A Case Study on Wild Birds: A Human Enteric Pathogens Transmission
Article Information
Mancini L1,*, Marcheggiani S1, D’angelo AM1, Chiudioni F1, Delibato E2, Dionisi AM3, Owczarek S3, De Medici D2, Ida Luzzi 4
1Department Environment and Health, Viale Regina Elena, 299 00161 Rome, Italy
2 Department of Veterinary Public Health and Food Safety, Viale Regina Elena, 299 00161 Rome, Italy
3Department of Infective Diseases, Viale Regina Elena, 299 00161 Rome, Italy
4 Former Department of Infective Diseases, Viale Regina Elena, 299 00161 Rome, Italy
Corresponding Author: Mancini L, Department Environment and Health, Viale Regina Elena, 299 00161 Rome, Italy
Received: 06 July 2020; Accepted: 14 July 2020; Published: 10 September 2020
Citation: Mancini L, Marcheggiani S, D’angelo AM, Chiudioni F, Delibato E, Dionisi AM, Owczarek S, De Medici D, Ida Luzzi. A Case Study on Wild Birds: A Human Enteric Pathogens Transmission. Journal of Environmental Science and Public Health 4 (2020): 267-281.
View / Download Pdf Share at FacebookAbstract
Wild birds have been shown to be vectors for enteric pathogens, they may constitute an environmental carrier of pathogens representing a source of human infection. The study aims to evaluate the intestinal carriage of Salmonella spp, Campylobacter spp and Yersinia enterocolitica in wild birds in the Italian Regional Natural Park of “Lagolungo and Ripasottile” lakes. A total of 276 samples from 16 bird species were sampled and tested by classical cultural methods and the presence of Salmonella and Y. enterocolitica was detected, using real time PCR as screening methods. None of the fecal samples were positive for Y. enterocolitica. Campylobacter spp was isolated from 4 fecal specimens and Salmonella positive results were obtained from three avian species. Four different colonies of S. Napoli isolated from Cettia cetti were submitted to molecular typing by PFGE, all showing the same genetic pattern and sharing 70-80% homology with the 3 representative human isolates. One strain of S. Typhimurium from Nycticorax nyctcorax and one strain of S. Livingstone from Sylvia atricapilla were also isolated. Campylobacter spp. were isolated from Phylloscopus collybita, Cyanistes caeruleus, Prunella modularis, Carduelis chloris chloris. Wild birds, might cause the contamination of vegetables crops either directly with faecal material, or indirectly, with pollution of irrigation water. The study demonstrated that the molecular platforms to detect Salmonella and Y. enterocolitica seem to be very appealing as screening methods. Enteric pathogen circulation associated to wild fauna strictly linked to water ecosystems open new scenarios of management agricultural practices.
Keywords
Enteric pathogens; S. Napoli; Wild birds; Bacteriology; Real-time PCR
Enteric pathogens articles, S. Napoli articles, Wild birds articles, Bacteriology articles, Real-time PCR articles
Enteric pathogens articles Enteric pathogens Research articles Enteric pathogens review articles Enteric pathogens PubMed articles Enteric pathogens PubMed Central articles Enteric pathogens 2023 articles Enteric pathogens 2024 articles Enteric pathogens Scopus articles Enteric pathogens impact factor journals Enteric pathogens Scopus journals Enteric pathogens PubMed journals Enteric pathogens medical journals Enteric pathogens free journals Enteric pathogens best journals Enteric pathogens top journals Enteric pathogens free medical journals Enteric pathogens famous journals Enteric pathogens Google Scholar indexed journals S. Napoli articles S. Napoli Research articles S. Napoli review articles S. Napoli PubMed articles S. Napoli PubMed Central articles S. Napoli 2023 articles S. Napoli 2024 articles S. Napoli Scopus articles S. Napoli impact factor journals S. Napoli Scopus journals S. Napoli PubMed journals S. Napoli medical journals S. Napoli free journals S. Napoli best journals S. Napoli top journals S. Napoli free medical journals S. Napoli famous journals S. Napoli Google Scholar indexed journals Wild birds articles Wild birds Research articles Wild birds review articles Wild birds PubMed articles Wild birds PubMed Central articles Wild birds 2023 articles Wild birds 2024 articles Wild birds Scopus articles Wild birds impact factor journals Wild birds Scopus journals Wild birds PubMed journals Wild birds medical journals Wild birds free journals Wild birds best journals Wild birds top journals Wild birds free medical journals Wild birds famous journals Wild birds Google Scholar indexed journals Bacteriology articles Bacteriology Research articles Bacteriology review articles Bacteriology PubMed articles Bacteriology PubMed Central articles Bacteriology 2023 articles Bacteriology 2024 articles Bacteriology Scopus articles Bacteriology impact factor journals Bacteriology Scopus journals Bacteriology PubMed journals Bacteriology medical journals Bacteriology free journals Bacteriology best journals Bacteriology top journals Bacteriology free medical journals Bacteriology famous journals Bacteriology Google Scholar indexed journals Real-time PCR articles Real-time PCR Research articles Real-time PCR review articles Real-time PCR PubMed articles Real-time PCR PubMed Central articles Real-time PCR 2023 articles Real-time PCR 2024 articles Real-time PCR Scopus articles Real-time PCR impact factor journals Real-time PCR Scopus journals Real-time PCR PubMed journals Real-time PCR medical journals Real-time PCR free journals Real-time PCR best journals Real-time PCR top journals Real-time PCR free medical journals Real-time PCR famous journals Real-time PCR Google Scholar indexed journals humans articles humans Research articles humans review articles humans PubMed articles humans PubMed Central articles humans 2023 articles humans 2024 articles humans Scopus articles humans impact factor journals humans Scopus journals humans PubMed journals humans medical journals humans free journals humans best journals humans top journals humans free medical journals humans famous journals humans Google Scholar indexed journals Control articles Control Research articles Control review articles Control PubMed articles Control PubMed Central articles Control 2023 articles Control 2024 articles Control Scopus articles Control impact factor journals Control Scopus journals Control PubMed journals Control medical journals Control free journals Control best journals Control top journals Control free medical journals Control famous journals Control Google Scholar indexed journals domestic animals articles domestic animals Research articles domestic animals review articles domestic animals PubMed articles domestic animals PubMed Central articles domestic animals 2023 articles domestic animals 2024 articles domestic animals Scopus articles domestic animals impact factor journals domestic animals Scopus journals domestic animals PubMed journals domestic animals medical journals domestic animals free journals domestic animals best journals domestic animals top journals domestic animals free medical journals domestic animals famous journals domestic animals Google Scholar indexed journals agricultural articles agricultural Research articles agricultural review articles agricultural PubMed articles agricultural PubMed Central articles agricultural 2023 articles agricultural 2024 articles agricultural Scopus articles agricultural impact factor journals agricultural Scopus journals agricultural PubMed journals agricultural medical journals agricultural free journals agricultural best journals agricultural top journals agricultural free medical journals agricultural famous journals agricultural Google Scholar indexed journals
Article Details
1. Introduction
Enteric illnesses are the second largest source of communicable diseases worldwide (Scallan et al ., [1]); Batz, Hoffmann and Morris, [2]). Wild-living animals might play a significant role in the dispersion of pathogenic microorganisms and the contamination of vegetables (Havelaar et al ., [3]; Smith et al., [4]). Particularly, wild-living birds are suspected to play an effective and important role in this dispersion and in the possible transmission of pathogens to other birds, livestock and humans, because they are characterized to fly freely covering long distances during annual movements (migration), colonize new areas and withstand a range of environments (Reed et al., [5] ).
The main way of enteric pathogens transmission includes physical contact with domestic animals, person-to-person spread, and consumption of contaminated food and water (Raül Ramos et al., [6]). Vegetables contaminated by zoonotic enteric pathogens have been also reported as vehicle of foodborne outbreaks. European Rapid Alert for Food and Feed System (RASFF) triggered alerts on contaminated vegetables, including Italian fresh products contaminated by Salmonella, Yersinia enterocolitica (Y. enterocolitica) and Campylobacter (European Commission, [7]).
Wild bird infections can reflect contamination of the environment by humans or livestock however it cannot be excluded that some avian species can act as reservoir for pathogens, direct or indirect contamination of water sources and be responsible for environmental contamination including food crops (Elmberg et al., [8]). The prevalence of bacterial human pathogens in wild birds has been reported (Benskin et al., [9]) and pathogens maintained by wild birds occasionally jump to human hosts, by the consumption of contaminated water and food, resulting in a considerable impact of human health (Fuller et al., [10]).
Among enteric pathogens, Salmonella and Campylobacter are the most frequent zoonotic pathogens responsible for foodborne infections over the world. In European Union, 246,571 cases of campylobacteriosis and 91,857 cases of salmonellosis have been reported in 2018 (EFSA, [11]). Yersiniosis remains the third most commonly reported bacterial food-borne zoonosis in the EU in 2018.
Food of animal origin are the most frequent vehicles for Salmonella and Campylobacter infections, however, vegetables contaminated by zoonotic enteric pathogens have been also reported as vehicle of foodborne outbreaks. European Rapid Alert for Food and Feed System (RASFF) triggered alerts on contaminated vegetables, including Italian fresh products contaminated by Salmonella enterica subsp. enterica serovar Napoli (S. Napoli). In 2013, a pilot study was carried out with the aim to evaluate the possible role of wild birds in the diffusion of Salmonella and a strain belonging to the S. Napoli was isolated from stool of a nightingale sampled in a natural area of Central Italy (Mancini et al., [12]). S. Napoli is among the most frequent serovars causing human infections in Italy, although it is relatively uncommon in other European countries (Luzzi et al., [13]; Graziani et al., [14]); it is mainly isolated from humans and the environment, but neither the reservoir nor its route of infection is clearly defined (Sabbatucci et al., [15]).
The results of the previous studies prompted us to extend the study and evaluate the intestinal carriage of Salmonella, Campylobacter spp and Y. enterocolitica, in free living birds in the Regional Park Latium region (Regional Natural Park “Lagolungo e Ripasottile), a pristine area without strong pressures and human impacts. The monitoring, on wild birds faecal samples, was performed using classical cultural methods to detect Campylobacter, Salmonella and Y. enterocolitica. Moreover, the presence of Salmonella and Y. enterocolitica was detected, using real time PCR as screening methods for evaluating the effectiveness of this rapid method for its use as screening methods for large monitoring (Delibato et al., [16]).
Human infections caused by S. Napoli are relatively rare in Europe (EFSA, [11]); however, in Italy, this serovar has been responsible for 2- 4% of all cases of human salmonellosis each year, for 10 years (Graziani et al., [14]; Sabbatucci et al., [15]). Moreover, several outbreaks related to this serovar outside of Italy have been linked to the consumption of exported Italian food products (e.g. chocolate bars (Greenwood and Hooper, [17]; Gill et al., [18] ) and rocket salad (Graziani et al., [19]). The Rapid Alert System for Food and Feed (RASFF) reported twelve notifications regarding S. Napoli to date, all but one involving fresh vegetable products exported from Italy to other European countries (as of July 2019) (https://webgate.ec.europa.eu/rasff-window/portal/)
S. Napoli is a particular serovar frequently isolated from environmental sources, mainly surface waters, but it is rarely found in animal and foodstuff of animal origin (Sabbatucci et al., [15]). On the basis of these findings, environmental sources and fresh and leafy vegetables should be considered important for the transmission of this serovar to humans. The circulation of Napoli in wild is well known and studied. Isolation of S. Napoli from several reservoirs needs to set up a plan to manage the cycle of infection (wild bird-water-plant-man), a highly complex cycle that cannot be referred to common human pressure but which necessarily must address water management agricultural irrigation. The aim of this work was performed a “pilot study on wild birds” in order to identify the possible environmental reservoirs of S. Napoli and other bacteria pathogens such as Y. enterocolitica and Campylobacter spp.
figure 1: Study area of Lungo and Ripasottile Lakes in Latium Region (Rieti) (adapted from Franceschini et al., [20])
2. Materials and Methods
2.1 Study area
The Regional Nature Reserve of Lungo and Ripasottile Lakes, in central Italy locations, is part of the international system of wetlands of Rhamsar convention, Sites of Community Interest and Special Protection Areas and is included in the "Natura 2000" network. The particular location, within the Italian territory makes it an important site for migrating, wintering and breeding for birds. The species were sampled by the experts of Bird Observatory, near Lake Ripasottile, municipality of Colli Sul Velino (Rieti), nearby water body ecosystem during hot season of 2012
2.2 Sampling
Ten bird trapping campaigns have been carried out from March to December 2012 and fecal samples from migratory and resident bird species were collected. The sampling methods used, was those reported by Mancini et al., [12] ). Each faecal sample was aanalyzed for the presence of Salmonella spp, Y. enterocolitica and Campylobacter spp. within 24 hours from collection.
3.Microbiological Analysis
Cultural and molecular methods adopted for recovery and identification of targeting microrganisms have been elaborated from those currently used in our laboratories for water and food analysis.
3.1 Cultural methods
3.1.1 Salmonella: One g of fecal sample was re-hydrated in 1 mL of Peptone Water (MERCK, Italy) and incubated for about 2 hours at 36 ± 1 °C. 1 mL of the suspension was transferred into 10 mL of Buffered Peptone Water (BPW) as pre-enrichment medium and incubated at 37 ± 1 °C for 24 hours. After incubation, samples were inoculated into Rappaport-Vassiliadis Broth (Oxoid, Basingstoke, UK) as enrichment medium and incubated at 42°C for 18 h. The cultures obtained were plated onto MacConkey agar selective medium (MERCK, Italy) incubated at 37°C and examined after 24 h. Suspected colonies were inoculated onto TSA agar (Oxoid, Basingstoke, UK). All isolates were biochemically identified by API20-E system (bioMe´rieux, Marcy l’Etoile, France), and serotyped according to Kauffman_ Le Minor scheme (Grimont and Weill, [21]).
3.1.2 Yersinia enterocolitica: One gram of faecal sample was transferred into 10 mL of Peptone Sorbitol and Bile (PSB) (Oxoid, Basingstoke, UK) broth as enrichment medium, and incubated at 25 ± 1 °C for 48 hours. After incubation samples were plated onto of Cefsulodin–Irgasan–Novobiocin (CIN) agar plates (Yersinia Selective Agar Base and Yersinia Selective Supplement, Oxoid, Basingstoke, UK) and examined after 24-48 hours incubation at 25 ± 1°C. Suspected colonies were inoculated onto TSA agar (Oxoid, Basingstoke, UK) and biochemically identified using the API 20E system (bioMerieux, Marcy l’Etoile, France).
3.1.3 Campylobacter spp.: One gram of fecal sample was suspended in 10 mL of Bolton broth (MERCK, Italy) and incubated at 37 ± 1 °C for 4 hours and then at 42 ± 1 °C for 48 hours under microaerobic conditions provided by CampyGen (Oxoid, Basingstoke, UK). After incubation the samples were streaked onto Campylobacter blood-free selective agar (mCCDA) (MERCK, Italy) and incubated in a microaereophilic conditions at 42 ± 1 °C for 48 hours. This particular atmosphere (5-7% O2 and 8-10% CO2) can be obtained by incubating the plates in anaerobic jar with envelopes of Anaerocult C (MERCK, Italy). five percent of the typical colonies were tested for oxidase production and motility by the method of the hanging drop and microscope under normal light. The colonies found to belong to the genus Campylobacter are inoculated onto TSA agar (Oxoid, Basingstoke, UK) and incubated to the same conditions described before and stored in cryo-banks at -20 °C.
3.1.4 Antimicrobial susceptibility assay: Salmonella isolates were subjected to the disk-diffusion assay. Isolates were grown in BHI (Oxoid, Basingstoke, UK) at 37 °C for 24 h, the inoculum was adjusted by the McFarland standard and a swab was dipped into the adjusted suspension, then spread onto the surface of plates containing Mueller Hinton agar (Oxoid, Basingstoke, UK), where disks of the following antimicrobials were added: nalidixic acid (NAL, 30), ampicillin (A, 10), cefotaxime (CTX, 5), ceftazidime (CAZ, 10), amoxicillin/clavulanic acid 2:1 (AMC, 30), meropenem (MEM, 10), chloramphenicol (C, 30), gentamicin (G, 10), kanamycin (K, 30), streptomycin (S, 10), sulfonamides (Su, 0.25), tetracycline (T, 30), trimethoprim (TMP, 5), and trimethoprim–sulfamethoxazole (SXT, 1.25/23.75). The plates were incubated at 35 °C for 18 h. All antimicrobials were purchased from Oxoid (Basingstoke, UK) and Escherichia coli ATCC 25922 was used as a pan-susceptible quality control. The diameters of the zones of inhibition were measured and the Salmonella isolates were characterized as resistant or susceptible according to EUCAST (EUCAST, [22]) For ciprofloxacin (CIP) the minimal inhibitory concentration (MIC) was determined using Etest strips (AB Biodisk, S-169 56 Solna, Sweden).
3.2 Molecular methods
3.2.1 DNA extraction method: Two mL of each pre-enrichment broth (BPW) (Oxoid, Basingstoke, UK) for Salmonella, PSB (Oxoid, Basingstoke, UK) for Y. enterocolitica) was transferred into a clean microcentrifuge tube, and centrifuged for 10 min at 10,000×g at 4°C. The supernatant was discarded carefully and the pellet was washed with 1 mL of Water for Molecular Biology (WMB) (MERCK, Italy) and centrifuged for 5 min at 10,000×g at 4ºC. Afterwards, the pellet was re-suspended in 200 µL of 6% Chelex 100 (Biorad, Hercules, CA, USA) by vortexing, and incubated for 20 min at 56°C and then for 8 min at 100°C. The suspension was immediately chilled on ice for 1 min, and centrifuged for 5 min at 10,000×g at 4°C.
3.2.2 Real-time PCR assay for Salmonella: The RT-PCR amplification protocol, used in Delibato et al., [16], is detailed below. The final volume of PCR mix reactions was 25 µL and prepared using: 12.5 µL of Mastermix (QuantiTecMultiplex PCR No Rox Master Mix – Qiagen, Hilden, Germany), 400 nM of each primer, 240 nM of ttr5 and IAC probe (labelled with FAM and HEX), 0.25 µL of IAC (about 1200 copies) and 4 µL of DNA template were added to each micro-well (of a 96-microwell plate). Negative and positive controls were prepared similarly (but avoiding the addition of the IAC) and adding 4 µL of WMB for negative control and 4 µL of DNA standard (extracted from a pure culture of S. Napoli 108 cfu/mL) for positive control. PCR 96-well-plate was spinned-down, and inserted into the RT-PCR platform (Stratagene Mx3005P–Agilent technologies, Santa Clara, USA). The amplification was performed using an initial hot-start step at 95 °C for 15min, followed by 40 cycles of a denaturation step at 95°C for 30 s, an annealing step at 65 °C for 60 s and an extension step at 72 °C for 30 s. The fluorescence was recorded (at 520 nm for FAM and at 556 nm for HEX) only at the end of the annealing step. Three PCR replicates were used for each sample and control. When an assay showed a quantification cycle (Cq) value ≤35 independently of the IAC Cq value, the result was interpreted as positive. When an assay showed a Cq value ≥35 with the IAC Cq value ≤40 was interpreted as negative. When an assay showed both the target and its corresponding IAC Cq values ≥37 the reaction was considered to have failed.
3.2.3 Real-time PCR assay for Y. enterocolitica: The RT-PCR amplification protocol, used in Made et al., [23] , is detailed below. The final volume of PCR mix reactions was 25 µL and prepared using: 12,5 µL Master mix (QuantiTecMultiplex PCR No Rox Master Mix – Qiagen, Hilden, Germany), 300 nM of each primer and 125 nM of ye-ail-tmp probe for Y. enterocolitca amplification, 250 nM of each primer and 100 nM of Tm-pUC 18 probe for IAC amplification (ye-ail-tmp and IAC probes were labelled with FAM and HEX, respectively), 1 µL of IAC (about 1000 copies) and 2,5 µl of DNA template were added to each micro-well (of a 96 microwell plate). Negative and positive controls were prepared similarly (but avoiding the addition of the IAC) and adding 2,5 µL of WMB for negative control and 2,5 µL of DNA standard (extracted from a pure culture of Y. enterocolitica 108 cfu/mL), for positive control.
PCR 96-well-plate was spinned-down, and inserted into the Real-Time PCR platform (Stratagene Mx3005P–Agilent technologies, Santa Clara, USA). The amplification was performed using an initial hot-start step at 95 °C for 10 min, followed by 45 cycles of a denaturation step at 95°C for 10 s, an annealing step at 60 °C for 30 s. The fluorescence was recorded (at 520 nm for FAM and at 556 nm for HEX) only at the end of the annealing step. Three PCR replicates were used for each sample and control. When an assay showed a quantification cycle (Cq) value ≤40 independently of the IAC Cq value, the result was interpreted as positive. When an assay showed a Cq value ≥40 with the IAC Cq value ≤32 was interpreted as negative. When an assay showed both the target and its corresponding IAC Cq values ≥40 the reaction was considered to have failed.
3.2.4 Pulsed-field Gel Electrophoresis for Salmonella: Analysis and comparison of PFGE types was performed according to the Pulsenet protocol (CDC, [24]) using a CHEF_Mapper (Bio-Rad Laboratories, Hercules, CA). Briefly, genomic DNA was digested with XbaI (New England Biolabs, Ipswich, MA, USA), and Salmonella enterica serovar Braenderup H9812 DNA was used as the molecular size marker. Electrophoresis conditions were an initial switch time of 2.16 sec, a final switch time of 63.8 sec, and a run time of 21 h. Pictures of PFGE gels were taken with Chemidoc system (Bio-Rad Laboratories, Hercules, CA). Dendrogram and cluster analyses were performed using algorithms included in the BioNumerics software package v.6.6 (Applied Maths, Sint-Martens-Latem, Belgium). The percentage similarity between different chromosomal fingerprints had been scored using the Dice coefficient. The unweighted pair group method with arithmetic means (UPGMA) with a 1.00% tolerance limit and 1.00% optimization was used to generate the dendrogram. Three S. Napoli strains representative of the most frequent PFGE patterns among human isolates were included in the analysis for comparison.
4. Results
A total of 92 specimens, corresponding to 16 species, were trapped and 276 faecal samples were collected. The most common sampled bird species were Gallinago gallinago Linnaeus, 1758, Nycticorax nycticorax, Linnaeus 1758, Erithacus rubecula (Linnaeus, 1758) and Sylvia atricapilla, Linnaeus 1758 (Table 1).
|
Bird species |
Common name |
Number of specimens |
|
Gallinago gallinago (Linnaeus, 1758) |
Stripe |
26 |
|
Nycticorax nycticorax, (Linnaeus, 1758) |
Night Heron |
13 |
|
Sylvia atricapilla, (Linnaeus, 1758) |
Blackcap |
8 |
|
Erithacus rubecula (Linnaeus, 1758) |
Robin |
10 |
|
Cettia cetti, (Temminck 1820) |
Nightingale or Cetti's Warblero |
6 |
|
Turdus merula (Linnaeus, 1758) |
Black birds |
3 |
|
Turdus philomelos (C.L.Brehm, 1831) |
Thrush |
1 |
|
Phylloscopus collybita (Vieillot, 1817) |
Common chiffchaff |
4 |
|
Cyanistes caeruleus (Linnaeus, 1758) |
Bluetit |
6 |
|
Prunella modularis (Linnaeus, 1758) |
Dunnock |
3 |
|
Passer montanus (Linnaeus, 1758) |
Eurasia tree sparrow |
2 |
|
Carduelis carduelis, (Linnaeus, 1758) |
Goldfinch |
3 |
|
Acrocephalus melanopogon (Temminck, 1823) |
Moustached warbler |
2 |
|
Parus major, (Linnaeus, 1758) |
Titmouse |
3 |
|
Alcedo atthis (Linnaeus, 1758) |
Kingfisher |
1 |
|
Carduelis chloris chloris (Linnaeus, 1758) |
Greenfinch |
1 |
|
Total |
92 |
|
Table 1: Number of specimens divided per bird species trapped in ten campaigns.
Microbiological results showed that all 92 specimens of wild bird were negative for Y. enterocolitica, five were positive for Campylobacter using classical methods and 3 were positive for Salmonella spp using Real Time PCR tests and cultural methods (Table 2). All Real Time PCR negative samples showed a positive IAC signal confirming that the negative results were not due to an inhibition during the amplifications. However, it was not possible to perform the RT-PCR on Campylobacter spp. because the sample stored has been corrupt due to damage to the refrigerator.
|
Microorganisms |
RT-PCR tests |
Cultural methods |
|
Yersinia enterocolitica |
negative |
negative |
|
Campylobacter |
Not performed |
5 positive samples |
|
Salmonella |
3 positive samples |
3 positive samples |
Table 2:Comparison between the results obtained by RT-PCR tests and Cultural methods.
Salmonella has been isolated in resident birds (Table 3). The 3 Salmonella isolates were found belonging to S. Typhimurium, S. Napoli and S. Livingstone. All these serovars, isolated in our study, belong to serovars reported as responsible for human infections, in Italy. In the last 5 years, S. Typhimurium is the most prevalent serovar accounting for approximately 54%; S. Livingstone is almost infrequent (0,3%) whereas S. Napoli accounts for 4 % of all human Salmonella serovars (Luzzi et al., [13]).
|
Species |
Salmonella spp. (serovar) |
|
Nycticorax nycticorax |
Typhimurium |
|
Sylvia atricapilla |
Livingstone |
|
Cettia cetti |
Napoli |
Table 3: The Enterobacteriaceae isolated from each bird species sampled are shown.
The results of antibiotic resistance assay performed on the three Salmonella isolated from fecal samples of three bird specimens are shown in table (Table 4)
*ampicillin (A, 10), cefotaxime (CTX, 5), ceftazidime (CAZ, 10), amoxicillin/clavulanic acid 2:1 (AMC, 30), meropenem (MEM, 10), chloramphenicol (C, 30), gentamicin (G, 10), kanamycin (K, 30), streptomycin (S, 10), sulfonamides (Su, 0.25), tetracycline (T, 30), trimethoprim (TMP, 5), and trimethoprim–sulfamethoxazole (SXT, 1.25/23.75).
Table 4: Antibiotic susceptibility of Salmonella serovars.
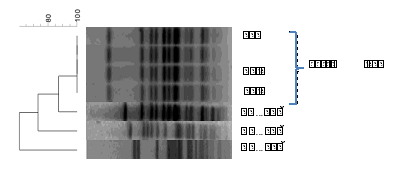
figure 2: Cluster analysis of PFGE profiles of S. Napoli isolated from Cettia cetti and from human infections.
PFGE analysis shows that the four different colonies of S. Napoli isolated from Cettia cetti had the same genetic pattern (figure 2). This result excludes the possible colonization of the same bird by different clones of this serovar. In addition, PFGE clustering shows a high genetic homology (>80%) with one of the three human isolates belonging to the most frequent PFGE patterns. A genetic relatedness less than 80% were detected between the bird’s strain and the two other human isolates. As described by Sabbatucci et al (Sabbatucci et al., [15] ) S. Napoli shows a high genetic variability among strains, however, the AA identified some main clusters which included both human, wild animals and environmental isolates thus suggesting an environmental dissemination of this serovar in Italy. In this respect, the results of this study add evidence on the role of the wild animals in the persistence in and spread through the environment of S. Napoli.
5. Discussion and Conclusion
This study analyzed the fecal carriage of Salmonella, Campylobacter and Y. enterocolitica in free living wild bird from a natural area in central Italy Salmonella spp. has been isolated from 3 out of 92 samples, whereas no sample was found positive for Y. enterocolitica. Several studies were performed on the presence of Salmonella in wild birds. In Spain, Reche et al., [25] ) quoted a prevalence of 4% in raptors, mainly S. Typhimurium DT 104. In another study, a higher Salmonella percentage (10%) is reported in wild raptors. In Italy, Botti et al., [26] ) reported a Salmonella positivity of 2% in all wild birds, even if half of the isolations occurred in raptors (n=12/24).
Napoli is an emerging serovar in Italy. Recent studies suggest that S. Napoli is mostly present in the environment even if they do not point out a specific source of exposure for humans [Graziani et al., [14] ; Sabbatucci et al., [15] ]. Wild birds, might cause the contamination of vegetables crops either directly with fecal material, or indirectly, with pollution of irrigation water.
Moreover, the present study demonstrates that the molecular platforms to detect Salmonella and Y. enterocolitica seem to be very appealing as screening methods, keeping in mind that the microbiological cultural methods require up to 7 days to produce final results. Despite this the isolation and serotyping of the Salmonella and Y. enterocolitica in samples remain important for epidemiological purpose. To this end the remaining pre-enrichment broths found positive to Real Time PCRs should be submitted to microbiological methods at the aim to isolate the strain, the Real-Time PCR assay for Salmonella is based on the co-amplification of a specific region of the Salmonella spp. ttr gene (Malorny et al., [27] ; Josefsen et al., [28] ) and an internal amplification control (IAC). The choice of the ttr locus as a target specific for Salmonella spp. over other published targets might have the advantage in the identification of all Salmonella spp. strains as the ability to respire tetrathionate is significant for Salmonella survival and out growth in anaerobic competitive environments (Winter et al., [29] ).
The duplex Real time PCR for Y. enterocolitica was based on amplification of a specific region of the Y. enterocolitica ail gene (Made et al., [23] ) and an internal amplification control (IAC). The chromosomal ail gene is stably inherited in contrast to the plasmid encoded virulence factors which may be lost during incubation due to instability of the virulence plasmid (Wannet et al., [30] ). The ail gene is found only in pathogenic strains of Yersinia, but is not present in non-pathogenic serotypes of Y. enterocolitica nor in non-pathogenic Yersinia species (Revell and Miller, [31] ). In addition, the use of an IAC in molecular microbiology diagnostics is necessary because indicates the presence of DNA polymerase inhibitors, errors caused by PCR components, or malfunction of the thermal cycler. The simultaneous use in a single reaction of two differently labelled fluorescent probes makes it possible to detect the target, and if negative results are obtained, the positive IAC signal can confirm that the negative result is not due to an inhibition during the amplification (Hoorfar et al., [32]). In this context the development of rapid, cost-effective, and automated methods for the detection of Salmonella and Y. enterocolitica, integrated with environmental preventive strategies, could significantly improve safety throughout the food chain.
The circulation of enteric pathogens not associated with anthropic activities but with wildlife, closely linked to the aquatic ecosystem, opens up new scenarios for the management of aquatic ecosystems and agricultural practices.
The three species positive for Salmonella are all revolve around the lakes or the river and surrounding riparian environments closely linked to water environments. In particular Nycticorax nycticorax (Ardeide-Ciconiiformes) found positive for S. Typhimurium was sampled from a nesting colony near Lake Votone of about 50 couples (RNR The Lakes along Ripasottile). In Europe, the breeding area is very fragmented, with localized colonies, while the wintering range includes the sub-Saharan region of West Africa to the Equator. Less than 1% of individuals winter in southern Europe. Immediately after nesting, the colonies are abandoned, with a scattering short-range, and by September the southward migration takes place. The return in the reproductive areas occurs from mid-March. Recapture data show that the species is long-lived (more than 10 years of life in some cases). Italy is a crossroads in the night heron migration system; particularly relevant are the numerically recaptures of individuals ringed in France, Czech Republic, Hungary and Yugoslavia. They can, however, make the movements of some significance to our country during migration, then cross directly wide arms of the sea and reach the wintering areas Sylvia atricapilla, found positive for S. Livingstone, is a species with different migratory strategies including sedentariness and erratic movements. The majority of individuals present in Italy are resident even if the nominal subspecies makes complete migration and Italy represents a simple stop site. In the ornithology station of Ripasottile approximately 3000 individuals have been ringed, mainly during the spring migration; the captures of the last 20 years show the presence of a resident population, a great faithfulness to the stop site of migration (individuals re-captured in 5 consecutive years) and to the breeding site. In addition, the particular microclimate of Ripasottile allows the breeding of the nominal subspecies S. atricapilla, with typical northern breeding habits.
The individual found positive for S. Livingstone showed morphologic features of the external flight apparatus and weight clearly indicating that the individual belonged to the migratory subspecies. Cettia cetti found positive for S. Napoli, is diffused overall Europe and in Italy it is distributed on the whole territory from the lowlands to the mountains given its pronounced adaptation to heterogeneous and degraded habitats. This species shows sedentariness and very low erratic movements and for this reason there are few data on the movements through our Country obtained from re-captures with the ring techniques (Brunelli et al. [33]). However, data obtained from the systematic captures of the same individuals in different period of the year and the re-captures in different years allowed to estimate a a resident breeding population of approximately 300 couples in the Riserva Naturale of Lungo and Ripasottile lakes.
Our dataset was composed of many different species, but the number of tested individual birds for each species was low in many cases. Earlier studies have pointed to certain species (gulls and corvids) in which the prevalence of Salmonella is sometimes high (2% to 20%), and argued that concern should be strong about epidemiologic disease transmission with these birds (Hubálek et al, [34] ; Palmgren, [35] ). These species have the capability to live in an opportunistic manner in close proximity to humans and can base their diet on waste products and garbage. Most bird species, however, have little or no niche overlap with humans or domesticated animals; virtually no data exists on the occurrence of Salmonella in this major group of migrating birds during a non-epizootic situation. Our results suggest that the natural occurrence of Salmonella in healthy birds during migration in Italy may be low. Therefore, the Salmonella incidence is probably also low for most wild bird species. We suggest that researchers consider analysing the non-epizootic natural occurrence of Salmonella in wild birds. Accumulated knowledge from many different regions, over many years, is a prerequisite for thorough risk assessment of the importance of Salmonella carriage in wild birds.
The overall proportion of Salmonella-positive samples was 3.20%. Salmonella isolation was positively associated with samples collected from birds in the proximity of water body. To our knowledge this is the first description of isolation of S. Napoli from wild birds and this finding provides an additional confirmation that S. Napoli is circulating in the wild and consequently in different natural habitats in Italy and justify the wide presence of this strains in surface water. In this respect, suitable public health interventions have to be applied by the Public Authority to avoid a possible diffusion of this serovar in vegetables irrigated using contaminated water as demonstrated by different outbreaks. Further, it should be to take in to account an important additional concern represented by the possible transmission of these strains in the farmed animals, because since to manage the contamination of surface waters and crops is extremely difficult. Besides water quality requirements, the awareness of growers, farmers and eco-systemic service users in general, about risks deriving from wild, represents an important issue to prevent and control S. Napoli human infections.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States — major pathogens. Emerging Infectious Diseases 17 (2011): 7-15.
- Batz MB, Hoffmann S, Morris JG. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. Journal of Food Protection 75 (2012): 1278-1291.
- Havelaar AH, Kirk MD, Torgerson PR, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Medicine 12 (2015): e1001923.
- Smith OM, Snyder WE, Owen JP. Are we overestimating risk of enteric pathogen spillover from wild birds to humans? Biological Reviews (2020).
- Reed KD, Meece JK, Henkel JS, et al. Birds, migration and emerging zoonoses: west nile virus, lyme disease, influenza A and enteropathogens. Clin Med Res 1 (2003): 5-12.
- Ramos R, Cerdà-Cuéllar M, Ramírez F, et al. Influence of Refuse Sites on the Prevalence of Campylobacter spp. and Salmonella Serovars in Seagulls. Aem applied and environmental microbiology (2010): 3052-3056.
- European Commission. RASFF portal (2017).
- Elmberg J, Berg C, Lerner H, et al. Potential disease transmission from wild geese and swans to livestock, poultry and humans: a review of the scientific literature from a One Health perspective. Infect Ecol Epidemiol 7 (2017): 1300450.
- Clare McW, Benskin H, Kenneth W, et al. Hartley Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biol. Revn 84 (2009): 349-373.
- Fuller T, Bensch S, Muller I, et al. Waldestrom The ecology of emerging infectious diseases in migratory birds: an assessment of the role of climate change and priorities for future research. Ecohealth 9 (2012): 80-8.
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2019. The European Union One Health 2018 Zoonoses report. EFSA Journal 17 (2019): 276.
- Mancini L, Marcheggiani S, D’Angelo A, et al. first isolation of Salmonella enterica serovar Napoli from wild bird in Italy. Annali dell’Istituto Superiore di Sanità 50 (2014): 96-98.
- Luzzi I, Garcia-Fernaandez A, Dionisi AM, et al. Enter-Net Italia e Registro italiano della sindrome emolitico uremica: sorveglianza delle infezioni da Salmonella, Campylobacter, Escherichia coli produttore di Shiga-tossina e Listeria monocytogenes (2010-2015). Roma: Istituto Superiore di Sanita (2017). (Rapporti ISTISAN 17/34).
- Graziani C, Luzzi I, Owczarek S, et al. Salmonella enterica serovar Napoli infectin in Italy from 2000 to 2013: spatial and spatio-temporal analysis of cases distributin and the effect of human and animal on the risk infection. PLoS One 11 (2015): 11-10.
- Sabbatucci M, Dionisi AM, Pezzotti P, et al. Molecular and Epidemiologic Analysis of Reemergent Salmonella enterica Serovar Napoli, Italy, 2011-2015.Emerg Infect Dis 24 (2018): 562-565.
- Delibato E, Rodriguez-Lazaro D, Gianfranceschi M, et al. European validation of Real-Time PCR method for detection of Salmonella spp. in pork meat. Int J Food Microbiol 184 (2014): 134-138.
- Greenwood MH, Hooper WL. Chocolate bars contaminated with Salmonella Napoli: an infectivity study. Br Med J (Clin Res Ed) (1983).
- Gill ON, Sockett PN, Bartlett CL, et al. Outbreak of Salmonella Napoli infection caused by contaminated chocolate bars. Lancet Lond Engl 1 (1983): 574-577.
- Graziani C, Busani L, Dionisi AM, et al. Virulotyping of Salmonella enterica serovar Napoli strains isolated in Italy from human and nonhuman sources. Foodborne Pathog Dis 8 (2011): 997-1003.
- Franceschini S, Formichetti P, Damiani G, et al. Qualità ambientale della Riserva Naturale dei laghi Lungo e Ripasottile (Rieti). Roma: Istituto Superiore di Sanita (2004).
- Grimont P, Weill FX. Antigenic formulae of the salmonella servovars. Who collab. Cent. Ref. Res. Salmonella (2008): 167.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2016).
- Mäde D, Reiting R, Strauch E. et al. A real-time PCR for Detection of Pathogenic Yersinia enterocolitica in food combined with an Universal Internal Amplification Control System. J. Verbr. Lebensm 3 (2008): 141-151.
- Centers for Disease Control and Prevention (CDC). Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Atlanta: CDC; Apr (2013).
- Reche MP, Jiménez PA, Alvarez F, et al. Incidence of salmonellae in captive and wild free-living raptorial birds in central Spain. J Vet Med B Infect Dis Vet Public Health 50 (2003): 42-44.
- Botti V, Navillod FV, Domenis L, et al. Salmonella spp. and antibiotic-resistant strains in wild mammals and birds in north-western Italy from 2002 to 2010. Vet. Ital 49 (2013): 195-202.
- Malorny B, Paccassoni E, Fach P, et al. Diagnostic real-time PCR for detection of Salmonellain food. Appl Environ Microbiol 70 (2004): 7046-7052.
- Josefsen MH, Krause M, Hansen F, et al. Optimization of a 12-hour TaqMan PCR-based method for detection of Salmonella bacteria in meat. Appl Environ Microbiol 73 (2007): 3040-3048.
- Winter S, Koerbler M, Stein B, et al. Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. Journal of General Virology 91 (2010): 1365-1372.
- Wannet WJ, Reessink M, Brunings LIA, et al. Detection of pathogenic Yersinia enterocolitica by a rapid and sensitive duplex pcr assay. J Clin Microbiol 39 (2001): 4483-4486.
- Paula A Revell, Virginia L Miller. Yersinia virulence: more than a plasmid. FEMS Microbiology Letters 205 (2001): 159-164.
- Hoorfar J, Cook N, Malorny B, et al. Making internal amplification control mandatory for diagnostic PCR. Letters in Applied Microbiology 38 (2004): 79-80
- Brunelli M, Sarrocco S, Corbi F, et al. Nuovo Atlante degli Uccelli Nidificanti nel Lazio. Edizioni ARP (Agenzia Regionale Parchi), Roma (2011): 464.
- Hubálek Z, Sixl W, Mikulásková M, et al. Salmonella in gulls and other free-living birds in the Czech republic. Cent Eur J Public Health 3 (1995): 21-24.
- Palmgren H, Sellin M, Bergström S, et al. Enteropathogenic bacteria in migrating birds arriving in Sweden. Scand J Infect Dis 29 (1997): 565-568.


 Impact Factor: * 3.6
Impact Factor: * 3.6 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 