671.
- Shuhang Wang, Stella T Tsui, Christina Liu, et al. EGFR C797S mutation mediates resistance to third-generation inhibitors in T790M-positive non-small cell lung cancer. J Hematol Oncol 9 (2016): 1-5.
- Puyu Shi, You-Take Oh, Guojing Zhang, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 380 (2016): 494-504.
- Zhe Yang, Nong Yang, Qiuxiang Ou, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non–small cell lung cancer patients Clin Cancer Res 24 (2018): 3097-3107.
- Attili I, Karachaliou N, Conte P, et al. Therapeutic approaches for T790M mutation positive non-small-cell lung cancer. Expert Rev Anticancer Ther 18 (2018): 1021-1030.
- Marzia Del Re, Stefania Crucitta, Giulia Gianfilippo, et al. Understanding the mechanisms of resistance in EGFR-positive NSCLC: from tissue to liquid biopsy to guide treatment strategy. Int J Mol Sci 20 (2019): 3951.
- Malapelle U, Rolfo C, Cancer I S. Liquid biopsy as a follow-up tool: Comment on longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor–tyrosine kinase inhibitors. Cancer 126 (2020): 22-25.
- Aparna R Parikh, Ignaty Leshchiner, Liudmila Elagina, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 25 (2019): 1415-1421.
- Chang L K, Chang Y L, Shih, et al. Primary resistance to osimertinib despite acquired T790M. Respirol Case Rep 8 (2020): e00532.
- Wang S, Song Y, Liu Delong. EAI045: the fourth-generation EGFR inhibitor overcoming T790M and C797S resistance. Cancer Lett 385 (2017): 51-54.
- Andersen J R, Steven K. Implantation metastasis after laparoscopic biopsy of bladder cancer. J Urol 153 (1995): 1047-1048.
- Mingwei Ma, Hongcheng Zhu, Chi Zhang, et al. “Liquid biopsy”—ctDNA detection with great potential and challenges. Ann Transl Med 3 (2015): 235.
- Leslie Calapre, Lydia Warburton, Michael Millward, et al. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett 404 (2017): 62-69.
- Kenneth S Thress, Roz Brant, Hedley Carr T, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 90 (2015): 509-515.
- Zheng D, Ye X, Zhang M Z, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 6 (2016): 1-9.
- Martin Reck, Koichi Hagiwara, Baohui Han, et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol 11 (2016): 1682-1689.
- Xin Yi, Jianhui Ma, Yanfang Guan, et al. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer 140 (2017): 2642-2647.
- Guillaume Herbreteau, Audrey Vallée, Sandrine Charpentier, et al. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): clinical application and future perspectives. J Thorac Dis 11 (2019): S113.
- Changcun Pan, Bill H Diplas, Xin Chen, et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol 137 (2019): 297-306.
- Zhou Z. et al. American Society of Clinical Oncology (2017).
- Suzanne Jenkins, James C-H Yang, Suresh S Ramalingam, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non–small cell lung cancer. J Thorac Oncol 12 (2017): 1061-1070.
- Jean-Charles Soria, Yuichiro Ohe, Johan Vansteenkiste, et al. Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer. N Engl J Med 378 (2018): 113-125.
- Geoffrey R Oxnard, Yuebi Hu, Kathryn F Mileham, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M–positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 4 (2018): 1527-1534.
- Benjamin P Brown, Yun-Kai Zhang, David Westover, et al. On-target resistance to the mutant-selective EGFR inhibitor osimertinib can develop in an allele-specific manner dependent on the original EGFR-activating mutation. 25 (2019): 3341-3351.
- Catherine A Eberlein, Daniel Stetson, Aleksandra A Markovets, et al. Acquired resistance to the mutant-selective EGFR inhibitor AZD9291 is associated with increased dependence on RAS signaling in preclinical models. Cancer Res 75 (2015): 2489-2500.
- Zofia Piotrowska, Hideko Isozaki, Jochen K Lennerz, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 8 (2018): 1529-1539.
- Adam J Schoenfeld, Joseph M Chan, Daisuke Kubota, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 26 (2020): 2654-2663.
- Shidai Jin, Chengzhi Zhou, Xue Hou, et al. A multicenter real-world study of tumor-derived DNA from pleural effusion supernatant in genomic profiling of advanced lung cancer. Transl Lung Cancer Res 9 (2020): 1507.
- Song Z, Cai Z, Yan J, et al. Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl Lung Cancer Res 8 (2019): 392.
- Jong Sik Lee, Jae Young Hur, In Ae Kim, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 18 (2018): 1236.
- Stanislav Volik, Miguel Alcaide, Ryan D Morin, et al. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 14 (2016): 898-908.
- Unger K.M, Raber M, Bedrossian C W, et al. Analysis of pleural effusions using automated flow cytometry. Cancer 52 (1983): 873-877.
- Qing-Bai She, David B Solit, Qing Ye, et al. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells 8 (2005): 287-297.
- Nicole M Davis, Melissa Sokolosky, Kristin Stadelman, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget 5 (2014): 4603.
- Srinivas Papaiahgari, Qin Zhang, Steven R Kleeberger, et al. Hyperoxia stimulates an Nrf2-ARE transcriptional response via ROS-EGFR-PI3K-Akt/ERK MAP kinase signaling in pulmonary epithelial cells. Antioxid Redox Signal 8 (2006): 43-52.
- Gallardo A, Lerma E, Escuin D, et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer 106 (2012): 1367-1373.
- Galatea Kallergi, Sofia Agelaki, Antonia Kalykaki, et al. Phosphorylated EGFR and PI3K/Akt signaling kinases are expressed in circulating tumor cells of breast cancer patients. Breast Cancer Res 10 (2008): R80.
- Bryan T Hennessy, Ana-Maria Gonzalez-Angulo, Katherine Stemke-Hale. et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69 (2009): 4116-4124.
- Eric J Allenspach, Ivan Maillard, Jon C Aster, et al. Notch signaling in cancer. Cancer Biol Ther 1 (2002): 466-476.
- Rizzo P, Osipo C, Foreman K, et al. Rational targeting of Notch signaling in cancer. Oncogene 27 (2008): 5124-5131.
- Victoria Bolós, Joaquín Grego-Bessa, José Luis de la Pompa. Notch signaling in development and cancer. Endocr Rev 28 (2007): 339-363.
- Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Current Opinion in Genetics & Development, 17 (2007): 52-59.
- Spyros Stylianou, Rob B Clarke, Keith Brennan. Aberrant activation of notch signaling in human breast cancer. Cancer Res 66 (2006): 1517-1525.
- Marina Pasca di Magliano, Matthias Hebrok. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer 3 (2003): 903-911.
- Jin Jiang, Chi-chung Hui. Hedgehog signaling in development and cancer. Dev Cell 15 (2008): 801-812.
- Sarah P Thayer, Marina Pasca di Magliano, Patrick W Heiser, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 425 (2003): 851-856.
- Huiting Wei, Xiaochun Yu. Proteomics & bioinformatics. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics 14 (2016): 131-139.
- Mike De Vos, Valérie Schreiber, Françoise Dantzer. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 84 (2012): 137-146.
- Huber A, Bai P, De Murcia J M, et al. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development 3 (2004): 1103-1108
- Xiaoting Lin, Dongshao Chen, Cheng Zhang, et al. Augmented antitumor activity by olaparib plus AZD1775 in gastric cancer through disrupting DNA damage repair pathways and DNA damage checkpoint. J Exp Clin Cancer Res 37 (2018): 129.


 Impact Factor: * 4.1
Impact Factor: * 4.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 