Screening for Mild Cognitive Impairment Using a Machine Learning Classifier and the Remote Speech Biomarker for Cognition: Evidence from Two Clinically Relevant Cohorts
Abstract
Background:
Modern prodromal Alzheimer’s disease (AD) clinical trials might extend outreach to a general population, causing high screen-out rates and thereby increasing study time and costs. Thus, screening tools that cost-effectively detect mild cognitive impairment (MCI) at scale are needed.
Objective:
Develop a screening algorithm that can differentiate between healthy and MCI participants in different clinically relevant populations.
Methods:
Two screening algorithms based on the remote ki:e speech biomarker for cognition (ki:e SB-C) were designed on a Dutch memory clinic cohort (N = 121) and a Swedish birth cohort (N = 404). MCI classification was each evaluated on the training cohort as well as on the unrelated validation cohort.
Results:
The algorithms achieved a performance of AUC 0.73 and AUC 0.77 in the respective training cohorts and AUC 0.81 in the unseen validation cohorts.
Conclusion:
The results indicate that a ki:e SB-C based algorithm robustly detects MCI across different cohorts and languages, which has the potential to make current trials more efficient and improve future primary health care.
INTRODUCTION
Recently, the focus of clinical trials in Alzheimer’s disease (AD) has shifted to the earlier phases of the disease [1]. To investigate early disease stages and avoid biased recruiting through memory-clinic recruitment funnels, some modern AD trials reach out to a general population. However, recruiting from a general population leads to a high number of screened out individuals who do not show AD pathology and are therefore no suitable subjects for drug development trials. The high number of screen-outs is accompanied by high costs [2] and timely expansion of AD clinical trials [3], which is considered to be a key barrier in modern AD trials [2].
An additional light-weight and scalable pre-screening step incorporated directly into the outreach funnel before the more costly onsite screening could help make those trials more efficient by improving patient selection and thus speeding up clinical development [3]. Traditional wet biomarkers (e.g., based on cerebrospinal fluid) reliably detect the pathologic hallmarks associated with AD but are not suitable for scaled frontline screening or remote scenarios as they are invasive and costly. Moreover, ethical issues might arise when applying traditional invasive wet biomarker procedures in patients who do not fulfill clinical indication yet. Therefore, AD clinical trials pose a clear opportunity for minimally invasive and especially digital biomarkers [4, 5].
Beyond biomarkers measuring biological processes, behavioral signs such as impaired cognition is an easy-to-observe hallmark of AD which is present throughout all phases of the disease trajectory including the early stage of mild cognitive impairment (MCI) [6]. Since the inclusion of an incipient dementia population is targeted in secondary prevention trials, the detection of the MCI state is of particular interest, as it is characterized by the onset of subtle cognitive impairments and is accompanied by a high conversion rate to dementia [7].
Speech biomarkers that detect cognition are especially promising because they allow for an automatic, non-invasive assessment, making them a low-burden digital tool, which offers an objective and scalable solution [2], optimal for screening scenarios in a broad population. Significant associations between speech and AD pathology, as assessed by cerebrospinal fluid and imaging biomarkers, were found even at a pre-symptomatic stage [8, 9], validating its usability for early disease detection. Moreover, speech can be captured remotely, which lowers the burden especially for the older adults target group [10] and allows application as a pre-screening tool before the on-site screening visits. Due to the automatic analysis, speech biomarkers allow for an extended outreach, making them attractive when trials recruit from a general population.
To address the needs of modern AD clinical trials, we set out to develop a screening algorithm that differentiates between MCI and healthy control participants. The algorithm is developed based on the validated ki:e SB-C (ki:e speech biomarker for cognition) [11], which measures cognition, composed of subscores for episodic memory, processing speed, and executive function. In this paper, we present results about the diagnostic accuracy of the MCI screening algorithm from two different cohorts, a representative birth cohort born 1944–1945 as well as a conventional memory clinic population.
MATERIALS AND METHODS
Training and validation of the screening algorithm is based on two clinical studies: the Dutch Deep Speech Analysis for cognitive assessment in clinical trials (DeepSpA) study [12, 13] and the Swedish H70 data set (subset of the Gothenburg H70 cohort; [14]). Both studies collected the ki:e SB-C over a mobile application.
Subjects
DeepSpA
Participants (N = 140) were recruited at the memory clinic in Maastricht as part of the Maastricht University Medical Center+(MUMC+) study (subjective cognitive impairment (SCI), MCI, and dementia). We excluded 6 subjects due to poor audio quality and operational issues and the dementia participants (N = 13) from the analysis, so that 121 participants remained for the analysis. Participants underwent an in-person assessment at the clinic at baseline. A diagnosis of MCI was based on DSM-5 criteria (minor neurocognitive disorder [15]). Participants without cognitive disorders were classified as SCI. The ki:e SB-C was collected using a mobile application. Mini-Mental State Examination (MMSE) and Clinical Dementia Rating (CDR) data were available.
H70 baseline
75-year-old participants (N = 404) were recruited 2019–2021 at the University of Gothenburg as part of the ongoing epidemiological H70 Birth Cohort Study. Eligible participants were initially obtained from the Swedish Tax Agency Population Registry. Participants were classified as either MCI or cognitively intact controls (CI). MCI was classified based on the criteria of Petersen (1999) [16]: Subjects’ performances were transformed into z-scores; subjects whose z-score was smaller or equal to –1.5 in the Rey Auditory Verbal Learning Test delayed recall (RAVLT) [17] OR smaller or equal to –1 on both the Phonemic Verbal Fluency (PVF) AND the Stroop task [18]. See pseudocode underneath for clarification:
The RAVLT delayed recall score acted as our amnestic criterion to define MCI, however we chose –1.5 as threshold for the z-score and not just –1 like one would typically do when applying the Petersen criteria. This is due to the fact that all subjects go through an extensive psychometric protocol when they do their cohort annual visits and this protocol features a lot of different word list learning tests. It can be assumed that the interference that is created by all those word lists systematically underestimates the memory performance of those participants. Hence we applied a more liberal criteria to categorize them as MCI.
The ki:e SB-C was collected using a mobile application.
Ethics
Both studies that provided data to this research have been conducted in compliance with the Ethical Principles for Medical Research Involving Human Subjects, as defined in the Declaration of Helsinki and the European General Data Protection Regulation. For the Dutch DeepSpA study, the local Medical Ethical Committee (METC MUMC/UM) approved the study (MEC 15-4-100). For the Swedish H70 Study, the National Ethical Review Board (Etikprövningsmyndigheten) approved the study (Dnr 2019-0158). All participants provided informed consent before completing any study-related procedures.
Measures
The ki:e SB-C takes speech recordings from two standard neuropsychological assessments as input: RAVLT and the Semantic Verbal Fluency (SVF) paradigm (the SVF version of the CERAD [20] is used). Speech from both tests is automatically processed using the proprietary speech analysis pipeline from ki:elements that involves automatic speech recognition to transcribe speech and extract features, which capture processing of task stimuli by analyzing subjective, serial or semantic groupings as well as semantic and temporal aspects of the verbal output of participants. The ki:e SB-C is built of more than 50 automatically extracted speech features from which 27 were selected. No features derived from the RAVLT delayed recall trial were selected, since this was used for MCI diagnosis in the H70 cohort. The features compose three distinct neurocognitive subdomain scores (learning and memory, executive function and processing speed), which cover the most frequently impaired cognitive domains in MCI [19]. From the three subdomain scores, one aggregated global score for cognition is derived.
Data analysis
A machine learning model was built and evaluated on each datasets. The best performing model for each of the two dataset was then evaluated on the respective other dataset (see Table 1). A support vector machine model, extra trees, and a random forest model were trained on the four features derived from the ki:e SB-C: the three neurocognitive subscores and the global composite score. Models were trained using leave-one-out cross validation and grid search for hyper parameter tuning. We used class weights to account for imbalanced diagnostic groups in some scenarios (i.e., model built on H70). For each dataset, the best performing model was selected based on the balanced accuracy.
Table 1
Overview of model training and validation
| DeepSpA-model | H70-model | |
| Training | DeepSpA | H70 |
| Validation | DeepSpA, H70 | H70, DeepSpA |
RESULTS
The two models that were built on the H70 and the DeepSpA sample based on the four ki:e SB-C scores (general cognition & subscores: processing speed, semantic and episodic memory) were evaluated on each sample.
Demographic and clinical information
Demographic and clinical information of the subjects of the two samples are presented in Tables 2 and 3.
Table 2
Descriptive statistics of the DeepSpA sample
| DeepSpA baseline | ||
| SCI | MCI | |
| N | 69 (24 F) | 52 (19 F) |
| Age | 62.20±10.71 | 70.29±9.83 |
| ki:e SB-C | 0.49±0.12 | 0.32±0.12 |
| CDR | 0.37±0.28 | 0.48±0.20 |
| MMSE | 28.71±1.23 | 26.85±2.04 |
Table 3
Descriptive statistics of the H70 sample
| H70 baseline | ||
| CI | MCI | |
| N | 356 | 48 |
| Age | 75 | 75 |
| ki:e SB-C | 0.44±0.12 | 0.29±0.11 |
| RAVLT delayed recall | 6.90±3.00 | 3.02±3.27 |
| PVF | 45.03±13.05 | 32.60±16.97 |
| Stroop | 36.42±10.88 | 24.98±12.64 |
In both samples, the groups differed significantly in their ki:e SB-C value (DeepSpA: Kruskal-Wallis Test, χ2(2) = 60.2, p < 0.001, d = 1.79, SCI > MCI; H70: Kruskal-Wallis Test, χ2(1) = 36.99, p < 0.001, d = 0.78, CI > MCI; see Tables 2 and 3).
Screening performance
The model that best differentiated among MCI and cognitively intact control participants in the H70 dataset and between SCI and MCI participants in the DeepSpA dataset based on the four ki:e SB-C scores was a support vector machine model. Performance metrics and confusion matrices for both models on both datasets are depicted in Tables 4 and 5.
Table 4
Performance metrics for both models
| DeepSpA model | H70 model | |||
| Performance | Validation | Validation | Validation | Validation |
| metric | on DeepSpA | on H70 | on H70 | on DeepSpA |
| F1 | 0.68 | 0.43 | 0.43 | 0.73 |
| Accuracy | 0.75 | 0.75 | 0.71 | 0.78 |
| Balanced accuracy | 0.74 | 0.77 | 0.76 | 0.77 |
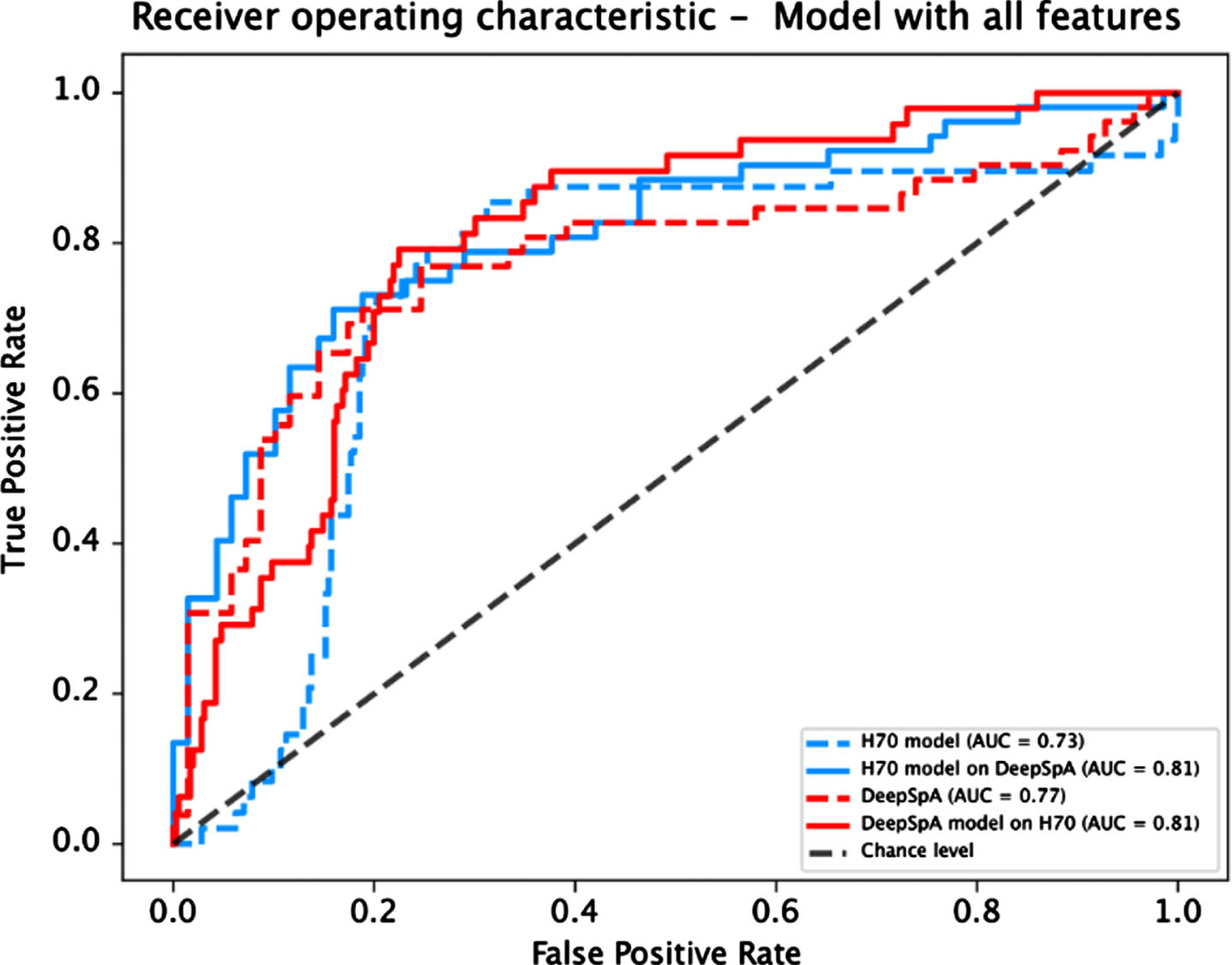
| ROC AUC | 0.77 | 0.81 | 0.73 | 0.81 |
| Sensitivity | 0.62 | 0.79 | 0.79 | 0.71 |
| Specificity | 0.86 | 0.75 | 0.72 | 0.83 |
| PPV | 0.76 | 0.30 | 0.28 | 0.76 |
| 1-NPV | 0.25 | 0.04 | 0.04 | 0.21 |
Table 5
Confusion matrices for both models on each dataset
| DeepSpA model | |||||
| Validation on DeepSpA | Validation on H70 | ||||
| Predicted SCI | Predicted MCI | Predicted CI | Predicted MCI | ||
| True SCI | 59 | 10 | True CI | 267 | 89 |
| True MCI | 20 | 35 | True MCI | 10 | 38 |
| H70 model | |||||
| Validation on DeepSpA | Validation on H70 | ||||
| Predicted SCI | Predicted MCI | Predicted CI | Predicted MCI | ||
| True SCI | 57 | 12 | True CI | 256 | 100 |
| True MCI | 15 | 37 | True MCI | 10 | 38 |
The “F1”-score is a way of combining the precision and recall of a model, which is defined as the harmonic mean of the model’s precision and recall, and measures a model’s accuracy on a dataset. “Accuracy” is the number of correctly predicted data points out of all the data points and is defined as the number of true positives and true negatives divided by the number of true positives, true negatives, false positives, and false negatives. “Balanced accuracy” score is a further development on the standard accuracy metric where it’s adjusted to perform better on imbalanced datasets. The way it does this is by calculating the average accuracy for each class, instead of combining them as is the case with standard accuracy. The “ROC-AUC” score is the measure of the ability of a classifier to distinguish between classes. “Sensitivity” is the ability of a test to correctly classify an individual as positive. Specificity is the ability of a test to correctly classify an individual as negative. The “positive predictive value” (PPV) is the probability that following a positive test result, that individual will truly be positive. The “negative predictive value” (NPV) is the probability that following a negative test result, that individual will truly be negative.
Fig. 1
Receiver operating characteristic curve for both models evaluated on each dataset.
DISCUSSION
This paper presents results of a screening algorithm that differentiates between MCI and subjectively impaired or cognitively intact control participants. We present results of the MCI screening algorithm from two different cohorts, a representative birth cohort recruited from a general population based on specific birth dates as well as a conventional memory clinic population.
Overall models trained based on the ki:e SB-C features, demonstrate good performance results (Balanced accuracy 0.74–0.77; AUC 0.73–0.81) in the discrimination of cognitively intact controls and SCI against MCI participants in both datasets. In the pre-screening setting described above, high specificity is particularly relevant, since the number of healthy subjects (True SCI/CI) is to be minimized to save study costs. That high specificity values (0.72–0.86) were achieved indicates that the two models are suitable for the given context. The results of specificity and sensitivity are comparable with other screening tools for MCI [21]. Moreover, we show that the performance of the algorithm reaches similar performances even when applied to a completely new, unknown data set (i.e., when trained on one data set the algorithm reaches good comparable performance when evaluated on the other data set and vice versa). Slightly different values were obtained for sensitivity and specificity depending on the evaluated dataset: the sensitivity is comparatively lower for each algorithm when the validation was performed on the DeepSpA dataset. This difference might be explained by different grouping criteria. Since the sample in the H70 dataset was divided into cognitively impaired and healthy participants based on test results, it is likely that the performance of an algorithm that uses similar tests as speech input is high. In contrast, the overlap between predicted and true groupings is expected to be lower when the grouping criteria differ. In total, the results demonstrate that a screening algorithm based on the ki:e SB-C is able to robustly detect MCI in a broad population and could be used to speed up recruitment for clinical trial enrichment.
One main strength of the study is that the algorithm was evaluated on two different populations that are typically used for recruitment in AD clinical trials: The DeepSpA sample including approximately 49% MCI participants represents a memory clinic population and the H70 sample, including 13% MCI participants, represents a general outreach population, since the prevalence of MCI in the general population is approximately 18.4% in the 60-year-old and 29.9% in the 70-year-old group [22]. Since the prevalence of MCI differs in the DeepSpA and the H70 population, the prevalence-dependent performance metrics (PPV and 1-NPV) differ between the validations on the two datasets (see Table 2). Hence, when specifically considering the case of general outreach, the results obtained when evaluating the H70 dataset must be considered. The PPV an 0.3, which means that approximately 30% of all included participants are actual MCI patients, implies a successful enrichment of the sample and thus demonstrates the suitability as a pre-screening tool when recruiting from a general population as conducted in early AD trials. Thus, the algorithm potentially supports recruitment in AD trials, helping to make them more efficient by saving time and money.
Moreover, the evaluation of two different populations is accompanied by two different languages (DeepSpA: Dutch, H70: Swedish), also evaluating a cross-over design (i.e., screening algorithm that has been built on Dutch cohort gets evaluated on the Swedish cohort). Since the algorithm shows good performance even when the model was evaluated on the other dataset, we conclude that the screening algorithm is robust and rather language-agnostic.
Finally, screening tools such as the evaluated algorithm have potential in future primary care scenarios. Since three new disease-modifying anti-amyloid drugs are at the border of receiving approval in 2022 [23], scalable and non-invasive screening tools that detect the disease in an early phase of the AD trajectory will be in demand more than ever. Based on the successful discrimination of a cognitively intact and a mildly impaired status based on an automatic and remote assessment, the presented screening algorithm is a particular convenient option for the early identification of individuals at risk and in rural areas. This closes an upcoming health care gap, which arises since contemporarily dementia diagnoses are usually not given by a primary care physician but instead with delay at a memory clinic. This will subsequently lead to a delayed provision of the right care and treatment for the patient. Thus, convenient screening tools play a relevant role in future care scenarios, as they enable identification of people at increased risk, a faster referral to specialized clinics for further multidisciplinary diagnostics, and in the end might enable earlier provision of relevant care and therapies, such as medications when these become available.
However, one limitation of the study is the definition of MCI. Although defining criteria, such as Petersen’s criteria, and standardized assessment procedures for MCI have been proposed [24], no established consensus on the definition of MCI criteria has prevailed [25]. Thus, a broad range of definitions and criteria are used in contemporary research and clinical practice. This is reflected in this study since MCI diagnosis was assigned differently in the two cohorts. The diverging diagnostic criteria introduced some heterogeneity in the data, which potentially impeded machine learning performance. Therefore, it is likely that the performance metrics reported here are an underestimation of the model performance that could be achieved for two cohorts with the same diagnostic criteria. However, since there is no consensus for diagnostic criteria in clinical practice either, it can be understood as a strength of this study that the model performance is still high when the model is applied to new data in which the clinical groups were defined differently. Another limitation concerning the MCI diagnosis in this study is that no biomarker information was taken into account, so no conclusion can be drawn about the etiology of cognitive impairment.
Conclusion
In summary, this work demonstrates that a screening algorithm based on the ki:e SB-C, measuring general cognition, processing speed, episodic memory, and executive functions can differentiate between healthy and MCI participants. Importantly, no matter on which of the two cohorts the screening algorithm was trained, it still performed well both on the training cohort during cross-validation as well as on the other cohort from a different language. This result is specifically notable, as the two data sets are in two different languages (Dutch and Swedish) and represent truly unrelated cohorts (one general population birth-cohort the other a memory clinic cohort). These results have two main implications: A screening algorithm like the one developed here can be used in clinical trials to screen out unsuitable candidates at low cost and over the phone reducing the costs and time lost in a general outreach recruitment funnel from a broad population. Moreover, the algorithm can improve primary healthcare by offering general practitioners a low-burden phone-based screening tool, to refer patients to memory clinics and experts.
ACKNOWLEDGMENTS
The authors have no acknowledgments to report.
FUNDING
SK was financed by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-965923, ALFGBG-81392, ALF GBG-771071), the Alzheimerfonden (AF-842471, AF-737641, AF-929959, AF-939825), the Swedish Research Council (2019-02075), and Psykiatriska Forskningsfonden.
DeepSpA has been funded by EIT-Health project grant agreement number 19249, as well as supported by Janssen Pharmaceutica NV through a collaboration agreement (Award/Grant number is not applicable).
CONFLICT OF INTEREST
For ki:elements, S. Schäfer, JT, AK, EM, LS, JZ and NL are employed by the digital biomarker company ki:elements. NL, AK and JT own shares of the company.
SK has served at SK scientific advisory boards and / or as consultant for Geras Solutions and Biogen.
AZ, IR, IS, JS have no conflict of interest to report.
DATA AVAILABILITY
The data supporting the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
[1] | Khoury R , Ghossoub E ((2019) ) Diagnostic biomarkers of Alzheimer’s disease: A state-of-the-art review. Biomark Neuropsychiatry 1: , 100005. |
[2] | Malzbender K , Lavin-Mena L , Hughes L , Bose N , Goldman D , Patel D (2020) Key barriers for clinical trials for Alzheimer’s disease. Schaeffer Center White Paper Series, Leonard D. Schaeffer Center for Health Policy &Economics, University of Southern California. |
[3] | Cummings J , Aisen PS , DuBois B , Frölich L , Jack CR Jr , Jones RW , Morris JC , Raskin J , Dowsett SA , Scheltens P ((2016) ) Drug development in Alzheimer’s disease: The path to 2025. Alzheimers Res Ther 8: , 39. |
[4] | Mattsson N , Carrillo MC , Dean RA , Devous MD Sr , Nikolcheva T , Pesini P , Salter H , Potter WZ , Sperling RS , Bateman RJ , Bain LJ , Liu E ((2015) ) Revolutionizing Alzheimer’s disease and clinical trials through biomarkers. Alzheimers Dement (Amst) 1: , 412–419. |
[5] | Dorsey ER , Papapetropoulos S , Xiong M , Kieburtz K ((2017) ) The first frontier: Digital biomarkers for neurodegenerative disorders. Digit Biomark 1: , 6–13. |
[6] | Porsteinsson AP , Isaacson RS , Knox S , Sabbagh MN , Rubino I ((2021) ) Diagnosis of early Alzheimer’s disease: Clinical practice in 2021. J Prev Alzheimers Dis 8: , 371–386. |
[7] | Geslani DM , Tierney MC , Herrmann N , Szalai JP ((2005) ) Mild cognitive impairment: An operational definition and its conversion rate to Alzheimer’s disease. Dement Geriatr Cogn Disord 19: , 383–389. |
[8] | Mueller KD , Van Hulle CA , Koscik RL , Jonaitis E , Peters CC , Betthauser TJ , Christian B , Chin N , Hermann BP , Johnson S ((2021) ) Amyloid beta associations with connected speech in cognitively unimpaired adults. Alzheimers Dement (Amst) 13: , e12203. |
[9] | Mazzon G , Ajcević M , Cattaruzza T , Menichelli A , Guerriero M , Capitanio S , Pesavento V , Dore F , Sorbi S , Manganotti P , Marini A ((2019) ) Connected speech deficit as an early hallmark of CSF-defined Alzheimer’s disease and correlation with cerebral hypoperfusion pattern. Curr Alzheimer Res 16: , 483–494. |
[10] | Skirrow C , Meszaros M , Meepegama U , Lenain R , Papp KV , Weston J , Fristed E ((2022) ) Validation of a novel fully automated story recall task for repeated remote high-frequency administration. JMIR Aging 5: , e37090. |
[11] | Tröger J , Baykara E , Zhao J , ter Huurne D , Possemis N , Mallick E , Schäfer S , Schwed L , Mina M , Linz N , Ramakers I , Ritchie C ((2022) ) Validation of the remote automated ki:e speech biomarker for cognition in mild cognitive impairment: Verification and validation following DiME V3 Framework. Digit Biomark 6: , 107–116. |
[12] | Ter Huurne DBG , Ramakers IHGB , Linz N , König A , Langel K , Lindsay H , Verhey FRJ , de Vugt M ((2021) ) Clinical use of deep speech parameters derived from the semantic verbal fluency task. Alzheimers Dement 17: (Suppl 8), e050380. |
[13] | Rydberg Sterner T , Ahlner F , Blennow K , Dahlin-Ivanoff S , Falk H , Havstam Johansson L , Hoff M , Holm M , Hörder H , Jacobsson T , Johansson B , Johansson L , Kern J , Kern S , Machado A , Mellqvist Fässberg M , Nilsson J , Ribbe M , Rothenberg E , Ryden L , Sadeghi A , Sacuiu S , Samuelsson J , Sigström R , Skoog J , Thorvaldsson V , Waern M , Westman E , Wetterberg H , Zetterberg H , Zetterberg M , Zettergren A , Östling S , Skoog I ((2019) ) The Gothenburg H70 Birth cohort study 2014–16: Design, methods and study population. Eur J Epidemiol 34: , 191–209. |
[15] | American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders (5th ed.). |
[16] | Petersen RC , Smith GE , Waring SC , Ivnik RJ , Tangalos EG , Kokmen E ((1999) ) Mild cognitive impairment: Clinical characterization and outcome. Archn Neurol 56: , 303–308. |
[17] | Rey A (1958) Memorisation d’une serie de 15 mots en 5 repetitions. L’examen Clinique en Psychologie. |
[18] | Stroop JR ((1935) ) Studies of interference in serial verbal reactions. J Exp Psychol 18: , 643–662. |
[19] | Nordlund A , Rolstad S , Hellström P , Sjögren M , Hansen S , Wallin A ((2005) ) The Goteborg MCI study: Mild cognitive impairment is a heterogeneous condition. J Neurol Neurosurg Psychiatry 76: , 1485–1490. |
[20] | Luck T , Riedel-Heller SG , Wiese B , Stein J , Weyerer S , Werle J , Kaduszkiewicz H , Wagner M , Mösch E , Zimmermann T , Maier W , Bickel H , van den Bussche H , Jessen F , Fuchs A , Pentzek M , AgeCoDe Study Group ((2009) ) CERAD-NP battery: Age-, gender- and education-specific reference values for selected subtests. Results of the German Study on Ageing, Cognition and Dementia in Primary Care Patients (AgeCoDe) [German]. Z Gerontol Geriatr 42: , 372–384. |
[21] | Zhuang L , Yang Y , Gao J ((2021) ) Cognitive assessment tools for mild cognitive impairment screening. J Neurol 268: , 1615–1622. |
[22] | Overton M , Pihlsgård M , Elmståhl S ((2019) ) Prevalence and incidence of mild cognitive impairment across subtypes, age, and sex. Dement Geriatr Cogn Disord 47: , 219–232. |
[23] | Jahnke H (2022) Alzheimer’s disease: An overview and current phase 3 disease-modifying biologic treatments. Biology and Microbiology Graduate Students Plan B Research Projects, 43. https://openprairie.sdstate.edu/biomicroplan-b/43. |
[24] | Foster NL , Bondi MW , Das R , Foss M , Hershey LA , Koh S , Logan R , Poole C , Shega JW , Sood A , Thothala N , Wicklund M , Yu M , Bennett A , Wang D ((2019) ) . Quality improvement in neurology: Mild cognitive impairment quality measurement set. Neurology 93: , 705–713. |
[25] | Anderson ND ((2019) ) State of the science on mild cognitive impairment (MCI). CNS Spectrums 24: , 78–87. |



