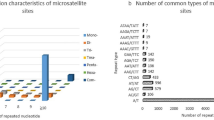
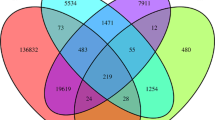
Abstract
We developed a set of SSR markers to investigate the genetic diversity and evolutionary history of Psammochloa villosa (Trin.) Bor, a dominant species of the Inner Mongolian Plateau and adjacent regions. We used sequenced transcriptomes of P. villosa to develop primer pairs for 97 SSR markers. Of these, we successfully amplified 46 (47%) SSRs and found that eleven were polymorphic among individuals. We tested these eleven SSRs in 122 accessions of P. villosa representing ten populations and determined that the number of alleles per locus ranged from one to seven, with an average of 2.727. The expected and observed heterozygosity per locus varied from 0 to 0.840 (mean: 0.471) and from 0 to 1.000 (mean: 0.679), respectively. The new SSR markers described in this study will be valuable for investigating the population genetic structure and history of P. villosa throughout its range and may help to develop hypotheses for understanding its evolutionary divergence from its closest relatives in the genus Stipa L.
Similar content being viewed by others
REFERENCES
Agarwal, M., Shrivastava, N., and Padh, H., Advances in molecular marker techniques and their applications in plant sciences, Plant Cell Rep., 2008, vol. 27, no. 4, pp. 617–631.
Apweiler, R., Bairoch, A., Wu, C.H., et al., The UniProt Consortium. UniProt: the universal protein knowledgebase, Nucleic Acids Res., 2017, vol. 45, no. D1, pp. D158–D169.
Ashburner, M., Ball, C. A., Blake, J. A., et al., Gene Ontology: tool for the unification of biology, The Gene Ontology Consortium, Nat. Genet., 2000, vol. 25, pp. 25–29.
Dong, M. and Alaten, B., Clonal plasticity in response to rhizome severing and heterogeneous resource supply in the rhizomatous grass Psammochloa villosa in an Inner Mongolian dune, China, J. Plant Ecol., 1999, vol. 141, no. 1, pp. 53–58.
Dong, M., Alaten, B., Xing, X.R., et al., Genet features and ramet population features in the rhizomatous grass species Psammochloa villosa, Chin. J. Plan. Ecol., 1999, vol. 23, no. 4, pp. 302–310.
Esselman, E.J., Li, J.Q., Crawford, D.J., et al., Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random applied polymorphic DNA (RAPD) and inter simple sequence repeat (ISSR) markers, Mol. Ecol., 1999, vol. 8, no. 3, pp. 443–451.
Finn, R.D., Bateman, A., Clements, J., et al., Pfam: the protein families database, Nucleic Acids Res., 2014, vol. 42, no. D1, pp. D222–D230.
Ghangal, R., Raghuvanshi, S., and Sharma, P.C., Isolation of good quality RNA from a medicinal plant sea buckthorn, rich in secondary metabolites, Plant Physiol. Biochem., 2009, vol. 47, no. 11–12, pp. 1113–1115.
Gomes, S., Martins-Lopes, P., Lopes, J., et al., Assessing genetic diversity in Olea europaea L. using ISSR and SSR markers, Plant Mol. Biol. Rep., 2009, vol. 27, pp. 365–373.
Haas, B.J., Papanicolaou, A., Yassour, et al., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis, Nat. Protoc. 2013. vol. 8, no. 8, pp. 1494–1512.
Hayden, M.J., Nguyen, T.M., Waterman, A., et al., Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat, Mol. Breeding, 2008, vol. 21, pp. 271–281.
Huang, Z.Y., Dong, M., and Zhang, S.M., Strategies of seed germination on sand dune and seedling desiccation tolerance of Psammochloa villosa (Poaceae), Acta Ecol. Sin., 2005, vol. 25, no. 2, pp. 298–303.
Izzah, N.K., Lee, J., Jayakodi, M., et al., Transcriptome sequencing of two parental lines of cabbage (Brassica oleracea L. var. capitata L.) and construction of an EST-based genetic map, BMC Genomics, 2014, vol. 15, p. 149.
Kanehisa, M. and Goto, S., KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 1999, vol. 27, no. 1, pp. 29–34.
Kim, B.Y., Park, H.S., Kim, S., et al., Development of microsatellite markers for Viscum coloratum (Santalaceae) and their application to wild populations, Appl. Plant Sci., 2017, vol. 5, no. 1, p. 1600102.
Langmead, B. and Salzberg, S., Fast gapped-read alignment with Bowtie 2, Nat. Methods, 2012, vol. 9, pp. 357–359.
Li, A. and Ge, S., Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers, Ann. Bot., 2001, vol. 87, no. 5, pp. 585–590.
Liu, Y. X., Desert Flora of China, Beijing: Science Press, 1985, vol. 1, p. 34.
Liu, B.B., Tian, B., Ma, H., et al., Development and characterization of EST-SSR markers in Ostryopsis (Betulaceae), Appl. Plant Sci., 2014, vol. 2, p. 1300062.
Liu, J.T., Zhou, Y.L., Luo, C.X., et al., De novo transcriptome sequencing of desert herbaceous Achnatherum splendens (Achnatherum) seedlings and identification of salt tolerance genes, Gene, 2016, vol. 7, no. 4, p. 12.
Liu, Y.P., Su, X., Luo, W.C., et al., Development of SSR markers from transcriptomes for Orinus (Poaceae), an endemic of the Qinghai–Tibetan Plateau, Appl. Plant Sci., 2017, vol. 5, no. 7, p. 1700029.
Lu, S.L. and Kuo, P.C., Psammochloa Hitchc. In: Kuo, P.C., ed. Flora Republicae Popularis Sinicae, Beijing: Science Press, 1987, pp. 307–309.
Lv, T., Liu, Y.P., Zhou, Y.H., et al., Germplasm collection and preliminary studies on genealogical differentiation of a desert species—Psammochloa villosa, Acta Agrest. Sin., 2018, vol. 26, no. 3, pp. 733–740.
Ma, Y.Q., Flora of Inner Mongolia, Hohhot: Inner Mongolia People’s Publishing House, 1994, vol. 5, pp. 115–152.
Rossetto, M., Jezierski, G., Hopper, S.D., et al., Conservation genetics and clonality in two critically endangered eucalypts from the highly endemic south-western Australian flora, Biol. Conserv., 1999, vol. 88, pp. 321–331.
Rousset, F., GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux, Mol. Ecol. Resour., 2008, vol. 8, no. 1, pp. 103–106.
Rozen, S. and Skaletsky, H., Primer3 on the WWW for general users and for biologist programmers, in Bioinformatics Methods and Protocols, Misener, S. and Krawetz, S.A., Eds., Methods in Molecular Biology, Totowa, New Jersey, USA: Humana Press, 2000, vol. 132, pp. 365–386.
Serra, I.A., Procaccini, G., Intrieri, M.C., et al., Comparison of ISSR and SSR markers for analysis of genetic diversity in the seagrass Posidonia oceanica, Mar. Ecol. Prog. Ser., 2007, vol. 338, pp. 71–79.
Simon, S.A., Zhai, J., Nandety, R.S., et al., Short-read sequencing technologies for transcriptional analyses, Annu. Rev. Plant Biol., 2009, vol. 60, no. 1, pp. 305–333.
Tani, N., Tomaru, N., Tsumura, Y., et al., Genetic structure within a Japanese stone pine (Pinus pumila Regel) population on Mt. Aino-dake in Central Honshu, Japan, J. Plant Res., 1998, vol. 111, no. 1101, pp. 7–15.
Tatusov, R.L., Fedorova, N.D., Jackson, J.D., et al., The COG database: an updated version includes eukaryotes, BMC Bioinformatics, 2003, vol. 4, p. 41.
Wang, K.Q., Ge, S., and Dong M., Allozyme variance and clonal diversity in the rhizomatous grass Psammochloa villosa (Gramineae), Acta Bot. Sin., 1999, vol. 41, no. 5, pp. 534–540.
Wheeler, D.L., Church, D.M., Federhen, S., et al., NCBI Resource Coordinators, Database resources of the National Center for Biotechnology Information, Nucleic Acids Res., 2014, vol. 42, no. D1, pp. D7–D17.
Yang, J.B., Dong, Y.R., Wong, K.M., et al., Genetic structure and differentiation in Dendrocalamus sinicus (Poaceae: Bambusoideae) populations provide insight into evolutionary history and speciation of woody bamboos, Sci. Rep., 2018, vol. 8, pp. 16933.
Zhang, Z.S., Li, L.L., and Chen, W.L., Ptilagrostis contracta (Stipeae, Poaceae), a new species endemic to Qinghai-Tibet Plateau, PLoS One, 2017, vol. 12, no. 1, e0166603.
ACKNOWLEDGMENTS
The authors thank anonymous reviewers for their comments on the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (41761009, 31960052), the Natural Science Foundation of Qinghai Province (2019-ZJ-7011), “The Dawn of West China” Talent Training Program of the Chinese Academy of Sciences (2019_1_4), the Key Laboratory of Medicinal Animal and Plant Resources of the Qinghai-Tibet Plateau in Qinghai Province (2020-ZJ-Y40), which supported international collaboration with AJ Harris, the “High-end Innovative Talents Thousands of People Plan” in Qinghai Province and the “135 High-level Personnel Training Project” in Qinghai Province. All grants were made to Su.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
DATA ACCESSIBILITY
We deposited raw sequencing reads in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject no. PRJNA730046). We also deposited sequence information for the developed primers in NCBI’s GenBank, and accession numbers are provided in Table 1.
About this article
Cite this article
Liu, YP., Liang, RF., Lv, T. et al. Development of SSR Markers for Psammochloa villosa (Trin.) Bor (Poaceae), a Dominant Species in the Inner Mongolian Plateau. Cytol. Genet. 55, 576–582 (2021). https://doi.org/10.3103/S0095452721060086
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452721060086