Genetic Study in Argentinian Patients with Clinical Long QT Syndrome Diagnosis
Article Information
Leonardo Dionisio1,2*, Sofía Stupniki1,2, Eugenio Aztiria1,2, Ezequiel Rías1,2, Leandro Dye1, Leonardo Onetto3, Franco Gregorietti3, Roberto Keegan3, Guillermo Spitzmaul1,2
1Laboratorio de Canales Iónicos, INIBIBB-CONICET-UNS, Bahía Blanca, Argentina
2Departamento de Biología Bioquímica y Farmacia, Universidad Nacional del Sur, Bahía Blanca, Argentina
3Servicio de Electrofisiología Cardíaca, Hospital Privado del Sur, Bahía Blanca, Argentina
*Corresponding Author: Leonardo Dionisio, Laboratorio de Canales Iónicos, INIBIBB-CONICET-UNS, Bahía Blanca, Argentina.
Received: 11 March 2023; Accepted: 18 March 2024; Published: 25 March 2024
Citation: Leonardo Dionisio, Sofía Stupniki, Eugenio Aztiria, Ezequiel Rías, Leandro Dye, Leonardo Onetto, Franco Gregorietti, Roberto Keegan, Guillermo Spitzmaul. Genetic Study in Argentinian Patients with Clinical Long QT Syndrome Diagnosis. Journal of Pediatrics, Perinatology and Child Health. 8 (2024): 41-52.
View / Download Pdf Share at FacebookAbstract
Long QT Syndrome (LQTS) is a genetic cardiac condition in which varying degrees of severity and treatment response. Three are primarily affected by mutations that cause cardiac ion channel dysfunction. In Argentina, most of the LQTS diagnoses are made by clinical examination and ECG analysis. In this study, we evaluated a group of individuals to correlate their clinical diagnosis of LQTS with genetic variations.
Using gDNA isolation, PCR, and exome sequencing, we screened the coding sequences of the KCNQ1, KCNH2, and SCN5A genes. We identified several changes in these genes, most of them previously described in the literature, but also a novel variation. We found an alteration in the sequence of KCNQ1 exon 16 which did not allow us to amplify it. This is the first analysis of genetic variations in LQTS in Argentina conducted by a national research laboratory. The combination of the detected variations may explain the prolongation of the QT interval observed in the ECG of some of the individuals and may help to improve the treatment making it more rational as well as provide genetic counselling to first-degree relatives.
Keywords
Long QT syndrome; Genetic screening; Argentina; Exon sequencing
Article Details
Abbreviations:
LQTS: long QT syndrome; ECG: Electrocardiogram; QTc: Corrected QT interval; gDNA: Genomic DNA; PVT: Polymorphic ventricular tachycardia
1. Introduction
Long QT syndrome (LQTS) is a genetic disorder that can cause lethal cardiac arrhythmia and sudden cardiac death [1]. Patients with LQTS display a prolonged corrected QT interval (QTc) on resting ECG with a world prevalence of 1:2,000 persons [2]. About 17 genes encoding ion channels or associated proteins have been implicated in this disease [3,4], and a pathogenic or likely pathogenic variant is considered a diagnostic criterion of LQTS regardless of clinical and/or ECG findings. The genes most frequently involved in LQTS are KCNQ1, KCNH2, and SCN5A [5]. The KCNQ1 gene encodes the KCNQ1 potassium channel subunit responsible for slow rectifying K+ current (IKs) [6,7]. Mutations in the KCNQ1 gene reduce IKs by preventing subunit assembly, interruption of membrane trafficking, or by impairment of channel opening. The KCNH2 gene encodes the hERG channel (Kv11.1), responsible for the rapid rectifying K+ current (IKr) [8]. The majority of mutations reported in this gene interfere with proper folding, assembly, or trafficking to the membrane [9]. The SCN5A gene encodes the sodium channel Nav1.5. Gain-of-function mutations in this channel lead to increased sodium entry into the heart cells, prolonging the QT interval and loss-of-function mutations, result in lower expression levels or translation into aberrant proteins [10]. Approximately 80% of genotype-positive patients have mutations in one of these 3 genes: KCNQ1 (40-45%), KCNH2 (30-35%), and SCN5A (10%) [2] allowing the disease to be classified into three major subtypes: LQT1, LQT2, and LQT3, respectively. In addition, between 5-10% of the patients with LQTS carry several alterations in more than one of these genes simultaneously. These patients generally have a more severe phenotype and earlier onset [11].
QT interval prolongation is a pathognomonic sign of the disease, but up to one-third of mutation carriers may have normal QT intervals on resting ECGs [5]. Therefore genetic testing for mutations that lead to cardiac channelopathies is essential for an accurate diagnostic evaluation and familial screening. Molecular identification of the causes of this disease contributes to better diagnosis, risk stratification, genetic counseling, and treatment of affected individuals. We aimed to correlate clinical LQTS diagnosis with genetic variants by analyzing LQTS cases in Argentinian subjects and their relatives. We examined the three most common LQTS genes (KCNQ1, KCNH2, and SCN5A) using exon amplification and direct DNA sequencing.
Methods
2.1 Ethics statement
Written informed consent for genetic explorations and authorization for publication of the results of this study were obtained from patients and family members. This study was conducted under the approval of the ethics committee of the Hospital Municipal de Agudos L. Lucero de Bahía Blanca, (Argentina) (expedient Nº 43-30154-2017) following the Declaration of Helsinki.
2.2 Cases selection
The Cardiac Electrophysiology Service of the Hospital Privado del Sur de Bahía Blanca, (Argentina) selected the cases with a clinical diagnosis of LQTS to perform the molecular analysis, according with the Schwartz criteria [12]. We investigated 6 Argentinian individuals (men and women) with an age of 0-62 years. 5 out of the 6 individuals, showed a prolonged QTc interval (i.e. >460 ms) while one of them (case #1 in Table 1) was a first-degree relative presenting a normal QTc interval (<450 ms). For case #5, we added 2 first-degree relatives to evaluate the same variation. As a control population, we studied 5 healthy unrelated individuals, with same age range.
|
Case |
Sex |
Age |
Presentation |
Family history |
Arrhythmia |
QTc (ms) |
Therapy |
|
1 |
F |
0.3 |
asymptomatic |
None |
None |
<450 |
None |
|
2 |
M |
27 |
asymptomatic |
None |
None |
510 |
BB |
|
3 |
F |
33 |
Presyncope and PVT |
None |
Yes |
460 |
BB + CA |
|
4 |
M |
45 |
palpitations |
None |
Yes |
495 |
BB |
|
5 |
M |
8 |
asymptomatic |
Yes |
None |
500 |
BB |
|
6 |
F |
62 |
bradycardia |
None |
None |
480 |
BB |
|
M: male, F: female, BB: beta-adrenergic receptor blocker, PVT: polymorphic ventricular tachycardia, CA: catheter ablation, ICD: Implantable cardioverter-defibrillator |
|||||||
Table 1: Clinical features of individuals with clinical LQTS diagnosis.
2.3 PCR amplification
Genomic DNA (gDNA) was extracted from whole blood cells using a commercial kit (Inbio Highway, Argentina). gDNA was used for PCR amplification of coding exons of KCNQ1, KCNH2, and SCN5A. The KCNQ1 gene has 16 coding exons, the KCNH2 gene has 15 and the SCN5A gene has 28. Specific primers were designed to amplify each exon (Tables 2, 3, and 4). Each primer pair was intended to hybridize ~100 bp before and after the coding region. For the KCNH2 gene, exon 1 codifies for 25 aminoacids however few polymorphisms were reported in it, so it was not analyzed. For the SCN5A gene, exon 1 is not a codifying region, so it was also not tested either. PCR products were separated by agarose gel electrophoresis and purified using a commercial kit (PB-L, Argentina).
|
Target |
Forward (5`-3`) |
Reverse (5`-3`) |
Product (bp) |
|
Exon 1 |
GGCTAAGCAGGTGGGCTCG |
CAGAGCTCCCCCACACCAG |
853 |
|
Exon 2 |
TCGAAGCACTGTCTGTTCCT |
GTTCCCCTCAGTCCTTGGTC |
433 |
|
Exon 3 |
GCATGGCTGGGTTCAAACAGG |
TGCTGAGGGCTGCCAATGC |
380 |
|
Exon 4 |
AGACGAGAGCAGGGTGTATG |
GCATCTGAGCAAGGTGGATGG |
261 |
|
Exon 5 |
CCTGTCGGGATGGACATATAC |
CTTGGGCTTGCTCTGAGTC |
418 |
|
Exon 6 |
TTAGGCGTCTGCACAGGAG |
CAAGCACAGGTTTGTGGACAG |
361 |
|
Exon 7 |
ATCAGAGTGGTGGGTTTGG |
CTCTGGAGTATAGCACCTTC |
340 |
|
Exon 8 |
TTCCAGCACTGACCATACC |
CAATGATGGTTCTGACAGGT |
304 |
|
Exon 9 |
CATGTCAAGCCTGTGACTCTG |
GGACATTGGGATGGCAGGAAC |
459 |
|
Exon 10 |
TGTGTGAAGACACTGGAGCTGG |
GAAGGCACCTGGAAGGTTTAC |
473 |
|
Exon 11 |
ATTGTCAGGGCTGGAGCTTC |
GCACTAGGCGAGTAGATAGCAC |
479 |
|
Exon 12 |
TGAACACTCTCCTTGTTTCTGG |
CCTTGCAACCCTCCACTATG |
306 |
|
Exon 13 |
AACCAGGCTTATGCCATCAC |
GGTTGAGAGGCAAGAACTCAG |
359 |
|
Exon 14 |
GAGGAAGTCTGAGAGGCAGC |
TTTCCACACCTAGAGCCTAACC |
565 |
|
Exon 15 |
TTTGACTCTCAGCTACCTCC |
CAGGAGCTTCACGTTCACAC |
266 |
|
Exon 16A |
TGCACTTGCAGAGACGGTTG |
GAAGAGGTGGCCTTGCTGAG |
548 |
|
Exon 16B |
TAGTGGTGTCCCCGCTAGG |
CCTGTCCTGTGTAGGAACCG |
1820 |
Table 2: List of primers used to amplify KCNQ1 coding exons.
2.4 DNA sequencing and analysis
PCR products were sequenced in both strands (5`→3`and 3`→5`) using an external service (Macrogen Inc., Korea). DNA alignments and polymorphism detection were carried out online using SnapGene (V5.2.4) and Benchling software. The sequences of the complete human KCNQ1 (NG_008935.1), KCNH2 (NG_008916.1), and SCN5A (NG_008934.1) genes described in GenBank were used as reference.
2.5 Structural analysis of human KCNQ1, hERG, and Nav1.5
Homology modeling was performed using the SWISS-MODEL server (https://swissmodel.expasy.org) [13]. The 3D-protein models were generated employing as target protein the FASTA amino acid sequences for human KCNQ1 (NP_000209.2), hERG (NP_000229.1), and Nav1.5 (NP_932173.1).
3. Results
3.1 Clinical features
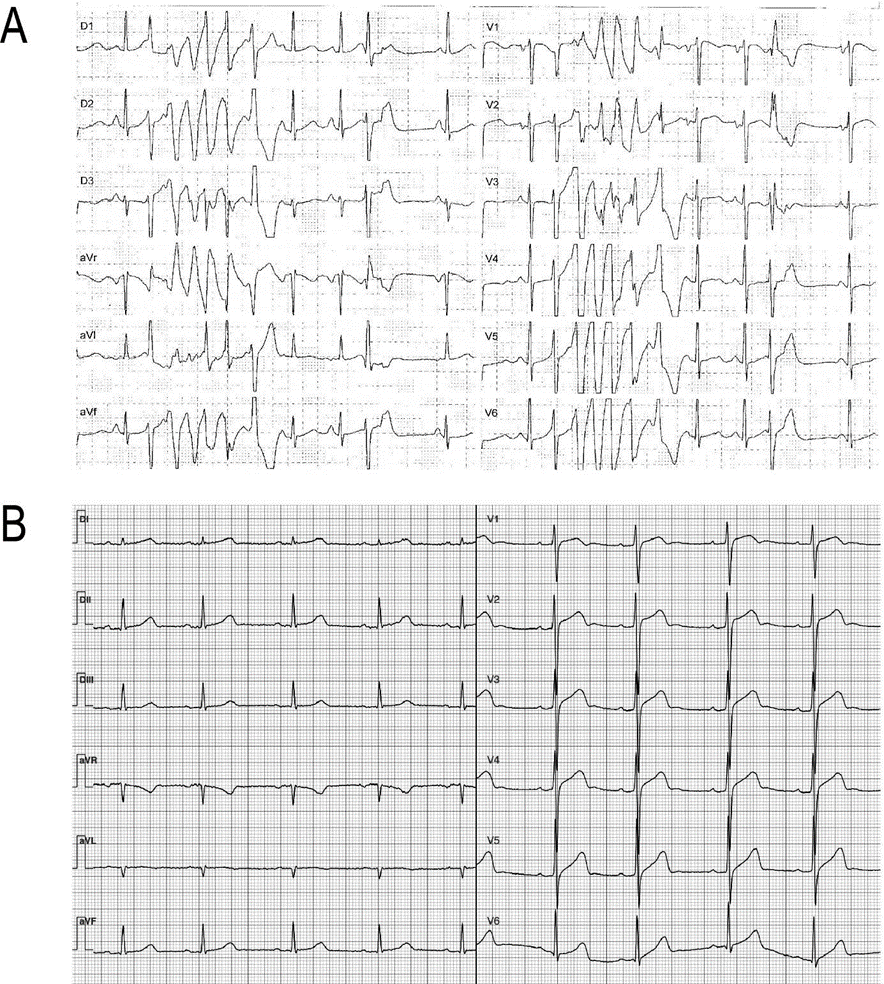
Out of the six individuals, five exhibited a prolonged QTc interval on their ECG while the remaining one was a first-degree relative presenting a normal QTc interval (case #2, Table 1). One case had a history of sudden cardiac death before the age of 40, while another experienced presyncope and documented polymorphic ventricular tachycardia (Table 1 and Figure 1A). Most subjects are currently under pharmacological treatment with β-adrenergic receptor blockers, while one of them has undergone catheter ablation and has an implantable defibrillator (Table 1).
3.2 KCNQ1 sequencing analysis
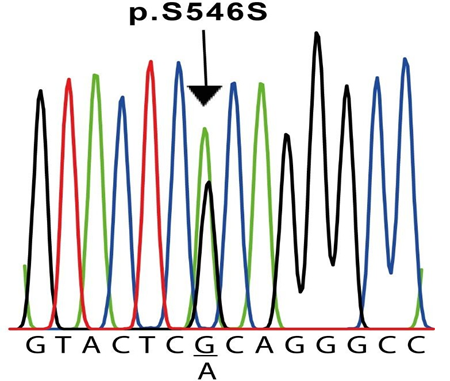
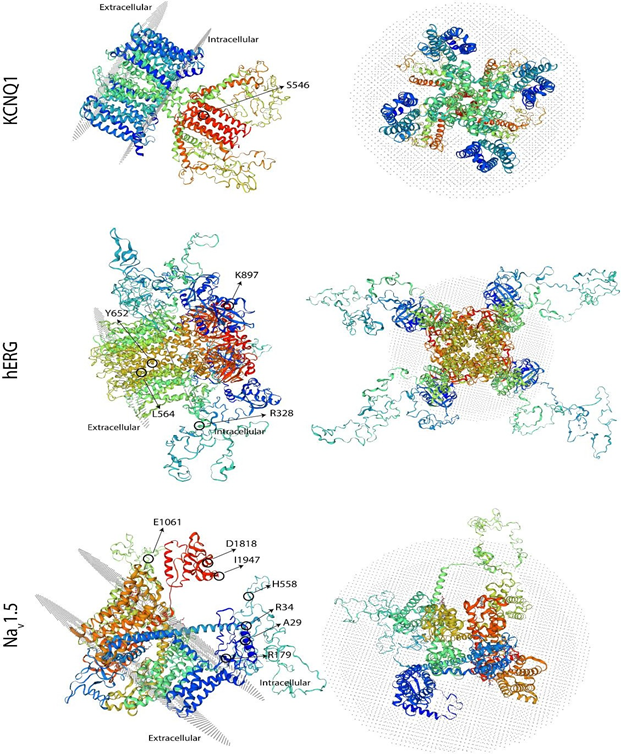
The results of the molecular screening of the KCNQ1 gene are presented in Table 5 and Figure 2. Exons 2 to 16 of this gene were analyzed in all six individuals and the control subjects. Exon 1, on the other hand, was not possible to be amplified in any sample. For this gene, we detected the synonymous NM_000218.3(KCNQ1):c.1638G>A (p.Ser546=) variant in exon 13 of cases #2 and #6 (Table 5 and Figure 2), located at the intracellular domain near the C-terminal (Figure 3). In both cases, this variant was found in heterozygosis. Additionally, in our experimental conditions, we were unable to amplify exon 16 of case #5 but on the contrary, we were able to do it in all other five cases and the five control samples. To verify this result, two different samples from the patient were used. To exclude the possibility that the primers would not anneal correctly on the sequence, we designed a second pair located far before and after in the sequence (Table 1). Also, in this case, we were unable to amplify this exon in this individual. To verify the same variant in 2 first-degree relatives (father and sister), we performed the same PCR reaction for exon 16 in both, resulting positive for them. The modeling shown in Figure 3 does not include this area due to resolution limitations.
Case #3: resting 12-lead ECG. Polymorphic ventricular tachycardia. B. Case #5: resting 12-lead ECG showing bradycardia (54 bpm) and a mild QTc prolongation (ECG after BB treatment).
Both patients underwent a c.1638G>A exchange in exon 13 in a heterozygous configuration, resulting in a silent mutation (p.S564S). The sequences shown correspond to the sense strand of patient #2 gDNA.
3.3 KCNH2 sequencing analysis
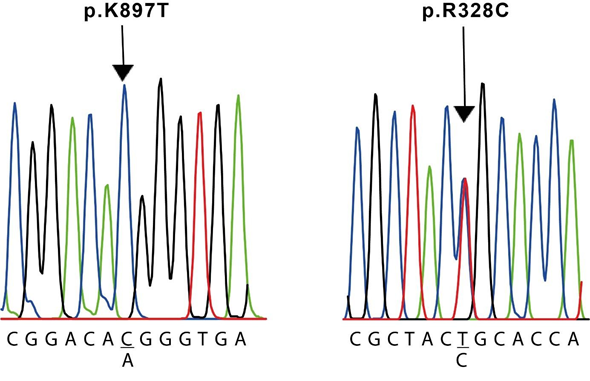
The results of the molecular screening of the KCNH2 gene are presented in Table 5. We identified the missense NM_000238.4(KCNH2):c.982C>T (p.Arg328Cys) variant in exon 5 of case #3, which is located at the proximal domain (Figure 3 and 4); the synonymous NM_000238.4(KCNH2):c.1692A>G (p.Leu564=) variant in exon 7 of case #1, located at the transmembrane domain (TM) 5 (Figure 3); the missense NM_000238.4(KCNH2):c.2690A>C (p.Lys897Thr) variant in exon 11 of case #2 and #6, located at the intracellular region near the C-terminal (Figure 3 and 4); and the synonymous NM_000238.4(KCNH2):c.1956T>C (p.Tyr6520) variant, located at the TM 6 of the channel, in all cases except case #3 (Table 5 and Figure 3).
3.4 SCN5A sequencing analysis
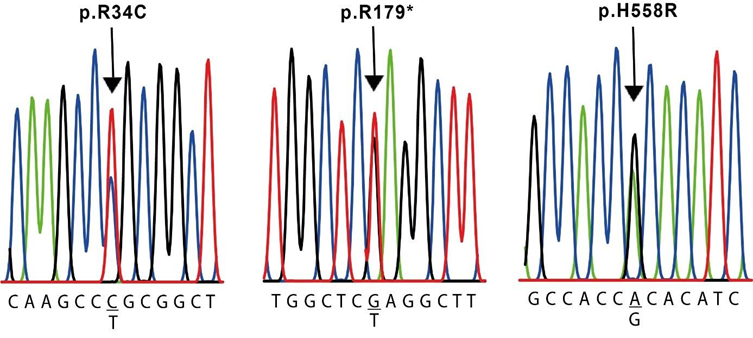
The results of the molecular evaluation of the SCN5A gene are presented in Table 5. We were able to identify 7 polymorphisms of the SCN5A gene in our population (Table 2): the synonymous NM_000335.5(SCN5A):c.87A>G (p.Ala29=) in exon 2 of cases #2, #4, #5 and #6, and the missense NM_000335.5(SCN5A):c.100C>T (p.Arg34Cys) in exon 2 of case #4, both located in the N-terminal region (Figure 3 and 5). The nonsense NM_000335.5(SCN5A):c.535C>T (p.Arg179Ter) in exon 5 of case #6, located in the TM2 of the domain I ((Figure 3 and 5), the missense NM_000335.5(SCN5A):c.1673A>G (p.His558Arg) in exon 12 of cases #3 and #4, located in the interdomain I-II (Figure 3 and 5); the synonymous NM_000335.5(SCN5A):c.5454T>C (p.Asp1818=) in exon 28 of case #2 and #4 and the synonymous NM_000335.5(SCN5A):c.5841C>T (p.Ile1947=) in exon 28 of case #4, both located after domain IV (Figure 3). The synonymous NM_000335.5 (SCN5A):c.3183A>G (p.Glu1061=) in exon 17 was found in all cases except case #4, in the interdomain III-IV (Figure 3). All of these changes are located in the intracellular side of the channel (Figure 3).
Left panel: side view, and right panel: extracellular view. The structures are oriented in space in such a way that the location of all the changes found can be observed.
R238C was founded in case #3 (left) and K897T was found in cases #2 and #6 (right). The sequences shown correspond to the sense strand of gDNA.
The sequences shown correspond to the sense strands of gDNA.
4. Discussion
Cases #1 and #2
Case #1 is a first-degree relative of case #2 (proband). We only found silent variants in it. We detected the L564L variant in exon 7, and the Y652Y variant located in exon 8, both of the KCNH2 gene classified as benign (Table 5) [14-16]. For the SCN5A gene we detected the benign silent variant E1061E, in exon 17. Case #1 shows a normal ECG, so it seems that those detected changes do not affect the QT interval.
Case #2
Case #2 is a young male with a considerably prolonged QT interval but asymptomatic and under treatment with BB. We found the infrequent benign S546S variant in exon 13 of the KCNQ1 gene (Table 5). It has been associated with LQT syndrome in European individuals [16,17] and with alterations in plasmatic lipid levels [18] in the Chinese population. The other silent variants were Y652Y, for KCNH2 gene, and A29A, E1061E, and D1818D, for SCN5A, all classified as benign [14,19-21]. Despite all of them are benign, some authors reported that synonymous mutations can affect the thermodynamic stability of mRNA secondary structures, having consequences in protein functional expression [22].
We also detected the K897T variant in exon 11 of KCNH2 gene (Figure 3), classified as benign but also associated with channel-kinetic changes and reduced hERG expression [23-25]. This variant acquires a pathogenic profile when it is associated with KCNQ1 polymorphisms [3,23]. hERG channels are regulated by thyroid hormone through PI3K signaling. This change, creates a phosphorylation site for Akt which reverses hormonal regulation through the PI3K signaling and inhibits channel activity, prolonging the QT Interval [26].
Considering that some of the silent variants are shared with Case 1 (normal QT interval), possibly, the combination of the silent variants with the K897T variant might be responsible for the prolongation of the QT interval observed in this case. In particular, silent variant in KCNQ1 gene that is not shared with Case #1.
Case #3
This is a young adult female with a medical history of presyncope and PVT, exhibiting a mild prolongation of QT interval (Table 1 and Figure 1). We identified two changes in the protein sequence: the R328C in KCNH2 gene and the H558R in the SCN5A gene, along with a silent variant E1031E. Despite they are not associated with a pathogenic status, these variants are reported in individuals with cardiac alterations [23,27]. Although the R328C variant does not reduce the IKr or alters channel function [28], it was reported in a case of Torsades de Pointes with respiratory distress, left ventricular dysfunction, and a prolonged QT interval [29]. Additionally, it has been associated with changes in conductance when is co-expressed with the KCNQ1 mutation R591H [30].On the other hand, it has been demonstrated that residues 326–345 located in the proximal domain (Figure 3) might be required for hormonal modulation of hERG [31,32]. This case also presents the H558R variant in exon 12 of SCN5A gene. It has been associated with alterations in intracellular trafficking [23] and reported as a pathogenic variant [33,34] (Table 3). R558-containing sodium channels are wild-type–like except when co- expressed with alternatively spliced transcripts [35].This change is located at the linker between DI and DII of the NaV1.5 channel (Figure 3), which was reported to be a hot-spot for Arginine methylation, affecting channel properties [36]. The combination of all these changes may act synergistically contributing to the severe clinical presentation observed in this individual.
|
Target |
Forward (5`-3`) |
Reverse (5`-3`) |
Product (bp) |
|
Exon 2 |
CACGCACAGCTGCGTTCG |
GCCCTCCTGAAAACCATCTC |
626 |
|
Exon 3 |
ATTGAGGGGAGCCATAAGGG |
CAAGCCACATCCTCAGGGTA |
524 |
|
Exon 4 |
TGGCGGTTTATGATGACTGGA |
GCACCCAGGACGTAGTGAAAA |
746 |
|
Exon 5 |
GTTTCTTGTGACTCCCCTGG |
AGCCCTTTACCAGACCTCTC |
584 |
|
Exon 6 |
GGGTGGGCATTCTGATGGAA |
GAGCATAGGTTTGCTGGGGT |
756 |
|
Exon 7 |
CAGTGTGGGCTTCACCTCTT |
CCCCCAAACCATGTCACGAT |
678 |
|
Exon 8 |
TGGAGCGCAGATGTACAAGG |
CATGGGCAAAAAGGGGCAAC |
488 |
|
Exon 9 |
CAGGCCTGGAGGTTGAGATTT |
AAGGAGAATGTGGGAACCCC |
586 |
|
Exon 10 |
ATTGCTTCCCCGGTTGTGTG |
TCCCTGCCCCCAATGTGATT |
538 |
|
Exon 11 |
TAGAGCAGCCTACAAAAGTCCC |
GGAAGGGATGGGAAGGTCTGA |
402 |
|
Exon 12 |
ATAGAACAAAGGAGGGCCAGG |
AGCTGAAAATGTTGGACACTCC |
682 |
|
Exon 13 |
CGAGAAGAGCAGCGACACTTG |
GAATGGAAGAAGGGGATCCAGC |
437 |
|
Exon 14 |
TCCCCTTCTTCCATTCCTAGCC |
GCAGGAACAAGGTTCAGGGAG |
466 |
|
Exon 15 |
TACTTCCCACCTTGGTGCCT |
GCTGTGCTTTCGAGTTCCTC |
485 |
Table 3: List of primers used to amplify KCNH2 coding exons.
|
Target |
Forward (5`-3`) |
Reverse (5`-3`) |
Product (bp) |
|
Exon 2 |
CTCTCTGCAAATGGTGTCCC |
CACCAGTGACTCATTTCCCC |
579 |
|
Exon 3 |
TGAGTCTACTGACCTGCCAA |
GGAATCAGCGCTACTCTCAC |
408 |
|
Exon 4 |
TCTCCTTGGAGACCCTGTTT |
GGACTGGGAAAGGCAAAAGA |
390 |
|
Exon 5 |
TTGATGGCCTCTGTTGAAGG |
CTCTCCCCACCAGGATGAG |
423 |
|
Exon 6 |
TAAGATGCCCAGGTTTGCCT |
TCTGGTGACAGGCACATTCG |
341 |
|
Exon 7 |
GTCTCAAAGCCCAGGAGAAG |
TCTAGCCTGGGAAGTCACAA |
572 |
|
Exon 8 |
CAGAGGAACAGAAGGAAGGC |
CTCCAGAAGCTGTCTCCTCT |
455 |
|
Exon 9 |
CTGTGGGGCATAAACTGGGT |
TGCTGATCCCTTCTCCCTCA |
407 |
|
Exon 10 |
TACCCTCCTCCCTAGGCTAT |
TCAGCGATACCACATTCACA |
539 |
|
Exon 11 |
TTGGGGTAGGTGTGCAAGTC |
AGGCCATGGGAAACAGGAAG |
435 |
|
Exon 12 |
GCCCTCAATGCTCTGAGAAG |
TCTGTCTGTCCCCATTAGCA |
667 |
|
Exon 13 |
AGGGAGTTGGGAACAGAGAA |
AGGCCAGATGTGGGAGTATT |
547 |
|
Exon 14 |
CACCTAGCAGCCCTGTCATC |
GACCCTGAGATTCCCTCCCT |
525 |
|
Exon 15 |
CCAAGCAAACCCCTACTGG |
GGTGAAGGCATGACAGATCA |
431 |
|
Exon 16 |
TAGTGGGTGCTCTGGGAGAA |
TGGACGGATGGGTAGATGGA |
620 |
|
Exon 17 |
ATAGCCAAACCTTCCACATT |
CATGAGTGGTGGATAGCAG |
883 |
|
Exon 18 |
CCTTGAGGGAGGAGTCTTCA |
AGAATTTCCCATTGGCCCTG |
527 |
|
Exon 19 |
TGACAGGCAAAAGTGGCTCT |
TCTAAGGCAGGGTGTTGGTG |
396 |
|
Exon 20 |
CACCCCCATCATCGTAGCTC |
CCGTGGGGTTGAGAGTTTGT |
447 |
|
Exon 21 |
GCAACAGAGCAAGACTGTCT |
TACGTCCTCCTTCCTCTCTG |
448 |
|
Exon 22 |
GCCAGGATACTCTTGGGCTT |
ACGGCCATAGGACATCAGAA |
455 |
|
Exon 23 |
CTCCCTTGAGTGTGGGATCT |
GGCACTGTGATCCTCCTATG |
557 |
|
Exon 24 |
CTCTGACCACCCAGGCATTT |
ACGAGATCTTGCCCTTGTGG |
335 |
|
Exon 25 |
TGGGCTAGTGACCTTCCTCT |
TACATCCCTGGACACACCCT |
452 |
|
Exon 26 |
GAGAAAGCCAGGAGGTGGTC |
AGGCTGGGACCTCTCTTCAT |
380 |
|
Exon 27 |
GGCTTTGGGCTCACTAGAGG |
GAGAGGTGTGTGTGCGTGTA |
562 |
|
Exon 28A |
TGGCTCCTTGCCATATAGAGA |
ATACGGAGTGGCTCAGACAG |
777 |
|
Exon 28B |
TGAGTGAGGACGACTTCGAT |
GAACTCTGCCTGGTTGATCC |
845 |
Table 4: List of primers used to amplify SCN5A exons.
|
Case |
Gene |
Region |
Nucleotide Change |
Amino acid change |
Zygosity |
ClinVar |
|
#1 |
KCNQ1 |
- |
- |
- |
||
|
KCNH2 |
Exon 7 |
c.1692A>G |
p.L564L |
Htz |
B |
|
|
Exon 8 |
c.1956T>C |
p.Y652Y |
Htz |
B |
||
|
SCN5A |
Exon 17 |
c.3183A>G |
p.E1061E |
Hmz |
B |
|
|
#2 |
KCNQ1 |
Exon 13 |
c.1638G>A |
p.S546S |
Htz |
B |
|
KCNH2 |
Exon 8 |
c.1956T>C |
p.Y652Y |
Htz |
B |
|
|
Exon 11 |
c.2690A>C |
p.K897T |
Htz |
B |
||
|
SCN5A |
Exon 2 |
c.87A>G |
p.A29A |
Htz |
B |
|
|
Exon 17 |
c.3183A>G |
p.E1061E |
Hmz |
B |
||
|
Exon 28 |
c.5454C>T |
p.D1818D |
Htz |
B/LB |
||
|
#3 |
KCNQ1 |
- |
- |
- |
- |
- |
|
KCNH2 |
Exon 5 |
c.982C>T |
p.R328C |
Htz |
B/LB/US |
|
|
SCN5A |
Exon 12 |
c.1673A>G |
p.H558R |
Htz |
B/LB |
|
|
Exon 17 |
c.3183A>G |
p.E1061E |
Hmz |
B |
||
|
#4 |
KCNQ1 |
- |
- |
- |
- |
- |
|
KCNH2 |
Exon 8 |
c.1956T>C |
p.Y652Y |
Hmz |
B |
|
|
SCN5A |
Exon 2 |
c.87A>G |
p.A29A |
Hmz |
B |
|
|
c.100C>T |
p.R34C |
Htz |
B |
|||
|
Exon 12 |
c.1673A>G |
p.H558R |
Htz |
B/LB |
||
|
Exon 28 |
c.5454T>C |
p.D1818D |
Htz |
B |
||
|
c.5841C>T |
p.I1947I |
Htz |
B |
|||
|
#5 |
KCNQ1 |
Exon 16 |
n. a. |
- |
- |
n. r. |
|
KCNH2 |
Exon 8 |
c.1956T>C |
p.Y652Y |
Hmz |
B |
|
|
SCN5A |
Exon 2 |
c.87A>G |
p.A29A |
Htz |
B |
|
|
Exon 17 |
c.3183A>G |
p.E1061E |
Hmz |
B |
||
|
#6 |
KCNQ1 |
Exon 13 |
c.1638G>A |
p.S546S |
Htz |
B |
|
KCNH2 |
Exon 8 |
c.1956T>C |
p.Y652Y |
Hmz |
B |
|
|
Exon 11 |
c.2690A>C |
p.K897T |
Hmz |
B |
||
|
SCN5A |
Exon 2 |
c.87A>G |
p.A29A |
Htz |
B |
|
|
Exon 5 |
c.535C>T |
p.R179* |
Htz |
P/LP |
||
|
Exon 17 |
c.3183A>G |
p.E1061E |
Hmz |
B |
||
|
n. a: not amplified, n. r.: not reported, Hmz: homozygous, Htz: heterozygous, ClinVar: ClinVar variant interpretation category, B: benign, LB: likely benign, P: pathogenic, LP likely pathogenic,US: uncertain significance. |
||||||
Table 5: Detected variants for 6 cases of the studied cohort.
Case #4
This individual is a middle-aged male with arrhythmia, palpitations and a prolonged QT interval (Table 1). He presents two changes in the protein sequence coded by the SCN5A gene: R34C and H558R. R34C variant is a frequent polymorphism in Asian and black population, although it has also been detected among Hispanics [34]. It is located at the N-terminal (Figure 3) and is associated with a loss-of-function change. H558R, as mentioned before, is associated with cardiac electrical alterations. The co-expression of these two changes in the same individual has not been reported before. A synergistic negative effect of both changes combined, could be present in this individual. Moreover, he exhibits several silent mutations in KCNH2 (Y652Y) and SCN5A genes (A29A, D1818D, and I1947I). Altogether these changes may contribute to the clinical manifestations that this individual shows.
Case #5
This is an asymptomatic pediatric individual with a family history of sudden death in a sports setting and exhibits a prolonged QTc interval (Figure 1). Gene analysis revealed several silent mutations in KCNH2 (Y652Y) and SCN5A (A29A and E1061E) genes but also an unidentified change in exon 16 of KCNQ1 gene. This change could result from a deletion or an insertion big enough to not be amplified by our PCR conditions. Either of these possibilities will affect the functional properties of the channel [37]. Young patients with deletions in exon 15 and 16 showed prolonged QTc interval [38,39]. Additionally, a small domain between residues 589 and 620 in the C-terminal region may function as an assembly domain for KCNQ1 subunits [37]. Without it, KCNQ1 would fail to assemble and functional channels will not be produced. Besides, this exon encodes the site of interaction between AKAP9 and KCNQ1, which is required for the functional channel regulation through phosphorylation [40]. Therefore, in this individual, the partial or complete lack of exon 16 may affect channel function through these mechanisms. A similar case was found in an adolescent patient who experienced QT lengthening following physical exercise [38]. In our case, a young individual with a family history of sudden death during sports activities, this exon 16 alteration could have similar implications.
Case #6
This case is a female who presents bradycardia and prolonged QT interval and shows several silent mutations: S546S in KCNQ1, Y652Y in KCNH2 and A29A and E1061E in SCN5A. Moreover, she also shows two variants that change the protein sequence: K897T in KCNH2 and the R179* in SCN5A. In this case, the K897T variant was found in homozygosity, so 100% of the channels will have the mutant subunit. More drastically, the R179* inserts a stop codon in exon 5 of SCN5A gene, generating a truncated protein that lacks all pore domains (Figure 3). Heterologous expression of the SCN5A-R179* alone results in non-functional channels [27]. This variant has been classified as pathogenic/likely pathogenic and also reported in individuals with Brugada Syndrome and LQTS [27,35,41]. As this mutation was found in heterozygosity, not all Nav1.5 channels are affected. Probably, the combination of both mutations leads to the development of an intermediate syndrome with characteristics of both, BS and LQTS.
5. Conclusion
This study conducted by a national research laboratory in Argentina represents the first public report of genetic variations in LQTS within this population. It marks the beginning of a systematic survey of the Argentinian population, which is known for its diverse genetic background.
The analysis of KCNQ1, KCNH2, and SCN5A genes in the studied individuals allowed the detection of both benign and pathological genetic variants. Among the cases, five individuals exhibited variations in these genes that may be associated with prolonged QT intervals and clinical symptoms.
Our study highlights the importance of detecting variants associated with alterations in the physiology and function of cardiac ion channels, since they can create a vulnerable substrate that, in the presence of specific triggers such as IKr blockers, can precipitate life-threatening ventricular arrhythmias.
Furthermore, the results demonstrate that the genetic classification of LQTS is not solely determined by a single pathogenic mutation in a gene. It is more likely that a combination of silent and protein-altering changes in different genes contributes to the phenotype observed in LQTS patients.
Consequently, genetic analysis of suspected LQTS cases, as well as their close relatives, can provide valuable information for accurate diagnosis, genetic counseling, and determining the most appropriate treatment strategies.
6. Acknowledgments
We would like to thank the laboratory of the Hospital Privado del Sur for their support in obtaining DNA samples.
Funding
This work was supported by the VT38-UNS8614 grant from the Ministry of Education to G.S. and a grant from Universidad Nacional del Sur (PGI 24/B269, PGI 24/B296) to E.A & G.S.
Competing interests
The authors declare that they have no competing interests.
Ethics declarations
Ethics approval and consent to participate. This study was conducted under the approval of the ethics committee of the Hospital Municipal de Agudos L. Lucero de Bahía Blanca, (expedient Nº 43-30154-2017) following the Declaration of Helsinki.
Authors' contributions
LD, SS, EA, ER and LD, perform molecular biology experiments, analyzed, and interpreted the patient data. LO, FG and RK selected the patients for the study. LD, EA, GS and RK were a major contributors in writing the manuscript. All authors read and approved the final manuscript.
References
- Bokil NJ, Baisden JM, Radford DJ, et al. Molecular genetics of long QT syndrome. Mol Genet Metab 101 (2010): 1-8.
- Schwartz PJ, Ackerman MJ, George AL, et al. Impact of Genetics on the Clinical Management of Channelopathies. J Am Coll Cardiol 62 (2013): 169-180.
- Bohnen MS, Peng G, Robey SH, et al. Molecular pathophysiology of congenital long QT syndrome. Physiological Reviews 97 (2017): 89-134.
- Adler A, Novelli V, Amin AS, et al. An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation 141 (2020): 418- 428.
- Wallace E, Howard L, Liu M, et al. Long QT Syndrome: Genetics and Future Perspective. Pediatr Cardiol 40 (2019): 1419-1430.
- Loussouarn G, Park K, Bellocq C, et al. KCNQ1 / KCNE1 voltage-gated potassium channels : a functional homology between voltage-gated and inward recti ® er K + channels. EMBO J 22 (2003) 20.
- Li Y, Zaydman MA, Wu D, et al. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc Natl Acad Sci USA 108 (2011): 9095-9100.
- Vandenberg JI, Perry MD, Perrin MJ, et al. hERG K(+) channels: structure, function, and clinical significance. Physiol Rev 92 (2012): 1393-1478.
- Ono M, Burgess DE, Schroder EA, et al. Long QT syndrome type-2: Emerging Strategies for correcting class 2 KCNH2 (hERG) mutations and identifying new patients. Biomolecules 10 (2020): 1144.
- Wilde AAM, Amin AS. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. JACC Clin. Electrophysiol 4 (2018): 569-579.
- Tester DJ, Will ML, Haglund CM, et al. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Hear Rhythm 2 (2005): 507-517.
- Schwartz PJ, Ackerman MJ, Wilde AAM. Channelopathies as Causes of Sudden Cardiac Death. Card Electrophysiol Clin 9 (2017): 537-549.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res 46 (2018): W296–W303.
- Iwasa H, Itoh T, Nagai R, et al. Twenty single nucleotide polymorphims (SNPs) and their allelic frequencies in four genes that are responsible for familial long QT syndrome in the Japanese population. Journal of Human Genetics 45 (2000): 182-183.
- Song MK, Bae EJ, Baek JS, et al. QT prolongation and life threatening ventricular tachycardia in a patient injected with intravenous meperidine (Demerol®). Korean Circ J 41 (2011): 342-345.
- Aydin A, Bähring S, Dahm S, et al. Single nucleotide polymorphism map of five long-QT genes. J Mol Med 83 (2005): 159-165.
- He FZ, McLeod HL, Zhang W, Current pharmacogenomic studies on hERG potassium channels. Trends Mol Med 19 (2013): 227-238.
- Chen X, Yang Y, Li S, et al. Several polymorphisms of KCNQ1 gene are associated with plasma lipid levels in general chinese populations. PLoS ONE 7 (2012): e34229.
- Qureshi SF, Ali A, John P, et al. Mutational analysis of SCN5A gene in long QT syndrome. Meta Gene 6 (2015): 26-35.
- Millat G, Chanavat V, Rodriguez-Lafrasse C, et al. Rapid, sensitive and inexpensive detection of SCN5A genetic variations by high resolution melting analysis. Clin Biochem 42 (2009): 491-499.
- Magnani JW, Brody JA, Prins BP, et al. Sequencing of scn5a identifies rare and common variants associated with cardiac conduction: Cohorts for heart and aging research in genomic epidemiology (charge) consortium. Circ Cardiovasc Genet 7 (2014): 365-373.
- Carlini DB, Chen Y, Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr. Genetics 159 (2001): 623-633.
- Gouas L, Nicaud V, Berthet M, et al. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. Eur J Hum Genet 13 (2005): 1213-1222.
- Paavonen KJ, Chapman H, Laitinen PJ, et al. Functional characterization of the common amino acid 897 polymorphism of the cardiac potassium channel KCNH2 (HERG). Cardiovasc Res 59 (2003): 603-611.
- Sinner MF, Pfeufer A, Akyol M, et al. The non-synonymous coding IKr-channel variant KCNH2- K897T is associated with atrial fibrillation: Results from a systematic candidate gene-based analysis of KCNH2 (HERG). European Heart Journal 29 (2008): 907-914.
- Gentile S, Martin N, Scappini E, et al. The human ERG1 channel polymorphism, K897T, creates a phosphorylation site that inhibits channel activity. Proc Natl Acad Sci USA 105 (2008): 14704-14708.
- Kawamura M, Ozawa T, Yao T, et al. Dynamic change in ST-segment and spontaneous occurrence of ventricular fibrillation in Brugada syndrome with a novel nonsense mutation in the SCN5A gene during long-term follow-up. Circ J 73 (2009): 584-588.
- Anderson CL, Delisle BP, Anson BD, et al. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation 113 (2006): 365-373.
- Hinterseer M, Irlbeck M, Ney L, et al. Acute respiratory distress syndrome with transiently impaired left ventricular function and Torsades de Pointes arrhythmia unmasking congenital long QT syndrome in a 25-yr-old woman. Br J Anaesth 97 (2006): 150-153.
- Grunnet M, et al. Functional assessment of compound mutations in the KCNQ1 and KCNH2 genes associated with long QT syndrome. Hear Rhythm 2 (2005): 1238-1249.
- Alonso-Ron C, Barros F, Manso DG, et al. Participation of HERG channel cytoplasmic structures on regulation by the G protein-coupled TRH receptor. Pflugers Arch Eur J Physiol 457 (2009): 1237-1252.
- Barros F, Domínguez P, de la Peña P. Cytoplasmic domains and voltage- dependent potassium channel gating. Front Pharmacol 3 (2012): 1-15.
- Darbar D, Kannankeril J, Donahue S, et al. Cardiac Sodium Channel (SCN5A) Variants Associated with Atrial Fibrillation. Enhanced Reader, Circulation 117 (2008): 1927-35.
- Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: Implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Hear Rhythm 1 (2004): 600-607.
- Kapplinger JD, Tester DJ, Salisbury BA, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION® long QT syndrome genetic test. Heart Rhythm 6 (2009): 1297-1303.
- Matsumura H, Nakano Y, Ochi H, et al. H558R, a common SCN5A polymorphism, modifies the clinical phenotype of Brugada syndrome by modulating DNA methylation of SCN5A promoters. J Biomed Sci 24 (2017): 1-10.
- Schmitt N, Schwarz M, Peretz A, et al. A recessive C-terminal Jervell and Lange-Nielsen mutation of the KCNQ1 channel impairs subunit assembly. EMBO J 19 (2000): 332-340.
- Ohno MHS, Fukuyama M, Itoh H, et al. Copy number variations in KCNQ1 gene were frequently identified in the pediatric patients of long QT syndrome and caused exercise related QT prolongation. Eur Heat J 34 (2013): P2291.
- Torrado M, Fernández G, Ganoza CA, et al. A cryptic splice-altering KCNQ1 variant in trans with R259L leading to Jervell and Lange-Nielsen syndrome. npj Genomic Med 6 (2021): 1-14.
- Chen L, Marquardt ML, Tester DJ, et al. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci USA 104 (2007): 20990-20995.
- Scouarnec S Le, Karakachoff M, Gourraud J, et al. Testing the burden of rare variation in arrhythmia- susceptibility genes provides new insights into molecular diagnosis for brugada syndrome. Human Molecular Genetics 24 (2015): 2757-2763.


 Impact Factor: * 3.8
Impact Factor: * 3.8 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 