Abstract
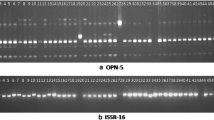
Festuca species have been used for forage, turf, soil conservation and ornamental purpose, which highlighted the importance of these species. This study was conducted to determine genetic variability among the 48 accessions of fine-leaved fescue from different regions of Iran using ISSR markers. To understand genetic relationships among these accessions, unweight pair-group method (UPGMA) were applied to the ISSR marker data set. The ten ISSR primers amplified in total 189 loci for 48 Festuca ovina accessions. All of the amplified loci were polymorphic. A high level of genetic diversity was detected in F. ovina accessions with an average genetic diversity index of 1.522. The expected heterozygosity (He) for the primers varied from 0.153 to 0.347. According to the ISSR data, accessions grouped in two main and seven sub clusters. Results indicated that each of the main clusters include plants that demonstrate Green and Silver phenotypes. Little congruence between the groupings patterns of accessions of F. ovina based on ISSR markers data and categorization based on phenotypes as a Green and Silver, was observed. In conclusion results reveal the high genetic diversity among F. ovina accessions, which can be used for alternative conservation and breeding programs.




Similar content being viewed by others
REFERENCES
Staneik, D., The genus Festuca (Poaceae: Loliinae) in Venezuela, Nord. J. Bot., 2003, vol. 23, pp. 191–205.
Harberd, D.J., Some observations on natural clones in Festuca ovina,New Phytol., 1962, vol. 61, no. 1, pp. 85–100.
Weibull, P., Ghatnekar, L., and Bengtsson, B.O., Genetic variation in commercial varieties and natural populations of sheep’s fescue, Festuca ovina L, Plant Breed., 1991, vol. 107, no. 3, pp. 203–209.
Wilkinson, M.J. Stace, C.A., A new taxonomic treatment of the Festuca ovina L. aggregate (Poaceae) in the British Isles, Bot. J. Linn. Soc., 2010, vol. 106, no. 4, pp. 347–397.
Mirhaji, M., Sanadgol, A., and Jafari, A.A., Evaluation of 16 accessions of Festuca ovina L. In the nursery of homand-abesard rangeland research station, Desalination, 2013, vol. 148, pp. 25–29.
Panahi, B. and Ghorbanzadeh Naghab, M., Genetic characterization of Iranian safflower (Carthamus tinctorius) using inter simple sequence repeats (ISSR) markers, Phys. Mol. Biol. Plant., 2013, vol. 19, no. 2, pp. 239–243.
Pirnajmedin, F., Majidi, M.M., Mirlohi, A., and Noroozi, A., Application of EST-derived microsatellite markers for analysis of genetic variation in tall fescue and its comparison with morphological markers, Biochem. Syst. Ecol., 2016, vol. 65, pp. 225–233.
Ghorbanzadeh Naghab, M. and Panahi, B., Molecular characterization of Iranian black cumin (Nigella sativa L.) accessions using RAPD marker, Biotechnologia, 2017, vol. 98, no. 2, pp. 97–102.
Ivanovych, Y.I., Udovychenko, K.M., Bublyk, M.O., and Volkov, R.A., ISSR-PCR fingerprinting of Ukrainian sweet cherry (Prunus avium L.) cultivars, Cytol. Genet., 2017, vol. 51, no. 1, pp. 40–47.
Campa, A. and Ferreira, J.J., Genetic diversity assessed by genotyping by sequencing (GBS) and for phenological traits in blueberry cultivars, PLoS One, 2018, vol. 13, no. 10, e0206361. https://doi.org/10.1371/journal.pone.0206361
Kölliker, R., Boller, B. and Widmer, F., Marker assisted polycross breeding to increase diversity and yield in perennial ryegrass (Lolium perenne L.), Euphytica, 2005, vol. 146, nos. 1–2, pp. 55–65.
Panahi, B., Afzal, R. Ghorbanzadeh Neghab, M., Mahmoodnia, M., and Paymard, B., Relationship among AFLP, RAPD marker diversity and agromorphological traits in safflower (Carthamus tinctorius L.), Prog. Biol. Sci., 2013, vol. 3, no. 1, pp. 90–99.
Xiang Zhang, Q., Shen, Y.K., Shao, R.X., Fang, J., He, Y.Q., Ren, J.X., Zheng, B.S., and Chen, G.J., Genetic diversity of natural Miscanthus sinensis populations in China revealed by ISSR markers, Biochem. Syst. Ecol., 2013, vol. 48, pp. 248–256.
Chtourou-Ghorbel, N., Mheni, N.B., Elazreg, H., Ghariani, S., Chakroun, M., and Trifi-Farah, N., Genetic diversity in Tunisian perennial forage grasses revealed by inter-simple sequence repeats markers, Biochem. Syst. Ecol., 2016, vol. 66, pp. 154–160.
Tiwari, A., Kumar, B., Tiwari, B., Kadam, G.B, Saha, T.N., Genetic diversity among turf grasses by ISSR markers, Indian J. Agri. Sci., 2017, vol. 87, no. 2, pp. 251–256.
Azizi, H., Sheidai, M., Mozaffarian, V., and Noormohammadi, Z., Genetic and morphological diversity in Tragopogon graminifolius DC (Asteraceae) in Iran., Cytol. Genet., 2018, vol. 52, no. 1, pp. 75–79.
Fjellheim, S. and Rognli, O.A., Genetic diversity within and among Nordic meadow fescue (Festuca pratensis Huds.) cultivars determined on the basis of AFLP markers, Crop Sci., 2005, vol. 45, no. 5, pp. 2081–2086.
Elazreg, H., Ghariani, S., Chtourou-Ghorbel, N., Chakroun, M., and Trifi-Farah, N., SSRs transferability and genetic diversity of Tunisian Festuca arundinacea and Lolium perenne,Biochem. Syst. Ecol., 2011, vol. 39, no. 2, pp. 79–87.
Rohlf, F.J., NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System Version 2.1, New York: Exeter Publishing Setauket, 2000.
Peakall, R. and Smouse, P.E., Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research, Mol. Ecol. Notes, 2006, vol. 6, no. 1, pp. 288–295.
Mahmoudi, B., Panahi, B., Mohammadi, S.A., Daliri, M., Babayev, M.S., Microsatellite based phylogeny and bottleneck studies of Iranian indigenous goat populations, Anim. Biotech., 2014, vol. 25, no. 3, pp. 210–222.
Majidi, M.M., Mirlohi, A.F., and Sayed-Tabatabaei, B.E., AFLP analyses of genetic variation in Iranian fescue accessions, Pak. J. Biol. Sci., 2006, vol. 9, pp. 1869–1876.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
About this article
Cite this article
Reza Mohammadi, Panahi, B. & Amiri, S. ISSR Based Study of Fine Fescue (Festuca ovina L.) Highlighted the Genetic Diversity of Iranian Accessions. Cytol. Genet. 54, 257–263 (2020). https://doi.org/10.3103/S0095452720030123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452720030123