Concurrent TERT Amplifications in EGFR Mutated Lung Adenocarcinoma are Associated with Higher Levels of Copy Number Aberrations - A Hypothesis-Generating Study
Article Information
Su Ir Lyu1#, Steffen Hamm1#, Pierce Heiden2, Michael Schultheis3, Christina Alidousty1, Sabine Merkelbach-Bruse1,4, Reinhard Büttner1,4, Philipp Lohneis1#, Anne Maria Schultheis1,4#*
1Institute of Pathology, University Hospital Cologne, 50937 Cologne, Germany
2Institute of Medical Statistics and Computational Biology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany
3Department of Dermatology, University Medical Center of the Johannes Gutenberg University Mainz, 55131 Mainz, Germany
4Medical Faculty, University of Cologne, 50931 Cologne, Germany
*Corresponding author: Anne M. Schultheis, Institute of Pathology, University Hospital Cologne, 50937 Cologne, Germany
#contributed equally
Received: February 07, 2024; Accepted: February 14, 2024; Published: March 11, 2024
Citation: Su Ir Lyu, Steffen Hamm, Pierce Heiden, Michael Schultheis, Christina Ali-dousty, Sabine Merkelbach-Bruse, Reinhard Büttner, Philipp Lohneis, Anne Maria Schultheis. Concurrent TERT Amplifications in EGFR Mutated Lung Adenocarcinoma are Associated with Higher Levels of Copy Number Aberrations - A Hypothesis-Generating Study. Journal of Cancer Science and Clinical Therapeutics 8 (2024): 95-101.
View / Download Pdf Share at FacebookAbstract
Objectives: EGFR mutated lung cancer is a molecular subtype of lung cancer that can be treated using EGFR targeting tyrosine kinase inhibitors. Despite improved survival rates of patients treated with these inhibitors, relapse and resistance remains a major concern. Concurrent mutations have recently been identified to drive disease outcome in EGFR mutated lung cancer. Here we sought to investigate the frequency and characteristics of TERT amplifications in EGFR L858R mutated lung cancer.
Materials and Methods: We performed a re-analysis of nine publicly available datasets concerning lung cancer.
Results: In this hypothesis generating study, we could show that tumors with TERT amplifications exhibited higher numbers of copy number alterations, indicating higher levels of genomic instability, potentially driving tumor biology in addition to EGFR mutations.
Conclusion: Taken together, our hypothesis-generating study indicates that the presence of TERT amplifications in EGFR mutated lung cancer represents a specific molecular subset of EGFR mutated carcinomas that need to be further analyzed to better understand their biology.
Keywords
Lung cancer, EGFR L858R, TERT amplification, CAN
Article Details
Abbreviations:
AKT - Protein kinase B
CNAs - Copy number alterations
EGFR - Epidermal growth factor receptor mTOR - Mammalian target of rapamycin
mTORC - Mammalian target of rapamycin complex
PI3K - Phosphoinositide 3-kinase
RICTOR - Rapamycin-insensitive companion of mammalian target of rapamycin TERT - Telomerase reverse transcriptase
TERTamp - Telomerase reverse transcriptase amplification
TERTwt - TERT wild type
TKB - Tyrosine kinase inhibitor TMB - Tumor mutational burden WGD - Whole-genome doubling
Introduction
EGFR mutated lung cancer is a molecular subtype of lung cancer characterized by activating mutations in the epidermal growth factor receptor (EGFR) gene. Potent EGFR targeting tyrosine kinase inhibitors have dramatically changed the way lung cancer patients with activating EGFR mutations are treated and their outcome has improved significantly [1, 2]. So far, three generations of EGFR inhibitors have been developed, differing by their time of development, effectiveness to bind EGFR and ability to overcome resistance mechanisms that emerged with earlier inhibitors. However, resistance mechanisms can still occur, even in response to third-generation inhibitors. This underscores the ongoing requirement for research aimed at deepening our understanding and improving treatment strategies for patients with EGFR-mutated lung cancer [3]. The TERT (telomerase reverse transcriptase) gene encodes the catalytic subunit of the telomerase enzyme. Telomerase plays a crucial role in maintaining the integrity and length of telomeres by synthesizing and appending repetitive DNA sequences (TTAGGG) to the ends of chromosomes, which counters the progressive shortening that occurs with each instance of cell division. This process ensures the stability and correct replication capacity of the genome [4]. Genetic alterations, such as amplifications, promoter mutations, or rearrangements within the TERT gene, as well as epigenetical events such as methylation of the promoter, all contribute to the subsequent dysregulation or elevation of TERT expression and of the telomerase activity [5, 6]. Thus, aberrant activation or dysregulation of TERT can lead to excessive telomerase activity, resulting in abnormal telomere lengthening, cellular immortality and cancer development [7-9]. Importantly, TERT alterations are associated with a poor prognosis and aggressive tumor behavior across a wide range of cancer types [10, 11]. Studying the role of concurrent mutations has yielded important insights into the mechanisms underlying tumor development, progression and resistance to targeted therapies in tumors. In cases of EGFR- mutant lung cancers, several reports have suggested that concurrent genetic alterations are associated with a decreased probability of responding to EGFR TKIs and a shorter overall survival [12-14]. Here we sought to investigate the frequency of TERT amplifications in EGFR L858R mutated lung cancer and to ascertain whether tumors with concurrent TERT amplifications would exhibit differences based on histological, genomic or clinical parameters.
Materials and Methods
Patient selection
We performed a re-analysis of nine publicly available datasets concerninglungcancer, encompassingcomprehensive genetic and clinical data (Lung Adenocarcinoma, Broad, Cell 2012; Lung Adenocarcinoma, CPTAC, Cell 2020;
Lung Adenocarcinoma, MSK, 2020; Lung Adenocarcinoma, MSK, J Thorac Oncol 2020; Lung Adenocarci-noma, MSK, NPJ Precision Oncology 2021; Lung Adenocarcinoma, TCGA, Pan-Cancer Atlas; Non-Small Cell Cancer; MSK, Cancer Discov 2017; Lung Adenocar-cinoma; MSK, Nature Cancer 2022; MSK MetTropism; MSK, Cell 2021) using the openly accessible cbioportal platform (www.cbioportal. org). Clinicopathological and molecular data from cases of EGFR L858R mutated lung adenocarcinoma were retrieved from the cbioportal website, distinguishing between those with TERT amplification (TERTamp) and those without (TERTwt) (www.cbioportal.org; accessed May 2023).
To ensure consistent comparisons, our study specifically focused on EGFR L858R mutations and TERT amplifications exclusively. We intentionally excluded all other EGFR mutations and TERT aberrations from this explorative, hypothesis-generating study.
Histologic and genomic stratification
Information regarding histologic subtypes, tumor grade and stage were retrieved from the individual datasets, whenever available. We also extracted details about co- existing pathogenic mutations (as classified by cbioportal), total number of mutations, tumor mutational burden (TMB), as well as the total count of copy number alterations (CNAs) along with all documented pathogenic amplifications and deletions present in the datasets (Supplementary Table 1).
Statistical analysis
All statistical analyses were performed using the SPSS statistical software package (IBM SPSS Statistics, Version
29.0.0.0. (241) IBM). Chi-Square tests and Fisher’s exact test were employed to test for equal distributions of qualitative variables in the TERTamp and TERTwt cohort. The Mann- Whitney-U-test was applied on quantitative data to test for possible differences in the central tendency of both cohorts. Kaplan-Meier curves and the log-rank test were utilized for survival-time analyses. Hypotheses were tested based on an alpha level of 0.05.
Results
Frequency of TERT amplifications in EGFR L858R mutated lung cancer
Across the nine datasets included in our project, a total of n=4091 primary lung adenocarcinomas were accounted for. Among these 4091 primary adenocarcinomas, n=486 (11.9%) exhibited an EGFR L858R mutation. Within the subset of adenocarcinomas harboring the EGFR L858R mutation, n=45 (9.2%) displayed a concurrent TERT amplification.
We included all n=45 cases characterized by both EGFR L858R mutation and TERT amplification (TERTamp), and also randomly selected n=89 non-TERT amplified cases harboring the EGFR L858R mutation (TERTwt) (Table 1).
Table 1: Clinico-pathologic and genomic parameters extracted from the cbioportal platform.
|
Total (n) |
Status |
Statistics |
|||||
|
TERTwt |
TERTamp |
median |
mean |
p-value |
|||
|
Total |
134 |
(n=89) 66% |
(n=45) 34% |
||||
|
Age at Diagnosis (years) |
TERTwt |
88 |
67 |
66.24 |
0.191** |
||
|
TERTamp |
45 |
65 |
64.13 |
||||
|
Gender |
female |
90 |
(n=55) 61% |
(n=35) 39% |
0.080* |
||
|
male |
44 |
(n=34) 77% |
(n=10) 23% |
||||
|
Pathological Stage at Diagnosis |
1 |
43 |
(n=30) 70% |
(n=13) 30% |
0.494*a |
||
|
2 |
8 |
(n=7) 88% |
(n=1) 13% |
||||
|
3 |
3 |
(n=2) 67% |
(n=1) 33% |
||||
|
4 |
10 |
(n=9) 90% |
(n=1) 10% |
||||
|
Smoking History |
non-smoker |
26 |
(n=19) 73% |
(n=7) 27% |
0.704* |
||
|
smoker |
27 |
(n=21) 78% |
(n=6) 22% |
||||
|
former smoker |
8 |
(n=7) 88% |
(n=1) 13% |
||||
|
Pack Years |
TERTwt |
36 |
2 |
10.15 |
0.915** |
||
|
TERTamp |
13 |
0.05 |
19.47 |
||||
|
Relapse-Free-Status(months) |
TERTwt |
27 |
16.6 |
23.42 |
0.122** |
||
|
TERTamp |
13 |
7 |
14.12 |
||||
|
Relapse Status |
No Relapse |
16 |
(n=6) |
(n=10) |
0.007* |
||
|
38% |
63% |
||||||
|
Relapse |
34 |
(n=26) |
(n=8) |
||||
|
77% |
24% |
||||||
|
Kaplan-Meier Estimate of Relapse- Free-Status (in months) |
TERTwt |
22 |
35.5 |
28.68 |
0.199*** |
||
|
TERTamp |
13 |
/ |
41.68 |
||||
|
Overall Survival (months) |
TERTwt |
28 |
23 |
22.05 |
0.994** |
||
|
TERTamp |
11 |
17 |
22.09 |
||||
|
Survival Status |
living |
53 |
(n=25) |
(n=28) |
0.018* |
||
|
47% |
53% |
||||||
|
deceased |
39 |
(n=28) |
(n=11) |
||||
|
72% |
28% |
||||||
|
Kaplan-Meier Estimate of Overall Survival (months) |
TERTwt |
52 |
40 |
48.96 |
0.410*** |
||
|
TERTamp |
37 |
56 |
42.64 |
||||
|
Mutation Count |
TERTwt |
89 |
7 |
22.73 |
0.648** |
||
|
TERTamp |
45 |
5 |
51.69 |
||||
|
Tumor Mutation Burden (nonsynonymous) |
TERTwt |
87 |
3.32 |
4.58 |
0.334** |
||
|
TERTamp |
42 |
3.9 |
5.57 |
||||
|
Copy Number Variations Count |
TERTwt |
89 |
1 |
36.8 |
0.001** |
||
|
TERTamp |
45 |
6 |
70.89 |
||||
*Chi-square test
**Mann-Whitney-U-Test
***Log Rang test (Mantel-Cox)
a4 cells (50.0%) had less than the five expected data, which was a limitation of the test results.
TERT amplification and clinicopathological parameters
There was no statistically significant difference in age at diagnosis between the two groups (TERTwt: median age: 67.00 years; TERTamp median age: 65.00 years; Mann- Whitney-U-test p=0.191; Table1). We noticed a tendency towards a higher proportion of female patients in the TERT- amp cohort (female with TERTamp/all females: (35/90) 39% vs. male with TER-Tamp/all male: (10/44) 23%; p-value =0.080; Table 1). However, a statistically significant difference in frequencies could not be detected. No significant difference was observed concerning TERTamp in relation to tumor stage at the time of diagnosis (for limitations of the test results see Table 1). However, there was inadequate available data for further analysis regarding stage-specific survival and stage-specific relapse. Analysis of overall-survival- time based on histological subtypes was not possible due to insufficient data. The distribution of histological subtypes as classified in the cbioportal dataset is presented in Table 2.
Both groups exhibited a comparable number of smokers and non-smokers (Table 1). A trend towards earlier relapses in TERTamp cases was observed, although the number of cases with comprehensive clinical data was limited (Table1).
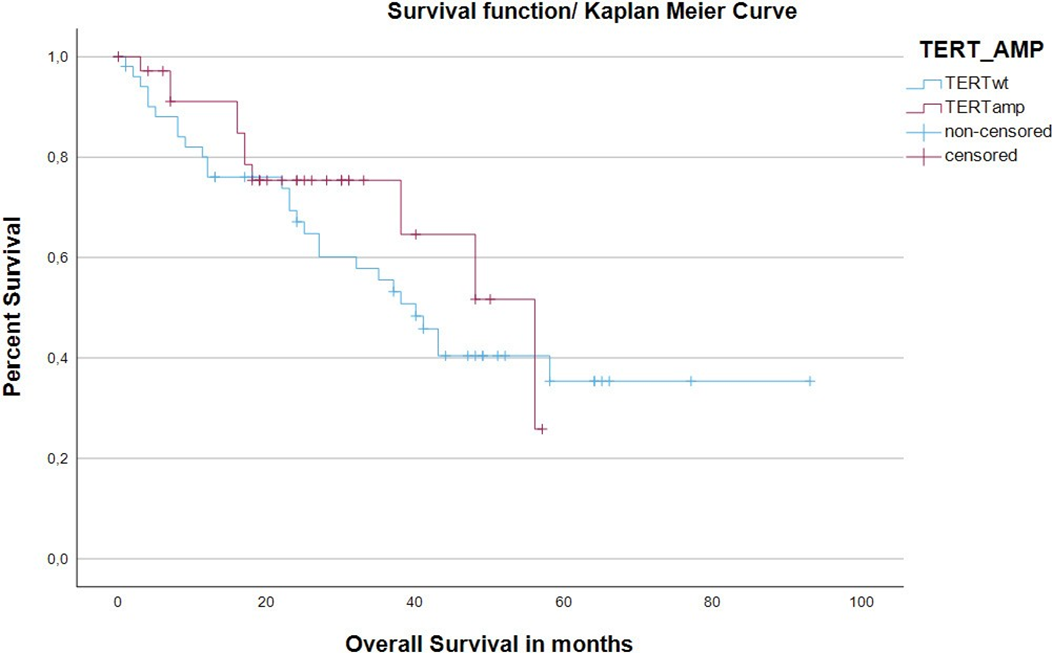
There were significantly more relapses in the TERTwt cohort (TERTwt: relapses: (26/34) 76.5% vs. TERTamp: relapses: (8/34) 23.5%; Chi-square test p=0.007; Table1). The Log-rank test with respect to relapse-free-status was not statistically significant (p=0.199). The TERTamp cohort showed a statistically significantly higher rate of survival status “living” (Chi-square test p=0.018; Table1). There was no statistically significant difference between the two cohorts in terms of overall survival (Kaplan-Meier estimate of overall survival (months), thus event at ‘deceased=yes’ and censoring in patients that were lost to follow up; TERTwt median: 40.00 vs. TERTamp median: 56.00; Log-rank test p= 0.410). The corresponding Kaplan-Meier estimates are graphically plotted in Figure 1.
Kaplan-Meier estimate of overall survival (months), thus event at ‘deceased=yes’ and censoring in patients that were lost to follow up. TERTwt showed a median of 40.00months vs. TERTamp showed a median of 56.00. (Log-rank test p= 0.410).
TERT amplification and mutation count/concurrent mutations
The two groups exhibited no significant difference in terms of mutation count (TER-Twt: median: 7.00 mutations
Kaplan-Meier estimate of overall survival (months), thus event at ‘deceased=yes’ and censoring in patients that were lost to follow up. TERTwt showed a median of 40.00months vs. TERTamp showed a median of 56.00. (Log-rank test p= 0.410).
Table 2: The distribution of histological subtypes as classified in the cbioportal dataset is presented in the table
|
Histology/Subtype/Tumor Grade |
||||
|
Histology/Subtype/Tumor Grade |
TERTwt |
TERTamp |
Total |
TERTamp/Total in % |
|
Acinar |
16 |
2 |
18 |
11.1 |
|
Papillary |
0 |
2 |
2 |
100 |
|
Acinar/ Papillar |
5 |
3 |
8 |
37.5 |
|
Solid CT |
7 |
1 |
8 |
12.5 |
|
Nonsolid CT |
5 |
1 |
6 |
16.7 |
|
Lepidic |
1 |
2 |
3 |
66.7 |
|
Mixed |
2 |
1 |
3 |
33.4 |
|
Poorly differentiated |
1 |
1 |
2 |
50 |
|
Moderately differentiated |
8 |
1 |
9 |
11.1 |
|
Well differentiated |
0 |
1 |
1 |
100 |
|
Total |
45 |
15 |
60 |
25 |
TERTamp: median: 5.00; Mann-Whitney-U-test p-value=0.648; Table1). Consistent with the aforementioned data, there was no statistically significant difference in tumor mutational burden (TERTwt median: 3.32 vs. TERTamp median 3.90; Mann-Whitney-U-test p-value=0.334; Table1). We did not identify any statistically significant recurrent mutations in either of the groups (Supplementary Table 1).
TERT amplifications and copy number alterations (CNAs)
TERTamp adenocarcinomas exhibited significantly higher numbers of CNAs compared to the TERTwt group. (TERTwt: median: 1.00 copy number variations vs TER- Tamp: median 6.00 copy number variations; Mann-Whitney- U-test: p-value=0.001; Table1). In addition, among copy number alterations, we observed the presence of recurrent RICTOR amplifications in the TERTamp cohort (TERTwt (n=0/89) 0% vs TERTamp (n=5/45) 11%; Fisher`s exact test p-value=0.004; Supplementary Table 1).
Discussion
Synergistic mutations pertain to genetic alterations wherein the combined effect of two or more mutations surpasses the individual effect of each mutation alone. Grasping the interaction among synergistic mutations holds paramount importance in deciphering the intricacies of cancer biology. This comprehension could facilitate a deeper understanding of tumor evolution, mechanisms of resistance, and potentially reveal novel therapeutic vulnerabilities. In recent years, a multitude of studies have demonstrated that concurrent, potentially synergistic mutations in EGFR-mutant lung cancer influence patients´ outcome [12-14]. In one of our previous studies, we could show that concurrent TERT amplifications in ALK translocated lung adenocarcinomas, another molecular subtype of lung cancer, were associated with particularly high numbers of genetic amplifications and deletions, indicating the prevalence of high numbers of copy number changes [15]. Copy number changes have been a focal point of extensive research in recent years. They hold a crucial role in evolutionary processes, population diversity [16], the onset of specific diseases [17] and influence host microbiome interactions [18]. Importantly, copy number changes causing alterations in the genome can increase the likelihood of survival, even in challenging or unfavorable environments. Copy number changes can arise from diverse biological processes. So called copy number alterations (CNAs) comprise deletions or amplifications of genomic material fragments, with a size as low as a few kilobases up to entire chromosomes and complex signatures of copy number alterations in cancer (CNAs) have been recently described [19]. Genomic instability, characterized by an increased propensity of genetic material to undergo alterations and mutations—ranging from substitutions, insertions, and deletions to more extensive structural changes such as duplications or rearrangements—at an elevated rate, can play a significant role in fostering the emergence of CNAs [20, 21]. At the early stages of human tumorigenesis, the rapid proliferation of cancer cells instigates an ongoing decline in telomere length. This diminishing of telomeres prompts a response of either cellular senescence or apoptosis when a small fraction of telomeres loses the ability to suppress DNA damage signaling pathways. Telomere exhaustion can lead to a critical state of “telomere crisis”, which includes phenomena like ‘chromothripsis’, or whole-genome doubling (WGD). These events, in turn, collectively contribute to the aforementioned genomic instability [22].
The result of this sequence of events is a transformed cell with a genome that has undergone extensive rearrangements yet achieved stability, all the while harboring novel genetic mutations with the potential to drive tumorigenesis, possibly among others including CNAs [19, 23-27]. Amplifications of oncogenes, potentially stemming from genomic instability related to CNAs, correlate with heightened aggressive behavior in tumors and possess the capability to foster resistance against targeted therapies [12-14, 28, 29]. In our analysis of the data, we detected a significantly more frequent occurrence of RICTOR amplifications within the TERTamp cohort (in comparison to the TERTwt cohort), among other copy number alterations. Rapamycin-Insensitive Companion of mTOR (RICTOR) assumes a pivotal function in overseeing cellular activities associated with growth, metabolism, and survival. It holds a position within the protein ensemble known as mammalian target of rapamycin complex 2 (mTORC2). The two entities, mTORC1 and mTORC2, represent discrete protein complexes that together constitute the mechanistic target of rapamycin (mTOR). One of the most disrupted pathways in human tumors is the signaling cascade governed by phosphoinositide 3-kinase (PI3K), AKT, and mTOR. This pathway assumes a significant role in events leading to resistance against epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs). Clinical investigations have substantiated that patients harboring EGFR mutations with activated PI3K pathway experience diminished progression- free survival and inferior overall survival. It is hypothesized that the amplification of RICTOR contributes to the constant activation of AKT, a kinase that becomes active downstream of PI3K. Remarkably, this mechanism operates irrespective of upstream signaling cues, culminating in both tumor advancement and the development of resistance to therapeutic drugs [30-35].
Taken together, our findings unveil a potential hypothetical sequence of occurrences that drive the emergence of a more aggressive subtype of EGFR-mutated lung adenocarcinoma. This process commences with an initial state of genomic instability, caused or accompanied by heightened telomerase activity due to TERT amplification, fostering the emergence of novel genetic mutations, including the amplification of oncogenes such as RICTOR, which could conceivably play a role in drug resistance. To validate this hypothesis and ascertain its potential impact on patient outcomes, a broader and more extensive cohort analysis is imperative. Collectively, within the scope of this retrospective and hypothesis- generating study, we could provide additional substantiation for the concept that EGFR mutated adenocarcinoma of the lung constitutes a heterogeneous group of tumors on a molecular plane. However, it is imperative to acknowledge the limitations of this investigation. The pool of cases furnished with comprehensive clinical data remains relatively small and the total number of TERT amplified, EGFR mutated lung carcinomas remains restricted. Nonetheless, the discoveries unveiled through this inquiry pave the way for subsequent investigations that aim to glean a more profound comprehension of the ramifications posed by TERT amplification and genomic instability in the realm of EGFR mutated lung cancer.
Conclusions
Our analysis underscores the importance to better understand the role of TERT amplifications in EGFR- mutated lung cancer, in particular the link between TERT amplifications and significantly higher levels of CNAs, potentially facilitating disease progression via genomic evolution and genetic events such as additional amplifications of oncogenes or deletions of tumor suppressor genes.
Declarations
Author Contributions
Su Ir Lyu: Writing - Original Draft, Investigation. Steffen Hamm: Methodology, Writ-ing - Original Draft, Investigation. Pierce Heiden: Statistical analyses and visualiza-tion. Michael Schultheis, Philip Lohneis: Methodology, Validation. Christina Ali-dousty: Writing - Review & Editing. Sabine Merkelbach-Bruse: Writing - Review & Editing. Reinhard Büttner: Supervision.
Anne Maria Schultheis, Philip Lohneis: Methodology, Supervision, Writing - Re-view & Editing, Project administration.
Funding and/or conflicts of interests/competing interests.
The authors declare no conflicts of interest. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of generative AI and AI-assisted technologies in the writing pro-cess
During the preparation of this work the authors used ChatGPT in order to edit English language and phrasing. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
References
- Lynch TJ, DW Bell, R Sordella, et Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med, 2004 350 (2004): 2129-2139.
- Paez JG, PA Jänne, JC Lee, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib Science 304 (2004): 1497-1500.
- Mok TS, YL Wu, S Thongprasert, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361 (2009): 947-957.
- Gomez DE, RG Armando, HG Farina, et al. Telomere structure and telomerase in health and disease (review). Int J Oncol 41 (2012): 1561-1569.
- Chiba K, FK Lorbeer, AH Shain et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step Science 357 (2017): 1416-1420.
- Guilleret I, P Yan, F Grange, et al. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase Int J Cancer 101 (2002): 335-341.
- Yuan X, C Larsson, and D Xu. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38 (2019): 6172-6183.
- Vinagre J, A Almeida, H Pópulo, et Frequency of TERT promoter mutations in human cancers. Nat Commun 4 (2013): 2185.
- Daniel M, GW Peek, and TO Tollefsbol. Regulation of the human catalytic subunit of telomerase (hTERT). Gene 498 (2012): 135-146.
- Huang FW, E Hodis, MJ Xu, et Highly recurrent TERT promoter mutations in human melanoma. Science 339 (2013): 957-959.
- Xing M, R Liu, X Liu, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest J Clin Oncol 32 (2014): 2718-2726.
- Yu HA, K Suzawa, E Jordan, et Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res 24 (2018): 3108-3118.
- Gini B, N Thomas, and CM Impact of concurrent genomic alterations in epidermal growth factor receptor (EGFR)-mutated lung cancer. J Thorac Dis 12 (2020): 2883-2895.
- Guo Y, J Song, Y Wang, et Concurrent Genetic Alterations and Other Biomarkers Predict Treatment Efficacy of EGFR-TKIs in EGFR-Mutant Non-Small Cell Lung Cancer: A Review. Front Oncol 10 (2020): 610923.
- Alidousty C, Nicolai D, Svenja WR, et Prevalence and potential biological role of TERT amplifications in ALK translocated adenocarcinoma of the lung. Histopathology 78 (2021): 578-585.
- Redon R, Shumpei I, Karen RF, et al. Global variation in copy number in the human genome. Nature 444 (2006): 444-454.
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease Trends Genet 14 (1998): 417-422.
- Mohajeri MH, Robert JMB, Robert AR, et al. The role of the microbiome for human health: from basic science to clinical Eur J Nutr 57 (2018): 1-14.
- Steele CD, Ammal A, SM Ashiqul Islam, et Signatures of copy number alterations in human cancer. Nature 606 (2022): 984-991.
- Hovhannisyan G, Tigran H, Rouben A, et al. DNA Copy Number Variations as Markers of Mutagenic Int J Mol Sci 20(2019): 4723.
- Moon JJ, A Lu, and C Moon. Role of genomic instability in human carcinogenesis. Exp Biol Med (Maywood) 244 (2019): 227-240.
- Artandi S, and RA Telomeres and telomerase in cancer. Carcinogenesis 31 (2010): 9-18.
- Maciejowski J, and T de Lange. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18 (2017): 175-186.
- Artandi SE, S Chang, S L Lee, et Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406(2000): 641-645.
- Maciejowski J, Yilong L, Nazario B, et Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 163 (2015): 1641-1654.
- Gupta S, Vanderbilt CM, Lin YT, et al. A Pan-Cancer Study of Somatic TERT Promoter Mutations and Amplification in 30,773 Tumors Profiled by Clinical Genomic J Mol Diagn 23 (2021): 253-263.
- Stephens PJ, Greenman CD, Fu B, et Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144 (2011): 27-40.
- Peters TL, Le AT, Davies KD, et al. Evolution of MET and NRAS gene amplification as acquired resistance mechanisms in EGFR mutant NSCLC. NPJ Precis Oncol 5 (2021): 91.
- Nishikawa S, Kimura H, Kobaet H, et al. Selective gene amplification to detect the T790M mutation in plasma from patients with advanced non-small cell lung cancer (NSCLC) who have developed epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) J Thorac Dis 10 (2018): 1431-1439.
- Laugier F, Finet-Benyair A, André J, et RICTOR involvement in the PI3K/AKT pathway regulation in melanocytes and melanoma. Oncotarget 6 (2015): 28120-28131.
- Fang W, Huang Y, Gu W, et PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl Lung Cancer Res 9 (2020): 1258-1267.
- Kim HR, Cho BC, Shim HS, et Prediction for response duration to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma. Lung Cancer 83 (2014): 374-382.
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung Nat Genet 49 (2017): 1693-1704.
- Yang G, Murashige DS, Humphrey SJ, et al. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Cell Rep 12 (2015): 937-943.
- Zhao D, Jiang M, Zhang X, et al. The role of RICTOR amplification in targeted therapy and drug Mol Med 26 (2020): 20.
|
Supplementary Table 1 Concurrent Copy Number Alterations |
||||||
|
CNAs: |
Total |
TERTamp |
TERTwt |
TERTamp/ Total |
Total/134 |
Significance* |
|
EGFR AMP |
29 |
10 |
19 |
34% |
21.6% |
no |
|
CDKN2A DeepDel |
23 |
6 |
17 |
26% |
17.2% |
no |
|
CDKN2B DeepDel |
22 |
6 |
16 |
27% |
16.4% |
no |
|
MDM2 AMP |
14 |
6 |
8 |
43% |
10.4% |
no |
|
NKX2-1 AMP |
12 |
4 |
8 |
33% |
9.0% |
no |
|
CDK4 AMP |
10 |
5 |
5 |
50% |
7.5% |
no |
|
MTAP DeepDel |
9 |
2 |
7 |
22% |
6.7% |
no |
|
FOXA1 AMP |
8 |
1 |
7 |
13% |
6.0% |
no |
|
ETV1 AMP |
7 |
2 |
5 |
29% |
5.2% |
no |
|
CCNE1 AMP |
6 |
3 |
3 |
50% |
4.5% |
no |
|
MYC AMP |
6 |
4 |
2 |
67% |
4.5% |
no |
|
CCND3 AMP |
5 |
3 |
2 |
60% |
3.7% |
no |
|
MET AMP |
5 |
1 |
4 |
20% |
3.7% |
no |
|
RICTOR AMP |
5 |
5 |
0 |
100% |
3.7% |
0.004* |
|
CARD11 AMP |
4 |
1 |
3 |
25% |
3.0% |
no |
|
FGF19 AMP |
4 |
2 |
2 |
50% |
3.0% |
no |
|
PTPRD DeepDel |
4 |
1 |
3 |
25% |
3.0% |
no |
|
RAC1 AMP |
4 |
1 |
3 |
25% |
3.0% |
no |
|
SDHA AMP |
4 |
3 |
1 |
75% |
3.0% |
no |
|
AKT2 AMP |
3 |
1 |
2 |
33% |
2.2% |
no |
|
MCL1 AMP |
3 |
2 |
1 |
67% |
2.2% |
no |
|
RHEB AMP |
3 |
2 |
1 |
67% |
2.2% |
no |
|
VEGFA AMP |
3 |
2 |
1 |
67% |
2.2% |
no |
|
AGO2 AMP |
2 |
2 |
0 |
100% |
1.5% |
no |
|
AURKA AMP |
2 |
2 |
0 |
100% |
1.5% |
no |
|
BAALC AMP |
2 |
1 |
1 |
50% |
1.50% |
no |
|
CDK6 AMP |
2 |
1 |
1 |
50% |
1.5% |
no |
|
FGF3 AMP |
2 |
1 |
1 |
50% |
1.5% |
no |
|
FGF4 AMP |
2 |
1 |
1 |
50% |
1.5% |
no |
|
FGFR4 AMP |
2 |
2 |
0 |
100% |
1.5% |
no |
|
MAPK1 AMP |
2 |
2 |
0 |
100% |
1.5% |
no |
|
NKX3-1 DeepDel |
2 |
0 |
2 |
0% |
1.5% |
no |
|
UBR5 AMP |
2 |
1 |
1 |
50% |
1.5% |
no |
|
ACTG1 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
AKT1 AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ATP6V1B2 DeepDel |
1 |
0 |
1 |
0% |
0.70% |
no |
|
AXL AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
CRBN DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
DUSP22 DeepDel |
1 |
1 |
0 |
100% |
0.7% |
no |
|
ELF3 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ERAS AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ESCO2 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
EZH2 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
GAB2 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
GLI1 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
HDAC6 AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
JUN AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
KEAP1 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
KRAS AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
LGR5 AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
MAG DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
MSH2 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
NSD1 DeepDel |
1 |
1 |
0 |
100% |
0.7% |
no |
|
PAK1 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
PAX5 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PCSK1N AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PMAIP1 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PPM1D AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
PPP2R2A DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PRKAR1A AMP |
1 |
0 |
1 |
0% |
0.70% |
no |
|
PTEN DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PTPRS DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
RAD51 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
RUNX1 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
SMAD4 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
SOX17 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
SPRED1 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
SRC AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
STK11 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
TP53BP1 DeepDel |
1 |
0 |
1 |
0% |
0.7% |
no |
|
YAP1 AMP |
1 |
1 |
0 |
100% |
0.7% |
no |
|
YES1 AMP |
1 |
0 |
1 |
0% |
0.7% |
no |
|
TERT AMP |
45 |
89 |
0.0% |
|||
|
* Fisher’s exact test p-value |
||||||
|
Supplementary Table 2: Concurrent pathogenic mutations |
||||||
|
Mutation |
Total |
TERTamp |
TERTwt |
TERTamp/ Total |
Total/ 134 |
Significance |
|
EGFR L858R |
134 |
45 |
89 |
34% |
100.0% |
|
|
TP53 |
75 |
27 |
48 |
36% |
56.0% |
no |
|
RBM10 |
14 |
8 |
6 |
57% |
10.4% |
no |
|
EGFR T790M |
13 |
5 |
8 |
38% |
9.7% |
no |
|
CTNNB1 |
6 |
2 |
4 |
33% |
4.5% |
no |
|
PTEN |
6 |
2 |
4 |
33% |
4.5% |
no |
|
CDKN2A |
5 |
0 |
5 |
0% |
3.7% |
no |
|
PIK3CA |
5 |
3 |
2 |
60% |
3.7% |
no |
|
RB1 |
4 |
0 |
4 |
0% |
3.0% |
no |
|
APC |
3 |
2 |
1 |
67% |
2.2% |
no |
|
MED12 |
3 |
2 |
1 |
67% |
2.2% |
no |
|
SMARCA4 |
3 |
0 |
3 |
0% |
2.2% |
no |
|
ARID1A |
2 |
1 |
1 |
50% |
1.5% |
no |
|
ATRX |
2 |
1 |
1 |
50% |
1.5% |
no |
|
CBFB |
2 |
2 |
0 |
100% |
1.50% |
no |
|
CDKN2C |
2 |
2 |
0 |
100% |
1.5% |
no |
|
ERBB2 |
2 |
2 |
0 |
100% |
1.5% |
no |
|
KDM5C |
2 |
1 |
1 |
50% |
1.5% |
no |
|
MAG |
2 |
1 |
1 |
50% |
1.5% |
no |
|
PIK3R2 |
2 |
0 |
2 |
0% |
1.5% |
no |
|
RASA1 |
2 |
2 |
0 |
100% |
1.5% |
no |
|
TERT |
2 |
0 |
2 |
0% |
1.5% |
no |
|
ANKRD11 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ARID2 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ARID4B |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ARID5B |
1 |
0 |
1 |
0% |
0.7% |
no |
|
ATM |
1 |
1 |
0 |
100% |
0.7% |
no |
|
B2M |
1 |
0 |
1 |
0% |
0.7% |
no |
|
BAP1 |
1 |
1 |
0 |
100% |
0.7% |
no |
|
BARD1 |
1 |
1 |
0 |
100% |
0.7% |
no |
|
BRCA2 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
BRIP1 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
CBFD |
1 |
1 |
0 |
100% |
0.7% |
no |
|
FANCE |
1 |
0 |
1 |
0% |
0.7% |
no |
|
KEAP1 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
KMT2C |
1 |
1 |
0 |
100% |
0.7% |
no |
|
LATS1 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
MEN1 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
NOTCH2 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
NRAS |
1 |
0 |
1 |
0% |
0.7% |
no |
|
PDGFRA |
1 |
1 |
0 |
100% |
0.7% |
no |
|
PRDM1 |
1 |
1 |
0 |
100% |
0.7% |
no |
|
PTPRT |
1 |
1 |
0 |
100% |
0.,7% |
no |
|
RB1 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
SETD2 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
SMAD4 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
STAT3 |
1 |
0 |
1 |
0% |
0.7% |
no |
|
TGFBR1 |
1 |
1 |
0 |
100% |
0.7% |
no |
|
TRX |
1 |
0 |
1 |
0% |
0.7% |
no |


 Impact Factor: * 4.1
Impact Factor: * 4.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 