Spatial-timely Quantitative Network Analysis for TGF-β pathway of Tumor Infiltrating Lymphocytes
Article Information
Yun Yang1,4#, Wenqin Li2,5#, Shuyi Chen3,Yuan Yuan4, Biaoru Li5*
1BS Undergraduate Student in Guangdong Medical University, Dongguan, Guangdong, 523808, China
2BS, Department of Chemistry, University of California, Irvine, CA 92697, USA
3Shuyi Chen, MD candidate, Geisinger Commonwealth School of Medicine, Scranton, PA 18510, USA
4Institute of Aging Research, Guangdong Medical University, Dongguan, Guangdong 523808, China
5Senior Faculty of Principal Research Scientist, Department of Pediatrics and GA Cancer Center, Children Hospital at GA, Augusta, GA 30913, USA.
#Yan Yang and Wenqin Li are both co-first authors.
*Corresponding Author: Biaoru Li, Senior Faculty of Principal Research Scientist and Full Member, Clinical Bioinformatics Specialist, Pediatric Oncology and Hematology Division, Children Hospital of Georgia, Augusta, GA 30912, USA.
Received: 25 October 2023; Accepted: 03 November 2023; Published: 10 November 2023.
Citation:
Yun Yang, Wenqin Li, Shuyi Chen,Yuan Yuan, Biaoru Li. Spatial-timely Quantitative Network Analysis for TGF-β pathway of Tumor Infiltrating Lymphocytes. Journal of Cancer Science and Clinical Therapeutics 7 (2023): 194-203.
View / Download Pdf Share at FacebookAbstract
Tumor-infiltrating lymphocytes (TILs) are to be subject to clinical applications by cultured TIL infusion in vivo for adoptive cell therapy (ACT) and by ex vivo TIL analysis for determining immune characteristics to kill autologous tumor cells so that TIL has been administrated tumor’s patients to immune-cell therapy and analyze patients’ immune characteristics for tumor diseases. To study the TIL features, we have established quantitative network modeling by TIL’s TCR signaling pathway, IL2 pathway, and TGF-β pathway for personalized immunotherapy about more than fifteen years. However, machine-learning analysis still has some challenges under the traditional quantitative pathway for network configurations to apply for patient treatment. For example, multiple protein complexes competing for downstream DNA binding-site or protein-protein complex will generate different effects. To address this question, we report here a temporal-spatial quantification network, termed a spatial-timely quantification network, to address the spatial-timely competition of complex proteins binding to downstream proteins or DNA in network analysis. After studying spatial-timely quantitative network modeling by TGF-β pathway activity in spatial-timely order, we discover that multiple protein complexes using spatial-timely quantitative networks are much better than traditional quantitative networks. Once the new system modeling is established, we can further analyze all pathways, such as the TCR signaling pathway and IL2 pathway from TIL, for different immunotherapy.
Keywords
Tumor-infiltrating lymphocytes (Tils), Pathway and network, Quantitative pathway and network, Spatial-timely quantitative pathway and network, Personalized therapy.
Tumor-infiltrating lymphocytes (Tils) articles; Pathway and network articles; Quantitative pathway and network articles; Spatial-timely quantitative pathway and network articles; Personalized therapy articles.
Tumor-infiltrating lymphocytes (Tils) articles Tumor-infiltrating lymphocytes (Tils) Research articles Tumor-infiltrating lymphocytes (Tils) review articles Tumor-infiltrating lymphocytes (Tils) PubMed articles Tumor-infiltrating lymphocytes (Tils) PubMed Central articles Tumor-infiltrating lymphocytes (Tils) 2023 articles Tumor-infiltrating lymphocytes (Tils) 2024 articles Tumor-infiltrating lymphocytes (Tils) Scopus articles Tumor-infiltrating lymphocytes (Tils) impact factor journals Tumor-infiltrating lymphocytes (Tils) Scopus journals Tumor-infiltrating lymphocytes (Tils) PubMed journals Tumor-infiltrating lymphocytes (Tils) medical journals Tumor-infiltrating lymphocytes (Tils) free journals Tumor-infiltrating lymphocytes (Tils) best journals Tumor-infiltrating lymphocytes (Tils) top journals Tumor-infiltrating lymphocytes (Tils) free medical journals Tumor-infiltrating lymphocytes (Tils) famous journals Tumor-infiltrating lymphocytes (Tils) Google Scholar indexed journals Pathway and network articles Pathway and network Research articles Pathway and network review articles Pathway and network PubMed articles Pathway and network PubMed Central articles Pathway and network 2023 articles Pathway and network 2024 articles Pathway and network Scopus articles Pathway and network impact factor journals Pathway and network Scopus journals Pathway and network PubMed journals Pathway and network medical journals Pathway and network free journals Pathway and network best journals Pathway and network top journals Pathway and network free medical journals Pathway and network famous journals Pathway and network Google Scholar indexed journals Quantitative pathway and network articles Quantitative pathway and network Research articles Quantitative pathway and network review articles Quantitative pathway and network PubMed articles Quantitative pathway and network PubMed Central articles Quantitative pathway and network 2023 articles Quantitative pathway and network 2024 articles Quantitative pathway and network Scopus articles Quantitative pathway and network impact factor journals Quantitative pathway and network Scopus journals Quantitative pathway and network PubMed journals Quantitative pathway and network medical journals Quantitative pathway and network free journals Quantitative pathway and network best journals Quantitative pathway and network top journals Quantitative pathway and network free medical journals Quantitative pathway and network famous journals Quantitative pathway and network Google Scholar indexed journals Spatial-timely quantitative pathway and network articles Spatial-timely quantitative pathway and network Research articles Spatial-timely quantitative pathway and network review articles Spatial-timely quantitative pathway and network PubMed articles Spatial-timely quantitative pathway and network PubMed Central articles Spatial-timely quantitative pathway and network 2023 articles Spatial-timely quantitative pathway and network 2024 articles Spatial-timely quantitative pathway and network Scopus articles Spatial-timely quantitative pathway and network impact factor journals Spatial-timely quantitative pathway and network Scopus journals Spatial-timely quantitative pathway and network PubMed journals Spatial-timely quantitative pathway and network medical journals Spatial-timely quantitative pathway and network free journals Spatial-timely quantitative pathway and network best journals Spatial-timely quantitative pathway and network top journals Spatial-timely quantitative pathway and network free medical journals Spatial-timely quantitative pathway and network famous journals Spatial-timely quantitative pathway and network Google Scholar indexed journals Personalized therapy articles Personalized therapy Research articles Personalized therapy review articles Personalized therapy PubMed articles Personalized therapy PubMed Central articles Personalized therapy 2023 articles Personalized therapy 2024 articles Personalized therapy Scopus articles Personalized therapy impact factor journals Personalized therapy Scopus journals Personalized therapy PubMed journals Personalized therapy medical journals Personalized therapy free journals Personalized therapy best journals Personalized therapy top journals Personalized therapy free medical journals Personalized therapy famous journals Personalized therapy Google Scholar indexed journals heterogeneous T-cells articles heterogeneous T-cells Research articles heterogeneous T-cells review articles heterogeneous T-cells PubMed articles heterogeneous T-cells PubMed Central articles heterogeneous T-cells 2023 articles heterogeneous T-cells 2024 articles heterogeneous T-cells Scopus articles heterogeneous T-cells impact factor journals heterogeneous T-cells Scopus journals heterogeneous T-cells PubMed journals heterogeneous T-cells medical journals heterogeneous T-cells free journals heterogeneous T-cells best journals heterogeneous T-cells top journals heterogeneous T-cells free medical journals heterogeneous T-cells famous journals heterogeneous T-cells Google Scholar indexed journals autologous tumor cells articles autologous tumor cells Research articles autologous tumor cells review articles autologous tumor cells PubMed articles autologous tumor cells PubMed Central articles autologous tumor cells 2023 articles autologous tumor cells 2024 articles autologous tumor cells Scopus articles autologous tumor cells impact factor journals autologous tumor cells Scopus journals autologous tumor cells PubMed journals autologous tumor cells medical journals autologous tumor cells free journals autologous tumor cells best journals autologous tumor cells top journals autologous tumor cells free medical journals autologous tumor cells famous journals autologous tumor cells Google Scholar indexed journals personalized immunotherap articles personalized immunotherap Research articles personalized immunotherap review articles personalized immunotherap PubMed articles personalized immunotherap PubMed Central articles personalized immunotherap 2023 articles personalized immunotherap 2024 articles personalized immunotherap Scopus articles personalized immunotherap impact factor journals personalized immunotherap Scopus journals personalized immunotherap PubMed journals personalized immunotherap medical journals personalized immunotherap free journals personalized immunotherap best journals personalized immunotherap top journals personalized immunotherap free medical journals personalized immunotherap famous journals personalized immunotherap Google Scholar indexed journals multiple protein complexes articles multiple protein complexes Research articles multiple protein complexes review articles multiple protein complexes PubMed articles multiple protein complexes PubMed Central articles multiple protein complexes 2023 articles multiple protein complexes 2024 articles multiple protein complexes Scopus articles multiple protein complexes impact factor journals multiple protein complexes Scopus journals multiple protein complexes PubMed journals multiple protein complexes medical journals multiple protein complexes free journals multiple protein complexes best journals multiple protein complexes top journals multiple protein complexes free medical journals multiple protein complexes famous journals multiple protein complexes Google Scholar indexed journals immunotherapy articles immunotherapy Research articles immunotherapy review articles immunotherapy PubMed articles immunotherapy PubMed Central articles immunotherapy 2023 articles immunotherapy 2024 articles immunotherapy Scopus articles immunotherapy impact factor journals immunotherapy Scopus journals immunotherapy PubMed journals immunotherapy medical journals immunotherapy free journals immunotherapy best journals immunotherapy top journals immunotherapy free medical journals immunotherapy famous journals immunotherapy Google Scholar indexed journals
Article Details
1. Introduction
Tumor-infiltrating lymphocytes (TILs, a group of heterogeneous T-cells) have made use of both (1) administrating TIL adoptive cell therapy (ACT) [1] and (2) studying immune characteristics regarding T-cell, B-cell, and NK cells, macrophage and neutrophil within the tumor tissues [2] so that TIL have been engaged to effectively treat different tumors and discover characteristics of immune response to tumor cells from the patients with tumor diseases to guide other immune therapy under the TIL characteristics from patients [3-4]. In the early periods between 1989 and 1994, we discovered that TIL could be used to treat solid tumors under more than three hundred TIL treatments for patients with solid tumors, but TIL ACT was found to have variable efficacy in treating solid tumor diseases during the early periods [5-10]. In order to study the efficacy of treating solid tumor diseases, we began to set up tumor tissue inventory [11] and study TIL genomic profiles, including setting up single-cell genomics profiles from TILs (including single-cell mRNA differential display from TIL published in 2007 [12] and single-cell RNA-seq from TIL published in 2013 [13]), establishing TIL quantitative network published in 2015 [14], and studying machine-learning analysis by artificial intelligence (AI) from TIL in 2022 [15]. Now, we know at least three quantitative networks, including the TCR signaling pathway, IL2 pathway, and TGF-β pathway, for guidance in immune diagnosis and treatment. Moreover, some physicians and clinical scientists have also applied genomic information such as SNPs from TIL and tumor cells to treat diseases such as CAR-T and TCR-T-cell or tumor vaccines [16-20]. Suppose a gene expression pattern using network configurations with or without SNP from TIL or tumor cells can be employed. In that case, they can significantly support personalized T-cell therapy and other immune treatments. However, many reports found that protein-protein complexes bind downstream DNA in spatial-timely orders, producing different effects for the network analysis, such as competitive inhibition or competitive activation to regulate downstream pathways [21]. To address this issue, in the manual, we are subject to study a new model to explain the regulation of protein-protein complexes for further downstream DNA binding analysis. Moreover, we compare the new module to previous methods published during 2014-2022. The current module focuses on the TGF-β pathway with protein-protein complexes bound to downstream DNA by a spatial-timely quantification network. This manual concentrates majorly on SMAD complexes and downstream transcription factor binding sites for spatial-timely quantitative modeling. The purpose of the manual is to discover more accurate evaluations for personalized immune therapy. After building the new model, finally, we shall use the modules for all pathways within TIL, such as the TCR signaling pathway and the IL2 pathway.
2. Materials and Methods
2.1 Mimic complex for the spatial-timely quantitative network:
To study the spatial-timely quantitative network from the complex proteins, quiescent TIL-cell pathway hubs derived from our previous published genomic profile [22-23] and targeting TGF-β pathway observed from the published STKE database. The new configurative hubs in the network platform focus on SMAD complex and downstream combined transcriptional factor binding sites [24-25]. The spatial-timely quantitative network modelling from complex regulation is SMAD complex (Smad-2, Smad-3, and Smad-4) with combination ratio (0%, 25%, 50%, 75%, and 100%) and downstream transcriptional factor complex binding sites with combination ratio (0%, 25%, 50%, 75%, and 100%) under our Python script plugin into Cytoscape (http://www.cytoscape.org/) which we have reported [26-27]. We mimic SMAD complex regulation and transcriptional factor complex binding sites and focus on spatial-timely simulation under TGF-β/SMAD signalling-dependent activation of the TGF-β pathway.
2.2 Comparing the new model to a traditional quantitative network in silico:
To compare the new spatial-timely quantitative network to traditional quantitative network, the conventional topology analysis is used by http://www.cytoscape.org as our previous reports or other publications, including “Betweenness Centrality, BC” and low “Connectivity Centrality, DC” and other values, indicating a significant targeting node. BC is the shortest path that goes through a given node over all pairs of nodes of the fraction, and DC is connectivity or degree centrality [28-30]. According to the research goal, the final mimic included a TCR signal, IL2 activating pathway with CTL for TNF-α, INF-γ, GrB, and PRF1, and inhibiting quiescent TGF-β pathway. In order to set up the comparison research, the analysis in silico only studies a TGF-β/SMAD pathway in a T-cell network. The combination values of the activities and inhibition begin with (1) zero and additional levels of activation (0%, 25%, 50%, 75%, and 100%), (2) the timely regulation of complex activity in quiescent TGF-β pathway and (3) spatial-timely simulation in quiescent TGF-β pathway.
2.3 Experimental study on the new module:
According to TIL CD8 immune networks, at least three pathways are majorly involved in T-cell immune response to tumor cells: CD8+T-cell receptor (TCR) signal from TCR antigen associated with MHC class I; T-cells reactivated with IL-2 to produce TNF-α (Tumor necrosis factor alpha), IFN-γ (Interferon-gamma), GrB or GZMB (Granzyme-B) and PRF1 (perforin-1) to kill tumor cells; TGF-β pathway to keep quiescent TIL-cell in tumor tissues. In order to confirm the spatial-timely pathway, we in silico mine inhibiting transcriptional factors by TF binding site by http://bioinfo.wilmer.jhu.edu/PDI/index.html. After discovering downstream DNA binding sites in promoters within TNF-α, IFN-γ, GrB, and PRF1, and SMAD complex in the TGF-β pathway related to CTL, we used TF to experiment to study the new modules. Briefly, cloned TIL from liver tumor tissues were cultured for 21 days, which were isolated and cloned as in the previous report. After day-0 to day-12 culture, the growth medium was replaced with Opti-MEM Invitrogen, Carlsbad, CA). For each transfection, 100nm and 200nm targeting ERF siRNA (sc-43754, because we found that ERF bound all four genes related with CTL) with scrambled control using DharmaFECT (Dharmacon, Lafayette, CO) according to the manufacturer’s protocol [31-32]. Each experimental condition was performed in triplicate. After cells were transfections for 72 hours, the cells were harvested and assay mRNA, protein, and CTL assay. Briefly, mRNA expression was measured ERF, TNF-α, INF-γ, GrB, and PRF1 by Q-rtPCR assay, protein level measurement was determined for ERF (sc-398269), TNF-α, INF-γ by Western blot. Our previous publications have reported all the methods [33-40]. To study functional TIL cytotoxic activity, cytotoxic T lymphocyte (CTL) assays were defined by MTT as our previous reports [41-45]. Briefly, Cytotoxic T lymphocyte (CTL) assays Cytotoxic T lymphocyte (CTL) was assayed for the HepG2 cell line as previous report. TIL was first measured for effector: target ratio by 1:25 and 1:50 at culture from day-12 to day-15.
3. Results
3.1 Results for mimicking complex for spatial-timely quantitative network:
To study spatial-timely quantitative network from the complex proteins, in silico experiment is SMAD complex regulation from TGF-β/SMAD signaling-dependent activation of quiescent TIL-cell pathway. The establishment of the network hubs is SMAD complex (Smad-2, Smad-3, and Smad-4) and transcriptional factors binding sites under our Python script. After the spatial-timely quantitative network modeling with SMAD complex performed combination-regulation ratio (0%, 25%, 50%, 75%, and 100%), we can harvest combination regulation activity. In order to visualize the spatial-timely quantitative network from the complex proteins, as Figure-1A showed in silico results with SMAD complex regulation (Smad2, Smad3, and Smad4). The results demonstrated SMAD complex regulation ratio (0%, 25%, 50%, 75%, and 100%), including spatial-timely combination shown in Figure-1B. Eventually, we can harvest maximal spatial-timely combination-regulation with the activity of SMAD complex including 15,625 combination regulation (data omitted), including maximal reactivity values (spatial- timely order) for SMAD complex, so that results in silico can discover maximal regulation from a TGF-β/SMAD signaling-dependent activation of the TGF-β pathway as Figure-1C.
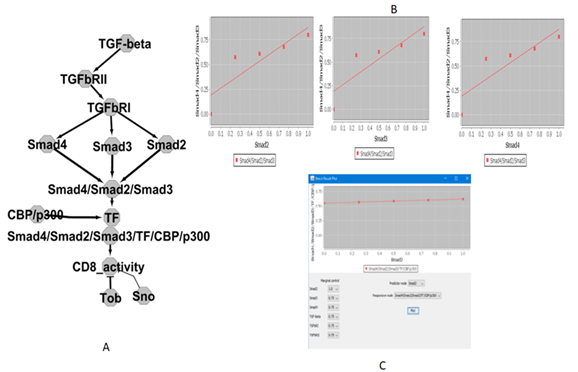
Figure 1: SMAD complex regulation ratio.
It showed in silico results with SMAD complex regulation (Smad2, Smad3 and Smad4). (0%, 25%, 50%, 75%, and 100%) including timely and spatial combination. B) It shows Smad2, Smad3 and Smad4 activity of SMAD complex including 15,625 combination regulation. C) Maximal reactivity values (timely and spatial order) for SMAD complex.
3.2 Comparing the new modelling to previous quantitative analysis:
To compare the new spatial-timely quantitative network to traditional quantitative network, the conventional topology analysis included BC, DC, and other values supported by the Cytoscape platform, which we have largely applied for, and in silico results demonstrated that the quantitative pathways had all values as possibility eleven liner-regulation (non-combination-regulation) (data omitted). All hubs and edges of the traditional quantitative pathway visualized TGF-β/SMAD signaling-dependent activation of the TGF-β pathway are shown in Figure 2A. To compare the new spatial-timely quantitative network from the complex proteins to the previous modelling, the mimic complex combines with the maximal spatial-timely with complex regulation including (1) activated SMAD2 and SMAD3 and SMAD4 with 0%, 25%, 50%, 75%, and 100% combination-regulation and other reaction regulation ratio as 0%, 25%, 50%, 75%, and 100%; (2) timely reactive regulation under the ratio and (3) spatial-timely reaction with the ratio. Finally, we can achieve maximal repeat regulation, including 390,625 combination regulation and activity. The visualized reactivity with spatial-timely complex regulation is demonstrated in Figure-2B. The complex regulations are much larger than those from traditional quantitative methods, even if hubs are also coming from similar TGF-β/SMAD signaling-dependent activation of the TGF-β pathway.
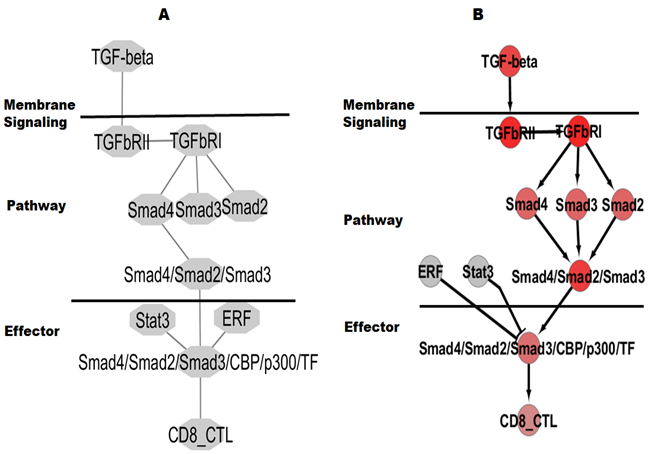
Figure 2: Traditional network and spatial-timely quantitative network.
A) Shown is a traditional network with 11 reactivity and B) Maximal reactivity values (timely and spatial order) for SMAD complex.
3.3. Experimental Results on the new module:
Three pathways are majorly involved in T-cell immune CTL to tumor cells: CD8+T-cell receptor (TCR) signal from TCR antigen associated with MHC class I; T-cells reactivated with IL-2 to induce TNF-α, INF-γ, GrB and PRF1; TGF-β pathway for quiescent TIL-cell in tumor tissues. The experiment focuses on TGF-β pathway for quiescent TIL-cell who’s cloned TIL have reported for several quiescent genes our previous reports [51]. The ERF and STAT3 (marking yellow) of transcriptional factors are shown at Table-1 which had also been discovered and reported by our previous publication [51].
Table 1: TF prediction for CTL with Quiescent T-cells
|
Genes |
TNF-alpha |
IFN-gamma |
Granzyme-B |
Perforin-1 |
|
Tob |
No |
No |
No |
No |
|
KLF2 |
No |
No |
No |
No |
|
Ski |
No |
No |
No |
No |
|
Sno-A |
No |
No |
No |
No |
|
ERF |
Yes |
Yes |
Yes |
Yes |
|
REST |
Yes |
No |
No |
No |
|
TCF-1 |
No |
No |
No |
No |
|
Bach-2 |
Yes |
No |
Yes |
Yes |
|
FOXO1 |
Yes |
No |
Yes |
No |
|
STAT3 |
Yes |
Yes |
Yes |
Yes |
We also performed a Transfac and TESS (Transcription Element Search System) in silico search and discovered Smad complex (Smad-2, Smad-3, and Smad-4) predicted binding site in promoter binding sites of TNF-α, INF-γ, GrB, and PRF1 as shown in Table-2.
Table 2: TF Prediction CTL within Smad pathway
|
Genes |
TNF-alpha |
IFN-gamma |
Granzyme-B |
Perforin-1 |
|
CBP (CITED1) |
Yes |
Yes |
Yes |
Yes |
|
p300 (HAT) |
Yes |
No |
No |
No |
|
Smad-2 |
Yes |
Yes |
Yes |
Yes |
|
Smad-3 |
Yes |
Yes |
Yes |
Yes |
|
Smad-4 |
Yes |
Yes |
Yes |
Yes |
|
MSG1 |
No |
No |
No |
No |
|
SMF |
No |
No |
No |
No |
|
ARC105 (MED15) |
No |
No |
No |
No |
|
AP-1 |
Yes |
Yes |
No |
Yes |
|
RUNX |
No |
No |
No |
No |
|
bZIP |
Yes |
Yes |
Yes |
Yes |
|
bHLH |
Yes |
Yes |
Yes |
Yes |
After ERF and STAT3 were mined by transcriptional binding in promoters of TNF-α, INF-γ, GrB, and PRF1 for whole T-cell function such as CTL while SMAD2 and SMAD3 and SMAD4 related to TGF-β pathway to inhibit CTL discovered by TF binding in promoters of TNF-α, INF-γ, GrB, and PRF1. As a research goal, ERF knockdown to confirm T-cell CTL reactivity under SMAD related to TGF- β pathway, two-step cloned TIL was cultured for these research. That is, normal TIL culture in the first step, and TIL was cultured in the functional induction and inhibition at the second step as in the previous report [14, 46]. The results of targeting protein demonstrated in Figure-3A indicate ERF expressing level 69% in the 100nm group and 45% in the 200nm group. TNF-α mRNA is 1.90 and 4.9 fold higher in transfected TILs than those in the scramble control group, respectively, to compare the 100nm and 200nm scramble TIL group (significantly increased on 200nm group, P=0.01513). The IFN-γ level is 1.1 and 3.9 fold higher in transfected TILs than those in 100nm and 200nm scramble TIL groups (significantly increased on 200nm group, P=0.0094). GrB mRNA is 0.66 and 3.11 fold higher in transfected TILs than those in the scramble control group, respectively, to compare the 100nm and 200nm scramble TIL group (significantly increased on 200nm group, P=0.00473). PRF levels are 2 and 4.93 fold higher in transfected TILs than those in 100nm and 200nm scramble TIL groups (significantly increased on 200nm group, P=0.00016). As Figure-3B, the results of the Western blot further confirmed for TNF-α and IFN-γ expression. Figure-3C, indicate ERF expressing level 111.47% in the 100nm group and 45.184% in the 200nm group. TNF-α protein is 1.029 and 1.802 fold higher in transfected TILs than those in the scramble control group, respectively, to compare the 100nm and 200nm scramble TIL group (increased on 200nm group although P=0.0837). The IFN-γ level is 1.040 and 2.060 fold higher in transfected TILs than those in 100nm and 200nm scramble TIL groups (significantly increased on 200nm group, P=0.0058). The results of CTL killing activity showed 32% and 56% in the transfected TILs group to compare 29% in the scrambled group at 200 nm, shown in Figure-3D. Cytotoxic T lymphocyte of knockdown ERF TIL was much higher than scrambled groups (significantly increased on 200nm group, P=0.0117).
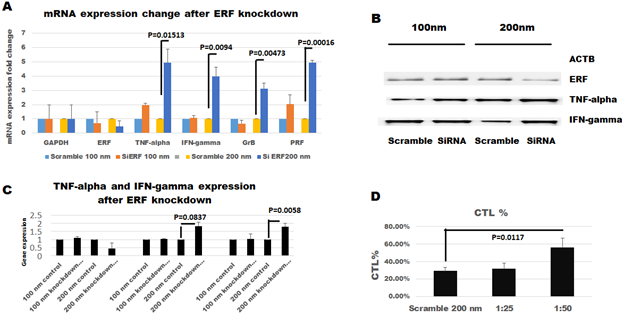
Figure 3: Experimental results from TIL.
A) Day-15 RT-qPCR quantification of targeting mRNA. B) Confirmation of ERF down regulation by Western blot at day-15. C) Confirmation with increase expression of TNF-α and IFN-γ under quantitative Western blot after ERF down regulation at day-15. D) Confirmation CTL increases with ERF down regulation on day-15.
4. Discussion
TGF-β is a pleiotropic cytokine from the tumor microenvironment (TME) that suppresses CD8+ cells in TILs, although it can show different regulation under some particular situations [47]. Active TGF-β binds to the dimeric TGF-β receptor type 2 (TGFbRII), a serine/threonine kinase activating downstream dimeric receptor type 1 (TGFbRI), forming a tetrameric body structure complex finally processing TGF-β pathway. TGF-β pathway has two signaling systems, either SMAD signaling pathway or the non-SMAD signaling pathway. TβRI first will be activated by phosphorylation and further phosphorylating SMAD2 and SMAD3 and then binding SMAD4 with the phosphorylated SMAD2 and SMAD3 to produce a heterotrimeric transcription complex, which transfers the signal to the nucleus to perform transcription function called as TGF-β/SMAD signaling [48] while SMAD-independent pathway of the TGF-β pathway process through RHO GTPases, RAS pathway, P38 pathway, and mTOR pathway according to current research [49]. Some scientists have studied the dynamic TGF-β-inducing phosphorylation of Smad2 and Smad3 to perform IP experiments with mass spectrometry [50]. These measurements can define non-phosphorylated Smad2 and Smad3, one phosphorylated Smad2 and Smad3 at the most C-terminal serine residue (pSmad2 at Ser467; pSmad3 at Ser425), and two phosphorylated Smad2 and Smad3 at the C-terminal serine residues (ppSmad2 at Ser465 and Ser467; ppSmad3 at Ser423 and Ser425). According to their research, they can measure the activity procedures mass spectrometry by predicting 84 possible combination regulations, timely and spatial [50]. In order to address a complex regulation, as the results are shown above, we set up a spatial-timely quantitation pathway; our results can demonstrate maximal spatial-timely combination-regulation with activity of SMAD complex including 15,625 combination regulation and activity so that maximal reactivity values can be found from 15,625 combination regulation so that it will more powerful than IP with mass spectrometry analysis. Although we have used the traditional quantitative network to analyze clinical specimens for more than fifteen years, in silico results demonstrated that the quantitative pathways had all values as possibility eleven reactivity for non-combination regulation to SMAD signaling pathway while the mimic complex combining with complex regulation, spatial-timely can be achieved maximal regulation including 390,625 combination regulation and activity from similar TGF-β/SMAD signaling-dependent activation of the TGF-β pathway. The combination of spatial-timely quantitative pathways from 390.625 activities can predict more powerful than those from traditional quantitative networks as above or those from IP with mass spectrometry analysis [50]. To confirm the spatial-timely quantitative network, we finally use experiment ex vivo to study the experiment in silico. As we all know, three pathways are majorly involved in T-cell immune CTL to tumor cells: CD8+T-cell receptor (TCR) signal from TCR antigen associated with MHC class I; T-cells reactivated with IL-2 to induce TNF-α, INF-γ, GrB, and PRF1; TGF-β pathway for quiescent TIL-cell in tumor tissues. After we found that ERF has a binding site in promoters in CTL genes such as TNF-α, INF-γ, GrB, and PRF1 while SMAD complex also involves the four promoters related to TGF-β/SMAD signaling-dependent activation of the TGF-β pathway, ERF down-regulation was used to study spatial-timely pathway for T-cell CTL. The mRNA results demonstrated TNF-α, IFN-γ GrB, and PRF1 are better than our previous results from traditional pathway configuration [51-55]. Western blot also supported mRNA results. The cytotoxic T lymphocyte (CTL) assays CTL of knockdown ERF TIL was much higher. The experiment data still support the spatial-timely quantitative pathway related to TIL function. Cancer immunotherapy mainly exerts either active stimulation of the immune system by vaccination or adoptive immunotherapy by cell therapy such as TIL, nature killer cells, cytokine-induced peripheral blood mononuclear cells (lymphokine-activated killer, LAK; cytokine-induced killer cells, CIK; dendritic cells and cytokine-induced killer cells, DC-CIK), neutrophil and macrophage [56-60]. According to published evidence, TILs obtained from tumor tissues with a higher population of CD8 cells, but they also have other immune cells, such as T-cell, B-cell, NK cells, macrophage, and neutrophil [61-68]. Because we have spent about 30 years of efforts to culture TIL and primary tumor cells [69], we found that TIL has a variable effect. In order to study TIL efficacy, we have studied TIL characteristics for (1) TIL infusion in vivo for personalized T-cell therapy under personalized genomic analysis and (2) TIL personalized genomic analysis ex vivo for determining all immune characteristics to kill autologous tumor cells. Based on the R&D, we have been researching quantitative network modeling to study TIL's TCR signaling pathway, IL2 pathway, and TGF-β pathway to individual subjects for personalized immunotherapy. After we set up tumor tissue inventory, single-cell genomics analysis, T-cell cloning and culture techniques, and primary tumor cell culture technique into system modeling [70], we can now use the information to activate and inhibit the genes by targeting in vivo for combination therapy or ex vivo for analysis of other immune cells. The manual can accurately evaluate quantitative networks to address multiple immune treatments such as TIL immunotherapy and other personalized immunotherapy.
Conclusion:
TIL have both clinical applications by TIL infusion in vivo for immune therapy and by TIL analysis ex vivo for determining the immune characteristics of tumor patients. Although the TIL ACT in vivo has been largely extended into clinics, TIL ACT had variable efficacy in treating solid tumor diseases. In order to study the efficacy of treating solid tumor diseases and analyse other immune treatments for all tumor patients, we have been studying personalized TIL therapy and analyzing other immune therapy from genomics profiles from both TILs and primary tumor cells. The TIL spatial-timely quantitative network with machine-learning analysis can support protein-protein complex binding downstream DNA. Because the spatial-timely quantitative network is a more powerful evaluation than current mass spectrometry, we can use the new model to address the regulation of protein-protein complexes, eventually, greatly support personalized T-cell therapy and other immune diagnosis and treatment.
Acknowledgments:
Under the support of Dr. H. D. Preisler, we have set up a method to analyze genomic profiles of CD3, CD4, and CD8 from TIL. This work was supported by National Cancer Institute IRG-91-022-09, USA (to BL). YY(1) and YY(4) working by YY(4) for Medical Research Foundation of Guangdong Province (A2020173) and Funds for PHD researchers of Guangdong Medical University in 2023 (GDMUB2023005) with Ruineng Yi Individualized Medical Technology Co., Ltd. The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation.
Author Contributions:
YY (1) and WQL perform experimental fields and in silico experiments, including TIL isolation, culture, and measurement. SYC perform biostatistics; YY (4) teaching YY (1) rtPCR and Western blot. BL conceived and designed the experiments.
Competing Interests Statement:
The authors declare competing financial interests.
References
- Rosenberg SA, Spiess P, Lafreniere R. A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science 233 (1986): 1318-13121.
- Paijens ST, Vledder A, de Bruyn M, et al. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol 18 (2021): 842-859.
- Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med 319 (1988): 1676-1680.
- Kumar A, Watkins R, Vilgelm AE. Cell Therapy with TILs: Training and Taming T Cells to Fight Cancer. Front Immunol 12 (2021): 690499.
- Li B, Tong SQ, Zhang XH, et al. A new experimental and clinical approach of combining usage of highly active tumor-infiltrating lymphocytes and highly sensitive antitumor drugs for the advanced malignant tumor, Chin Med J (Engl) 107 (1994): 803-807.
- Li B, Tong SQ, Zhang XH, et al. Development of adoptive T-cell immunotherapy-Future of personalized immunotherapy. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 137-154.
- Hu BC, Li GW, Wei C, et al. Clinical application of infiltrating lymphocytes in malignant brain tumors, Journal of Immunology (Chinese) 2 (1997): 1-2.
- Hua ZD, Lu J, Li HF, et al. Clinical study of tumor infiltrating lymphocytes in ovarian cancer. Chinese Journal of Obstetrics and Gynecology (Chinese) 31 (1996): 55-57.
- Lu J, Hu LW, Hua ZD, et al. Analysis of the therapeutic effects of different therapeutic approaches for TIL; Chinese Journal Cancer Biotherapy (Chinese) 3 (1996): 127-129.
- Cai XM, Lu J, Hua ZD, et al. Clinical Application of TIL from Different Sources; Journal of Immunology (Chinese) 12 (1996): 251-254.
- Li B, Ding J, Larson A, et al. Tumor Tissue Recycling - A New Combination Treatment for Solid Tumors: Experimental and Preliminarily Clinical Research. Anticancer (IN VIVO) 13 (1999): 1-6.
- Zhang W, Ding J, Qu Y, et al. Genomic expression analysis of quiescent CD8 T-cells from tumor-infiltrating lymphocytes of in vivo liver tumor by single-cell mRNA differential display, Immunology 127 (2009): 83-90.
- Xu YB, Hu HL, Zheng J, et al. Feasibility of whole RNA sequencing from single-TIL cell mRNA amplification. Genetics of Research International 4 (2013): 1.
- Li B, Hu HL, Ding J, et al. Functional cell-proliferation and differentiation by system modelling for cell therapy. IJLSRST (2015): 1-11.
- Ying XN and Li B. Machine-learning Modeling for Personalized Immunotherapy- An Evaluation Module. Biomedical Journal of Scientific and Technical Research (2022): 38211-38216.
- Li S, Perabekam S, Devemy E, et al. genetically modified T-cells affinity to tumor cells. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 174-179.
- Li B. Breakthrough of 2015-Personalized immunotherapy. iMedPub Journals 1 (2015): 1-2.
- Li B, Liu G, Hu HL, et al. Biomarkers Analysis for Heterogeneous Immune Responses of Quiescent CD8+cells -A Clue for Personalized Immunotherapy. iMedPub Journals 1 (2015): 1-12.
- Lu YC, Yao X, Crystal JS, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 20 (2014): 3401-3410.
- Jeon EY, Choi DS, Choi S, et al. Enhancing adoptive T-cell therapy with fucoidan-based IL-2 delivery microcapsules. Bioeng Transl Med 8 (2022): 10362.
- Chaudhary A, Raza SS, Haque R. Transcriptional factors targeting in cancer stem cells for tumor modulation. Semin Cancer Biol 88 (2023): 123-137.
- Li B, Perabekam S, Liu G, et al. Experimental and bioinformatics comparison of gene expression between T cells from TIL of liver cancer and T Cells From UniGene. J Gastroenterol 37 (2002): 275-282.
- Li B. Identification of mRNAs expressed in tumor-infiltrating lymphocytes by A strategy for rapid and high throughput screening. GENE 255 (2000): 273-279.
- Nguyen TH, Han TH, Newfeld SJ, et al. Selective Disruption of Synaptic BMP Signaling by a Smad Mutation Adjacent to the Highly Conserved H2 Helix. Genetics 216 (2020): 159-175.
- Kaminska B, Cyranowski S. Recent Advances in Understanding Mechanisms of TGF Beta Signaling and Its Role in Glioma Pathogenesis. Adv Exp Med Biol 1202 (2020): 179-201.
- Wang ZH, Hu HL, Zheng J, et al. Gene Expression and Pathway Analysis of Quiescent CD8+ T Cells from Liver Cancer, Liver Sinusoid and Peripheral Blood - Study on Toxicogenomics and Prevention Targeting, BIBE (2011): 72-76.
- Zheng J, Zhang D, Przytycki PF, et al. SimBoolNet--a Cytoscape plugin for dynamic simulation of signaling networks. Bioinformatics 26 (2010): 141-142.
- Zhang W, Qu Y, Lin MH, et al. Immune cells signaling-pathway and genomic profiles for personalized immunotherapy. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 20-32.
- Liu G, Zheng J and Li B. Bioinformatics of T-cell and primary tumor cells. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 118-136.
- Wang NX, Zheng JJ. Computational studies of H5N1 influenza virus resistance to oseltamivir. Protein Sci 18 (2009): 707-715.
- Oseghale AR, Zhu X, Li, B, et al, Conjugate prodrug AN-233 induces fetal hemoglobin expression in sickle erythroid progenitors and β-YAC transgenic mice. Blood Cells, Molecules, and Diseases 79 (2019): 102345.
- Starlard-Davenport A, Smith A, Vu L, et al. (2019). MIR29B mediates epigenetic mechanisms of HBG gene activation. Br J Haematol (2019): 1-10.
- LI B, Preisler HD, Gao XZ, et al. Poor prognosis AML. II. Biological and molecular biological characteristics. Leukemia Research 24 (2000): 777-789.
- LI B, Gao XZ, Tao M, et al. Telomerase activity in preleukemia and AML. Leukemia and Lymphoma 36 (2000): 579-587.
- Min Tao, LI B. SCF, IL-1 beta, IL-1ra and GM-CSF in the bone marrow and serum of normal individuals and of AML CML patients. Cytokine 12 (2000): 699-707.
- Preisler HD and LI B. Suppression of telomerase activity and cytokines mRNA levels in AML in vivo in patients by Amifostine and IL-4. Cancer Research 6 (2000): 807-812.
- Mundle SD, BY Mativi, JD Cartlidge, et al. Signal antonymy unique to MDS correlates with altered expression of E2F1. British Journal of Hematology 109 (2000): 376-381.
- Yang J and LI B. Tyrosine phosphorylation of Shc proteins in normal CD34- progenitor cells and leukemia cells. Blood 94 (1999): 373-374.
- Preisler HD, Gao XZ, Tao M, et al. Marrow cytokine transcripts and the secondary hematologic disorders. Leukemia and lymphoma 35 (1999): 297-302.
- Perambakam S, LI B. Quantitation of Interferon regulatory factor transcripts in patients with AML. Cancer Investigation 19 (2001): 346-351.
- Wang JH, Tong SQ, Li B, et al. Immunological Character of TIL in Ovarian Carcinoma. Chinese Journal of Cancer Research (Chinese) 12 (2000): 99-104.
- Gu QL, Lin YQ, Yin HR, et al. Preliminary study on cryopreservation of tumor infiltrating lymphocytes. Journal of Immunology (Chinese) 04 (1195): 251-252.
- Tong SQ, Wang JH, Li L, et al. Biological characteristics of invasive lymphocytes from ovarian cancer; Chinese Journal of Immunology (Chinese) 16 (2000): 131-134.
- Hu BY, Tong SQ, Wang JH, et al. Immunological characteristics of TIL from ovarian cancer; Chinese Journal of Cancer (Chinese) 4 (1999): 1-5.
- Li B, Tong SQ, Zhu YM, et al. Establishment of a method for separation of tumor infiltrating lymphocytes with high vitality; Journal of Immunology (Chinese) 10 (1994): 44-47.
- Li B, Perabekam S, Larson A, et al. Primary cell culture and T-cell cloning. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 97-114.
- Salmond RJ. Regulation of T-Cell Activation and Metabolism by Transforming Growth Factor-Beta. Biology (Basel) 12 (2023): 297.
- Abarca-Buis RF, Mandujano-Tinoco EA, Cabrera-Wrooman A, et al. The complexity of TGFβ/activin signaling in regeneration. J Cell Commun Signal 15 (2021): 7-23.
- Li X, Fang Y, Chen L, et al. Bone morphogenetic protein 4 inhibits rat stem/progenitor Leydig cell development and regeneration via SMAD-dependent and SMAD-independent signaling. Cell Death Dis 13 (2022): 1039.
- Lucarelli P, Schilling M, Kreutz C, et al. Resolving the Combinatorial Complexity of Smad Protein Complex Formation and Its Link to Gene Expression. Cell Syst 6 (2018): 75-89.
- Li B. Why will TIL produce different efficacy to treat solid tumor. Frontier in Immunology 3 (2022): 1-13.
- Li B. A strategy to identify genomic expression profiles at single-T-cell level and a small number of cells (review paper). Journal of Biotechnology (2005): 71-82.
- Li B, Zheng J and Liu G. (2020). Chapter-12: System modeling of T-cell function. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 197-223.
- Kongkaew T, Thaiwong R, Tudsamran S, et al. TIL expansion with high dose IL-2 or low dose IL-2 with anti-CD3/anti-CD28 stimulation provides different quality of TIL-expanded T cell clones.J Immunol Methods 503 (2022): 113229.
- Knochelmann HM, Rivera-Reyes AM, Wyatt MM, et al. 1.Modeling ex vivo tumor-infiltrating lymphocyte expansion from established solid malignancies. Oncoimmunology 10 (2021): 1959101.
- Schoenberg MB, Li X, Li X, et al. The predictive value of tumor infiltrating leukocytes in Hepatocellular Carcinoma: A systematic review and meta-analysis. Eur J Surg Oncol 47 (2021): 2561-2570.
- González-Tablas Pimenta M, Otero Á, Arandia Guzman DA, et al. Tumor cell and immune cell profiles in primary human glioblastoma: Impact on patient outcome. Brain Pathol 31 (2021): 365-380.
- Kim SR, Chun SH, Kim JR, et al. The implications of clinical risk factors, CAR index, and compositional changes of immune cells on hyperprogressive disease in non-small cell lung cancer patients receiving immunotherapy. BMC Cancer 21 (2021): 19.
- Qiu P, Guo Q, Lin J, et al. An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer. Sci Rep 12 (2022): 22322.
- Li Y, Chen S, Li X, et al. CD247, a Potential T Cell-Derived Disease Severity and Prognostic Biomarker in Patients With Idiopathic Pulmonary Fibrosis. Front Immunol 12 (2021): 762594.
- Chen Y, Klingen TA, Aas H, et al. CD47 and CD68 expression in breast cancer is associated with tumor-infiltrating lymphocytes, blood vessel invasion, detection mode, and prognosis. J Pathol Clin Res 9 (2023): 151-164.
- López-Janeiro Á, Villalba-Esparza M, Brizzi ME, et al. The association between the tumor immune microenvironments and clinical outcome in low-grade, early-stage endometrial cancer patients. J Pathol 258 (2022): 426-436.
- Ding K, He Y, Wei J, et al. A score of DNA damage repair pathway with the predictive ability for chemotherapy and immunotherapy is strongly associated with immune signaling pathway in pan-cancer. Front Immunol 13 (2022): 943090.
- Wang F, Cathcart SJ, DiMaio DJ, et al. Comparison of tumor immune environment between newly diagnosed and recurrent glioblastoma including matched patients. J Neurooncol 159 (2022): 163-175.
- Darvishian F, Wu Y, Ozerdem U, et al. Macrophage density is an adverse prognosticator for ipsilateral recurrence in ductal carcinoma in situ. Breast 64 (2022): 35-40.
- Lund MC, Ellman DG, Nissen M, et al. The Inflammatory Response after Moderate Contusion Spinal Cord Injury: A Time Study. Biology (Basel) 11 (2022): 939.
- Hanamura T, Kitano S, Kagamu H, et al. Immunological profiles of the breast cancer microenvironment represented by tumor-infiltrating lymphocytes and PD-L1 expression. Sci Rep 12 (2022): 8098.
- Yoshida C, Kadota K, Yamada K, et al. Tumor-associated CD163+ macrophage as a predictor of tumor spread through air spaces and with CD25+ lymphocyte as a prognostic factor in resected stage I lung adenocarcinoma. Lung Cancer 167 (2022): 34-40.
- Li B. Biobank for personalized immunotherapy. Personalized Immunotherapy for Tumor Diseases and Beyond (2020): 224-247.
- Li B. Personalized Immunotherapy of Patients: Defining by Single-cell RNA-seq with Artificial Intelligence. Medical Research Archives 11 (2023): 1-14.


 Impact Factor: * 4.1
Impact Factor: * 4.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 