The Immune Microenvironment of the Hydatidiform Mole and Invasive Mole
Article Information
Yiqun Yanga, Jia Weia, Xiaoxue Zhanga,*, Juncheng Weia,*
a Department of Obstetrics & Gynecology, Tongji Hospital, Tongji Medical College, Huazhong University of Science & Technology, Hubei Province, China.
*Corresponding Author’s: Xiaoxue Zhang, Juncheng Wei, Department of Obstetrics & Gynecology, Tongji Hospital, Tongji Medical College, Huazhong University of Science & Technology, Hubei Province, China.
Received: 18 May 2023; Accepted: 22 May 2023; Published: 25 May 2023
Citation:
Yiqun Yang, Jia Wei, Xiaoxue Zhang, Juncheng Wei. The immune microenvironment of the hydatidiform mole and invasive mole. Obstetrics and Gynecology Research. 6 (2023): 171-180.
View / Download Pdf Share at FacebookAbstract
Aim:
To investigate the expression profiles of the cytotoxic T-lymphocyte protein 4 (CTLA-4), programmed cell death protein 1(PD-1), programmed cell death-Ligand 1(PD-L1), Twist1, Ki-67, CD4, CD8 and CD11b in hydatidiform mole (HM) and invasive mole (IM).
Methods:
In this study, 19 cases of complete hydatidiform mole (CHM), 9 cases of partial hydatidiform mole (PHM) and 10 cases of IM were selected based on histopathological criteria. All cases were confirmed by the P57 immunohistochemistry (IHC). The expression of CTLA-4, PD-1, PD-L1, Twist1, Ki-67, CD4, CD8 and CD11b were detected by IHC using paraffin-embedded tissue sections. Expression level of these markers was scored semi-quantitatively according to the staining intensity, percentage of positive cells and immunoreactivity score (IRS).
Results:
As for HM, CD11b staining in decidual cells were predominant, followed by CD4+ cells, PD-L1+ cells, Ki-67+ cells, CTLA-4+ cells and CD8+ cells. PD-L1+ cells were present in 14/15 cases. In the villi, the major immune cells were PD-L1+ cells, followed by Ki-67+ cells. Noteworthy, CTLA-4 and CD8 did not express in the villi. Specifically, in the decidual compartment, Twist1 expression was stronger in PHM compared with CHM (p=0.039). While in the villi, Ki-67 was significantly expressed in CHM compared with PHM (p=0.0175). PD-L1 and CD4 immunostaining were higher in PHM than CHM (p=0.0153 and p=0.0127, respectively). In addition, HM has an increased IRS of PD-1, PD-L1, Ki-67and CD11b compared with IM.
Conclusions:
This study demonstrated that PD-1/PD-L1 and CTLA-4 pathways collectively contribute to the immune tolerance of GTD and evidenced that a more prominent immunosuppressive microenvironment is present in PHM than CHM.
Keywords
CTLA-4; hydatidiform mole; immune Checkpoint Proteins; PD-1; PD-L1
CTLA-4 articles CTLA-4 Research articles CTLA-4 review articles CTLA-4 PubMed articles CTLA-4 PubMed Central articles CTLA-4 2023 articles CTLA-4 2024 articles CTLA-4 Scopus articles CTLA-4 impact factor journals CTLA-4 Scopus journals CTLA-4 PubMed journals CTLA-4 medical journals CTLA-4 free journals CTLA-4 best journals CTLA-4 top journals CTLA-4 free medical journals CTLA-4 famous journals CTLA-4 Google Scholar indexed journals hydatidiform mole articles hydatidiform mole Research articles hydatidiform mole review articles hydatidiform mole PubMed articles hydatidiform mole PubMed Central articles hydatidiform mole 2023 articles hydatidiform mole 2024 articles hydatidiform mole Scopus articles hydatidiform mole impact factor journals hydatidiform mole Scopus journals hydatidiform mole PubMed journals hydatidiform mole medical journals hydatidiform mole free journals hydatidiform mole best journals hydatidiform mole top journals hydatidiform mole free medical journals hydatidiform mole famous journals hydatidiform mole Google Scholar indexed journals immune Checkpoint Proteins articles immune Checkpoint Proteins Research articles immune Checkpoint Proteins review articles immune Checkpoint Proteins PubMed articles immune Checkpoint Proteins PubMed Central articles immune Checkpoint Proteins 2023 articles immune Checkpoint Proteins 2024 articles immune Checkpoint Proteins Scopus articles immune Checkpoint Proteins impact factor journals immune Checkpoint Proteins Scopus journals immune Checkpoint Proteins PubMed journals immune Checkpoint Proteins medical journals immune Checkpoint Proteins free journals immune Checkpoint Proteins best journals immune Checkpoint Proteins top journals immune Checkpoint Proteins free medical journals immune Checkpoint Proteins famous journals immune Checkpoint Proteins Google Scholar indexed journals PD-1 articles PD-1 Research articles PD-1 review articles PD-1 PubMed articles PD-1 PubMed Central articles PD-1 2023 articles PD-1 2024 articles PD-1 Scopus articles PD-1 impact factor journals PD-1 Scopus journals PD-1 PubMed journals PD-1 medical journals PD-1 free journals PD-1 best journals PD-1 top journals PD-1 free medical journals PD-1 famous journals PD-1 Google Scholar indexed journals PD-L1 articles PD-L1 Research articles PD-L1 review articles PD-L1 PubMed articles PD-L1 PubMed Central articles PD-L1 2023 articles PD-L1 2024 articles PD-L1 Scopus articles PD-L1 impact factor journals PD-L1 Scopus journals PD-L1 PubMed journals PD-L1 medical journals PD-L1 free journals PD-L1 best journals PD-L1 top journals PD-L1 free medical journals PD-L1 famous journals PD-L1 Google Scholar indexed journals Gestational trophoblastic disease articles Gestational trophoblastic disease Research articles Gestational trophoblastic disease review articles Gestational trophoblastic disease PubMed articles Gestational trophoblastic disease PubMed Central articles Gestational trophoblastic disease 2023 articles Gestational trophoblastic disease 2024 articles Gestational trophoblastic disease Scopus articles Gestational trophoblastic disease impact factor journals Gestational trophoblastic disease Scopus journals Gestational trophoblastic disease PubMed journals Gestational trophoblastic disease medical journals Gestational trophoblastic disease free journals Gestational trophoblastic disease best journals Gestational trophoblastic disease top journals Gestational trophoblastic disease free medical journals Gestational trophoblastic disease famous journals Gestational trophoblastic disease Google Scholar indexed journals pregnancy-related disorders articles pregnancy-related disorders Research articles pregnancy-related disorders review articles pregnancy-related disorders PubMed articles pregnancy-related disorders PubMed Central articles pregnancy-related disorders 2023 articles pregnancy-related disorders 2024 articles pregnancy-related disorders Scopus articles pregnancy-related disorders impact factor journals pregnancy-related disorders Scopus journals pregnancy-related disorders PubMed journals pregnancy-related disorders medical journals pregnancy-related disorders free journals pregnancy-related disorders best journals pregnancy-related disorders top journals pregnancy-related disorders free medical journals pregnancy-related disorders famous journals pregnancy-related disorders Google Scholar indexed journals trophoblastic disease articles trophoblastic disease Research articles trophoblastic disease review articles trophoblastic disease PubMed articles trophoblastic disease PubMed Central articles trophoblastic disease 2023 articles trophoblastic disease 2024 articles trophoblastic disease Scopus articles trophoblastic disease impact factor journals trophoblastic disease Scopus journals trophoblastic disease PubMed journals trophoblastic disease medical journals trophoblastic disease free journals trophoblastic disease best journals trophoblastic disease top journals trophoblastic disease free medical journals trophoblastic disease famous journals trophoblastic disease Google Scholar indexed journals chemotherapy articles chemotherapy Research articles chemotherapy review articles chemotherapy PubMed articles chemotherapy PubMed Central articles chemotherapy 2023 articles chemotherapy 2024 articles chemotherapy Scopus articles chemotherapy impact factor journals chemotherapy Scopus journals chemotherapy PubMed journals chemotherapy medical journals chemotherapy free journals chemotherapy best journals chemotherapy top journals chemotherapy free medical journals chemotherapy famous journals chemotherapy Google Scholar indexed journals reproductive immunology articles reproductive immunology Research articles reproductive immunology review articles reproductive immunology PubMed articles reproductive immunology PubMed Central articles reproductive immunology 2023 articles reproductive immunology 2024 articles reproductive immunology Scopus articles reproductive immunology impact factor journals reproductive immunology Scopus journals reproductive immunology PubMed journals reproductive immunology medical journals reproductive immunology free journals reproductive immunology best journals reproductive immunology top journals reproductive immunology free medical journals reproductive immunology famous journals reproductive immunology Google Scholar indexed journals
Article Details
INTRODUCTION
Gestational trophoblastic disease (GTD), a set of pregnancy-related disorders, consists a spectrum of premalignant to malignant disorders. Hydatidiform moles(HM), the most prevalent histologic type, are divided into complete hydatidiform mole (CHM) and partial hydatidiform mole (PHM). CHM develops when the monosperm or disperm fertilized with an empty ovum. In contrast, PHM arises from fertilization of a normal ovum with disperm, usually resulting in a triploid karyotype[1]. It is of great importance to differentiate CHM from PHM since the risk of persistent trophoblastic disease is higher in CHM patients (10%–30%) than PHM (0.5%–5%)[2]. Most are sensitive to chemotherapy, but others are ineffective. Although the immunotherapy has been confirmed effective in GTD, the immunology is not fully understood[3,4]. Tumorigenesis is closely related to immune escape and suppression, by leveraging a plethora of pathways to disguise themselves and suppress T cell function, such as the immune checkpoints. The most researched are cytotoxic T-lymphocyte protein 4 (CTLA-4) and programmed cell death protein 1 pathway (PD-1/PD-L1). So far, it demonstrated promising results by inhibiting the CTLA-4 and PD-1/PD-L1 and some have been approved for cancer treatment by Food and Drug Administration[5]. Like malignant cells, human trophoblastic cells are able to invade blood vessels and the host immune system do not exclude, which indicate immunology play an important role in the regulation[6]. There are revelations that many immune cells and immune pathways are changed in GTD[7–9]. It has been reported that PD-L1 expression in premalignant and malignant trophoblasts from GTD is ubiquitous and independent of clinical outcomes[6,10]. Furthermore, the trophoblast cells of gestational trophoblastic neoplasia(GTN) show high expression of PD-L1, B7-H3 and VISTA in a manner that is independent of clinical outcomes[11]. In addition, TIM-3 and PD-1 pathways get involved in regulating decidual CD8+ T-cell function and maintain normal pregnancy[12]. Furthermore, immune checkpoint molecules, such as CTLA-4, TIM-3, PD-1, involve in reproductive immunology[13]. Thus, we inferred that immune checkpoints get involved in the occurrence and development of GTD and a deeper understanding of the exact immune checkpoints engaging in the development of GTD should be further explored. In this work, we examined the expression of CTLA-4, PD-1/PD-L1, Twist1, Ki67, CD4, CD8 and CD11b in the CHM, PHM and invasive mole (IM), and to determine their potential role in the pathogenesis of the GTD[6].
Methods
Case Selection
In total, formalin-fixed paraffin-embedded tissues were retrieved from Tongji Hospital, Tongji Medical College of Huazhong University of Science & Technology between January 2020 and June 2022 after Tongji Hospital Institutional Review Board approval with permit number TJ-IRB20230389. Haematoxylin and eosin-stained sections of the specimens were reviewed independently with no knowledge of the specimens’ clinical information and were classified according to the main morphological findings. There were 19 CHM, 9 PHM, and 10 IM and clinical data of each case were retrieved from medical records.
Immunohistochemistry
Immunohistochemistry was performed in 4-mm-thick full-tumor sections. The sections were deparaffinized in xylene and dehydrated through a series of graded ethanol. Antigen retrieval was done by microwave treatment for 15 minutes using pH 6.0 boiled citrate. The slides were left to cool about room temperature for 60 minutes. Sections were incubated using General SP Kit (Mouse/Rabbit Streptavidin-Biotin Detection Systems; catalog number SP9001; ZSGB-BIO, Beijing, China) following the manufacturer’s protocol. Antibodies used are outlined in supplementary table 1. Pre-titrated dilutions of primary antibodies were incubated overnight at +4°C in 5% skim milk (1:20; catalog number G5002; Servicebio, Wuhan, China). Diaminobenzidine was used as the chromogen for the immunostaining. Finally, sections were counterstained with hematoxylin and mounted. Staining was imaged using an upright microscope (CX23LEDRFS1C; Olympus, Guangzhou, China) and a slide scanner (SQS-40R; Shengqiang, Shenzhen, China) with a software Image Viewer. Five high power fields were selected randomly at the decidual compartment (DC) or the villi for each case. Appropriate positive controls were performed according manufacturer’s instruction. Negative controls were obtained by substitution of the primary antibody by phosphate buffered saline.
Scoring of Immunostaining
We adopted the definition of P57 expression from the previous study, as shown in the supplementary figure 1 [14]. PD-1, PD-L1, CTLA-4, CD4, CD8 and CD11b expressions were assessed as cell membrane staining while nuclear staining were used for Ki-67 and Twist1. The expression patterns of all markers were scored semi-quantitatively using the immunoreactive score formula: immunoreactivity score (IRS) = staining intensity score × positive cell proportion score,as described elsewhere[15]. Visual scoring method was completed blindly with cases scored based on intensity and frequency of expression from 0 to 4 as outlined in supplementary table 2. After all relevant slides had been examined, staining intensity was scored as follows: negative = 0, weak (1+) = 1, moderate (2+) = 2, and strong (3+) = 3, as shown in the supplementary figure 2. Positive cell proportion was scored as follows: <5% = 0, 5-25% = 1, 25-50% =2, 50-75% = 3, >75% = 4.
Statistical Analysis
IBM Statistical Product and Service Solutions 26.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 9.0 software (GraphPad Software Inc., San Diego, CA) were used for statistical analysis. The semi-quantitative data were represented by median (P25, P75) or mean ± SD and p value <0.05 was significant. The t-test or one-way analysis of variance test was used when the normality assumption was met, whereas the Mann-Whitney test or the Kruskal-Wallis test and median was calculated for variables that violated normality. Post hoc comparisons were conducted using the Mann-Whitney U-test with Bonferroni correction.
Results
Demographic Data
According to the histopathologic diagnosis of the total selected cases (n=38), 19 (50%) cases were identified as CHM, 9 cases (23.7%) as PHM, and 10 cases (26.3%) as IM. P57 was positive in all PHM, whereas no P57 immunostaining was found in all CHM (supplementary figure 1). Invasive mole describes the condition where a CHM or PHM invades the myometrium and there were 7/10 (70%) who had surgery of hysterectomy and 3/10(30%) who had an operation of lesion removal by hysteroscope. The age of patients with CHM ranged from 18 to 48 years (30.260 ± 6.556 years). The mean age were 32.0 ± 6.0 years in the PHM group and 42.3 ± 8.354 years in the IM group. There was a significant difference in the mean age of the patients between CHM and IM group (P<0.0001) and PHM and IM group (P=0.008). The median initial β-human chorionic gonadotropin(β-HCG) was 98705.3 U/L (108542) for the CHM group and 51497.0 U/L (85073) for the PHM group. There was no significant difference between the two groups (P = 0.105).
Immunoreactivity in HM
Description of the immune microenvironment of the HM is shown in Table 1. In the DC compartment, PD-L1+ cells were present in 14/15 cases, followed by CD11b (87.5%), CD4 (82.35%), Ki-67 (68.75%), CTLA-4 (57.14%), CD8 (50%), PD-1 (33.33%) and Twist1 (31.25%). CD11b+ cells [median (IQR): 3 (2.4, 4.05)] were the predominant immune cell population, followed by CD4+ cells [median (IQR): 2.2 (1.1, 2.9)], PD-L1+ cells [median (IQR): 1.4 (1.2, 2.8)], Ki-67 + cells [median (IQR): 1 (0, 2.4)], CTLA-4 [median (IQR): 0.7 (0.3, 1.55)] and CD8 [median (IQR): 0.3 (0, 1.2)] (Figure 1).
Other immune cell populations were rarely detected. In the villi compartment, PD-L1 was expressed in all samples, followed by Ki-67 (89.29%), CD11b (55.56%), CD4 (51.85%), Twist1 (34.62%) and PD-1 (26.92%). Noteworthy, CTLA-4 and CD8 did not express in the villi. The predominant immune cells were PD-L1+ cells [median (IQR): 3 (2.8, 4)], followed by Ki-67+ cells [median (IQR): 2.7 (1.5, 5.15)] (Figure 2).
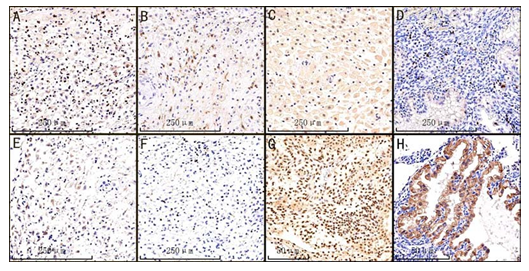
Figure 1: Representative IHC staining of CD11b, CD4, PD-L1, KI-67, CTLA-4, CD8, Twist1 and PD-1 in the ecidual of HM samples, irrespective of CHM or PHM, ×10 magnification.(A)Membrane expression of CD11b.
(B) Membrane expression of CD4. (C) Membrane expression of PD-L1. (D) Nuclear KI-67 expression. (E) Membrane expression of CTLA-4. (F) Membrane expression of CD8. (G) Nuclear Twist1 expression. (H) Membrane expression of PD-1.
|
Parameter |
Total cases |
Positive cases |
positive percentage* |
Median (P25,P75) |
|
PD-1 |
||||
|
-DC |
15 |
5 |
33.33% |
0(0, 1.20) |
|
-Villi |
26 |
7 |
26.92% |
0(0,3.60) |
|
PD-L1 |
||||
|
-DC |
15 |
14 |
93.33% |
1.4(1.2,2.8) |
|
-Villi |
27 |
27 |
100.00% |
3(2.8,4) |
|
CTLA-4 |
||||
|
-DC |
14 |
8 |
57.14% |
0.70(0.30 1.55) |
|
-Villi |
26 |
0 |
0 |
NA* |
|
Twist1 |
||||
|
-DC |
16 |
5 |
31.25% |
0(0,6.55) |
|
-Villi |
26 |
9 |
34.62% |
0(0,9.25) |
|
Ki-67 |
||||
|
-DC |
16 |
11 |
68.75% |
1(0,2.4) |
|
-Villi |
28 |
25 |
89.29% |
2.70(1.50 5.15) |
|
CD4 |
||||
|
-DC |
17 |
14 |
82.35% |
2.2(1.1,2.9) |
|
-Villi |
27 |
14 |
51.85% |
0.6(0,1.2) |
|
CD8 |
||||
|
-DC |
14 |
7 |
50.00% |
0.3(0,1.2) |
|
-Villi |
26 |
0 |
0 |
NA* |
|
CD11b |
||||
|
-DC |
16 |
14 |
87.50% |
3(2.40,4.05) |
|
-Villi |
27 |
15 |
55.56% |
0.6(0,1.2) |
positive cases means immunoreactivity score>0; HM, hydatidiform mole ; NA, no
Table 1: Description of the immune microenvironment of the HM, including decidual compartment and villi
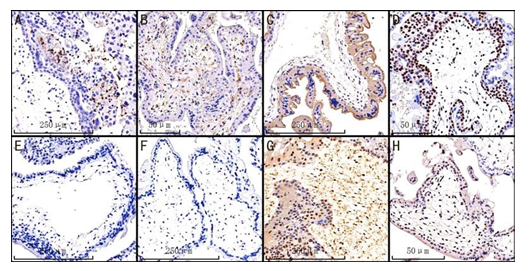
Figure 2: Representative IHC staining of CD11b, CD4, PD-L1, KI-67, CTLA-4, CD8, Twist1 and PD-1 in the villi of HM samples, irrespective of CHM or PHM, ×10 magnification. (A) CD11b; (B) CD4; (C) PD-L1; (D) KI-67; (E) CTLA-4; (F) CD8; (G) Twist1; (H) PD-1
Comparison Between CHM and PHM Groups
Comparison of the immune microenvironment characteristics between CHM and PHM is shown in Table 2. In the decidual compartment, Twist1 infiltrated in 2 CHM case (2/12) and 3 PHM cases (3/4). Twist1 expression was stronger in PHM compared with CHM (median = 6.6 versus 0, p=0.039). While in the villi, Ki-67 was significantly expressed in CHM compared with PHM (mean = 3.78 versus 2.02, p=0.0175). Furthermore, PD-L1 (median = 4.6 versus 2.8, p=0.0153) and CD4 (median = 1.2 versus 0, p=0.0127) were higher in PHM compared with CHM. Furthermore, the differences of others among CHM and PHM groups were not significant.
|
Protein |
CHM (n=19) |
PHM(n=9) |
P |
|
PD-1 |
|||
|
-Decidua |
0(0,0.4) |
2(0.3,4.45) |
0.0908 |
|
-Villi |
0(0,0) |
3.4(0,4.3) |
0.0578 |
|
PD-L1 |
|||
|
-Decidua |
1.75±0.89 |
2.6±1.44 |
0.2075 |
|
-Villi |
2.8(2.4,3.6) |
4.6(2.9,5.6) |
0.0153* |
|
CTLA-4 |
|||
|
-Decidua |
0.4(0,1.4) |
1(0.8,-) |
0.2225 |
|
-Villi |
|||
|
Twist1 |
|||
|
- Decidua |
0(0,0) |
6.6(1.3,9.65) |
0.0390* |
|
-Villi |
0(0,7.8) |
0(0,10.7) |
0.3268 |
|
Ki-67 |
|||
|
- Decidua |
1(0,2.7) |
1.5(0.15,2.4) |
0.9863 |
|
- Villi |
3.78±2.48 |
2.02±1.19 |
0.0175* |
|
CD4 |
|||
|
-Decidua |
2.05±0.92 |
1.72±1.75 |
0.6121 |
|
-Villi |
0(0,1) |
1.2(0.5,2.1) |
0.0127* |
|
CD8 |
|||
|
-Decidua |
0(0,1.2) |
0.6(0.6,-) |
0.1648 |
|
-Villi |
|||
|
CD11 |
|||
|
-Decidua |
3.1±1.73 |
2.55±1.79 |
0.5937 |
|
- Villi |
0.3(0,0.75) |
0.6(0,1.2) |
0.4548 |
expression; Values were given as median(P25,P75) or mean ± SD ( standard deviation ); CHM, complete hydatidiform mole; PHM, partial hydatidiform mole;
*: difference statistically significant (p < 0.05).
Table 2: Comparison of the characteristics of the microenvironment between CHM and PHM
Comparison Between HM and IM Groups
We compared the immune microenvironment characteristics between HM and IM groups (Table 3) and concluded that HM had an increased score of PD-1 ([median(IQR): 0 (0, 2.825)] VS [median(IQR): 0 (0, 0)], p=0.034), PD-L1 ([median (IQR): 2.8 (2.3, 3.6)] VS [median(IQR): 0 (0, 0.4)], p=0.0001), Ki-67 ([median(IQR): 2.6 (1.2, 3.525)] VS [median(IQR): 0 (0, 0.375)], p=0.0001) and CD11b ([median(IQR): 1.2 (0.6, 1.8)] VS [median(IQR): 0 (0, 0.5)], p=0.002). For details, the expression levels between CHM, PHM and IM are shown in Figure 3. PHM had higher IRS of PD-1 ([median(IQR): 2.7 (0, 4.45)] VS 156 [median(IQR): 0 (0, 0)],p=0.0037) and PD-L1([median(IQR): 3.6 (2.8, 5.25)] VS [median(IQR): 0 (0, 0.4)],p<0.0001) than IM, and CHM had a higher IRS of PD-L1 ([median(IQR): 2.45 (2, 3.25)] VS [median(IQR): 0 (0, 0.4)],p=0.0008), Ki-67 ([median(IQR): 2.7 (1.2, 4.2)] VS [median(IQR): 0 (0, 0.375)],p=0.0013) and CD11b ([median(IQR): 1.5 (0.6, 2.18)] VS [median(IQR): 0 (0, 0.5)],p=0.0056) than IM.
|
Protein |
HM(n=28) |
IM(n=10) |
P |
|
PD-1 |
0(0,2.825) |
0(0,0) |
0.034* |
|
PD-L1 |
2.8(2.3,3.) |
0(0,0.4) |
0.0001* |
|
CTLA-4 |
0(0,0.4) |
0(0,0) |
0.062 |
|
Twist1 |
0(0,9.05) |
0(0,5.1) |
0.827 |
|
Ki-67 |
2.6(1.2,3.525) |
0(0,0.375) |
0.0001* |
|
CD4 |
1.096±0.7778 |
1.18±0.8917 |
0.781 |
|
CD8 |
0(0,0.3) |
0.2(0,0.75) |
0.121 |
|
CD11b |
1.2(0.6,1.8) |
0(0,0.5) |
0.002* |
HM, hydatidiform mole; IM, invasive mole; *: difference statistically significant (p < 0.05).
Table 3: Comparison of the characteristics of the microenvironment between HM and IM, irrespective of DC or villi
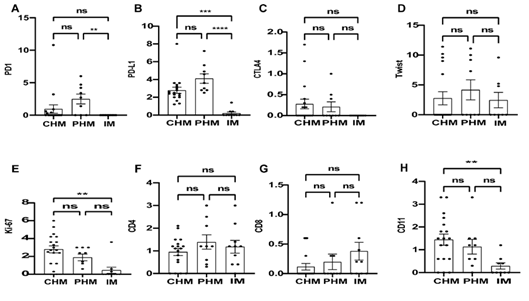
Figure 3. The expression levels of PD-1, PD-L1, CTLA-4, Twist1,Ki-67, CD4,CD8 and CD11b in the whole section ( DC and villi) of CHM, PHM and IM. ns: difference not statistically significant (p > 0.05); * : p < 0.05; ** : p < 0.01; *** :p < 0.001;****: p < 0.0001.
Discussion
Compelling evidence indicates that CTLA-4 and PD-1/PD-L1 immune check-points play an essential role in immune regulation, leading to immune evasion and tumor growth. This makes immune checkpoints as an attractive target for cancer immunotherapy. In the current study, we determined the expression profiles of CTLA-4, PD-1, PD-L1, Twist1, Ki67, CD4, CD8 and CD11b in HM and IM. This is the first evaluation of CD11b proteins in GTD. We detected that CD11b+ cells are predominant in the DC, instead of NK cells in the normal pregnancy. At the same time, previous studies were mainly focused on immune cells, but in this study, we investigated immune checkpoints in the GTD. To the best of our knowledge the first evaluation of CTLA-4 protein expression of GTD, our results demonstrate that, similar to PD-1/PD-L1, CTLA-4 pathway takes part in the immune tolerance to the development of GTD. In this study, the expression level of markers in the DC as well as villi was evaluated quantitatively (percentage of positive cells) and qualitatively (staining intensity) separately and as IRS. In our study, we showed that CD11b+ cells were verified as the predominant cell population in the DC compartment, followed by CD4+ cells, PD-L1+ cells,Ki-67+ cells,CTLA-4+ cells and CD8+ cells. Other immune biomarkers were rarely detected. In a first trimester pregnancy, the decidua displays an inflammatory infiltrate composed of 70% of NK cells, 20% of macrophages, the remaining immune cells being T lymphocytes. Among the T lymphocyte population, 30-45% are CD4+ lymphocytes and 45-75% are CD8+ lymphocytes[16]. Compared with literature data onto normal pregnancy, we observed that CD11b+ cells are abundant, which was in line with the results of recent studies that Maroa et al. reported that CD11c+ cells are dominant[8]. A previous report showed that CD11b+ DCs secreted high levels of IL-10 and had the specific function to induce type 1 regulatory T cells that induced antigen-specific tolerance[17]. The dendritic cells compartment is heterogeneous and CD11b+ dendritic cells are more potent at stimulating CD4+ T cells[18,19]. Therefore, the abundance of CD4+ cells, rather CD8+ cells, could largely result from the fact that CD11b+ DC2s are rich in DC. Previous studies have suggested that FoxP3+ regulatory T-cell infiltration was highest in the CHM[7], constitutively expressing immune checkpoint molecule CTLA-4, which play positive roles in the establishment and/or maintenance of maternal–fetal tolerance of normal pregnancy[20]. CTLA-4, competing with the T cell co-stimulatory checkpoint CD28 receptor, interacts with the ligands CD80/86 on antigen presenting cells (APCs) to inhibit T cell activation, but also depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on APCs[21].We found that HM, like normal pregnancy, harness CTLA-4 pathways to establish or maintain immune tolerance, but there is no expression in the villi and without difference between CHM and PHM. The presence of CTLA-4+ cells was not sought in GTD, allowing for a comparison. Furthermore, PD-1+ cells were uncommon, consistent with Emanuela Veras who observed PD-1+ immune cells were present in association with choriocarcinomas but density varied widely from 0 to 5 positive cells per high-power fields[6]. A recent study indicated that Expression of Twist-1 is higher and more intense in villous stromal cells of CHM when compared with PHM[22]. Claudio et al. discovered that Twist1 and Snail2 were highly expressed in stromal villi cells of molar disease. Particularly, Twist1 was highly expressed in CHM compared to PHM[23]. A research demonstrated that all CHM cases were stromal positive about Twist1 expression and CHM was significantly higher than PHM in stromal immunostaining score[14]. However, the study did not independently analyze DC. In this study, Twist1 expression in the DC was stronger in PHM than CHM with no difference in the villi. Twist1 protein induces epithelial-mesenchymal transition (EMT) by the reduction of cadherin,engaging in tumor invasion and metastasis[14]. EMT involves in the pathogenesis of the HM and we highlighted that in molar disease not only the trophoblast and villi, but even the DC, are involved. But, in 2019, Dubruc, E. et al. confirmed that GTN did not display epithelial-to-mesenchymal transition (EMT) features[24]. Besides, we observed no differences between PHM and CHM of CD4+ and CD8+ cells. However, Wongweragiat et al. analyzed 36 cases (10 first trimester normal pregnancy,13 PHM and 13 CHM) and reported that there are significantly increased numbers of CD3 + and CD4+ T cells in CHM compared to PHM[25]. In addition, Maroa et al. examined the composition of the immune cell microenvironment of 19 CHM and 17 PHM and found the CD4+ lymphocytes are more abundant than CD8+ lymphocytes in molar pregnancies and a higher number of CD4+ cells and CD8+ cells in PHM than in CHM[8]. Perhaps, it was caused by different evaluation standard.
In the villi compartment, Ki-67 was stronger in CHM than PHM, which corresponded with that CHM is more invasive. This is consistent with previous reports that Ki-67 is a good marker for differentiating CHM from PHM[14]. PD-L1 was expressed in the villous trophoblasts in all cases, which is in line with previous studies describing the constitutive presence of PD-L1 in the trophoblast in all subtypes of preneoplasia and trophoblastic malignant neoplasia[6,10,11]. PD-L1 was also expressed in the DC in the almost all cases. It can be seen that PD-1/PD-L1 pathways play an essential role in the establishment and/or maintenance of maternal–fetal tolerance like a normal pregnancy[12,26]. Maroa et al. found decidual immune cells in the DC showed no statistically significant difference of PD-L1 in PHM compared to CHM, which is in accordance with our results, although they did not compare the difference between villi[8]. Besides, in keeping with our results, CD8+ cells were not detected in the villi of CHM and PHM[27]. Finally, HM has an increased IRS of PD-1, PD-L1, Ki-67and CD11b compared with IM. Quite to the contrary, PD-L1 expression of premalignant and malignant trophoblasts from GTD is ubiquitous[10]. It could be due to insufficient materials and numbers of IM. Collectively, these findings indicate an immunosuppressive microenvironment in molar pregnancies, which is more prominent in PHM. In general, GTD are chemosensitive disorders with very high cure rates. However, individuals with chemoresistant diseases need to receive innovative medications[28]. Immune checkpoints therapy, targeting at PD-1/PD-L1, is a potential approach to GTD. The TROPHIMMUN trial assessed avelumab in women with chemotherapy-resistant GTN and demonstrated avelumab had a favorable safety profile and cured approximately 50% of patients[29]. Besides, a recent phase 2 trial reported Camrelizumab plus apatinib showed promising antitumor activity and acceptable toxicity and could be a salvage therapy option for the treatment of high-risk chemo-refractory or relapsed GTN[30]. We have reason to believe immunotherapy as a potential therapeutic approach will be an effective method to cure GTD. At the same time, there are limitations on this study. First, our sample size is insufficient due to the rarity the GTD and there were some samples which contained villi but no DC, so more samples are required to further verify the results. Second, our study is limited by its retrospective nature and I wish there will be a prospective study. Third, there is a lack of normal pregnancy control, which is indeed difficult to obtain. Although this study demonstrates the presence of PD-L1/PD-1 and CTLA-4 immune checkpoints in HM and IM, GTD still warrants further investigation. Future research is needed to determine whether other checkpoint pathways operate in human trophoblastic cells, such as negative immune regulation: indolaimine-2, 3-deoxygenase (IDO), T cell immunoglobulin mucin-3 (TIM-3), lymphocyte activation gene-3 (LAG-3) and B and T cell lymphocyte attenuator (BTLA or CD272), in addition, whether have deficiency in positive immune regulation: glucocorticoid-induced TNFR-related gene (GITR) and OX40[31]. In conclusion, this study reveals that PD-1/PD-L1 and CTLA-4 pathways collectively contribute to the immune tolerance of GTD and provides evidence that provides evidence that a more prominent immunosuppressive microenvironment is present in PHM than CHM.
Acknowledgements
The study was funded by the National Natural Science Foundation of China (NSFC:81772775).
Disclosure
All authors have no conflicts of interest to disclose.
REFERENCES
- Lok C, Frijstein M, van Trommel N. Clinical presentation and diagnosis of Gestational Trophoblastic Disease. Best Practice & Research Clinical Obstetrics & Gynaecology 2021; 74: 42-52.
- Crisp H, Burton J L, Stewart R, Wells M. Refining the diagnosis of hydatidiform mole: image ploidy analysis and p57 KIP2 immunohistochemistry: Refining the diagnosis of hydatidiform mole. Histopathology 2003; 43: 363-73.
- Chen S, Li T, Meng L, Liu K. Advances in immunotherapy and molecular targeted therapy of gestational trophoblastic tumor: current practice and future perspectives n.d.:11.
- Hoeijmakers YM, Gorris MAJ, Sweep FCGJ, Bulten J, Eysbouts YK, Massuger LFAG, et al. Immune cell composition in the endometrium of patients with a complete molar pregnancy: Effects on outcome. Gynecologic Oncology 2021; 160: 450-456.
- Darvin P, Toor S M, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018; 50: 1–11.
- Veras E, Kurman RJ, Wang T-L, Shih I-M. PD-L1 Expression in Human Placentas and Gestational Trophoblastic Diseases. Int J Gynecol Pathol 2017; 36: 146–153.
- Sundara Y T, Jordanova E S, Hernowo B S, Gandamihardja S, Fleuren G J. Decidual infiltration of FoxP3+ regulatory T cells, CD3+ T cells, CD56+ decidual natural killer cells and Ki-67 trophoblast cells in hydatidiform mole compared to normal and ectopic pregnancies. Mol Med Rep 2012; 5: 275-281.
- Dridi M, Papoudou-Bai A, Kanavaros P, Perard M, Clemenson A, Chauleur C, et al. The immune microenvironment of the hydatidiform mole. Human Pathology 2022; 120: 35-45.
- Tsonis O, Karpathiou G, Tsonis K, Paschopoulos M, Papoudou-Bai A, Kanavaros P. Immune cells in normal pregnancy and gestational trophoblastic diseases. Placenta 2020; 101: 90-96.
- Bolze P-A, Patrier S, Massardier J, Hajri T, Abbas F, Schott AM, et al. PD-L1 Expression in Premalignant and Malignant Trophoblasts From Gestational Trophoblastic Diseases Is Ubiquitous and Independent of Clinical Outcomes: International Journal of Gynecological Cancer 2017; 27: 554–561.
- Zong L, Zhang M, Wang W, Wan X, Yang J, Xiang Y. -PLD1, B7-H3 and VISTA are highly expressed in gestational trophoblastic neoplasia. Histopathology 2019; 75: 421-430.
- Sc W, Yh L, Hl P, Xw H, D Z, Yy X, et al. PD-1 and Tim-3 pathways are associated with regulatory CD8+ T-cell function in decidua and maintenance of normal pregnancy. Cell Death & Disease 2015;6.
- Miko E, Meggyes M, Doba K, Barakonyi A, Szereday L. Immune Checkpoint Molecules in Reproductive Immunology. Front Immunol 2019; 10: 846.
- Moussa R A, Eesa A N, Abdallah Z F, Abdelmeged A, Mahran A, Bahaa H. Diagnostic Utility of Twist1, Ki-67, and E-Cadherin in Diagnosing Molar Gestations and Hydropic Abortions. American Journal of Clinical Pathology 2018; 149: 442-455.
- Hussein M R. Analysis of p53, BCL-2 and epidermal growth factor receptor protein expression in the partial and complete hydatidiform moles. Experimental and Molecular Pathology 2009; 87: 63-69.
- Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The role of decidual immune cells on human pregnancy. J Reprod Immunol 2017; 124: 44–53..
- Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10–dependent ILT4/HLA-G pathway. Blood 2010; 116: 935-944.
- Duong E, Fessenden TB, Lutz E, Dinter T, Yim L, Blatt S, et al. Type I interferon activates MHC class I-dressed CD11b+ conventional dendritic cells to promote protective anti-tumor CD8+ T cell immunity. Immunity 2022;55:308-323.e9.
- Human dendritic cell subsets: an update - Collin - 2018 - Immunology - Wiley Online Library n.d. (accessed December 10, 2022).
- Wang S, Sun F, Li M, Qian J, Chen C, Wang M, et al. The appropriate frequency and function of decidual Tim-3+CTLA-4+CD8+ T cells are important in maintaining normal pregnancy. Cell Death Dis 2019; 10: 407.
- Tekguc M, Wing J B, Osaki M, Long J, Sakaguchi S. Treg-expressed CTLA-4 depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on antigen-presenting cells. Proc Natl Acad Sci U S A 2021; 118: e2023739118.
- Jahanbin B, Sarmadi S, Ghasemi D, Nili F, Moradi J-A, Ghasemi S. Pathogenic role of Twist-1 protein in hydatidiform molar pregnancies and investigation of its potential diagnostic utility in complete moles. Diagn Pathol 2023; 18: 40.
- Luchini C, Parcesepe P, Mafficini A, Nottegar A, Parolini C, Veronese N, et al. Specific expression patterns of epithelial to mesenchymal transition factors in gestational molar disease. Placenta 2015; 36: 1318-1324.
- Dubruc E, Allias F, Morel AP, Golfier F, Puisieux A, Devouassoux-Shisheboran M. Gestational trophoblastic neoplasms (GTNs) do not display epithelial-to-mesenchymal transition (EMT) features. Virchows Arch 2019; 475: 121-122.
- Wongweragiat S, Searle RF, Bulmer JN. Decidual T lymphocyte activation in hydatidiform mole. J Clin Pathol 1999; 52: 888-894.
- Meggyes M, Miko E, Szigeti B, Farkas N, Szereday L. The importance of the PD-1/PD-L1 pathway at the maternal-fetal interface. BMC Pregnancy Childbirth 2019; 19: 74.
- Nagymanyoki Z, Callahan M, Parast M, Fulop V, Mok S, Berkowitz R. Immune cell profiling in normal pregnancy, partial and complete molar pregnancy. Gynecologic Oncology 2007; 107: 292-297.
- Paydas S. Immune checkpoint inhibitor using in cases with gestational trophoblastic diseases. Med Oncol 2023; 40: 106.
- You B, Bolze P-A, Lotz J-P, Massardier J, Gladieff L, Joly F, et al. Avelumab in Patients With Gestational Trophoblastic Tumors With Resistance to Single-Agent Chemotherapy: Cohort A of the TROPHIMMUN Phase II Trial. JCO 2020; 38: 3129-3137.
- Cheng H, Zong L, Kong Y, Wang X, Gu Y, Cang W, et al. Camrelizumab plus apatinib in patients with high-risk chemorefractory or relapsed gestational trophoblastic neoplasia (CAP 01): a single-arm, open-label, phase 2 trial. Lancet Oncol 2021; 22: 1609-1617.
- G M, Ba H, P S, Ja W. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021; 184.
Supplementary Data
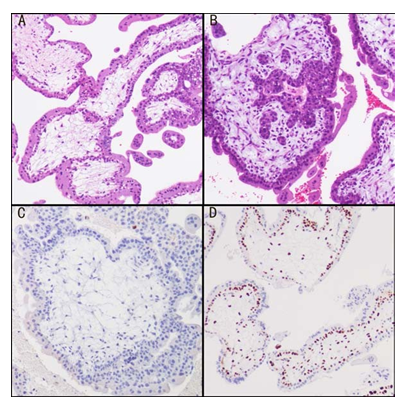
Supplementary Figure 1: (A)H&E staining of CHM;( B)H&E staining of PHM; (C) Complete loss of expression of p57 in villous cytotrophoblasts lining stroma and stromal cells in CHM; (D) Positive p57 expression in PHM
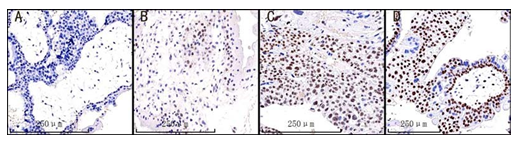
Supplementary Figure 2: Visual scoring system based on intensity and percentage of positive cells (browncolour) on the immunostained slide. (A) non-detectable staining and scored as 0; (B) weak staining and scored as 1; (C) moderate staining and scored as 2; (D) strong staining and scored as 3;
|
Antibody |
Clone, source |
Dilution |
|
PD-1 |
Mouse monoclonal ab52587, Abcam |
1/100 |
|
PD-L1 |
Rabbit monoclonal E1L3N, 13684T, CST |
1/100 |
|
CTLA-4 |
Rabbit monoclonal E2V1Z, 53560T, CST |
1/100 |
|
CD4 |
Rabbit monoclonal ab133616, Abcam |
1/200 |
|
CD8 |
Rabbit monoclonal D8A8Y, 85336S, CST |
1/100 |
|
CD11b |
Rabbit monoclonal ab133357, Abcam |
1/4000 |
|
Ki 67 |
Rabbit monoclonal D2H10, 9027S, CST |
1/200 |
|
Twist |
Mouse monoclonal ab175430, Abcam |
1/200 |
|
P57 |
Mouse monoclonal Kp10, ZSGB-BIO |
Ready-to-use |
Supplementary table 1: Immunohistochemical factors studied and evaluation.
|
score |
intensity |
Quantity* |
|
0 |
Non-detectable |
<5% |
|
1 |
Weak |
5-25% |
|
2 |
Moderate |
25-50% |
|
3 |
Strong |
50-75% |
|
4 |
>75% |
Supplementary table 2: Scoring of immunohistochemistry based on intensity and quantity. All IHC slides were blindly scored based on intensity and abundance. The total score was multiplied by the two individual scores. *: positive cell proportion.


 Impact Factor: * 3.2
Impact Factor: * 3.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 