Network Meta-analysis on the Molecular Mechanisms of Palmitic Acid in Liver Fibrosis
Article Information
Myeong Gil Jun1, Heping Zhou*
Department of Biological Sciences, Seton Hall University, South Orange, NJ 07079, USA
*Corresponding Author: Heping Zhou, Department of Biological Sciences, Seton Hall University, South Orange, NJ 07079, USA
Received: 25 October 2021; Accepted: 01 November 2021; Published: 29 November 2021
Citation:
Myeong Gil Jun, Heping Zhou. Network Meta-analysis on the Molecular Mechanisms of Palmitic Acid in Liver Fibrosis. Journal of Food Science and Nutrition Research 4 (2021): 286-302.
View / Download Pdf Share at FacebookAbstract
Elevated levels of free fatty acids have been suggested as the main factors contributing to the development of non-alcoholic steatohepatitis. This study conducted network meta-analysis using the QIAGEN Ingenuity Pathway Analysis (IPA) to examine the roles of saturated fatty acids (SFAs) and n-3 unsaturated fatty acids (UFAs) in the activation of Hepatic Fibrosis Signaling Pathway and collagen accumulation. Our analysis identified the shortest paths from palmitic acid (PA), a SFA, to the Hepatic Fibrosis Signaling Pathway, and found that elevated level of PA may increase the activities of transcription factors, such as CEBPβ, JUN/FOS, NFκB, and PPARγ, cytokines/chemokines/growth factors, such as TNF, IL1, CCL2, CCN2, LEP, and TGFβ1, oxidative stress mediators, such as NOX, and fibrinolysis regulators, such as SERPINE1, which may in turn activate the Hepatic Fibrosis Signaling Pathway. Our analysis also identified the shortest paths from PA to collagen accumulation and found that elevated level of PA may increase the activities of signaling mediators, such as ERK1/2, cytokines/chemokines/growth factors, such as TNF, IL6, CCL2, CCN2, LEP, and TGFβ1, which may in turn increase the accumulation of collagens. In contrast, the increased levels of n-3 UFAs inhibited the activities of PDGFA, PDGFB, TNF, IL1, and CCL2. Our analysis also identified seven PA- and liver fibrosis-associated molecules mapped to the shortest paths from PA the Hepatic Fibrosis Signaling Pathway and from PA to collagens. Mapping of these seven molecules, TNF, IL1A, CCL2, TGFβ1, CCN2, LEP, and SERPINE1, to the Hepatic Fibrosis Signaling Pathway showed that these molecules may lead to proinflammatory response in the liver, reduced ECM degradation, and enhanced ECM accumulation. Our studies shed light on the molecular mechanisms by which PA contributes to liver fibrosis and the key mediating molecules that may be used for further research and
Keywords
Liver fibrosis, Palmitic acid, n-3 unsaturated fatty acids, Collagen, Saturated fatty acids
Liver fibrosis articles; Palmitic acid articles; n-3 unsaturated fatty acids articles; Collagen articles; Saturated fatty acids articles
Liver fibrosis articles Liver fibrosis Research articles Liver fibrosis review articles Liver fibrosis PubMed articles Liver fibrosis PubMed Central articles Liver fibrosis 2023 articles Liver fibrosis 2024 articles Liver fibrosis Scopus articles Liver fibrosis impact factor journals Liver fibrosis Scopus journals Liver fibrosis PubMed journals Liver fibrosis medical journals Liver fibrosis free journals Liver fibrosis best journals Liver fibrosis top journals Liver fibrosis free medical journals Liver fibrosis famous journals Liver fibrosis Google Scholar indexed journals Palmitic acid articles Palmitic acid Research articles Palmitic acid review articles Palmitic acid PubMed articles Palmitic acid PubMed Central articles Palmitic acid 2023 articles Palmitic acid 2024 articles Palmitic acid Scopus articles Palmitic acid impact factor journals Palmitic acid Scopus journals Palmitic acid PubMed journals Palmitic acid medical journals Palmitic acid free journals Palmitic acid best journals Palmitic acid top journals Palmitic acid free medical journals Palmitic acid famous journals Palmitic acid Google Scholar indexed journals n-3 unsaturated fatty acids articles n-3 unsaturated fatty acids Research articles n-3 unsaturated fatty acids review articles n-3 unsaturated fatty acids PubMed articles n-3 unsaturated fatty acids PubMed Central articles n-3 unsaturated fatty acids 2023 articles n-3 unsaturated fatty acids 2024 articles n-3 unsaturated fatty acids Scopus articles n-3 unsaturated fatty acids impact factor journals n-3 unsaturated fatty acids Scopus journals n-3 unsaturated fatty acids PubMed journals n-3 unsaturated fatty acids medical journals n-3 unsaturated fatty acids free journals n-3 unsaturated fatty acids best journals n-3 unsaturated fatty acids top journals n-3 unsaturated fatty acids free medical journals n-3 unsaturated fatty acids famous journals n-3 unsaturated fatty acids Google Scholar indexed journals Collagen articles Collagen Research articles Collagen review articles Collagen PubMed articles Collagen PubMed Central articles Collagen 2023 articles Collagen 2024 articles Collagen Scopus articles Collagen impact factor journals Collagen Scopus journals Collagen PubMed journals Collagen medical journals Collagen free journals Collagen best journals Collagen top journals Collagen free medical journals Collagen famous journals Collagen Google Scholar indexed journals Saturated fatty acids articles Saturated fatty acids Research articles Saturated fatty acids review articles Saturated fatty acids PubMed articles Saturated fatty acids PubMed Central articles Saturated fatty acids 2023 articles Saturated fatty acids 2024 articles Saturated fatty acids Scopus articles Saturated fatty acids impact factor journals Saturated fatty acids Scopus journals Saturated fatty acids PubMed journals Saturated fatty acids medical journals Saturated fatty acids free journals Saturated fatty acids best journals Saturated fatty acids top journals Saturated fatty acids free medical journals Saturated fatty acids famous journals Saturated fatty acids Google Scholar indexed journals monounsaturated fatty acid articles monounsaturated fatty acid Research articles monounsaturated fatty acid review articles monounsaturated fatty acid PubMed articles monounsaturated fatty acid PubMed Central articles monounsaturated fatty acid 2023 articles monounsaturated fatty acid 2024 articles monounsaturated fatty acid Scopus articles monounsaturated fatty acid impact factor journals monounsaturated fatty acid Scopus journals monounsaturated fatty acid PubMed journals monounsaturated fatty acid medical journals monounsaturated fatty acid free journals monounsaturated fatty acid best journals monounsaturated fatty acid top journals monounsaturated fatty acid free medical journals monounsaturated fatty acid famous journals monounsaturated fatty acid Google Scholar indexed journals insulin articles insulin Research articles insulin review articles insulin PubMed articles insulin PubMed Central articles insulin 2023 articles insulin 2024 articles insulin Scopus articles insulin impact factor journals insulin Scopus journals insulin PubMed journals insulin medical journals insulin free journals insulin best journals insulin top journals insulin free medical journals insulin famous journals insulin Google Scholar indexed journals Obesity articles Obesity Research articles Obesity review articles Obesity PubMed articles Obesity PubMed Central articles Obesity 2023 articles Obesity 2024 articles Obesity Scopus articles Obesity impact factor journals Obesity Scopus journals Obesity PubMed journals Obesity medical journals Obesity free journals Obesity best journals Obesity top journals Obesity free medical journals Obesity famous journals Obesity Google Scholar indexed journals
Article Details
1. Introduction
Liver plays many important functions including metabolism, glycogen storage, detoxification, and bile production. Non-alcoholic fatty liver disease (NAFLD), characterized by excess accumulation of triglycerides in the liver, has become a very common chronic liver disease worldwide with a prevalence of 37.1% [1,2]. NAFLD includes a spectrum of chronic liver disorders without significant alcohol consumption, ranging from simple liver steatosis to non-alcoholic steatohepatitis (NASH), which may accumulate fibrosis and can evolve to cirrhosis [3,4].
Fasting serum levels of free fatty acids (FFAs) are reportedly considerably higher in NAFLD patients than controls [4]. Studies have suggested that FFAs, not triglycerides accumulated in lipid droplets, are the main factors contributing to the development of NASH [5-7]. Patients with advanced liver fibrosis also exhibit higher level of palmitic acid (PA), stearic acid (SA), monounsaturated fatty acid (MUFA) and insulin [8].
The number of people with obesity has nearly tripled worldwide since 1975 with 38% of adults aged 18 years or older being overweight and 13% being obese [9]. About two-thirds of obese patients have dyslipidemia with increased circulating levels of triglycerides, very-low-density lipoprotein (VLDL), and free fatty acids (FFAs) including saturated fatty acids (SFAs) [10,11]. Obesity is strongly associated with increased risk of insulin resistance, diabetes, and NAFLD [1,12]. Obesity stimulates de novo lipogenesis in the liver [13]. Insulin resistance may increase lipolysis from the adipose tissue, thereby increasing the amount of FFAs delivered to the liver [14,15]. Hepatic FFAs may also come from the diet [16]. The serum levels of total fatty acids are significantly higher in mice fed a high fat diet [17]. Elevated levels of FFAs may increase the formation of ceramides, which may lead to oxidative stress, inflammation, liver injury, and development of NAFLD [14,18-20]. Excessive SFAs, especially PA, have been shown to cause cellular injury, promote cell death, and contribute to the development of liver fibrosis, NAFLD [21] and Type 2 diabetes [22]. PA-enriched diet reportedly induces hepatic steatosis and injury in adult Zebrafish [23]. In contrast, n-3 UFAs such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) are reportedly inversely associated with NAFLD risk in US adults [24], and attenuate NAFLD by inhibiting oxidative stress and inflammation [5,25].
Previously we conducted Canonical Pathway Analysis of biological molecules associated with PA using Ingenuity Pathway Analysis (IPA), the bioinformatic tool from QIAGEN, and found that PA was strongly associated with the Hepatic Fibrosis Signaling Pathway (p < 2.48E-40), only second to the Neuroinflammation Signaling Pathway (p < 3.59E-46) [26], which suggests that besides neuroinflammation, PA may be strongly associated with liver fibrosis. However, the molecular mechanisms by which different FFAs affect hepatic fibrosis are not clearly defined. Therefore, this study used IPA to examine the molecular mechanisms underlying PA-induced hepatic fibrosis. Molecules affected by PA and liver fibrosis were obtained from QIAGEN Knowledge Base (QKB), a repository of over 13 million manually curated biological findings and more than 7 million curated relationships between different entities (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis), and used for IPA Core Expression Analysis. Differential impacts of PA and n-3 UFAs on the Hepatic Fibrosis Signaling Pathway and collagen accumulation were also examined using the “Path Explorer” tool and the “Molecule Activity Predictor (MAP)” tool in IPA.
2. Materials and Methods
2.1. Ingenuity Pathway Analysis (IPA) Software
The license for Ingenuity Pathway Analysis (IPA), a web-based bioinformatics tool, was purchased from QIAGEN (Germantown, MD, USA). IPA provides insightful data analysis building on comprehensive, manually curated information in the QIAGEN Knowledge Base (QKB). QKB includes more than 13 million biological findings and is constantly updated with newest findings.
2.2. IPA Analysis
IPA Core Expression Analysis was used to identify the most significant biological Canonical Pathways that the molecules in the dataset were involved in. Molecules associated with PA and liver fibrosis were retrieved from QKB with chemicals trimmed from the dataset. The dataset of molecules associated with PA was expanded using the “Grow” tool in IPA with chemicals trimmed to focus on biological molecules [26]. The “Path Explorer” tool was used to examine the relationships between different molecules including PA and n-3 UFAs and liver fibrosis or collagen production. After these terms were added to the new pathway, “Path Explorer” tool was then selected, and chemicals were trimmed to focus on biological molecules, and then the shortest +1 paths were selected to examine the paths from PA or n-3 UFAs to liver fibrosis or collagens production via one intermediate molecule [26]. The “Molecule Activity Predictor (MAP)” tool was used to simulate the increased or decreased levels of PA or n-3 UFAs and predict their effects on the intermediate molecules and, in turn, on liver fibrosis or collagen production [26]. Data retrieval and analysis was conducted between November 15, 2020 and July 31, 2021.
3. Results
3.1 Overlapping of molecules associated with PA and those associated with liver fibrosis
Molecules associated with PA were retrieved from QIAGEN Knowledge Base (QKB) using IPA’s “Grow” tool. After chemicals were trimmed from the dataset, 400 biological molecules and complexes were identified. Molecules associated with liver fibrosis were retrieved from QKB. After chemicals were trimmed from the dataset, 449 biological molecules and complexes were identified. These two datasets were then compared, and 61 molecules were found to be overlapped between PA- and liver fibrosis-associated molecules (Figure 1A). Among these 61 molecules, 11 molecules were involved in liver function; 18 molecules were involved in transcription regulation; 9 molecules were involved in signaling; 6 molecules were involved in stress response; 12 molecules were cytokines/chemokines/growth factors/hormones; and 5 molecules were enzymes or enzyme regulators. The 6 molecules in stress response include DNA damage inducible transcript 3 (DDIT3), glutamate-cysteine ligase catalytic subunit (GCLC), heme oxygenase 1 (HMOX1), neutrophil cytosolic factor 1 (NCF1), nitric oxide synthase 2 (NOS2), and X-box binding protein 1 (XBP1), suggesting that excess PA may increase oxidative stress and endoplasmic reticulum (ER) stress in liver. The overlapped molecules also include 6 cytokines, namely interleukin (IL)10, IL1A, IL6, transforming growth factor beta 1 (TGFβ1), tumor necrosis factor (TNF), and TNF superfamily member 10 (TNFSF10); 3 chemokines, namely C-C motif chemokine ligand 2 (CCL2), CCL4, and C-X-C motif chemokine ligand 8 (CXCL8); 1 growth factor, cellular communication network factor 2 (CCN2)/connective tissue growth factor (CTGF); and 2 hormones, namely insulin (INS) and leptin (LEP), suggesting that PA may affect inflammation and hormonal regulation of metabolism. The 5 enzymes/enzyme regualtors include alanine aminotransferase (ALT), serine protease inhibitor Family E member 1 (SERPINE1), sirtuin 1 (SIRT1), patatin-like phospholipase domain-containing protein 3 (PNPLA3), and stearoyl-CoA desaturase (SCD) (Figure 1B).
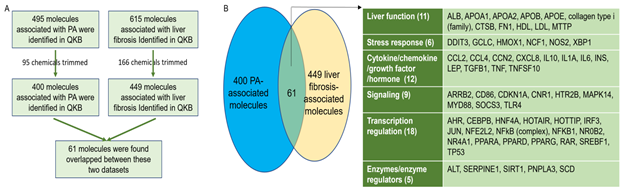
Figure 1: Identification and comparison of PA-associated and liver fibrosis-associated molecules. A. Flowchart for the identification and comparison of PA- and liver fibrosis-associated molecules. B. 61 biological molecules were found to be overlapped between PA- and liver fibrosis-associated molecules, including those involved in liver function, stress response, cytokine/chemokine/growth factor, signaling, transcription regulation, and enzyme/enzyme regulation.
3.2 Shortest paths from PA to hepatic fibrosis signaling pathway
Since the second most significant pathway associated with molecules impacted by PA is the Hepatic Fibrosis Signaling Pathway, we examined the potential paths by which PA may affect the Hepatic Fibrosis Signaling Pathway. Using the “Path Explorer” tool in IPA, 68 shortest paths were identified from PA to Hepatic Fibrosis Signaling Pathway (Figure 2). 19 of the 61 molecules associated with both PA and liver fibrosis were identified on the shortest paths, including CCL2, CCN2, CCAAT/enhancer-binding protein beta (CEBPβ), cannabinoid receptor 1 (CNR1), CXCL8, IL1A, INS, JUN, LEP, mitogen activated protein kinase (MAPK)14, MYD88, NCF1, nuclear factor kappa B (NFκB) complex, NFκB1, peroxisome proliferator-activated receptor gamma (PPARγ), SERPINE1, TGFβ1, toll-like receptor (TLR)4, and tumor necrosis factor (TNF). Then the “Molecule Activity Predictor (MAP)” tool was applied to predict the effects of increased PA level on the Hepatic Fibrosis Signaling Pathway. As shown in Figure 2, simulated increase in the level of PA enhanced the activities of transcription factors, such as activating transcription factor (ATF)4, CEBPβ, FOXO1, JUN/FOS, NFκB1, NFκB complex, RELA, SP1, and peroxisome proliferator activated receptor gamma (PPARγ) while attenuating the activities of cAMP response element-binding protein (CREB), hepatocyte nuclear factor (HNF)1A, mothers against decapentaplegic homolog (SMAD)4, signal transducer and activator of transcription (STAT)3, and yes-associated protein (YAP)1. Increased PA level also augmented the activities of cytokines/chemokines/growth factors, such as CCL2, CCL5, CCN2, CXCL8, IL1B, IL1RN, LEP, TGFβ1, and TNF while attenuating the activities of INS. Simulated increase of PA also increased the activities of oxidative stress mediators, such as NADPH oxidase (NOX) and NCF1 and augmented the activities of SERPINE1 (Figure 2). Moreover, simulated increase of PA level enhanced the activities of caspase 3 (CASP3) while attenuating the activities of anti-apoptotic B-cell lymphoma 2 (BCL2). Therefore, elevated level of PA may promote inflammation, oxidative stress, and apoptosis, leading to the activation of the Hepatic Fibrosis Signaling Pathway.
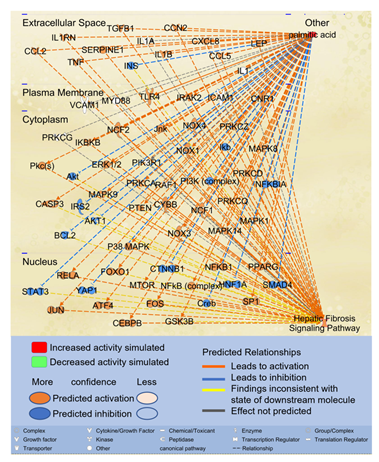
Figure 2: The shortest paths from PA to Hepatic Fibrosis Signaling Pathway were established using the “Path Explorer” tool in IPA, and the effects of increased level of PA was simulated and explored using the “MAP” tool in IPA.
3.3 Effects of n-3 UFAs on the shortest paths from PA to hepatic fibrosis signaling pathway
Considering that n-3 UFAs have been reported to exhibit protective activities against NAFLD, we examined the effects of n-3 UFAs on the Hepatic Fibrosis Signaling Pathway. Using the “Path Explorer” tool in IPA, 12 shortest paths from n-3 UFAs to Hepatic Fibrosis Signaling Pathway were identified. The “MAP” tool was then used to predict the effects of increased levels of n-3 UFAs on the Hepatic Fibrosis Signaling Pathway. As shown in Figure 3A, decreased levels of n-3 UFAs were predicted to increase the activities of IL1B, TNF, CCL2, NFkB, platelet-derived growth factor (PDGF)A, PDGFB, and protein kinase C beta (PRKCB), and decrease the activities of insulin receptor substrate (IRS)2 and phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1). Increased level of PA and decreased levels of n-3 UFAs altogether significantly augmented the activation of the Hepatic Fibrosis Signaling Pathway (Figure 3A). In contrast, increased levels of n-3 UFAs together with decreased level of PA significantly attenuated the activation of the Hepatic Fibrosis Signaling Pathway (Figure 3B).
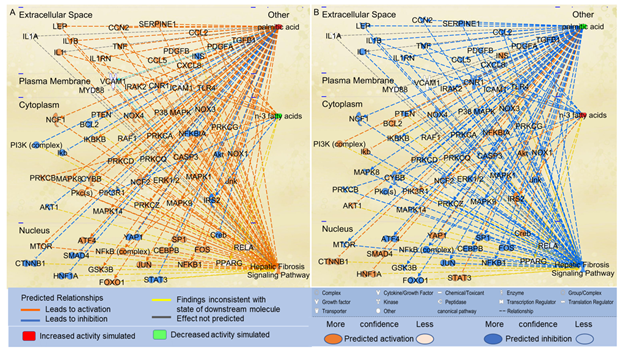
Figure 3: The shortest paths from PA and n-3 FAs to the Hepatic Fibrosis Signaling Pathway were established using the “Path Explorer” tool, and the effects of altered levels of n-3 UFAs and PA on these paths were simulated and explored using the “MAP” tool in IPA. (A) Simulated increase in the level of PA and decrease in the levels of n-3 UFAs were predicted to activate the Hepatic Fibrosis Signaling Pathway using the “MAP” tool in IPA. (B) Simulated decrease in the level of PA and increase in the levels of n-3 FAs were predicted to inhibit the Hepatic Fibrosis Signaling Pathway using the “MAP” tool in IPA.
3.4 PA modulation of collagen accumulation
Considering that liver fibrosis features excessive accumulation of extracellular matrix (ECM) proteins such as collagens [27], we examined the effects of excessive PA on collagen accumulation. Using the “Path Explorer” tool in IPA, 39 shortest paths were identified from PA to collagens (Figure 4). 17 of the 61 molecules associated with both PA and liver fibrosis were identified on the shortest paths, including apolipoprotein E (APOE), arrestin beta 2 (ARRB2), CCL2, CCN2, cathepsin B(CTSB), fibronectin (FN)1, IL10, IL1A, IL6, low density lipoprotein (LDL), LEP, nitric oxide synthase (NOS)2, nuclear receptor subfamily 4 group A member 1(NR4A1), SERPINE1, sirtuin (SIRT)1, TGFβ1, and TNF. Simulated increase in the level of PA was predicted to increase the activities of CCN2, CCL2, extracellular signal?regulated protein kinase (ERK)1/2, F2R Like Trypsin Receptor 1 (F2RL1), IL6, LEP, oxidized low density lipoprotein receptor (OLR)1, p38MAPK, SERPINE1, TGFβ1, and TNF, which in turn increased the accumulation of collagens. Simulated increase in the level of PA was also predicted to inhibit the activities of IL10, which may also, in turn, lead to increased accumulation of collagens (Figure 4).
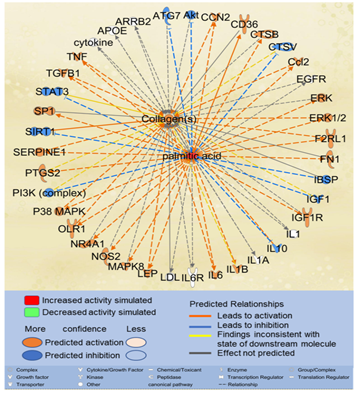
Figure 4: The shortest paths from PA to collagens were established using the “Path Explorer” tool in IPA, and the effects of increased level of PA were simulated and explored using the “MAP” tool in IPA.
3.5 Effects of n-3 UFAs on the shortest paths from PA to collagens
We also examined the effects of n-3 UFAs on the paths from PA to collagens. We used the “Path Explorer” tool in IPA and identified 8 shortest paths from n-3 UFAs to collagens, and then used the “MAP” tool to predict the effects of n-3 UFAs on collagen accumulation. Decreased levels of n-3 UFAs were predicted to increase the activities of CCL2, IL1, PDGFB, prostaglandin-endoperoxide synthase 2 (PTGS2), and TNF. Increased level of PA and decreased levels of n-3 UFAs altogether significantly augmented the accumulation of collagens (Figure 5A). In contrast, increased levels of n-3 UFAs together with decreased level of PA were predicted to significantly attenuate the accumulation of collagens (Figure 5B).
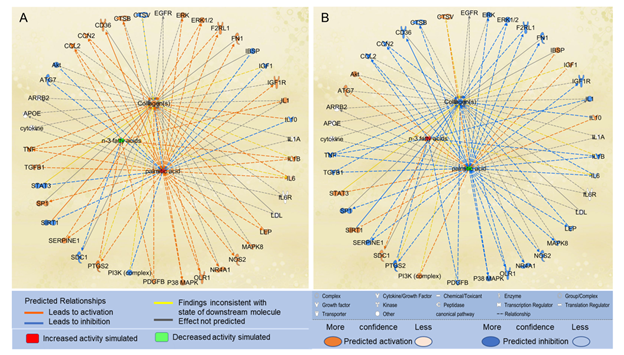
Figure 5: The shortest paths from PA to collagens were established using the “Path Explorer” tool, and the effects of altered levels of n-3 UFAs and PA on these paths were simulated and explored in IPA using the “MAP” tool. (A) Increased level of PA and decreased levels of n-3 UFAs were predicted to enhance collagen accumulation using the “MAP” tool in IPA. (B) Decreased level of PA and increased levels of n-3 UFAs were predicted to attenuate collagen accumulation using the “MAP” tool in IPA.
3.6 Mapping of key molecules onto the hepatic fibrosis signaling pathway
Among the 61 biological molecules that overlapped between PA- and liver fibrosis-associated molecules retrieved from QKB, 19 were mapped to the shortest paths from PA to Hepatic Fibrosis Signaling Pathway, and 17 were mapped to the shortest paths from PA to collagens with 7 molecules mapped to both. These 7 molecules were TNF, IL1A, CCL2, TGFβ1, CCN2, LEP, and SERPINE1. These molecules were then identified on the Hepatic Fibrosis Signaling Pathway, and the “MAP” tool was used to predict the effects of increased activities of these molecules on different processes involved in the Hepatic Fibrosis Signaling Pathway. As shown in Figure 6A, simulated increase in the activities of TNF and IL1 increased the activities of other inflammatory mediators, including ERK1/2, NFκB, and chemokines, and the production of collagen type I, leading to proinflammatory response and accumulation of ECM in the liver. Simulated increase in the activities of TGFβ increased the activities of ERK1/2, NOX4, and transcription factors including NFκB, CEBPβ, and SMADs, leading to enhanced production of collagen type I, collagen type III, chemokines, tissue inhibitor of metalloproteinase (TIMP)1, SERPINE1, CCN2, and proinflammatory response in the liver. Increased production of collagen type I and collagen type III increased the accumulation of ECM while increased activities of TIMP1 and SERPINE1 inhibit the degradation of ECM (Figure 6C). Simulated increase in the activities of LEP increased the activities of such molecules as ERK1/2, STAT3, TIMP1, and SERPINE1, leading to increased production of collagen type I alpha 1 chain (COL1A1), increased deposition of ECM and decreased degradation of ECM (Figure 6C). Simulated increase in the activities of CCN2 was predicted to increase the activities of such molecules as ERK1/2, JNK, and STAT3, leading to proliferation of hepatic stellate cells (HSC), increased production of COL1A1, and enhanced deposition of ECM (Figure 6D).
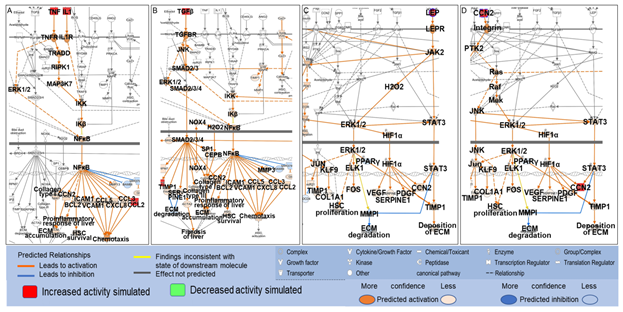
Figure 6: Mapping of TNF, IL1, TGFβ, CCL2, LEP, and CCN2 onto the Hepatic Fibrosis Signaling Pathway and the effects of simulated increase in the activities of TNF (A), IL1 (A), CCL2 (A), TGFβ (B), LEP (C) and CCN2 (D) on different processes in the Hepatic Fibrosis Signaling Pathway using the “MAP” tool.
4. Discussion
Liver plays a key role in the uptake of serum FFAs and the transport, metabolism, and storage of lipids [4]. Many chronic liver diseases including NASH may develop fibrosis characterized by excessive accumulation of collagens and other ECM proteins [28]. This study was undertaken to examine the molecular mechanisms by which PA affected liver fibrosis. We found that increased level of PA may activate the Hepatic Fibrosis Signaling Pathway by promoting inflammation via a number of mediators, including cell surface receptors, such as TLR4, signaling mediators, such as MAPKs, transcription factors, such as NFκB, JUN, CEBPβ, and PPARγ, and cytokines and chemokines, such as TNF and CCL2. Our analysis also showed that PA may increase the accumulation of collagens via mediators including CCN2, SERPINE1, cytokines and chemokines, such as IL1A and CCL2. We also found that n-3 UFAs may attenuate the activation of the Hepatic Fibrosis Signaling Pathway and the accumulation of collagens.
Obesity is well-recognized to be associated with chronic low-grade inflammation [29]. The increased production of chemokines, such as CCL2, and cytokines, such as TNF and IL1, during adipose tissue inflammation contributes to local insulin resistance and increased release of FAs from the adipose tissue, which, in turn, induce the production of inflammatory mediators and insulin resistance in other organs [30-33]. Elevated fasting serum levels of FFAs are strongly associated with advanced fibrosis in NAFLD patients [4]. Treatment with a mixture of PA/oleic acid (OA) at a 1:2 molar ratio increases the mRNA expression of IL6 and IL8 in hepatocytes [34]. PA has been reported to increase the production of IL1B and IL18 in mouse Kupffer cells [35], and stimulate the activation of NLRP3 inflammasome through the TLR4-NFkB signaling pathway in HSCs [36,37]. Activated HSCs represent a major source of collagens in fibrotic liver [27]. PA is also reported to cooperate with TLR2 to contribute to the development of NASH through inflammasome activation in mice [38]. In contrast, dietary supplementation of EPA, a n-3 UFA, has been reported to improve hepatic inflammation and fibrosis [39], which is consistent with our study that increased levels of n-3 UFAs attenuated the activities of IL, TNF, and PDGF, and inhibited the activation of Hepatic Fibrosis Signaling Pathway and collagen accumulation.
LEP is a pleiotropic hormone and an activator of inflammation. As a hormone, LEP plays a key role in the regulation of lipolysis and lipogenesis [40]. In vivo administration of LEP leads to suppression of lipogenesis, enhanced hydrolysis of triglycerides, and increased FFAs [41]. Obese patients have been reported to exhibit higher circulating level of LEP and LEP resistance [42]. PA treatment has been shown to increase LEP expression in 3T3-L1 adipocytes [43]. LEP also acts as a proinflammatory adipokine, stimulating the production of pro-inflammatory cytokines [40,44]. Indeed, intracerebroventricular administration of PA induces central LEP resistance as well as increased expression of TNF, IL1B, and IL-6 in the hypothalamus [45]. Consistently, IPA analysis of PA-associated molecules identified Neuroinflammation Signaling Pathway as the most significant Canonical Pathway associated with PA [26].
CCN2 is an important player in fibrogenic pathways and consistently upregulated in human liver cirrhosis of various etiologies. Serum level of CCN2 has been proposed as an indicator of fibrogenic progression [46]. Expression of CCN2 is directly correlated with fibrotic changes in the liver [47], and knockdown of CCN2 considerably attenuates experimental liver fibrosis [48]. PA treatment increases the mRNA expression of TGFβ1, CCN2, tissue inhibitor of metalloproteinase-1 and procollagen type I and activates HSCs [37]. Our study found that increased activities of TGFβ1 and CCN2 may contribute to PA-induced activation of the Hepatic Fibrosis Signaling Pathway and accumulation of collagens. Consistently, exogenous CCN2 promotes proliferation and collagen production in HSCs [49]. Chronic exposure to TGFβ1 increases the expression of collagen and induces cytoskeletal rearrangement that resembles the epithelial-mesenchymal transition of hepatocytes to collagen-producing mesenchymal cells [50]. Circulating level of TGFβ1 is associated with hepatic fibrosis severity [51]. Moreover, TGFβ has been shown to enhance the expression of CCN2 in HSCs [49,52].
Our analysis found that PA increased the activity of SERPINE1, which in turn contribute to PA-induced activation of Hepatic Fibrosis Signaling Pathway and accumulation of collagens. SERPINE1 codes for Plasminogen activator inhibitor 1 (PAI1), which inhibits the activities of tissue-type plasminogen activator (tPA), a serine protease that initiates fibrinolysis. PAI1 and tPA together help to maintain tissue homeostasis [28]. Obesity is associated with increased plasma levels of tPA and PAI1 with reduced fibrinolysis via inhibition of tPA by PAI1 [53]. Similarly, PA treatment leads to an increase in tPA and a larger increase in PAI1, leading to a net decrease in tPA activity in the culture medium of human primary hepatocytes [54]. Inflammatory cytokines such as TNFA has been shown to induce the expression of PAI in a number of cells including hepatocytes [55-57]. Administration of TGFB in mice causes activation of PAI-1 and progression of steatohepatitis [58]. These results, therefore, further support the significant roles of PAI-1 in PA-induced liver fibrogenesis by inhibiting the degradation of collagens.
Several limitations of this study may potentially affect the findings. First, as more progress is made on the understanding of liver fibrosis and actions of PA, more molecules and relationships will be identified, which will improve the comprehensiveness of liver fibrosis- and PA-associated molecules in QIAGEN’s QKB database for IPA analysis. Secondly, only the shortest paths with one intermediate molecule from FA to the Hepatic Fibrosis Signaling Pathway or collagen accumulation were explored. PA may affect the Hepatic Fibrosis Signaling Pathway or collagen accumulation via more than one intermediate molecules. The paths involving those molecules were beyond the scope of analysis of this study. Thirdly, IPA analysis attributes all the molecules in the pathway an equal weight even though different molecules make vary in their significance in the interactions and relationships [26,59]. Finally, this study focused on the analysis of PA and n-3 UFAs on liver fibrosis. Obesity-associated metabolic disturbances, oxidative stress, endoplasmic reticulum (ER) stress, insulin resistance, dyslipidemia, and pathologies in multiple tissues and organs [60-62] may further complicate liver fibrogenesis, are worthy of further studies.
5. Conclusions
Our study investigated the molecular mechanisms by which PA affects liver fibrosis using IPA. We found that increased level of PA, a SFA, activated the Hepatic Fibrosis Signaling Pathway and accumulation of collagens by multiple mechanisms, including enhancing inflammation, as indicated by increased activities of proinflammatory molecules such as NFkB, FOS, JUN, TNF, and CCL2, and increasing the production of growth factors, such as TGFB and CCN2, LEP, a proinflammatory adipokine, and SERPINE1, suggesting that PA may not only increase the production of collagens but also decrease the degradation of collagens. Furthermore, our study found that increased levels of n-3 UFAs inhibited the activities of PDGFA, PDGFB, TNF, IL1, and CCL2. Decreased level of n-3 UFAs augmented the activation of Hepatic Fibrosis Signaling Pathway and accumulation of collagens induced by increased level of PA. Our results shed light on the molecular mechanisms by which PA may contribute to liver fibrosis and provide key mediating molecules that may be used for further research and therapeutic intervention.
Author Contributions
- Z. conceived the study. H.Z. designed the study. H.Z. and M.G.J. performed the study. H.Z. and M.G.J. analyzed the data and drafted the manuscript. All authors provided input into and approved the final draft of the manuscript.
Acknowledgments
This study was supported by the research fund from Department of Biological Sciences at Seton Hall University. The authors also thank Dr. Eric Seiser for his consultation on IPA software.
Conflicts of interest
The authors declare no conflict of interest.
Data availability statement
The biological molecule datasets used in this study were retrieved from QIAGEN Knowledge Base within Ingenuity Pathway Analysis (IPA) (https://www.qiagen.com/qiagen-ipa (accessed during 15 November 2020 and 31 July 2021)) to allow exploration in the context of other datasets. IPA is commercially available from QIAGEN.
References
- Pais R, Maurel T. Natural History of NAFLD. Journal of clinical medicine 10 (2021).
- Ciardullo S, Perseghin G. Prevalence of NAFLD, MAFLD and associated advanced fibrosis in the contemporary United States population. Liver international: official journal of the International Association for the Study of the Liver 41 (2021): 1290-1293.
- Calzadilla Bertot L, Adams LA. The natural course of non-alcoholic fatty liver disease. International journal of molecular sciences 17 (2016).
- Zhang J, Zhao Y, Xu C. et al. Association between serum free fatty acid levels and nonalcoholic fatty liver disease: a cross-sectional study. Scientific reports 4 (2014): 5832.
- Musso G, Cassader M, Paschetta E. et al. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology 155 (2018): 282-302.
- Alkhouri N, Dixon LJ, Feldstein AE. Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert review of gastroenterology & hepatology 3 (2009): 445-451.
- Liu J, Han L, Zhu L. Free fatty acids, not triglycerides, are associated with non-alcoholic liver injury progression in high fat diet induced obese rats. Lipids in health and disease 15 (2016): 27.
- Cansancao K, Silva Monteiro L, Carvalho Leite N. et al. Advanced liver fibrosis is independently associated with palmitic acid and insulin levels in patients with non-alcoholic fatty liver disease. Nutrients 10 (2018).
- Organization WH. Obesity and overweight fact sheet (2017).
- Feingold KR Obesity and Dyslipidemia; Endotext (2020).
- Rumora AE, LoGrasso G, Hayes JM, et al. The divergent roles of dietary saturated and monounsaturated fatty acids on nerve function in murine models of obesity. The Journal of neuroscience : the official journal of the Society for Neuroscience 39 (2019): 3770-3781.
- Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology 51 (2010): 679-689.
- Donnelly KL, Smith CI, Schwarzenberg SJ, et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. The Journal of clinical investigation 115 (2005): 1343-1351.
- Morigny P, Houssier M, Mouisel E, et al. Adipocyte lipolysis and insulin resistance. Biochimie 125 (2016): 259-266.
- Bessone F, Razori MV, Roma MG. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cellular and molecular life sciences 76 (2019): 99-128.
- Alves-Bezerra M, Cohen DE. Triglyceride Metabolism in the Liver. Comprehensive Physiology 8 (2017): 1-8.
- Wang L, Xu F, Song Z, et al. A high fat diet with a high C18:0/C16:0 ratio induced worse metabolic and transcriptomic profiles in C57BL/6 mice. Lipids in health and disease 19 (2020): 172.
- Mota M, Banini BA, Cazanave SC, et al. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism: clinical and experimental 65 (2016): 1049-1061.
- Kasumov T, Li L, Li M, et al. Ceramide as a mediator of non-alcoholic fatty liver disease and associated atherosclerosis. Plos one 10 (2015): e0126910.
- Boden G. Fatty acid-induced inflammation and insulin resistance in skeletal muscle and liver. Current diabetes reports 6 (2006): 177-181.
- Ogawa Y, Imajo K, Honda Y, et al. Palmitate-induced lipotoxicity is crucial for the pathogenesis of nonalcoholic fatty liver disease in cooperation with gut-derived endotoxin. Scientific reports 8 (2018): 11365.
- Lytrivi M, Castell AL, Poitout V, et al. Recent Insights Into Mechanisms of beta-Cell Lipo- and Glucolipotoxicity in Type 2 Diabetes. Journal of molecular biology 432 (2020): 1514-1534.
- Park KH, Ye ZW, Zhang J, et al. Palmitic acid-enriched diet induces hepatic steatosis and injury in adult Zebrafish. Zebrafish 16 (2019): 497-504.
- Cui J, Li L, Ren L, et al. Dietary n-3 and n-6 fatty acid intakes and NAFLD: A cross-sectional study in the United States. Asia Pacific journal of clinical nutrition 30 (2021): 87-98.
- Geng Y, Faber KN, de Meijer VE, et al. How does hepatic lipid accumulation lead to lipotoxicity in non-alcoholic fatty liver disease? Hepatology international 15 (2021): 21-35.
- Joshi CJ, Zhou H. Molecular Mechanisms of palmitic acid augmentation in COVID-19 Pathologies. Int J Mol Sci 27 (2021).
- Bataller R, Brenner DA. Liver fibrosis. The Journal of clinical investigation 115 (2005): 209-218.
- Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. Journal of cellular physiology 227 (2012): 493-507.
- Zhou H, Urso CJ, Jadeja V. Saturated fatty acids in obesity-associated inflammation. Journal of inflammation research 13 (2020): 1-14.
- Greenberg AS, Obin MS. Obesity and the role of adipose tissue in inflammation and metabolism. The American journal of clinical nutrition 83 (2006): 461S-465S.
- Burhans MS, Hagman DK, Kuzma JN, et al. Contribution of Adipose Tissue Inflammation to the Development of Type 2 Diabetes Mellitus. Comprehensive Physiology 9 (2018): 1-58.
- Kawai T, Autieri MV, Scalia R. Adipose tissue inflammation and metabolic dysfunction in obesity. American journal of physiology. Cell physiology 320 (2021): C375-C391.
- Gutierrez DA, Puglisi MJ, Hasty AH. Impact of increased adipose tissue mass on inflammation, insulin resistance, and dyslipidemia. Current diabetes reports 9 (2009): 26-32.
- Chavez-Tapia NC, Rosso N, Uribe M, et al. Kinetics of the inflammatory response induced by free fatty acid accumulation in hepatocytes. Annals of hepatology 13 (2013): 113-120.
- Cai C, Zhu X, Li P, et al. NLRP3 Deletion inhibits the non-alcoholic steatohepatitis development and inflammation in kupffer cells induced by palmitic acid. Inflammation 40 (2017): 1875-1883.
- Dong Z, Zhuang Q, Ning M, et al. Palmitic acid stimulates NLRP3 inflammasome activation through TLR4-NF-kappaB signal pathway in hepatic stellate cells. Annals of translational medicine 8 (2020): 168.
- Duan NN, Liu XJ, Wu J. Palmitic acid elicits hepatic stellate cell activation through inflammasomes and hedgehog signaling. Life sciences 176 (2017): 42-53.
- Miura K, Yang L, Van Rooijen N, et al. Toll-like receptor 2 and palmitic acid cooperatively contribute to the development of nonalcoholic steatohepatitis through inflammasome activation in mice. Hepatology 57 (2013): 577-589.
- Tanaka N, Sano K, Horiuchi A, et al. Highly purified eicosapentaenoic acid treatment improves nonalcoholic steatohepatitis. Journal of clinical gastroenterology 42 (2008): 413-418.
- Martinez-Sanchez, N. There and back again: Leptin actions in white adipose tissue. International journal of molecular sciences 21 (2020).
- Harris RB. Direct and indirect effects of leptin on adipocyte metabolism. Biochimica et biophysica acta 1842 (2014): 414-423.
- Izquierdo AG, Crujeiras AB, Casanueva FF, et al. Leptin, Obesity, and Leptin Resistance: Where Are We 25 Years Later? Nutrients 11 (2019).
- Korczynska J, Czumaj A, Chmielewski M, et al. Increased Expression of the leptin gene in adipose tissue of patients with chronic kidney disease-the possible role of an abnormal serum fatty acid profile. Metabolites 10 (2020).
- Perez-Perez A, Sanchez-Jimenez F, Vilarino-Garcia T, et al. Role of leptin in inflammation and vice versa. International journal of molecular sciences 21 (2020).
- Cheng L, Yu Y, Szabo A, et al. Palmitic acid induces central leptin resistance and impairs hepatic glucose and lipid metabolism in male mice. The Journal of nutritional biochemistry 26 (2015): 541-548.
- Gressner OA, Fang M, Li H, et al. Connective tissue growth factor (CTGF/CCN2) in serum is an indicator of fibrogenic progression and malignant transformation in patients with chronic hepatitis B infection. Clinica chimica acta; international journal of clinical chemistry 421 (2013): 126-131.
- Hayashi N, Kakimuma T, Soma Y, et al. Connective tissue growth factor is directly related to liver fibrosis. Hepato-gastroenterology 49 (2002): 133-135.
- Gressner OA, Gressner AM. Connective tissue growth factor: a fibrogenic master switch in fibrotic liver diseases. Liver international : official journal of the International Association for the Study of the Liver 28 (2008): 1065-1079.
- Rachfal AW, Brigstock DR. Connective tissue growth factor (CTGF/CCN2) in hepatic fibrosis. Hepatology research : the official journal of the Japan Society of Hepatology 26 (2003): 1-9.
- Bi WR, Yang CQ, Shi Q. Transforming growth factor-beta1 induced epithelial-mesenchymal transition in hepatic fibrosis. Hepato-gastroenterology 59 (2012): 1960-1963.
- Kurabekova R, Tsirulnikova O, Pashkova I, et al. Transforming growth factor beta 1 levels in the blood of pediatric liver recipients: Clinical and biochemical correlations. Pediatric transplantation 24 (2020): e13693.
- Williams EJ, Gaca MD, Brigstock DR, et al. Increased expression of connective tissue growth factor in fibrotic human liver and in activated hepatic stellate cells. Journal of hepatology 32 (2000): 754-761.
- Tjarnlund-Wolf A, Brogren H, Lo E, et al. Plasminogen activator inhibitor-1 and thrombotic cerebrovascular diseases. Stroke 43 (2012): 2833-2839.
- Zheng Z, Nakamura K, Gershbaum S, et al. Interacting hepatic PAI-1/tPA gene regulatory pathways influence impaired fibrinolysis severity in obesity. The Journal of clinical investigation 130 (2020): 4348-4359.
- Swiatkowska M, Szemraj J, Cierniewski CS. Induction of PAI-1 expression by tumor necrosis factor alpha in endothelial cells is mediated by its responsive element located in the 4G/5G site. The FEBS journal 272 (2005): 5821-5831.
- Plomgaard P, Bouzakri K, Krogh-Madsen R, et al. Tumor necrosis factor-alpha induces skeletal muscle insulin resistance in healthy human subjects via inhibition of Akt substrate 160 phosphorylation. Diabetes 54 (2005): 2939-2945.
- Takeshita Y, Takamura T, Hamaguchi E, et al. Tumor necrosis factor-alpha-induced production of plasminogen activator inhibitor 1 and its regulation by pioglitazone and cerivastatin in a nonmalignant human hepatocyte cell line. Metabolism: clinical and experimental 55 (2006): 1464-1472.
- Chanda D, Lee CH, Kim YH, et al. Fenofibrate differentially regulates plasminogen activator inhibitor-1 gene expression via adenosine monophosphate-activated protein kinase-dependent induction of orphan nuclear receptor small heterodimer partner. Hepatology 50 (2009): 880-892.
- Huang W, Zhou H, Hodgkinson C, et al. Network Meta-Analysis on the Mechanisms Underlying Alcohol Augmentation of COVID-19 Pathologies. Alcoholism, clinical and experimental research 45 (2021): 675-688.
- Aburasayn H, Al Batran R, Ussher JR. Targeting ceramide metabolism in obesity. American journal of physiology. Endocrinology and metabolism 311 (2016): 423-435.
- Furukawa S, Fujita T, Shimabukuro M, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. The Journal of clinical investigation 114 (2004): 1752-1761.
- Kawasaki N, Asada R, Saito A, et al. Obesity-induced endoplasmic reticulum stress causes chronic inflammation in adipose tissue. Scientific reports 2 (2012): 799.


 Impact Factor: * 3.8
Impact Factor: * 3.8 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 