Abstract
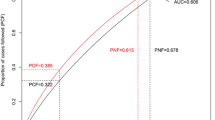
Multiple common variations discovered via genome-wide association studies (GWASs) were shown to have a minimal association with breast cancer (BC) risk in Vietnamese women. This study analyzed the cumulative effect in predicting BC risk of ten single nucleotide polymorphisms (SNPs) identified by previous GWAS and were common in Vietnamese. In this case-control research, 240 BC patients and 271 healthy controls were recruited to assess candidate SNPs’ association with BC risk. A polygenic risk score (PRS) was then created from SNPs strongly related to the risk of BC among the assessed population. The area under the receiver operating characteristic curve (AUC) was used to assess the effectiveness of the PRS model with BC risk. Logistic regression results showed seven individual SNPs (rs2155209, rs4784227, rs2605039, rs3817198, rs2981582, rs11614913, and rs12325489) were significantly associated with BC risk after multiple testing. These SNPs were then used to create the PRS model. Compared with women in the lowest quartile, women in the highest quartile of PRS had a considerably higher risk (odds ratio 2.65; 95% confidence interval (95% CI) 1.61–4.40) with AUC at 71%. These findings suggest that the 7-SNP PRS would effectively distinguish between women with high and low risk of BC, indicating the genetic marker for BC risk prediction in a Vietnamese population.

Similar content being viewed by others
REFERENCES
Allman, R., Dite, G.S., Hopper, J.L., et al., SNPs and breast cancer risk prediction for African American and Hispanic women, Breast Cancer Res. Treat., 2015, vol. 154, no. 3, pp. 583–589. https://doi.org/10.1007/s10549-015-3641-7
Bastami, M., Choupani, J., Saadatian, Z., et al., Evidences from a systematic review and meta-analysis unveil the role of miRNA polymorphisms in the predisposition to female neoplasms, Int. J. Mol. Sci., 2019, vol. 20, no. 20, art. ID 5088. https://doi.org/10.3390/ijms20205088
Black, M.H., Li, S., LaDuca, H., et al., Polygenic risk score for breast cancer in high-risk women, J. Clin. Oncol., 2018, vol. 36, pp. 1508-1508. https://doi.org/10.1200/JCO.2018.36.15_suppl.1508
Bradbury, A.R. and Olopade, O.I., Genetic susceptibility to breast cancer, Rev. Endocr. Metab. Disord., 2007, vol. 8, no. 3, pp. 255–267. https://doi.org/10.1007/s11154-007-9038-0
Cai, Q., Long, J., Lu, W., et al., Genome-wide association study identifies breast cancer risk variant at 10q21. 2: results from the Asia Breast Cancer Consortium, Hum. Mol. Genet., 2011, vol. 20, no. 24, pp. 4991–4999. https://doi.org/10.1093/hmg/ddr405
Cai, Q., Zhang, B., Sung, H., et al., Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1, Nature Genetics, 2014, vol. 46, no. 8, pp. 886–890. https://doi.org/10.1038/ng.3041
Campa, D., Kaaks, R., Le Marchand, L., et al., Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium, J. Natl. Cancer Inst., 2011, vol. 103, no. 16, pp. 1252–1263. https://doi.org/10.1093/jnci/djr265
Chan, C.H.T., Munusamy, P., Loke, S.Y., et al., Evaluation of three polygenic risk score models for the prediction of breast cancer risk in Singapore Chinese, Oncotarget, 2018, vol. 9, no. 16, pp. 12796–12804. https://doi.org/10.18632/oncotarget.24374
Chen, Q.H., Wang, Q.B., and Zhang, B., Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: a HuGE meta-analysis, Tumor Biol., 2014, vol. 35, no. 1, pp. 529–543. https://doi.org/10.1007/s13277-013-1074-7
Chen, Y., Fu, F., Lin, Y., et al., The precision relationships between eight GWAS-identified genetic variants and breast cancer in a Chinese population, Oncotarget, 2016, vol. 7, no. 46, art. ID 75457. https://doi.org/10.18632/oncotarget.12255
Chen, Y., Shi, C., and Guo, Q., TNRC9 rs12443621 and FGFR2 rs2981582 polymorphisms and breast cancer risk, World J. Surg. Oncol., 2016, vol. 14, no. 1, art. ID 50. https://doi.org/10.1186/s12957-016-0795-7
Choupani, J., Nariman-Saleh-Fam, Z., Saadatian, Z., et al., Association of mir-196a-2 rs11614913 and mir-149 rs2292832 polymorphisms with risk of cancer: an updated meta-analysis, Front. Genet., 2019, vol. 10, art. ID 186. https://doi.org/10.3389/fgene.2019.00186
Couch, F.J., Kuchenbaecker, K.B., Michailidou, K., et al., Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer, Nat. Commun., 2016, vol. 7, no. 1, art. ID 11375. https://doi.org/10.1038/ncomms11375
Dai, Z.J., Shao, Y.P., Wang, X.J., et al., Five common functional polymorphisms in microRNAs (rs2910164, rs2292832, rs11614913, rs3746444, rs895819) and the susceptibility to breast cancer: evidence from 8361 cancer cases and 8504 controls, Curr. Pharm. Des., 2015, vol. 21, no. 11, pp. 1455–1463. https://doi.org/10.2174/1381612821666141208143533
Dai, Z.M., Kang, H.F., Zhang, W.G., et al., The Associations of Single Nucleotide Polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-compliant observational study, Medicine (Baltimore), 2016, vol. 95, no. 7, art. ID e2826. https://doi.org/10.1097/MD.0000000000002826
Darabi, H., Czene, K., Zhao, W., et al., Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement, Breast Cancer Res., 2012, vol. 14, no. 1, art. ID R25. https://doi.org/10.1186/bcr3110
Dinger, M.E., Amaral, P.P., Mercer, T.R., et al., Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation, Genome Res., 2008, vol. 18, no. 9, pp. 1433–1445. https://doi.org/10.1101/gr.078378.108
Dite, G.S., MacInnis, R.J., Bickerstaffe, A., et al., Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: australian breast cancer family registry, Cancer Epidemiol. Biomarkers Prev., 2016, vol. 25, no. 2, pp. 359–365. https://doi.org/10.1158/1055-9965.Epi-15-0838
Dite, G.S., Mahmoodi, M., Bickerstaffe, A., et al., Using SNP genotypes to improve the discrimination of a simple breast cancer risk prediction model, Breast Cancer Res. Treat., 2013, vol. 139, no. 3, pp. 887–896. https://doi.org/10.1007/s10549-013-2610-2
Easton, D.F., Pooley, K.A., Dunning, A.M., et al., Genome-wide association study identifies novel breast cancer susceptibility loci, Nature, 2007, vol. 447, no. 7148, pp. 1087–1093. https://doi.org/10.1038/nature05887
Evans, D.G., Brentnall, A., Byers, H., et al., The impact of a panel of 18 SNPs on breast cancer risk in women attending a UK familial screening clinic: a case–control study, J. Med. Genet., 2017, vol. 54, no. 2, art. ID 111113. https://doi.org/10.1136/jmedgenet-2016-104125
Fernandes, G.C., Michelli, R.A., Scapulatempo-Neto, C., et al., Association of polymorphisms with a family history of cancer and the presence of germline mutations in the BRCA1/BRCA2 genes, Hered. Cancer Clin. Pract., 2016, vol. 14, no. 1, art.ID. 2. https://doi.org/10.1186/s13053-015-0042-1
Fernandez-Navarro, P., Pita, G., Santamarina, C., et al., Association analysis between breast cancer genetic variants and mammographic density in a large population-based study (Determinants of Density in Mammographies in Spain) identifies susceptibility loci in TOX3 gene, Eur. J. Cancer, 2013, vol. 49, no. 2, pp. 474–481. https://doi.org/10.1016/j.ejca.2012.08.026
Fletcher, O., Johnson, N., Orr, N., et al., Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study, J. Natl. Cancer Inst., 2011, vol. 103, no. 5, pp. 425–435. https://doi.org/10.1093/jnci/djq563
Fogarty, M.P., Emmenegger, B.A., Grasfeder, L.L., et al., Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells, Proc. Natl. Acad. Sci., 2007, vol. 104, no. 8, pp. 2973–2978. https://doi.org/10.1073/pnas.0605770104
Gapska, P., Scott, R.J., Serrano-Fernandez, P., et al., Vitamin D receptor variants and breast cancer risk in the Polish population, Breast Cancer Res. Treat., 2009, vol. 115, no. 3, pp. 629–633. https://doi.org/10.1007/s10549-008-0107-1
Ghosh, J.C., Dohi, T., Kang, B.H., et al., Hsp60 regulation of tumor cell apoptosis, J. Biol. Chem., 2008, vol. 283, no. 8, pp. 5188–5194. https://doi.org/10.1074/jbc.M705904200
Gibb, E.A., Brown, C.J., and Lam, W.L., The functional role of long non-coding RNA in human carcinomas, Mol. Cancer, 2011, vol. 10, no. 1, art. ID 38. https://doi.org/10.1186/1476-4598-10-38
Glubb, D.M., Maranian, M.J., Michailidou, K., et al., Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1, Am. J. Hum. Genet., 2015, vol. 96, no. 1, pp. 5–20. https://doi.org/10.1016/j.ajhg.2014.11.009
Gold, B., Kirchhoff, T., Stefanov, S., et al., Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33, Proc. Natl. Acad. Sci., 2008, vol. 105, no. 11, pp. 4340–4345. https://doi.org/10.1073/pnas.0800441105
Guttman, M., Amit, I., Garber, M., et al., Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals, Nature, 2009, vol. 458, no. 7235, pp. 223–227. https://doi.org/10.1038/nature07672
Haiman, C.A., Chen, G.K., Vachon, C.M., et al., A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer, Nat. Genet., 2011, vol. 43, no. 12, pp. 1210–1214. https://doi.org/10.1038/ng.985
Han, M.-R., Long, J., Choi, J.-Y., et al., Genome-wide association study in East Asians identifies two novel breast cancer susceptibility loci, 2016, Hum. Mol. Genet., vol. 25, no. 15, pp. 3361-3371. https://doi.org/10.1093/hmg/ddw164
Han, M.R., Deming-Halverson, S., Cai, Q., et al., Evaluating 17 breast cancer susceptibility loci in the Nashville breast health study, Breast Cancer, 2015, vol. 22, no. 5, pp. 544–551. https://doi.org/10.1007/s12282-014-0518-2
Harrison, R.E., Sikorski, B.A., and Jongstra, J., Leukocyte-specific protein 1 targets the ERK/MAP kinase scaffold protein KSR and MEK1 and ERK2 to the actin cytoskeleton, J. Cell Sci., 2004, vol. 117, no. 10, pp. 2151–2157.https://doi.org/10.1242/jcs.00955
Hein, A., Rack, B., Li, L., et al., Genetic breast cancer susceptibility variants and prognosis in the prospectively randomized SUCCESS a study, Geburtshilfe Frauenheilk., 2017, vol. 77, no. 6, pp. 651–659. https://doi.org/10.1055/s-0042-113189
Hill, K., The demography of menopause, Maturitas, 1996, vol. 23, no. 2, pp. 113–127. https://doi.org/10.1016/0378-5122(95)00968-x
Hsieh, Y.C., Tu, S.H., Su, C.T., et al., A polygenic risk score for breast cancer risk in a Taiwanese population, Breast Cancer Res. Treat., 2017, vol. 163, no. 1, pp. 131–138. https://doi.org/10.1007/s10549-017-4144-5
Huarte, M. and Rinn, J.L., Large non-coding RNAs: missing links in cancer?, Hum. Mol. Genet., 2010, vol. 19, no. R2, pp. R152–R161. https://doi.org/10.1093/hmg/ddq353
Hughes, E., Judkins, T., Wagner, S., et al., Development and validation of a residual risk score to predict breast cancer risk in unaffected women negative for mutations on a multi-gene hereditary cancer panel, J. Clin. Oncol., 2017, vol. 35, pp. 1579–1579. https://doi.org/10.1200/JCO.2017.35.15_suppl.1579
Hunter, D.J., Kraft, P., Jacobs, K.B., et al., A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer, Nat. Genet., 2007, vol. 39, no. 7, pp. 870–874. https://doi.org/10.1038/ng2075
Kim, H.-C., Lee, J.-Y., Sung, H., et al., A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study, Breast Cancer Res., 2012, vol. 14, no. 2, art. ID R 56. https://doi.org/10.1186/bcr3158
Kuchenbaecker, K.B., McGuffog, L., Barrowdale, D., et al., Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers, J. Natl. Cancer Inst., 2017, vol. 109, no. 7. https://doi.org/10.1093/jnci/djw302
Lakeman, I.M.M., Hilbers, F.S., Rodríguez-Girondo, M., et al., Addition of a 161-SNP polygenic risk score to family history-based risk prediction: impact on clinical management in non-BRCA1/2 breast cancer families, J. Med. Genet., 2019, vol. 56, no. 9, pp. 581–589. https://doi.org/10.1136/jmedgenet-2019-106072
Li, H., Feng, B., Miron, A., et al., Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab, Genet. Med., 2017, vol. 19, no. 1, pp. 30–35. https://doi.org/10.1038/gim.2016.43
Li, N., Zhou, P., Zheng, J., et al., A polymorphism rs12325489C>T in the LincRNA-ENST00000515084 exon was found to modulate breast cancer risk via GWAS-based association analyses, PLoS One, 2014, vol. 9, no. 5, art. ID e98251. https://doi.org/10.1371/journal.pone.0098251
Lin, Y., Fu, F., Chen, M., et al., Associations of two common genetic variants with breast cancer risk in a Chinese population: a stratified interaction analysis, PLoS One, 2014, vol. 9, no. 12, pp. 1–12. https://doi.org/10.1371/journal.pone.0115707
Lindström, S., Thompson, D.J., Paterson, A.D., et al., Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk, Nat. Commun., 2014, vol. 5, no. 1, art. ID 5303. https://doi.org/10.1038/ncomms6303
Lobrich, M. and Jeggo, P., The impact of a negligent G2/M checkpoint on genomic instability cancer induction, Nat. Rev. Cancer, 2007, vol. 7, no. 11, pp. 861–869. https://doi.org/10.1038/nrc2248
Long, J., Shu, X.O., Cai, Q., et al., Evaluation of breast cancer susceptibility loci in Chinese women, Cancer Epidemiol., Biomarkers Prev., 2010, vol. 19, no. 9, pp. 2357–2365. https://doi.org/10.1158/1055-9965.EPI-10-0054
Ma, L., Teruya-Feldstein, J., and Weinberg, R.A., Tumour invasion and metastasis initiated by microRNA-10b in breast cancer, Nature, 2007, vol. 449, no. 7163, pp. 682–688. https://doi.org/10.1038/nature06174
MacLachlan, T.K., Sang, N., and Giordano, A., Cyclins, cyclin-dependent kinases and Cdk inhibitors: implications in cell cycle control and cancer, Crit. Rev. Eukaryotic Gene Expression, 1995, vol. 5, no. 2, pp. 127–156. https://doi.org/10.1615/critreveukargeneexpr.v5.i2.20
Mavaddat, N., Pharoah, P.D., Michailidou, K., et al., Prediction of breast cancer risk based on profiling with common genetic variants, J. Natl. Cancer Inst., 2015, vol. 107, no. 5, art. ID djv036. https://doi.org/10.1093/jnci/djv036
Mealiffe, M.E., Stokowski, R.P., Rhees, B.K., et al., Assessment of clinical validity of a breast cancer risk model combining genetic and clinical information, J. Natl. Cancer Inst., 2010, vol. 102, no. 21, pp. 1618–1627. https://doi.org/10.1093/jnci/djq388
Michailidou, K., Beesley, J., Lindstrom, S., et al., Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer, Nat. Genet., 2015, vol. 47, no. 4, pp. 373–380. https://doi.org/10.1038/ng.3242
Möller, S., Mucci, L.A., Harris, J.R., et al., The heritability of breast cancer among women in the nordic twin study of cancer, Cancer Epidemiol. Prev. Biomarkers, 2016, vol. 25, no. 1, pp. 145–150. https://doi.org/10.1158/1055-9965.EPI-15-0913
Mu, K., Wu, Z.Z., Yu, J.P., et al., Meta-analysis of the association between three microRNA polymorphisms and breast cancer susceptibility, Oncotarget, 2017, vol. 8, no. 40, pp. 68809–68824. https://doi.org/10.18632/oncotarget.18516
Muranen, T.A., Mavaddat, N., Khan, S., et al., Polygenic risk score is associated with increased disease risk in 52 Finnish breast cancer families, Breast Cancer Res. Treat., 2016, vol. 158, no. 3, pp. 463–469. https://doi.org/10.1007/s10549-016-3897-6
Na, Li., Ping, Zhou., Jian, Zheng., et al., A polymorphism rs12325489C>T in the LincRNA-ENST00000515084 exon was found to modulate breast cancer risk via GWAS-based association analyses, PLoS One, 2014, vol. 9, no. 5, pp. e98251–e98251. https://doi.org/10.1371/journal.pone.0098251
Orr, N., Dudbridge, F., Dryden, N., et al., Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2, Hum. Mol. Genet., 2015, vol. 24, no. 10, pp. 2966–2984. https://doi.org/10.1093/hmg/ddv035
Özgöz, A., İçduygu, F.M., Yükseltürk, A., et al., Low-penetrance susceptibility variants and postmenopausal oestrogen receptor positive breast cancer, J. Genet., 2020, vol. 99, no. 1, art. ID 15. https://doi.org/10.1007/s12041-019-1174-2
Pace, A., Barone, G., Lauria, A., et al., Hsp60, a novel target for antitumor therapy: structure-function features and prospective drugs design, Curr. Pharm. Des., 2013, vol. 19, no. 15, pp. 2757–2764. https://doi.org/10.2174/1381612811319150011
Peto, J., Collins, N., Barfoot, R., et al., Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer, J. Natl. Cancer Inst., 1999, vol. 91, no. 11, pp. 943–949. https://doi.org/10.1093/jnci/91.11.943
Pharoah, P.D., Dunning, A.M., Ponder, B.A., et al., Association studies for finding cancer-susceptibility genetic variants, Nat. Rev. Cancer, 2004, vol. 4, no. 11, pp. 850–860. https://doi.org/10.1038/nrc1476
Qi, P., Wang, L., Zhou, B., et al., Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population, Genet. Mol. Res., 2015, vol. 14, no. 2, pp. 6289–6296. https://doi.org/10.4238/2015.June.11.2
Qian, B., Zheng, H., Yu, H., et al., Genotypes and phenotypes of IGF-I and IGFBP-3 in breast tumors among Chinese women, Breast Cancer Res. Treat., 2011, vol. 130, no. 1, pp. 217–226. https://doi.org/10.1007/s10549-011-1552-9
Ricol, D., Cappellen, D., El Marjou, A., et al., Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer, Oncogene, 1999, vol. 18, no. 51, pp. 7234–7243. https://doi.org/10.1038/sj.onc.1203186
Safari, S., Baratloo, A., Elfil, M., et al., Evidence based emergency medicine; part 5 receiver operating curve and area under the curve, Emergency (Tehran, Iran), 2016, vol. 4, no. 2, pp. 111–113. https://doi.org/10.22037/aaem.v4i2.232
Sawyer, S., Mitchell, G., McKinley, J., et al., A role for common genomic variants in the assessment of familial breast cancer, J. Clin. Oncol., 2012, vol. 30, no. 35, pp. 4330–4336. https://doi.org/10.1200/JCO.2012.41.7469
Schwartz, M.D., Isaacs, C., Graves, K.D., et al., Long-term outcomes of BRCA1/BRCA2 testing: risk reduction and surveillance, Cancer, 2012, vol. 118, no. 2, pp. 510–517. https://doi.org/10.1002/cncr.26294
Shan, J., Dsouza, S.P., Bakhru, S., et al., TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness, Cancer Res., 2013, vol. 73, no. 9, pp. 2840–2849. https://doi.org/10.1158/0008-5472.CAN-12-4313
Shan, J., Mahfoudh, W., Dsouza, S.P., et al., Genome-Wide Association Studies (GWAS) breast cancer susceptibility loci in Arabs: susceptibility and prognostic implications in Tunisians, Breast Cancer Res. Treat., 2012, vol. 135, no. 3, pp. 715–724. https://doi.org/10.1007/s10549-012-2202-6
Shi, J., Zhang, Y., Zheng, W., et al., Fine-scale mapping of 8q24 locus identifies multiple independent risk variants for breast cancer, Int. J. Cancer, 2016, vol. 139, no. 6, pp. 1303–1317. https://doi.org/10.1002/ijc.30150
Shieh, Y., Hu, D., Ma, L., et al., Breast cancer risk prediction using a clinical risk model and polygenic risk score, Breast Cancer Res. Treat., 2016, vol. 159, no. 3, pp. 513–525. https://doi.org/10.1007/s10549-016-3953-2
Shieh, Y., Hu, D., Ma, L., et al., Joint relative risks for estrogen receptor-positive breast cancer from a clinical model, polygenic risk score, and sex hormones, Breast Cancer Res. Treat., 2017, vol. 166, no. 2, pp. 603–612. https://doi.org/10.1007/s10549-017-4430-2
Stacey, S.N., Manolescu, A., Sulem, P., et al., Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer, Nat. Genet., 2007, vol. 39, no. 7, pp. 865–869. https://doi.org/10.1038/ng2064
Stacey, S.N., Manolescu, A., Sulem, P., et al., Common variants on chromosome 5p12 confer susceptibility to estrogen receptor–positive breast cancer, Nat. Genet., 2008, vol. 40, no. 6, pp. 703–706. https://doi.org/10.1038/ng.131
Starlard-Davenport, A., Allman, R., Dite, G.S., et al., Validation of a genetic risk score for Arkansas women of color, PLoS One, 2018, vol. 13, no. 10, art. ID e0204834. https://doi.org/10.1371/journal.pone.0204834
Tajbakhsh, A., Farjami, Z., Darroudi, S., et al., Association of rs4784227-CASC16 (LOC643714 locus) and rs4782447-ACSF3 polymorphisms and their association with breast cancer risk among Iranian population, EXCLI J., 2019, vol. 18, pp. 429–438. https://doi.org/10.17179/excli2019-1374
Tan, T., Zhang, K., Chen, W., Genetic variants of ESR1 and SGSM3 are associated with the susceptibility of breast cancer in the Chinese population, Breast Cancer, 2017, vol. 24, no. 3, pp. 369–374. https://doi.org/10.1007/s12282-016-0712-5
Turnbull, C., Ahmed, S., Morrison, J., et al., Genome-wide association study identifies five new breast cancer susceptibility loci, Nat. Genet., 2010, vol. 42, no. 6, pp. 504–507. https://doi.org/10.1038/ng.586
Vachon, C.M., Pankratz, V.S., Scott, C.G., et al., The contributions of breast density and common genetic variation to breast cancer risk, J. Natl. Cancer Inst., 2015, vol. 107, no. 5, art. ID dju397. https://doi.org/10.1093/jnci/dju397
Wacholder, S., Hartge, P., Prentice, R., et al., Performance of common genetic variants in breast-cancer risk models, N. Engl. J. Med., 2010, vol. 362, no. 11, pp. 986–993. https://doi.org/10.1056/NEJMoa0907727
Wang, J., Wang, Q., Liu, H., et al., The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies, Mutagenesis, 2012, vol. 27, no. 6, pp. 779–788. https://doi.org/10.1093/mutage/ges052
Wang, P.Y., Gao, Z.H., Jiang, Z.H., et al., The associations of single nucleotide polymorphisms in miR-146a, miR-196a and miR-499 with breast cancer susceptibility, PLoS One, 2013, vol. 8, no. 9, art. ID e70656. https://doi.org/10.1371/journal.pone.0070656
Wen, W., Shu, X.O., Guo, X., et al., Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry, Breast Cancer Res., 2016, vol. 18, no. 1, art. ID 124. https://doi.org/10.1186/s13058-016-0786-1
Wu, Z., Wang, P., Song, C., et al., Evaluation of miRNA-binding-site SNPs of MRE11A, NBS1, RAD51 and RAD52 involved in HRR pathway genes and risk of breast cancer in China, Mol. Genet. Genomics, 2015, vol. 290, no. 3, pp. 1141–1153. https://doi.org/10.1007/s00438-014-0983-5
Xu, M., Xu, Y., Chen, M., et al., Association study confirms two susceptibility loci for breast cancer in Chinese Han women, Breast Cancer Res. Treat., 2016, vol. 159, no. 3, pp. 433–442. https://doi.org/10.1007/s10549-016-3952-3
Xu, W., Xu, J., Liu, S., et al., Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis, PLoS One, 2011, vol. 6, no. 5, art. ID e20471. https://doi.org/10.1371/journal.pone.0020471
Yu, K., Xu, J., Liu, Z., et al., Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth, Development, 2003, vol. 130, no. 13, pp. 3063–3074. https://doi.org/10.1242/dev.00491
Zhang, H., Zhang, Y., Yan, W., et al., Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: a meta-analysis, Oncotarget, 2017, vol. 8, no. 1, pp. 393–407. https://doi.org/10.18632/oncotarget.13426
Zhang, Y., Zeng, X., Liu, P., et al., Association between FGFR2 (rs2981582, rs2420946 and rs2981578) polymorphism and breast cancer susceptibility: a meta-analysis, Oncotarget, 2017, vol. 8, no. 2, pp. 3454–3470. https://doi.org/10.18632/oncotarget.13839
Zheng, W., Wen, W., Gao, Y.-T., et al., Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women, J. Natl. Cancer Inst., 2010, vol. 102, no. 13, pp. 972–981. https://doi.org/10.1093/jnci/djq170
Zheng, Y., Ogundiran, T.O., Falusi, A.G., et al., Fine mapping of breast cancer genome-wide association studies loci in women of African ancestry identifies novel susceptibility markers, Carcinogenesis, 2013, vol. 34, no. 7, pp. 1520–1528. https://doi.org/10.1093/carcin/bgt090
Zhu, R.M., Lin, W., Zhang, W., et al., Modification effects of genetic polymorphisms in FTO, IL-6, and HSPD1 on the associations of diabetes with breast cancer risk and survival, PLoS One, 2017, vol. 12, no. 6, art. ID e0178850. https://doi.org/10.1371/journal.pone.0178850
Zuo, X., Wang, H., Mi, Y., et al., The association of CASC16 variants with breast Cancer risk in a northwest Chinese female population, Mol. Med., 2020, vol. 26, no. 1, pp. 1–10. https://doi.org/10.1186/s10020-020-0137-7
FUNDING
This research is funded by University of Science, VNU-HCM under grant number T2021-50.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement of compliance with standards of research involving humans as subjects. During research enrollment, participants signed informed consent. The Ethical Committee approved this study of Oncology Hospital Ho Chi Minh City (no. 177/ÐÐÐ-CÐT November 18th, 2014).
About this article
Cite this article
Thanh Thi Ngoc Nguyen, Nguyen, T.H., Phan, H.N. et al. Seven-Single Nucleotide Polymorphism Polygenic Risk Score for Breast Cancer Risk Prediction in a Vietnamese Population. Cytol. Genet. 56, 379–390 (2022). https://doi.org/10.3103/S0095452722040065
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452722040065