In vitro and In Vivo Evidences Propound Potential of Lipocalin 2 as a Therapeutic Target in Cervical Carcinoma
Article Information
Nehanjali Dwivedi1,2, Tahmina Mazumder3, Shivani Tihara3, Gayathri Veeraraghavan4, Ramanujam Siva5, Smitha P K6, Rohit Ranade7, Anil K M8, Sujan K Dhar3*, Manjula Das1*
1Molecular Immunology, MSMF, Narayana Health City, Bangalore 560099, India
2MAHE, Manipal, 576104, India
3Computational Biology, MSMF, Narayana Health City, Bangalore 560099, India
4Centre for Toxicology and Developmental research, CEFTE, SRIHER, Porur, Chennai 600116, India
5Centre for Toxicology and Developmental Research, Porur, Chennai - 600116, India
6Product Research Group, MSMF, Narayana Health City, Bangalore 560099, India
7Department of Gynaecology Oncology, Mazumdar Shaw Medical Centre, Narayana Health City, Bangalore 560099, India
8Beyond Antibody, Dodda Bommasandra, Bangalore 560097, India
*Corresponding Author: Dr. Manjula Das, Molecular Immunology and Dr Sujan K Dhar, Computational Biology, MSMF, Narayana Health City, Bangalore 560099, India
Received: 06 November 2023; Accepted: 14 November 2023; Published: 04 December 2023.
Citation: Nehanjali Dwivedi, Tahmina Mazumder, Shivani Tihara, Gayathri Veeraraghavan, Ramanujam Siva, Smitha P K, Rohit Ranade, Anil K M, Sujan K Dhar, Manjula Das. In vitro and In Vivo Evidences Propound Potential of Lipocalin 2 as a Therapeutic Target in Cervical Carcinoma. Journal of Cancer Science and Clinical Therapeutics 7 (2023): 233-248.
View / Download Pdf Share at FacebookAbstract
Aim: This study assessed efficacy of a novel α-Lipocalin 2 (LCN2) monoclonal antibody (MAb) employing in vitro and in vivo xenograft models of cervical cancer (CC).
Main methods: Anti-proliferative and anti-invasive effects of α-LCN2 MAb was assessed in vitro on STR-authenticated-HeLa cells. Gelatin zymography and qPCR were used to decipher molecular mechanism of the observed effects. To assess its anti-tumorigenic potential in vivo, Hela cell tumors grown as mouse model xenografts were treated with α-LCN2 MAb. Treatment efficacy was assessed by tumor palpability and volume regression followed by histopathological and image texture analysis of tumor. Molecular pathways were deciphered by analyzing differential RNASeq data separately for the injected human and recruited mouse cell-components in both MAb-treated and mock-treated xenografts.
Key findings: α-LCN2 MAb caused tumor regression and softening through necrosis of both tumor and stromal cells. Differential gene expression analysis of human component indicated LCN2 inhibition abolished tumorigenic pathways while significantly increasing necrosis and cell death pathways in tumor via reduction in LCN2 and a downfall of TNFα-IL17 axis leading to further decline in LCN2 expression, suggestive of a negative feedback loop. In mouse transcriptome component, T-cell activation and decline in M2 macrophage population upon α-LCN2 treatment underlies its potential as an immune sensitizer that can devour tumor cells through T-cell mediated cytotoxicity leading to an anti-tumorigenic milieu.
Significance: Results of this study strongly suggests LCN2 to be a potential therapeutic target for CC. α-LCN2 MAb used in this study can be further developed for clinical usage in CC using intra-tumoral delivery.
Keywords
Anti-LCN2 therapeutic-MAb, Cervical Cancer, Tumor softening, RNA-Seq; Xenograft, Texture-feature analysis.
Anti-LCN2 therapeutic-MAb articles; Cervical Cancer articles; Tumor softening articles; RNA-Seq articles; Xenograft articles; Texture-feature analysis articles.
Anti-LCN2 therapeutic-MAb articles Anti-LCN2 therapeutic-MAb Research articles Anti-LCN2 therapeutic-MAb review articles Anti-LCN2 therapeutic-MAb PubMed articles Anti-LCN2 therapeutic-MAb PubMed Central articles Anti-LCN2 therapeutic-MAb 2023 articles Anti-LCN2 therapeutic-MAb 2024 articles Anti-LCN2 therapeutic-MAb Scopus articles Anti-LCN2 therapeutic-MAb impact factor journals Anti-LCN2 therapeutic-MAb Scopus journals Anti-LCN2 therapeutic-MAb PubMed journals Anti-LCN2 therapeutic-MAb medical journals Anti-LCN2 therapeutic-MAb free journals Anti-LCN2 therapeutic-MAb best journals Anti-LCN2 therapeutic-MAb top journals Anti-LCN2 therapeutic-MAb free medical journals Anti-LCN2 therapeutic-MAb famous journals Anti-LCN2 therapeutic-MAb Google Scholar indexed journals Cervical Cancer articles Cervical Cancer Research articles Cervical Cancer review articles Cervical Cancer PubMed articles Cervical Cancer PubMed Central articles Cervical Cancer 2023 articles Cervical Cancer 2024 articles Cervical Cancer Scopus articles Cervical Cancer impact factor journals Cervical Cancer Scopus journals Cervical Cancer PubMed journals Cervical Cancer medical journals Cervical Cancer free journals Cervical Cancer best journals Cervical Cancer top journals Cervical Cancer free medical journals Cervical Cancer famous journals Cervical Cancer Google Scholar indexed journals Tumor softening articles Tumor softening Research articles Tumor softening review articles Tumor softening PubMed articles Tumor softening PubMed Central articles Tumor softening 2023 articles Tumor softening 2024 articles Tumor softening Scopus articles Tumor softening impact factor journals Tumor softening Scopus journals Tumor softening PubMed journals Tumor softening medical journals Tumor softening free journals Tumor softening best journals Tumor softening top journals Tumor softening free medical journals Tumor softening famous journals Tumor softening Google Scholar indexed journals RNA-Seq articles RNA-Seq Research articles RNA-Seq review articles RNA-Seq PubMed articles RNA-Seq PubMed Central articles RNA-Seq 2023 articles RNA-Seq 2024 articles RNA-Seq Scopus articles RNA-Seq impact factor journals RNA-Seq Scopus journals RNA-Seq PubMed journals RNA-Seq medical journals RNA-Seq free journals RNA-Seq best journals RNA-Seq top journals RNA-Seq free medical journals RNA-Seq famous journals RNA-Seq Google Scholar indexed journals Xenograft articles Xenograft Research articles Xenograft review articles Xenograft PubMed articles Xenograft PubMed Central articles Xenograft 2023 articles Xenograft 2024 articles Xenograft Scopus articles Xenograft impact factor journals Xenograft Scopus journals Xenograft PubMed journals Xenograft medical journals Xenograft free journals Xenograft best journals Xenograft top journals Xenograft free medical journals Xenograft famous journals Xenograft Google Scholar indexed journals Texture-feature analysis articles Texture-feature analysis Research articles Texture-feature analysis review articles Texture-feature analysis PubMed articles Texture-feature analysis PubMed Central articles Texture-feature analysis 2023 articles Texture-feature analysis 2024 articles Texture-feature analysis Scopus articles Texture-feature analysis impact factor journals Texture-feature analysis Scopus journals Texture-feature analysis PubMed journals Texture-feature analysis medical journals Texture-feature analysis free journals Texture-feature analysis best journals Texture-feature analysis top journals Texture-feature analysis free medical journals Texture-feature analysis famous journals Texture-feature analysis Google Scholar indexed journals histopathological articles histopathological Research articles histopathological review articles histopathological PubMed articles histopathological PubMed Central articles histopathological 2023 articles histopathological 2024 articles histopathological Scopus articles histopathological impact factor journals histopathological Scopus journals histopathological PubMed journals histopathological medical journals histopathological free journals histopathological best journals histopathological top journals histopathological free medical journals histopathological famous journals histopathological Google Scholar indexed journals anti-tumorigenic articles anti-tumorigenic Research articles anti-tumorigenic review articles anti-tumorigenic PubMed articles anti-tumorigenic PubMed Central articles anti-tumorigenic 2023 articles anti-tumorigenic 2024 articles anti-tumorigenic Scopus articles anti-tumorigenic impact factor journals anti-tumorigenic Scopus journals anti-tumorigenic PubMed journals anti-tumorigenic medical journals anti-tumorigenic free journals anti-tumorigenic best journals anti-tumorigenic top journals anti-tumorigenic free medical journals anti-tumorigenic famous journals anti-tumorigenic Google Scholar indexed journals α-LCN2 MAb articles α-LCN2 MAb Research articles α-LCN2 MAb review articles α-LCN2 MAb PubMed articles α-LCN2 MAb PubMed Central articles α-LCN2 MAb 2023 articles α-LCN2 MAb 2024 articles α-LCN2 MAb Scopus articles α-LCN2 MAb impact factor journals α-LCN2 MAb Scopus journals α-LCN2 MAb PubMed journals α-LCN2 MAb medical journals α-LCN2 MAb free journals α-LCN2 MAb best journals α-LCN2 MAb top journals α-LCN2 MAb free medical journals α-LCN2 MAb famous journals α-LCN2 MAb Google Scholar indexed journals cytotoxicity articles cytotoxicity Research articles cytotoxicity review articles cytotoxicity PubMed articles cytotoxicity PubMed Central articles cytotoxicity 2023 articles cytotoxicity 2024 articles cytotoxicity Scopus articles cytotoxicity impact factor journals cytotoxicity Scopus journals cytotoxicity PubMed journals cytotoxicity medical journals cytotoxicity free journals cytotoxicity best journals cytotoxicity top journals cytotoxicity free medical journals cytotoxicity famous journals cytotoxicity Google Scholar indexed journals
Article Details
Highlights
- α-LCN2 Intra-tumoral injection regressed and softened tumor in xenograft model
- Texture -features of both tumor & stroma revealed increase in disorder upon treatment
- RNASeq of the treated tumor revealed negative feedback loop of LCN2 via IL17 axis
- The treatment increased cytotoxic T cell and decreased M2 macrophage infiltration
- The novel biologic suggests reduced dosage and resistance and combination therapy
1. Introduction
Cervical cancer (CC), the fourth most frequent form of female cancer overall, second in developing nations and third in developed nations, has a significant impact on women's health all over the world. India sees about 95-100,000 new cases every year with 18% incidence and 11.4% mortality rate [1]. Unfortunately, majority of CC cases are diagnosed in advanced stages leading to a death every 8 minutes in India. Standard of treatment for early-stage cancers (I-IIA) is radical hysterectomy with bilateral pelvic node dissection followed by adjuvant radiation and chemotherapy according to associated risk factors, while for locally advanced (IIB-IIIB) tumors, preferred therapy is a combination chemo-radiation. Treatment strategies/modalities depend upon age, overall health of patient and cancer stage. Despite the approval of MAbs [2-4] (tisotumab, bevacizumab and pembrolizumab) as the second line of therapy in CC, the treatment options are not very encouraging due to escalated cost and poor efficacy [5]. Though CC is primarily caused by HPV infections [6], involvement of various cytokines [7], including lipocalins in the disease pathology have been reported [8, 9]. Overexpression of LCN2 was found to increase the invasive potential of human CC [10, 11]. LCN2 levels were reported to be higher in advanced clinical CC stages with HPV16, a high-risk genotype of the virus that accounts for around 61% of incidences [12, 13]. LCN2 levels were significantly increased post ≥ 8 weeks of treatment with external beam radiotherapy and supplementary radiotherapy. LCN2 is known to be a potential therapeutic target in a large number of solid human malignancies. A mesenchyme- like cell morphology has been reported to be induced by LCN2 overexpression, along with SNAI1, TWIST1, CDH1, FN1, and MMP9 that promote invasiveness [14]. On the contrary, Wang et al demonstrated that nm23-H1 knockdown mediated increase in LCN2 promoter activity resulted in decreased cell migration and invasion in SiHa uterine cervical cancer cells correlating with poor prognosis and overall survival of patients [15]. However, majority of the studies implicate correlation of LCN2 levels with the disease pathogenesis. LCN2 is known to have intracellular effect through its receptors in a cell-specific manner [16] and an extracellular effect by binding to and stabilizing the protease activity of MMP9 [17]. Thus, a correlation between increased MMP9 activity and LCN2 expression has been reported in CC cell line, C33A [18]. In this study, we evaluated the effect of abrogation of LCN2 activity by treatment with an α-LCN2 MAb. In vitro effects were assessed in HeLa, the first immortal human cell, “cancer in a test tube”, from a very aggressive glandular carcinoma of the cervix [19]. The molecular mechanism of action was predicted by qPCR on the cells and verified by end-point functional assays like migration, invasion and gelatin zymography. In vivo analysis using a HeLa cell xenografts included tumor size regression, histopathological and texture analysis and transcriptome analysis of the tumor. Our findings proclaim the tumor-regressive role of the α-LCN2 MAb and potentiates its therapeutic usage in cervical cancer to improve patient prognosis.
2. Materials and Methods
2.1 Analysis of public databases:
The Cancer Genome Atlas (TCGA) is a project that has produced thorough, multidimensional maps of the major genetic alterations in many forms of cancer. Expression of the genes of interest were assessed in TCGA Cervical and Endocervical Carcinoma (TCGA-CESC) dataset using RNA-seq normalized count values acquired from Broad Institute Firehose portal to compare their expression in tumor and normal samples.
2.2 Recombinant LCN2 protein and α-LCN2 MAb production:
Full-length, codon optimized human LCN2 cDNA (NM_005564) was synthesized and cloned by GeneScript, USA, between NdeI and XhoI restriction sites of pET-28b(+) vector, expressed in E.coli Rosetta cells as a tag-less protein. The recombinant protein was purified using DEAE ion exchange chromatography (Sigma, # DCL6B100). For hybridoma generation, BALB/c mice were injected with 10µg purified LCN2 protein with Freund's complete adjuvant (Sigma, # F5881) in 1:1 ratio, followed by two boosters with a gap of 21 days each with Freund's Incomplete Adjuvant (Sigma, # F5506) in 1:1 ratio and a final injection with 50µg of protein prior to fusing spleenocytes with Sp2/O cells. Monoclonal antibody was then isolated from the cell supernatant using Protein A beads (MagGenome, # MG17PA) from the single cell cloned antibody-producing colonies obtained through HAT/HT (Sigma, # H0262/HN0137) selection.
2.3 HeLa cell culture:
HeLa cells were cultured in DMEM medium, pH 7.2, (Thermo Fisher Scientific, #11995-065) supplemented with 10% FBS (Thermo Fisher Scientific, #10270106) and 1X penicillin-streptomycin solution (Thermo Fisher Scientific, #15140122) at 37°C. The cells were confirmed for their authenticity by STR profiling (Supplementary Table 1). performed by TheraCUES Innovations, Bangalore using GenePrint 10 (Promega Corporation), version 3.0.0 SoftGenetics. GeneMarker_HID was used to analyze the result and confirm profile referring to the ATCC and CLASTR STR databases. For assessing therapeutic potential of LCN2 as a target, tumorigenicity of HeLa cells was determined under the effect of a-LCN2 MAb as described in next sections.
2.4 RNA isolation, cDNA conversion and qPCR:
RNA isolation from the cells was performed using TRIzol method, as per manufacturer’s instructions. Briefly, 1´106 cells were resuspended and incubated for 10 minutes on ice in 1mL TRIzol (Ambion, #15596018) followed by addition of 200µL chloroform (Sigma, #496189) and centrifugation at 13,000 g for 15 minutes at 4°C. The aqueous layer was transferred into a fresh tube containing equal volume of 100% isopropanol (Qualigen, # Q26897) without disturbing either the middle interface or the lower organic phase. The tube was gently invert-mixed and incubated at room temperature for 10 minutes followed by centrifugation at 13,000 g for 10 minutes at 4°C. The supernatant was discarded and the pellet was washed with 75% ethanol (HiMedia, # MB228) followed by centrifugation at 7500 g for 5 minutes at 4 °C. Air-dried pellet was then dissolved in nuclease free water for further processing and 1μg of total RNA was used for cDNA conversion using AMV Reverse Transcriptase (NEB, # M0277L) in a 20μL reaction. A set of two reference genes - ACTB and GAPDH were used for expression analysis of LCN2 and its receptors by qPCR on a Roche LightCycler 480 II instrument using KAPA SyBr green Universal (Merck, # KK4601), with each sample analyzed in triplicates. In a total of 5µL reaction volume containing SyBr mix, 1:5 diluted cDNA template, primers (100nM each), and water were used. The reaction conditions were: pre-incubation at 95°C for 10 seconds followed by 45 cycles of amplification (95°C – 1 second; 95°C – 10 seconds; 60°C – 15 seconds; and 72°C – 15 seconds). List of primers are detailed in supplementary table 2.
2.5 Proliferation Assay:
HeLa cells were seeded at a concentration of 5´103 cells per well in a 96-well plate, overnight at 37°C in 5% CO2 followed by a 12 h treatment with α-LCN2 MAb and mIgG at indicated concentrations. Proliferative potential of the cells was calculated using Alamar Blue Reagent (Sigma, # R7017) at a final concentration of 0.2mg/mL, after 2 h of incubation at 37°C. Mean ± standard deviation of relative fluorescence intensities (excitation at 560 nm and emission at 590 nm) of three independent experiments were computed
2.6 Invasion Assay:
Transwell inserts (HiMedia, # TCP083) were coated with 100 µl of 1mg/mL ECM gel (Sigma, E1270) in serum free (SF) medium followed by incubation for 2 h at 37 °C. HeLa cells (5X103) were seeded in SF medium with indicated treatments of α-LCN2 MAb and mIgG and allowed to invade for 24 h. Following invasion, the inserts were stained with 2% crystal violet (HiMedia, # S012) and quantified by eluting the stain with 10% acetic acid (Qualigen, #Q21057) as described before [20]. The findings are presented as the mean ± standard deviation of three independent experiments.
2.7 Gelatin zymography:
Gelatin zymography was performed by co-polymerizing the gels (SDS-PAGE, 10%) with gelatin (1 mg/ml) (HiMedia, #GRM019). Ten microliters of gel-loading buffer and 20 microliters of concentrated conditioned medium of cultured HeLa cells (with and without treatment with α-LCN2 antibody) were loaded onto wells of the gel. Electrophoresis was carried out at a constant voltage of 125 V, until the dye reached bottom of the gel, in cold conditions. After electrophoresis, gels were washed in washing buffer [2.5% Triton X-100 (Fisher Scientific, #10655) in 50 mM Tris-HCl (pH 7.5) (Sigma, # T5941), 5mM CaCl2 (Sigma, # C1016), 1µM ZnCl2 (Fisher Scientific, #28785)] for 1 h at room temperature with gentle agitation. Gels were then incubated for 18 h at 37°C in developing buffer [5 mM CaCl2, 1µM ZnCl2 in 50 mM Tris-HCl (pH 7.5), 1% TritonX-100] followed by staining with Coomassie blue (HiMedia, # MB153) and de-staining with 30% methanol (HiMedia, #AS059) and 10% acetic acid. Gels were acquired and photographed by gel imaging system (Molecular Imager® Gel DocTM XR; Bio-Rad Laboratories).
2.8 Animal husbandry:
Male NCr nude mice (procured from Vivo Biotech, Hyderabad, India) were housed in individually ventilated cages covered with stainless steel grid top. Temperature and humidity were maintained in the range of 19-23°C and 30-70%, respectively with 12-15 air changes/h. Animal confinement was provided with 12h artificial light photoperiod and 12h darkness. Animal experiments were performed under the approval of Institutional Animal Ethics Committee.
2.9 Xenograft cervical tumor model:
HeLa cells (5x106) were injected using a needle of size 26 gauges subcutaneously into the dorsal flanks of animals maintained under sterile and controlled conditions. Tumor volume was determined using the formula 0.5 ´ LP2) where L,P are the largest dimension and and its perpendicular dimension lengths measured using a slide caliper. Once tumors grew to about 100 mm3 in volume, they were injected intra-tumor with 100µg α-LCN2 once every alternate day. PBS was used as vehicle control in no treatment group. Animals were sacrificed when the tumor grew to ~2,000 mm3 in volume or after 2 weeks of treatment whichever was earlier.
2.10 Histopathology analysis:
Half of the tumors harvested from sacrificed animals were fixed in 10% (v/v) formalin (Sigma, #HT501128) and embedded in paraffin (Qualigen, #Q19215) for sectioning (n=3 in each group). Sections of 5µm thickness were stained with hematoxylin and eosin. Slides were analyzed for the extent of tumor boundary and regions of necrosis and other cellular events before converting to digitized images using a Grundium ocus40 whole-slide scanner.
2.11 Texture analysis of histopathology images:
H&E-stained histopathology images of treated and untreated tumor samples were analyzed to extract some of the grey-level co-occurrence matrix (GLCM) features originally proposed by Haralick et al [21]. Whole-slide images were tiled into 16 ´ 16 pixel patches with minimum 90% tissue content and colour normalized. Three basic GLCM features including entropy, contrast and dissimilarity which are measures of randomness and disorder in the images were calculated for each image patch using python scikit image library. For a visual representation, feature value of each patch was converted to a colour map with red to green colour gradient indicating maximum to minimum value of each feature.
2.12 RNA sequencing:
RNA was isolated from remaining tumor using Qiagen RNA isolation kits (Qiagen, #74004) as per manufacturer’s instructions. Library for RNAseq was prepared using illumina TruSeq library preparation kit and sequencing was performed on a NovaSeq 6000 (illumina) platform using 2 ´ 150 bp chemistry at LifeCell International, Chennai.
2.13 Transcriptome analysis:
Sequence reads were filtered for quality using Trimmomatic and filtered reads were separated based on mapping to human (GRCh38) and mouse (GRCm39) reference assembly using FastQ Screen. Separated reads were aligned to respective genome using STAR pipeline and differentially expressed genes were determined using edgeR. Functional and pathway enrichment analysis of the differentially expressed genes were carried out using EnrichR web server and ClusterProfiler R library. Gene expression heatmaps were drawn using pathway signatures from MSigDB and published literature. Estimation of mouse immune cell infiltration in the tumor was estimated using seq-ImmuCC cell type deconvolution tool from mouse gene counts.
3. Results
3.1 Expression of LCN2, receptors and ligands in Cervical Cancer:
Analysis of expression of LCN2, its known ligand MMP9, and its receptors (SLC22A17, LRP2, MC1R, MC3R and MC4R) in TCGA-CESC dataset dataset revealed that LCN2 and MMP9 have a significantly higher expression in CC compared to normal samples (Figure 1a,b), whereas SLC22A17, the most widely known LCN2 receptor showed the opposite behaviour (Figure 1c). Other receptors (LRP2, MC1R, MC3R and MC4R) did not show any significant difference between normal and tumor samples, while MC1R remained undetected (data not shown). In-house analysis using qPCR in HeLa cells revealed the same pattern, wherein, LCN2 and MMP9 are overexpressed as compared to the receptors (Figure 1b), thereby confirming only MMP9-mediated activity of LCN2 in HeLa cells.
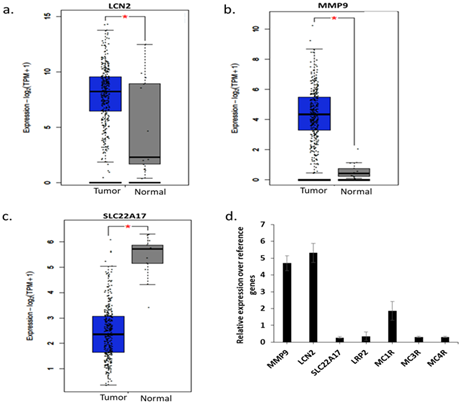
Figure 1: mRNA expression analysis. (a) LCN2 expression in TCGA database between tumor and normal samples, (b) MMP9 expression in TCGA database between tumor and normal samples, (c) SLC22A17 expression in TCGA database between tumor and normal samples , (d) qPCR analysis of expression of indicated genes on HeLa cells. *p<0.05
3.2 α-LCN2 antibody abrogates the tumorigenicity of cervical cancer cell line:
Treatment with α-LCN2 antibody decreased the proliferative ability of HeLa in a dose-dependent manner, starting as early as 12 hours (Figure 2a). Treatment with α-LCN2 antibody also reduced the invasive ability of the cells in a dose-dependent manner (Figure 2b, c). Control mIgG addition did not show any statistical significance at the equivalent highest molar concentration of α-LCN2 antibody, suggesting that the decrease in tumorigenicity is specifically due to the MAb. Expression of Claudin mRNA, a tight junction protein [22], increased upon treatment with α-LCN2 antibody suggesting restoration of cell-cell adhesion and restriction in migratory abilities of the cells (Figure 2d). HeLa cells upon treatment, showed a reduction in expression of active metalloproteases (known to form complex with LCN2 and increase invasion and matrix degradation [23]) in gelatin zymography, suggesting a quick degradation of these proteases and thus inhibiting migratory and invasive ability of HeLa cells (Figure 2e). All the data taken together suggests that LCN2 abrogation resulted in inhibition of migratory and invasive potential of HeLa cells via the reduction in MMP9 activity and not through other receptors of LCN2.
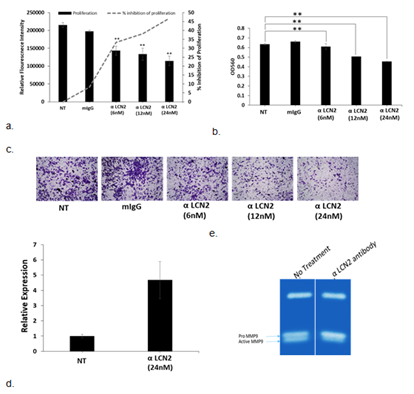
Figure 2: Effect of treatment of α-LCN2 antibody on HeLa cells. (a) Proliferative potential as measured by alamar blue indicated by solid bars (RFI), dashed line indicates the % inhibition of proliferation (b) Intensity of crystal violet measured at 590nm to quantitate the number of invaded cells with and without the treatment of α-LCN2 antibody, (c) Bright field microscopic examination of invaded cells with and without the treatment with α-LCN2 antibody, stained with crystal violet. (d) qPCR expression of Claudin 1 in HeLa cells upon treatment with α-LCN2 antibody (e) Gelatin zymography to check the MMP9 activity upon indicated treatments. All images represent 4X magnification. **p<0.05. NT – No treatment, mIgG – mouse IgG, RFI – Relative Fluorescent Intensity. α-LCN2 antibody treatments done for 12 h, error bars representing the standard deviation.
3.3 Cervical cancer xenograft tumor model and treatment:
Tumor formation was observed at the site of injection after approximately 7 days of subcutaneous HeLa cell line administration. During the time of treatment, animals appeared to be normal without any distress. After reaching a volume of 100mm3, the tumors were treated with α-LCN2 antibody at a concentration of 100µg/mL every alternate day, directly into the tumor along with the vehicle control in the no treatment group. After 2 weeks of treatment, once the control group’s tumor grew to ~200 mm3 in size, all the animals were sacrificed because of ethical considerations and tumors were resected for further analysis.
Effect of α-LCN2 on tumor morphology – Treatment with α-LCN2 antibody resulted in a significant 39% overall reduction in tumor volume compared to the volume observed on day 6 of the treatment (Figure 3a) which went upto 60% at the time of sacrifice of animals. Palpation assessment indicated tumor softening in the treatment group.
Effect of α–LCN2 on tumor histology – Blinded Histopathological analysis manifested tumor epithelial cells with clearly defined boundaries in control group (Figure 3b), whereas, tumor sections obtained post treatment revealed areas of necrosis and fibrosis demarcated by the tissue areas having shrivelled cellular morphology of both the tumor epithelia and stromal cells (Figure 3b). Treatment with α-LCN2 antibody thus showed tumor regression characterized by necrotic regions in the tumor, promulgating the therapeutic potential of LCN2. Necrosis of stromal cells is the likely cause for observed tumor softening. Texture features of whole-slide histopathology images such as entropy, dissimilarity and contrast have been correlated in literature with clinical end-points or molecular subtypes in multiple cancers including breast [24], oral [25] and multiple other types of cancers [26]. In this analysis, the three features - entropy, dissimilarity and contrast, presented with higher values in images from treated relative to untreated tumors (Figure 3c) indicating emergence of disorder due to necrosis of tumor and stromal cells in treated samples. Differences in each parameter between images from control and treatment group animals were found to be statistically significant (p << 0.001) using t-test, which corroborates with the qualitative observation of pathologist (Figure 3b).
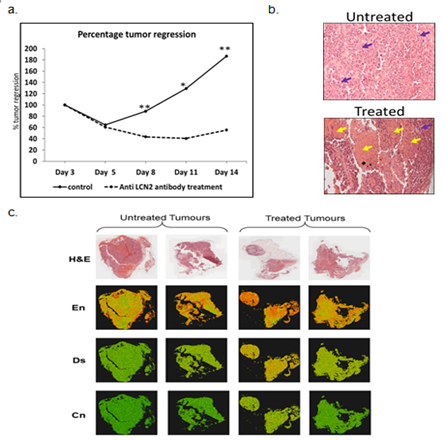
Figure 3: Effect of α-LCN2 antibody on tumor regression. (a) Percentage tumor regression post treatment with α-LCN2 antibody, (b) Histological analysis of the tumor obtained from control group, and the treatment group. Purple arrows show tumor cells, while the yellow arrows indicates the necrotic regions (c) H&E-stained histopathology images of untreated and treated tumors and corresponding visual representation of entropy (En), dissimilarity (Ds) and contrast (Cn) values for each 16´16 px in the images using a colour map with red to green colour gradient indicating maximum to minimum value of each feature. More red dots are seen in treated sample images signifying higher degree of disorder.
3.4 Transcriptome Analysis:
Since the tumor from human cells was formed in another host species, sequencing reads were segregated into mouse and human components based on mapping to reference assemblies (Figure S1a). Unique human fragments mapped were more in number as compared to the unique mouse fragments evidencing human origin of extracted tumor. Mouse genes differentially expressed in treated samples revealed activation of cytokine pathways due to immune activation (Figure S1b). Analysis of human gene expression indicated higher expression of LCN2 in control tumors compared to treated ones (Figure S2)
Mice gene expression – Treatment with α-LCN2 antibody resulted in an increase of CD8+ T cell-estimates in treated samples over control (Figure S3a). Overall activation of T-cells and T-cell mediated killing in treated samples was demonstrated by overexpression of lymphocyte activating 3 (Lag3), regenerating islet-derived 3 gamma (Reg3g) and chemokine Cxcr14 [27, 28] (Figure 4). On the other hand, overall macrophage proportions (Figure S3b) and the pro-tumorigenic, M2 mouse macrophage markers (Figure S3c) showed a decline post-treatment signifying tumor regression. This observed T-cell activation and debilitation of M2 mouse macrophages by α-LCN2 underlies its potential as an immune sensitizer that can potentially devour tumor cells through T-cell mediated cytotoxicity (Figure 4).
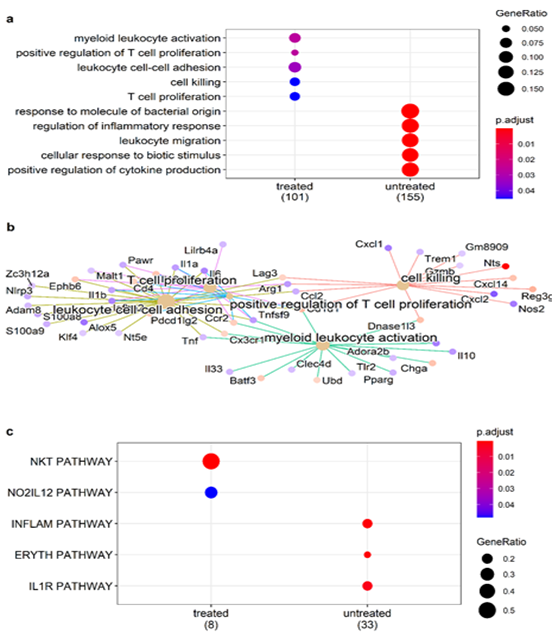
Figure 4: Role of immune cells in tumor regression. (a) Enrichment of T-cell activation related GO biological processes and (b) pathway-gene maps for pathways enriched by differentially expressed mouse genes in treated tumor; (c) enrichment of Biocarta NKT pathway involved in T-cell polarization in treated tumors.
Human gene expression – Enrichment analysis of differentially expressed human genes revealed suppression of pro-tumorigenic TNF-a and IL17 signalling pathways. Some of the major players of IL17 signalling pathway including NFkB, CXCLs and interestingly LCN2 were down regulated post-treatment (Figure 5) suggesting a negative feedback loop, in which action of α-LCN2 antibody abrogates LCN2 expression directly as well as via IL17 pathway. Further, pathway analysis of the differentially expressed genes revealed that the treated samples had higher expression of genes pertinent to cell death pathways (cell death pathway gene signature taken from GSEA mSigDB) (Figure 6a), which was again in concordance with histopathology findings. These results clearly articulate that α-LCN2 antibody treatment increased the cellular death pathways in the treated tumor, thus promoting fibrosis and necrosis of the same, a pre-requisite of tumor regression. Upon further analysis of the tumorigenicity pathways from literature, it was observed that the untreated samples had higher expression of genes involved in promoting invasion (Figure 6b), hypoxia (Figure 6c), and migration (Figure 6d).
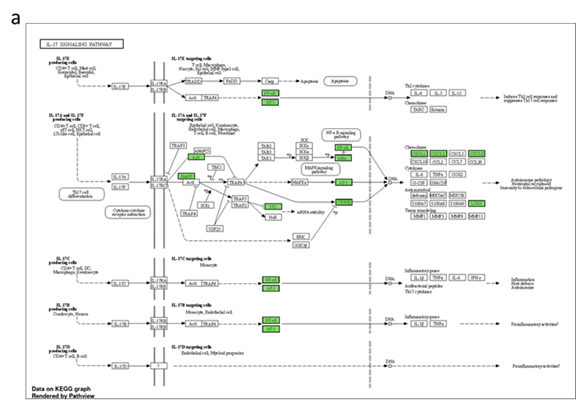
Figure 5: Pathways enriched by differentially expressed human genes post-treatment. Functional analysis of differentially expressed genes depicting (a) Genes (shaded in red) down regulated in the treatment group, (b) cancer hallmark gene sets enriched in treated and untreated samples and (c) genes associated with TNF-a and KRAS signalling pathways enriched only in untreated samples (and hence suppressed under treatment).
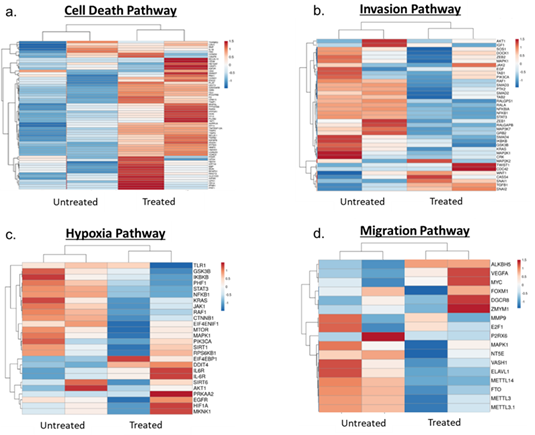
Figure 6: Heat maps showing pathway expression between untreated and treated samples. (a) Cell death pathway, (b) Invasion pathway, (c) Hypoxia pathway, (d) Migration pathway
4. Discussion
In the present study, inhibition of LCN2 with an α-LCN2 antibody decreased the activity of MMP9 in vitro, leading to inhibition of invasion, whereas the hIgG-treated cells remained very solidly packed. In the xenograft model, along with tumor regression, softening of tumors was observed in the treated group. It is important to delve deep into the molecular landscape of this process. Recent reports have suggested that matrix stiffness plays an important role in the prediction of aggressiveness and affects the morphology, drug sensitivity and mechanical properties of CC cells [29, 30]. Increased tumor stiffness is a widely approved characteristic of solid tumors [31] which impedes intra-tumoral injection. Piao et.al reported that substrate stiffness affected epithelial to mesenchymal transition of HeLa and SiHa CC cell lines [32]. Thus, α-LCN2 antibody might even act as a companion therapeutic to soften the tumor and make efficacious approved biologics act at a much lower dosage by direct administration to the tumor. Owing to the high sequence and domain structure similarity between mouse and human LCN2 [33], it can therefore be stated that treatment with α-LCN2 antibody decreased the stiffness of the tumor, thus helping in tumor regression. The softening of tumor by α-LCN2 administration opens an avenue for localized treatment by intra-tumoral injection. However, more mechanistic experiments have to be performed to correlate the involvement of LCN2 in decreasing tumor stiffness. Sequence analysis of these tumors established the presence of mouse transcripts in the human cell xenograft showing a transformation of mouse cells precipitated by human tumor cells. In the human transcript landscape, inhibiting LCN2 in vivo using α-LCN2 antibody resulted in suppression of major tumor-promoting signaling pathways like TNFa, IL17 and NFkB signaling, which ultimately resulted in tumor regression. Numerous reports suggest that all these pathways are linked to LCN2 in various human malignancies [34, 35]. For example, in PC-3 prostate cancer cells, it has been reported that TNF alpha stimulates LCN2, thus protecting the tumor cells from apoptotic cell death [36]. Identification of cytokine dysregulation has been of crucial significance in the development and spread of CC [37, 38]. Specifically IL17 has been regarded as a significant factor in the progression of various human malignancies, including CC [39]. Additionally, IL17 has been implicated in the progression of cervical carcinogenesis, not only with high-risk HPV infection [40], but also with HPV negative cervical cancer cells [40]. Since in the study we demonstrated down regulation of IL17 signaling pathway by α-LCN2 antibody treatment, the antibody can be effective irrespective of the HPV infection status. Down regulation of these tumors promoting pathways by the antibody in the xenograft model in this study proves conclusively that LCN2 can be a potential therapeutic target for abrogating tumor progression. Additionally, in the mouse component, sequencing analysis revealed that intervention with α-LCN2 antibody increased the cytotoxic T lymphocytes and reduced the M2 mouse macrophages suggesting the immunotherapeutic effect of α-LCN2 antibody in this system.
Conclusion:
In conclusion, it can be stated that LCN2 inhibition improves tumor regression properties both in vitro and in vivo. By encouraging cancer cell motility by activating EMT pathway components, LCN2 appears to be a potential therapeutic target. Our in-house developed a-LCN2 MAb emphatically abrogates the stiffness and invasive-migratory properties of tumor in both in vitro and in vivo context. We plan to develop this agent further for clinical usage both as mono and combination therapy in CC.
Declarations
Acknowledgements
The authors would like to acknowledge Dr. Anjali Karande from Indian Institute of Science for Sp2/O and HeLa cells used in the study.
Author’s contributions
Authors MD and SKD conceptualized the study. In vitro experiments were performed by ND while the bioinformatics and image texture analysis was performed by authors TM, ST under the supervision of SKD. In vivo experiments were performed under the supervision of GV. Authors ND and SPK prepared the recombinant protein and monoclonal antibody. Authors MD, SKD and ND wrote the manuscript. Authors MD and ND revised and finalized the manuscript. RR helped in obtaining the latest cervical cancer treatment regimen and contributed to writing of clinical aspects. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved [IAEC/66/SRIHER/782/2022] by the ethics committee of Sri Ramachandra Institute of Higher Education and Research.
Availability of data and materials
Transcriptome sequencing data generated in this study is available at NCBI Gene Expression Omnibus (Accession No GSE242667).
Competing interests
The authors declare that they have no competing interests
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Appendix A
Supplementary information
References
- H Sung, J Ferlay, RL Siegel, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA. Cancer J. Clin 71 (2021): 209-249.
- HC Chung, W Ros, JP Delord, et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Cervical Cancer: Results From the Phase II KEYNOTE-158 Study, J. Clin. Oncol 37 (2019): 1470-1478.
- M Frumovitz, MF Munsell, JK Burzawa, et al. Combination therapy with topotecan, paclitaxel, and bevacizumab improves progression-free survival in recurrent small cell neuroendocrine carcinoma of the cervix, Gynecol. Oncol 144 (2017): 46-50.
- RL Coleman, D Lorusso, C Gennigens, et al. Efficacy and safety of tisotumab vedotin in previously treated recurrent or metastatic cervical cancer (innovaTV 204/GOG-3023/ENGOT-cx6): a multicentre, open-label, single-arm, phase 2 study, Lancet Oncol 22 (2021): 609-619.
- A Adedimeji, R Ajeh, A Pierz, et al. Challenges and opportunities associated with cervical cancer screening programs in a low income, high HIV prevalence context, BMC Womens. Health 21 (2021).
- N Muñoz, FX Bosch, S de Sanjosé, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer, N. Engl. J. Med 348 (2003): 518-527.
- M Branca, M Ciotti, C Giorgi, et al. Predicting high-risk human papillomavirus infection, progression of cervical intraepithelial neoplasia, and prognosis of cervical cancer with a panel of 13 biomarkers tested in multivariate modeling, Int. J. Gynecol. Pathol 27 (2008): 265-273.
- S Xu, P Venge. Lipocalins as biochemical markers of disease, Biochim. Biophys. Acta 1482 (2000): 298-307.
- P Devarajan. Neutrophil gelatinase-associated lipocalin: new paths for an old shuttle, Cancer Ther 5 (2007): 463.
- IH Chung, TI Wu, CJ Liao, et al. Overexpression of lipocalin 2 in human cervical cancer enhances tumor invasion, Oncotarget 7 (2016): 11113-11126.
- PH Wang, LY Lin, S fa Yang, et al. The role of lipocalin 2 and its concernment with human nonmetastatic clone 23 type 1 and p53 in carcinogenesis of uterine cervix, Reprod. Sci 18 (2011): 447-455.
- A Vitkauskaite, J Celiešiute, S Paškauskas, et al. Associations among Serum Lipocalin-2 Concentration, Human Papilloma Virus and Clinical Stage of Cervical Cancer, Medicina (Kaunas) 55 (2019).
- S Syrjänen, P Naud, L Sarian, et al. Up-regulation of lipocalin 2 is associated with high-risk human papillomavirus and grade of cervical lesion at baseline but does not predict outcomes of infections or incident cervical intraepithelial neoplasia, Am. J. Clin. Pathol 134 (2010): 50-59.
- S Lee, JY Park, WH Lee, et al. Lipocalin-2 is an autocrine mediator of reactive astrocytosis, J. Neurosci 29 (2009): 234-249.
- PH Wang, JL Ko, SF Yang, et al. Implication of human nonmetastatic clone 23 Type 1 and its downstream gene lipocalin 2 in metastasis and patient’s survival of cancer of uterine cervix, Int. J. Cancer 129 (2011): 2380-2389.
- S Probst, B Scharner, R McErlean, et al. Inverse Regulation of Lipocalin-2/24p3 Receptor/SLC22A17 and Lipocalin-2 Expression by Tonicity, NFAT5/TonEBP and Arginine Vasopressin in Mouse Cortical Collecting Duct Cells mCCD(cl1): Implications for Osmotolerance, Int. J. Mol. Sci 20 (2019): 5398.
- M Feng, Y Wang, K Chen, et al. IL-17A Promotes the Migration and Invasiveness of Cervical Cancer Cells by Coordinately Activating MMPs Expression via the p38/NF-κB Signal Pathway 9 (2014): 108502.
- CY Lin, PH Tsai, CC Kandaswami, et al. Matrix metalloproteinase-9 cooperates with transcription factor Snail to induce epithelial-mesenchymal transition, Cancer Sci 102 (2011): 815-827.
- JR Masters. HeLa cells 50 years on: The good, the bad and the ugly, Nat. Rev. Cancer 2 (2002): 315-319.
- The Cancer Genome Atlas Program - NCI, (n.d.).
- Broad GDAC Firehose, (n.d.).
- ATCC: The Global Bioresource Center | ATCC, (n.d.).
- T Robin, A Capes-Davis, A Bairoch. CLASTR: The Cellosaurus STR similarity search tool - A precious help for cell line authentication, Int. J. Cancer 146 (2020): 1299-1306.
- N Dwivedi, C Gangadharan, V Pillai, et al. Establishment and characterization of novel autologous pair cell lines from two Indian non-habitual tongue carcinoma patients, Oncol. Rep 48 (2022): 1-12.
- RM Haralick, I Dinstein, K Shanmugam. Textural Features for Image Classification, IEEE Trans. Syst. Man Cybern 3 (1973): 610-621.
- AM Bolger, M Lohse, B Usadel. Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics 30 (2014): 2114-2120.
- SW Wingett, S Andrews. FastQ Screen: A tool for multi-genome mapping and quality control, F1000Research 7 (2018): 1338.
- A Dobin, CA Davis, F Schlesinger, et al.STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29 (2013): 15-21.
- MD Robinson, DJ McCarthy, GK Smyth. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26 (2010): 139-140.
- M V Kuleshov, MR Jones, AD Rouillard, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update, Nucleic Acids Res 44 (2016): 90-97.
- T Wu, E Hu, S Xu, et al. clusterProfiler 40: A universal enrichment tool for interpreting omics data, Innov 2 (2021): 100141.
- GSEA | MSigDB | Investigate Human Gene Sets, (n.d.).
- Z Chen, L Quan, A Huang, et al. seq-ImmuCC: Cell-centric view of tissue transcriptome measuring cellular compositions of immune microenvironment from mouse RNA-seq data, Front. Immunol 9 (2018): 1-11.
- M Geoffroy, A Kleinclauss, S Kuntz, et al. Claudin 1 inhibits cell migration and increases intercellular adhesion in triple-negative breast cancer cell line, Mol. Biol. Rep 47 (2020): 7643-7653.
- U Bahrun, W Wildana, H Rahmawati, et al. Lipocalin 2 could predict circulating MMP9 levels in patients with breast cancer, Breast Dis (2021): 1-3.
- Y Hao, S Qiao, L Zhang, et al. Breast Cancer Histopathological Images Recognition Based on Low Dimensional Three-Channel Features, Front. Oncol 11 (2021): 1-15.
- TY Rahman, LB Mahanta, AK Das, et al. Tissue and Cell Automated oral squamous cell carcinoma identi fi cation using shape , texture and color features of whole image strips, Tissue Cell 63 (2020): 101322.
- S Öztürk, B Akdemir. Application of Feature Extraction and Classification Methods for Histopathological Image using GLCM, LBP, LBGLCM, GLRLM and SFTA, Procedia Comput. Sci 132 (2018): 40-46.
- SR Woo, ME Turnis, M V Goldberg, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape, Cancer Res 72 (2012): 917-927.
- Cxcl14 chemokine (C-X-C motif) ligand 14 [Mus musculus (house mouse)] - Gene - NCBI, (n.d.).
- B Jin, W Kong, X Zhao, et al. Substrate stiffness affects the morphology, proliferation, and radiosensitivity of cervical squamous carcinoma cells, Tissue Cell 74 (2022): 101681.
- Y Zhuang, Y Huang, Z He, et al. Effect of substrate stiffness on the mechanical properties of cervical cancer cells, Arch. Biochem. Biophys 725 (2022): 109281.
- L Wullkopf, AK V West, N Leijnse, et al. Cancer cells’ ability to mechanically adjust to extracellular matrix stiffness correlates with their invasive potential, Mol. Biol. Cell 29 (2018): 2378.
- J Piao, K You, Y Guo, et al. Substrate stiffness affects epithelial-mesenchymal transition of cervical cancer cells through miR-106b and its target protein DAB2, Int. J. Oncol 50 (2017): 2033-2042.
- S Chakraborty, S Kaur, Z Tong, et al. Neutrophil Gelatinase Associated Lipocalin: Structure, Function and Role in Human Pathogenesis, in: Acute Phase Proteins - Regul. Funct. Acute Phase Proteins, InTech (2011).
- F Yao, Y Deng, Y Zhao, et al., A targetable LIFR-NF-κB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis, Nat. Commun 12 (2021).
- GS Santiago-Sánchez, V Pita-Grisanti, B Quiñones-Díaz, et al. Biological functions and therapeutic potential of lipocalin 2 in cancer, Int. J. Mol. Sci 21 (2020): 1-15.
- L Li, J Liu, C Liu, et al. The correlation between tnf-α-308 gene polymorphism and susceptibility to cervical cancer, Oncol. Lett 15 (2018): 7163-7167.
- PH Paradkar, JV Joshi, PN Mertia, et al. Role of cytokines in genesis, progression and prognosis of cervical cancer, Asian Pac. J. Cancer Prev 15 (2014): 3851-3864.
- B Li, L Zhang, J Zhao, et al. The value of cytokine levels in triage and risk prediction for women with persistent high-risk human papilloma virus infection of the cervix, Infect. Agent. Cancer 14 (2019): 1-12.
- Y Bai, H Li, R Lv. Interleukin-17 activates JAK2/STAT3, PI3K/Akt and nuclear factor-κB signaling pathway to promote the tumorigenesis of cervical cancer, Exp. Ther. Med 22 (2021): 1-11.
- Y Pan, X Luo, J Ma, et al. The dual role of interleukin-17 in the growth of human papillomavirus-negative cervical cancer cells, Eur. J. Inflamm 20 (2022): 1-8.
Supplementary Figures:
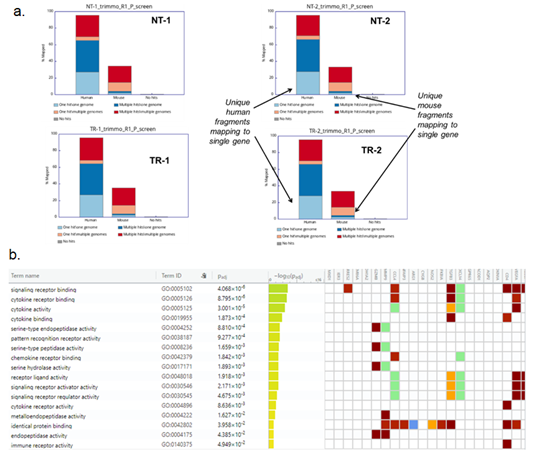
Figure S1: (a) Mapping of RNA sequencing reads to human and mouse genome (b) Pathway association of the differentially expressed genes mapped to mouse genome. NT – No treatment; TR – Treatment with α-LCN2 antibody
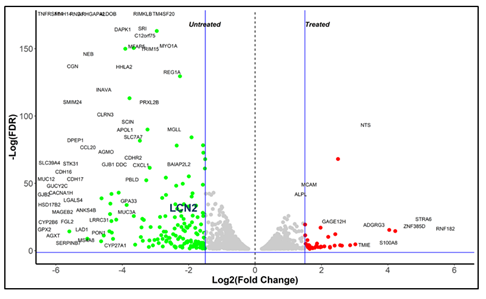
Figure S2: Differential expression analysis showing LCN2 expression in untreated samples.
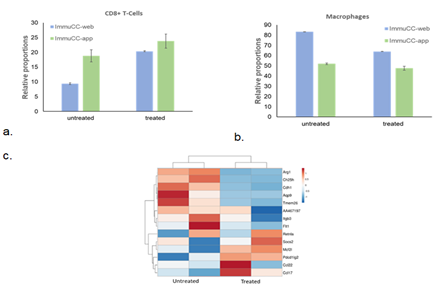
Figure S3: Role of mouse immune cells in tumor regression. (a) Estimated proportions of mouse CD8+ T-cells, (b) macrophages in untreated and treated samples determined using seq-ImmuCC web tool and R application. (c) Sequencing analysis showing decline in M2 macrophage gene signature in treated samples as compared to untreated samples.
Supplementary Tables:
Supplementary Table 1: STR analysis of HeLa cell line. The STR profile matched 100% to HeLa cell line from ATCC database [74] as authenticated by TheraCUES.
|
Marker |
Allele#1 |
Allele#2 |
|
|
HeLA |
TH01 |
7 |
7 |
|
D21S11 |
27 |
28 |
|
|
D5S818 |
11 |
12 |
|
|
D13S317 |
- |
- |
|
|
D7S820 |
8 |
12 |
|
|
D16S539 |
9 |
10 |
|
|
CSF1PO |
9 |
10 |
|
|
AMEL |
X |
X |
|
|
vWA |
16 |
18 |
|
|
TPOX |
8 |
12 |
Supplementary Table 2: Primer sequences used in the study
|
GAPDH |
Fw 5'-TCGACAGTCAGCCGCATCTTCTTT-3' |
|
Rv 5'-GCCCAATACGACCAAATCCGTTGA-3' |
|
|
ACTB |
Fw 5'- GCGCGATATTTCTTCTTGCAGG-3' |
|
Rv 5'- TTCGTACCTGGCATGACTGG-3' |
|
|
Claudin |
Fw 5'-AGCCATGTACGTTGCTATCCA-3' |
|
Rv 5'-ACCGGAGTCCATCACGATG-3' |
|
|
SLC22A17 |
Fw 5'- GTTACCCCGCAGACAGATTTG-3' |
|
Rv 5'- CCCAAGAGGAATCGGAGGG-3' |
|
|
LRP2 |
Fw 5'- GTTCAGATGACGCGGATGAAA-3' |
|
Rv 5'- TCACAGTCTTGATCTTGGTCACA-3' |
|
|
MC1R |
Fw 5'- CATCGCCAAGAACCGGAAC-3' |
|
Rv 5'- GGATGACGGCCGTCTCC-3' |
|
|
MC3R |
Fw 5'- GGGCATCGTCAGTCTGCTG-3' |
|
Rv 5'- AGGGCATTGGACACACTTACC-3' |
|
|
MC4R |
Fw 5'- ACACCCACTCCTCCACCTTT-3' |
|
Rv 5'- TGCTGTAGCCAAATTCGTTG-3' |
|
|
LCN2 |
Fw 5'- GAAGTGTGACTACTGGATCAGGA-3' |
|
Rv 5'- ACCACTCGGACGAGGTAACT-3' |
|
|
MMP9 |
Fw 5’ – TTGGTCCACCTGGTTCAACT – 3’ |
|
Rv 5’ – ACGACGTCTTCCAGTACCGA – 3’ |


 Impact Factor: * 4.1
Impact Factor: * 4.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 