Mechanisms of TLR4-Mediated Autophagy and Nitroxidative Stress
- 1State Key Laboratory of Livestock and Poultry Breeding, Guangdong Key Laboratory of Animal Breeding and Nutrition, Institute of Animal Science, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 3Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences and Comparative Medicine Center, Peking Union Medical College, Beijing, China
- 4Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding/Guangdong Provincial Research Center of Gene Editing Engineering Technology, Foshan University, Foshan, China
Pathogenic infections have badly affected public health and the development of the breeding industry. Billions of dollars are spent every year fighting against these pathogens. The immune cells of a host produce reactive oxygen species and reactive nitrogen species which promote the clearance of these microbes. In addition, autophagy, which is considered an effective method to promote the destruction of pathogens, is involved in pathological processes. As research continues, the interplay between autophagy and nitroxidative stress has become apparent. Autophagy is always intertwined with nitroxidative stress. Autophagy regulates nitroxidative stress to maintain homeostasis within an appropriate range. Intracellular oxidation, in turn, is a strong inducer of autophagy. Toll-like receptor 4 (TLR4) is a pattern recognition receptor mainly involved in the regulation of inflammation during infectious diseases. Several studies have suggested that TLR4 is also a key regulator of autophagy and nitroxidative stress. In this review, we describe the role of TLR4 in autophagy and oxidation, and focus on its function in influencing autophagy-nitroxidative stress interactions.
Introduction
Autophagy is a physiological metabolic compensatory process of eukaryotic cells that maintains homeostasis. However, autophagy is also a conserved defense mechanism that evolved during cell evolution and which has an important role in the process of pathogenic infection. Generally, autophagy degrades or digests intracellular aging or damaged organelles, protein, metabolin, and even pathogenic microorganisms by autophagosome encapsulation and lysosomal binding (Nakatogawa et al., 2012). Autophagy of immune cells resists the invasion of pathogens by regulating cellular functions. For example, promoting autophagy helps to clear Streptococcus, Helicobacter. pylori, and Pneumonia (Wang et al., 2009; Ye et al., 2015; Nozawa et al., 2017). However, some microorganisms have evolved the ability to inhibit, escape, and even use autophagy, allowing them to continue to survive in the host body (Starr et al., 2012; Sharma et al., 2021). Moreover, autophagy is involved in the regulation of inflammation. Moderate autophagy maintains homeostasis by negatively regulating inflammation (Liu et al., 2016b; Zhong et al., 2016). Incorrect autophagy can damage the body and even cause organ failure by triggering cytokine storms (Lu et al., 2019; Zhao et al., 2019). How to regulate and use autophagy to protect the body is a hot spot of current research.
Nitroxidative stress is a state of physiological imbalance mainly related to the excessive production of reactive oxygen species (ROS) and reactive nitrogen species (RNS). It is well-known that nitroxidative stress is a general factor in many pathological conditions, including autoimmune diseases, infectious diseases, and tumor (Su et al., 2017; Smallwood et al., 2018). ROS and RNS also have dual-roles in the processes of removing pathogenic microorganisms and maintaining homeostasis. Low concentrations of ROS/RNS help maintain normal physiological metabolism and protect against infectious pathogens (Valko et al., 2007). However, the excessive ROS/RNS leads to an imbalance of redox states, metabolic disorders, and even damage of tissues and organs (Islam, 2017; Di Meo and Venditti, 2020). In addition, a serious pathogenic infection can induce severe nitroxidative stress followed by cytokine storm, resulting in the acute injury of tissues and organs (Mrityunjaya et al., 2020). Therefore, understanding how the body regulates nitroxidative stress levels is important.
The recognition of pathogenic microorganisms by pattern recognition receptors (PRRs) is the initial step in host innate immune responses. The Toll like receptor (TLR) family is an important class of PRRs that recognize a variety of bacteria and viruses, and induces the secretion of inflammatory cytokines and chemokines. Moreover, the activation of TLRs directly enhances the phagocytosis and killing capacity of innate immune cells that promotes the elimination of pathogens (Vasselon and Detmers, 2002). TLR4 is mainly expressed on the membranes of macrophages, dendritic cells and neutrophils. Activation of TLR4 is closely related to inflammation, autophagy, and nitroxidative stress during a pathogenic infection (Deng et al., 2020). In this review, we discuss recent studies on the activities of TLR4, autophagy, and nitroxidative stress during pathogenic microorganism infections, with a particular focus on the mechanisms of TLR4-mediated autophagy and nitroxidative stress.
The Initiation of Autophagy and Nitroxidative Stress During Bacterial Infection
Initiation of Autophagy
Autophagy is also known as type II programmed cell death. Moderate autophagy can degrade damaged organelles, denaturated macromolecules and intracellular pathogens, and then provide raw materials for cell metabolism (Levine and Kroemer, 2019; Zhu et al., 2019). Excessive autophagy and insufficient autophagy lead to disease (Lavandero et al., 2015). It was confirmed that autophagy is closely associated with proliferation of pathogenic microorganisms (Wang et al., 2020b). The stage and site of pathogenic infection, type of infected cell, and physiological state of the host body all affect the activity and outcome of autophagy. Therefore, identifying the initiation process of autophagy can help us understand its multiple functions.
Mammalian target of rapamycin (mTOR) is a key factor of autophagy initiation. It is widely known that mTOR is composed of two protein complexes mTORC1 and mTORC2, which have different structures and functions and are involved in many physiological processes. Unc-51-like kinase 1 (ULK1) is an indispensable component in autophagy vesicles, which can form the ULK1 complex with autophagy related 13 (Atg13), 200-kDa FAK family kinase interaction protein (FIP200), and Atg101 to induce autophagy (Chen et al., 2014; Hurley and Young, 2017). Under normal circumstances, mTORC1 suppresses autophagy by directly inhibiting the ULK1 complex activity. However, under stress conditions, mTORC1 is phosphorylated leading to the rapid disinhibition of ULK1 and Atg13, which triggers autophagy. In addition, mTORC1 also negatively regulates autophagy by phosphorylating autophagy/Beclin-1 regulator 1 (Ambra1) at Ser52 and 4’,6-diamidino-2-phenylindole (DAP1) at Ser3 and Ser51 (Koren et al., 2010; Cianfanelli and Cecconi, 2015). Conversely, mTORC2 indirectly inhibits autophagy through activation of the AKT/mTORC1 signaling axis. The mechanism involves the AKT-dependent phosphorylation inhibition of tuberous sclerosis complex 1/2 (TSC1/2) which induces Rheb activity that promotes mTORC1 activation (Bernard et al., 2020). Another major mTOR related signaling pathway is the AMP-activated protein kinase (AMPK) -mTOR pathway. The regulation of AMPK is extremely complex. Activation of AMPK phosphorylates TSC2 at Ser792 of mTORC1. Furthermore, PIM2 directly phosphorylates TSC2 at Ser1798 to activate mTORC1 (Lee et al., 2010; Lu et al., 2013). Moreover, AMPK initiates autophagy by directly phosphorylating ULK1 at Ser317 and Ser777 in response to stress (Pagano et al., 2014). In addition, a novel signaling axis AMPK- E3 ligase S-phase kinase-associated protein 2 (SKP2)-co-activator-associated arginine methyltransferase 1 (CARM1), was found to regulate autophagy activation by nutrient starvation (Shin et al., 2016). A recent study indicated that PKCα phosphorylated ULK1 at Ser423, which prevented autosomal formation and inhibited autophagy (Wang et al., 2018a). Atg13, ULK1, and TFEB can be negatively regulated by mTORC1. The secretion of TFEB during stress helped regulate the expression of genes involved in lysosomal biogenesis and lipid catabolism (Settembre et al., 2013).
The processes of autophagy initiation induced by different pathogenic microorganisms are also different. It was shown that Salmonella typhimurium invasion breaks through vesicles and enters the cytoplasm to selectively activate mTOR and degrade AMPK to escape autophagy (Liu et al., 2018). Streptococcus pneumoniae PavA activates the AMPK signaling pathway to induce autophagy in alveolar epithelial cells by inhibiting the mTOR pathway (Kim et al., 2017). In H. pylori infected gastric epithelial cells, inactivated transforming growth factor-β (TGF-β)-activated kinase 1 (TAK1) inhibits AMPK phosphorylation, as well as autophagy and cell survival (Lv et al., 2014).
Formation of Nitroxidative Stress
Nitroxidative stress is a state of physiological imbalance mainly caused by excessive ROS and RNS. Under normal conditions, low levels of ROS/RNS trigger the hosts protective immune response, which is of great significance for anti-infection, anti-inflammatory, and tumor inhibition. However, excessive ROS/RNS directly leads to membrane rupture, DNA peroxidation damage, and cell dysfunction, such as loss of energy metabolism, changes in cell signal transduction, and gene mutation. During some pathogenic microorganisms infection, the host’s antioxidant system and metabolic balance of free radicals becomes disordered, resulting in nitroxidative stress.
ROS components include O2, H2O2 and –OH, and RNS components includes NO, NO2, and ONOO− (Stamler et al., 1992). ROS functions as a signaling molecule in a variety of intracellular processes, which lead to cell proliferation, apoptosis, and defense against microorganisms (Fratelli et al., 2005; Scherz-Shouval et al., 2007). Endogenous ROS is mainly induced by the mitochondrial respiratory chain (Zou et al., 2017). Specific enzymes of the NADPH oxidase (NOX) family and double oxidase family are involved in ROS production. These enzymes catalyze the conversion of intracellular O2 to O2-, and then O2- is converted to H2O2 in the presence of superoxide dismutase. Subsequently, H2O2 reacts with metal ions to form –OH (Si et al., 2015). For example, sodion, a second messenger, interacts with phospholipids and controls oxidase in mitochondria. Furthermore, sodion helps transfer electrons from the substrate to oxygen ions, and subsequently promotes ROS formation and causes oxidative stress (Hernansanz-Agustin et al., 2020). Studies reported that SoxRS and OxyR are the main redox response transcription factors involved in oxidative stress in bacteria. OxyR senses H2O2 and organic peroxide, and SoxRS regulates O2−-mediated oxidative stress. In addition, other stress factors including RpoS and PerR are also involved in regulating oxidative stress responses (Fei et al., 2020). ROS and RNS are produced through different processes including ultraviolet irradiation, metal-catalyzed reactions, electron transport reactions, and inflammation (Valko et al., 2006). Many studies have shown that a variety of pathogens induce nitroxidative stress (Li et al., 2019). The increase of ROS/RNS production was found during microbial infection, or infection with hepatitis C virus, Herpes simplex virus type 1, E. coli, and Salmonella (Molteni et al., 2014; Rhen, 2019). When pathogenic microorganisms infect a host, ROS and RNS can be beneficial and detrimental via anti-inflammatory, anti-pathogen, protective immunity, or cytotoxicity functions (Umezawa et al., 1997). Moderate insulin-like growth factor 1 promoted mitochondrial ROS synthesis and NO synthase gene expression which participated in the elimination of P. falciparum (Drexler et al., 2014). Activation of TLR4 helped clear invading pathogens through the production of NO which was induced by increasing the activity of iNOS via guanosine triphosphate cyclohydrolase (Deng et al., 2015). Inhibition of mitochondrial ROS production impaired the ability of immune cells to kill Salmonella typhimurium in mice (West et al., 2011). In addition, excessive ROS/RNS was closely associated with tissue and organ damage (Deng et al., 2012). For example, excessive ROS triggered apoptosis by inducing the expression of NLRP3, caspase-1, and ASC (Dai et al., 2019).
Antioxidant Systems
Because persistent and excessive nitroxidative stress can lead to host tissue damage, organisms have an antioxidant system that maintains redox homeostasis. There are two types of antioxidant systems: antioxidant enzymes and non-enzymatic antioxidants (Halliwell, 1996). Antioxidants help to mediate peroxidation by catalyzing the formation of active oxygen intermediates which lead to remove free radicals of oxygen and nitrogen. For example, the massive accumulation of ROS promotes the production of superoxide dismutase (SOD), which catalyzes the conversion of superoxide into O2 and H2O2 (Lipinski et al., 2010). Then, catalase scavenges peroxide by catalyzing the conversion of H2O2 into O2 and H2O (He et al., 2017). In addition, the GPx family and Trx system, endogenous antioxidants, also participate in the elimination of nitroxidative stress (Papp et al., 2007; Liu et al., 2014a). Moreover, several signaling pathways, of which nuclear factor erythroid 2-related factor 2 (NRF2) signaling is the most important, are involved in regulating nitroxidative stress. ROS directly oxidizes the cysteine residues on Kelch-like ECH-associated protein 1 (KEAP1) freeing NRF2 from the KEAP1-NRF2 complex. After free NRF2 translocates into the nucleus, it promotes the expression of multiple antioxidant genes by binding to their regulatory regions (Schieber and Chandel, 2014). Furthermore, NO- triggers NRF2 transcription and the expression of ferroportin which activate the antioxidant system and nutrition immunity (Nairz et al., 2013). In turns, inhibiting NRF2 signaling increased ROS levels by suppressing the expression of HO-1 during H. pylori infection (Ko et al., 2016).
Overview of TLR4: An Important Signal Transduction Protein
Studies over the past few decades have shown that the recognition of microbes is based on hosts genes encoding PRRs, which consist of TLRs, RIG-I-like receptors (RLRs), C-type lectin receptors (CLRs), and Nod-like receptors (NLRs) (Janeway, 2013). TLRs are Type I transmembrane proteins that recognize various microbial pathogen-associated molecular patterns (PAMPs). More than a dozen TLRs have been identified to date. Different TLRs can recognize different PAMPs in different cell compartments, including the cell membrane, endosomes, cytoplasm, and endosome (Akira et al., 2006). The correct localization of TLRs is crucial for the regulation of signal transduction. Furthermore, cell type specific signaling downstream of TLRs determines specific innate immune responses (Kawasaki and Kawai, 2014). Generally, TLR signaling is divided into two types: MyD88-dependent and the MyD88-independent signaling pathways. Both pathways activate downstream signaling molecules to promote the production of pro-inflammatory cytokines, chemokines, and type I interferon (IFN) which help remove pathogens (Yamashita et al., 2012). Here, we mainly focus on studies of TLR4 which is also involved in autophagy and nitroxidative stress.
TLR4 Signal Transduction
TLR4 is known for recognizes a broad variety of substances including bacterial lipopolysaccharide (LPS), viral structural protein, fungal mannan, and parasitic glycoinositolphospholipids and has a key role in activating innate immunity (Kawai and Akira, 2011; Mu et al., 2011). For example, when Gram-negative bacteria infect a host, bacterial LPS is directly recognized by TLR4 in the presence of LPS binding protein, CD14, and myeloid differentiation factor 2 (MD-2). Subsequently, TLR4 undergoes oligomerization and recruits downstream adaptor proteins through its Toll-interleukin-1 receptor domains, which initiates signal transduction (Lu et al., 2008). Similar to other TLRs, TLR4 also has two types of signal transduction. After TLR4 is combined with MyD88, phosphorylated IRAK-4 activates the TRAF6-TAK1-NF-κB/MAPK signaling axis which promotes the release of inflammatory factors, such as IL-1β, IL-6, and TNF-α (Monlish et al., 2016; Dajon et al., 2017). Theses cytokines are responsible for inflammation, autophagy and nitroxidative stress. In the MyD88-independent pathways, TRAM is selectively recruited to TLR4 to link TLR4 and TRIF, which induces the production of type I IFN and other proinflammatory cytokines through the activation of IRF3, NF-κB, and MAPK signaling (Kawai and Akira, 2011).
TLR4-Mediated Innate Immune Responses Have Dual Functions
However, when TLR4 is overactivated or its negative regulatory system is obstructed, TLR4 can induce endotoxic shock, autoimmune disease, and even cytokine storm where the immune system attacks the host. The SARS-CoV-2 virus has spread worldwide since 2020, infecting billions of people and killing millions of people (Harrison et al., 2020). Cytokine storm is thought to be an important cause of death in patients with severe and critical COVID-19, which is caused by the SARS-CoV-2 virus (Chen et al., 2020). Compared with a severe influenza group, PBMCs from COVID-19 patients showed a high inflammatory response, and a marked TNF/IL-1β driven inflammatory response. IFN, TNF-α, and IL-1β co-drive inflammatory responses in the monocytes of patients with severe COVID-19, but not in those of patients with mild COVID-19 (Lee et al., 2020). Further research showed that TLR4 is probably involved in recognizing the spike protein of SARS-CoV-2 and then inducing inflammatory responses (Bhattacharya et al., 2020; Choudhury and Mukherjee, 2020). After SARS-CoV-2 virus infects a host, the SARS-CoV-2 spike trimer directly binds to TLR4 and the trigger section of IL-1β. Inhibiting TLR4 completely blocks SARS-CoV-2-induced IL-1β. In contrast, ACE2-deficient or TMPRSS2-inhibition did not affect SARS-CoV-2-induced IL-1β (Zhao et al., 2021). This indicates that if TLR4 signals are not effectively controlled, they can seriously threaten health. Several tightly-regulated mechanisms regulate TLR4 signal transduction to avoid excessive immune response (Table 1) (Liu et al., 2016a).
Regulatory Relationships Among TLR4, Autophagy, and Nitroxidative Stress
In previous studies, researchers mainly investigated the functions of TLR4 with regard to the activation of innate immune responses and production of proinflammatory cytokines during infection. Recently studies have suggested that TLR4 is involved in regulating autophagy and nitroxidative stress during various physiological states (Wang et al., 2016; Wang et al., 2018b; Wang et al., 2020a). Here, we summarize signaling pathways that connect TLR4, autophagy, and nitroxidative stress, and discuss their triangular relationships that affect cellular homeostasis.
Key Signaling Nodes That Link TLR4, Autophagy and Nitroxidative Stress
mTOR Signaling Pathway
Identifying the signaling pathways shared by TLR4, autophagy and nitroxidative stress help us better understand the process of host resistance to pathogens. mTOR is involved in many cellular physiological processes, such as inflammation, energy metabolism, oxidation and autophagy (Zhou et al., 2018). The TLR4-MyD88-MAPK and TLR4/PI3K/Akt signaling pathways affect mTOR activity and influence autophagy (Huang et al., 2020). Generally, nitroxidative stress affects the phosphorylation of AMPK and PI3K signaling, which then influence autophagy (Hinchy et al., 2018; Mistry et al., 2019). Mechanistically, bacteria activate mTORC1 via the upstream TLR4-PI3K-Akt signaling cascade, which inhibits autophagy by suppressing autophagy initiating kinase ULK1 (Shariq et al., 2021). Down regulating TLR4 and MyD88 promoted autophagy by suppressing the phosphorylation of MAPK, mTOR and p65. However, activated TLR4-TRIF and TLR4-MAPK signaling pathways induced autophagy by promoting the dissociation of Beclin 1 and Bcl-2. The release of Beclin 1 from Bcl-2 biased cells toward autophagy (Shi and Kehrl, 2008). Then, mTOR-dependent autophagy precisely regulates downstream NF-κB activation, which leads to the production of nitroxidative stress (Zhou et al., 2018). ROS and RNS signaling were demonstrated to be related to TLR4-dependent NF-κB activation and inflammatory cytokines production (Weigert et al., 2018). The membrane-associated enzyme complex NADPH acts as a key intermediate in regulating the TLR4-mediated production of ROS. Furthermore, the inhibition of mTOR or mTORC1 down-regulated the TLR4-mediated production of ROS and RNS. mTOR-induced the expression of NOX2 and NOX4, which are essential for ROS formation (Sohrabi et al., 2018).
TRAF6 Signaling Pathway
TLR4-MyD88 signaling activates of TRAF6, another important factor that links autophagy and oxidation. TRAF6 promotes mtROS production during LPS stimulation or bacterial infection through the direct ubiquitylation of domains of evolutionarily conserved signaling intermediates in Toll pathways (ECSIT), which is a key mitochondrial respiratory chain assembly factor (West et al., 2011; Wi et al., 2014). Moreover, the inhibition of TRAF6 ubiquitin-ligase activity suppresses autophagy (Min et al., 2018). Mechanistically, activated TRAF6 promoted the K63-linked polyubiquitination of Beclin-1, which is essential for the initiation of autophagy (Lee et al., 2018). Furthermore, activated TRAF6 also induced the stabilization and activation of ULK1, which is the most important factor for the biogenesis of autophagosomes by inducing its Lys63 ubiquitination (Han et al., 2019). In addition, under non-autophagic conditions, mTOR induces the phosphorylation of Ambra1. Then, activated Ambra1 interacts with TRAF6 to mediate the ubiquitination of ULK1 that further inhibits the initiation of autophagy (Nazio et al., 2013). Furthermore, TRAF6 is necessary for the translocation of mTORC1 to lysosomes. The p62/TRAF6 complex induces the activation of mTORC1 via the K63-linked polyubiquitination of mTOR (Linares et al., 2013). These intriguing studies indicate that mTOR and TRAF6 are factors joints that connect TLR4 signaling, autophagy, and nitroxidative stress (Figure 1).
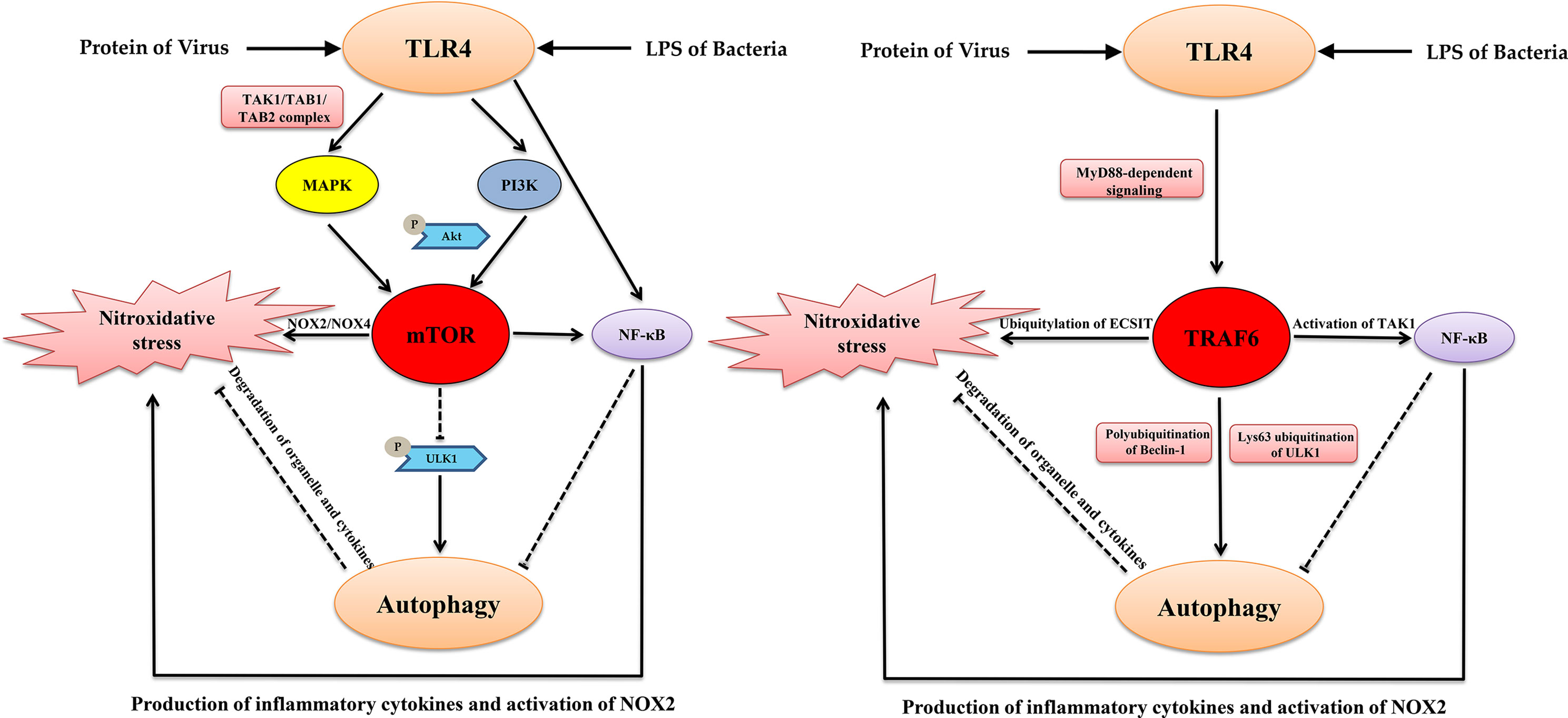
Figure 1 The key signaling nodes that link TLR4, autophagy and nitroxidative stress. TLR4, Autophagy, and Nitroxidative Stress: A Triangular Relationship That Affects Cellular Homeostasis.
TLR4 and Nitroxidative Stress Induce Autophagy
The process of TLR4 activation is always accompanied by autophagy and nitroxidative stress in host immune cells (Figure 2). For example, mycobacterium tuberculosis infection, recognized by TLR4, induced SIRT3-mediated autophagy and nitroxidative stress in mouse BMDM (Kim et al., 2019). E. coli LPS stimulation also induced autophagy and nitroxidative stress via MAPK signaling in macrophages (Wang et al., 2020a). The host recognition of bacteria is the first step for activation of the immune system. Generally, TLR4 recognizes PAMPs and induces the secretion of inflammatory cytokines, including type I IFN, which induces the production of ROS/RNS. Moderate intracellular oxidation is an important method for the host to eliminate pathogenic microorganisms. Evidence suggests that TLR4 interacts directly with NADPH oxidase. Indeed, the TIR-domain of TLR4 interacts directly with the carboxy-terminal region of Nox2/4 under LPS stimulation (Park et al., 2004). Activated NADPH oxidase induces the transmembrane transport of electrons and production of ROS. Furthermore, TLR4 promotes the expression of iNOS via NF-κB signaling. This promotes NO production that suppresses SOD activity and promotes MDA production, which aggravates nitroxidative stress (Li et al., 2019). In addition, TLR4-mediated pro-inflammatory cytokines, such as TNF-α and IL-1β, and also induces ROS (Sasaki et al., 2008). Moreover, inhibiting the expression of TLR4 significantly decreased the level of autophagy (Kandadi et al., 2012). The excessive accumulation of ROS/RNS is harmful to homeostasis and can cause body oxidative damage in the host. It is widely thought that ROS/RNS induces autophagy, which maintains oxidation intermediates at a low level. ROS directly phosphorylated the p65 subunit of NF-κB at Ser-536, which activated the autophagy receptor P62 (Song et al., 2017). In addition, many oxidation intermediates promote the transcription of BNIP3 and NIX, which dissociate Beclin-1from Bcl2 to induce autophagy (Mahalingaiah and Singh, 2014; Xu et al., 2020). Moreover, H2O2 mediated the binding of AMPK and GSH which helped phosphorylate the ULK1 complex leading to autophagy (Filomeni et al., 2015). Furthermore, excessive RNS and ROS cause DNA damage (Wiseman and Halliwell, 1996). Upon DNA damage, Ataxia telangiectasia mutated promoted AMPK phosphorylation and the p53-dependent expression of autophagic genes, including ATG4, ULK1 and UVRAG (Hurley and Bunz, 2007; Fullgrabe et al., 2014). Nrf2 sensed cellular ROS or RNS, and then activated AMPK, which suppressed mTOR signaling leading to autophagy (Kapuy et al., 2018). In addition, the inhibition of ROS by N-acetyl-l-cysteine abolished autophagic flux in porcine trophectoderm cells and ROS production activated MAPK and PI3K/Akt pathways, which were involved in this process (Luo et al., 2019). As mentioned above, TLR4 regulated autophagy via the TLR4-MyD88-MAPK/NF-κB and TLR4/PI3K/Akt/mTOR signaling pathways. However, whether TLR4 activation has an active role in the induction of autophagy remains controversial. Most studies have shown that LPS or infection can induce autophagy via TLR4 signaling, and that the knock-down/knock-out of TLR4 down-regulates autophagy (Chen et al., 2015; Zhao et al., 2016; Wang et al., 2020a). However, a study reported that the activation of TLR4 by bacterial LPS inhibited autophagy through the MyD88-TAK1-MAPK-dependent phosphorylation of mTOR (Zhou et al., 2018). Another recent study showed that the intraperitoneal injection of LPS promoted neuroinflammation by activating TLR4, inhibiting autophagic markers, and inducing excessive oxidation intermediates (Jamali-Raeufy et al., 2021a). We hypothesize that this opposite result is related to the different concentrations and duration of stimulation or different types of cells used. However, there is no doubt that TLR4 is involved in regulating autophagy.
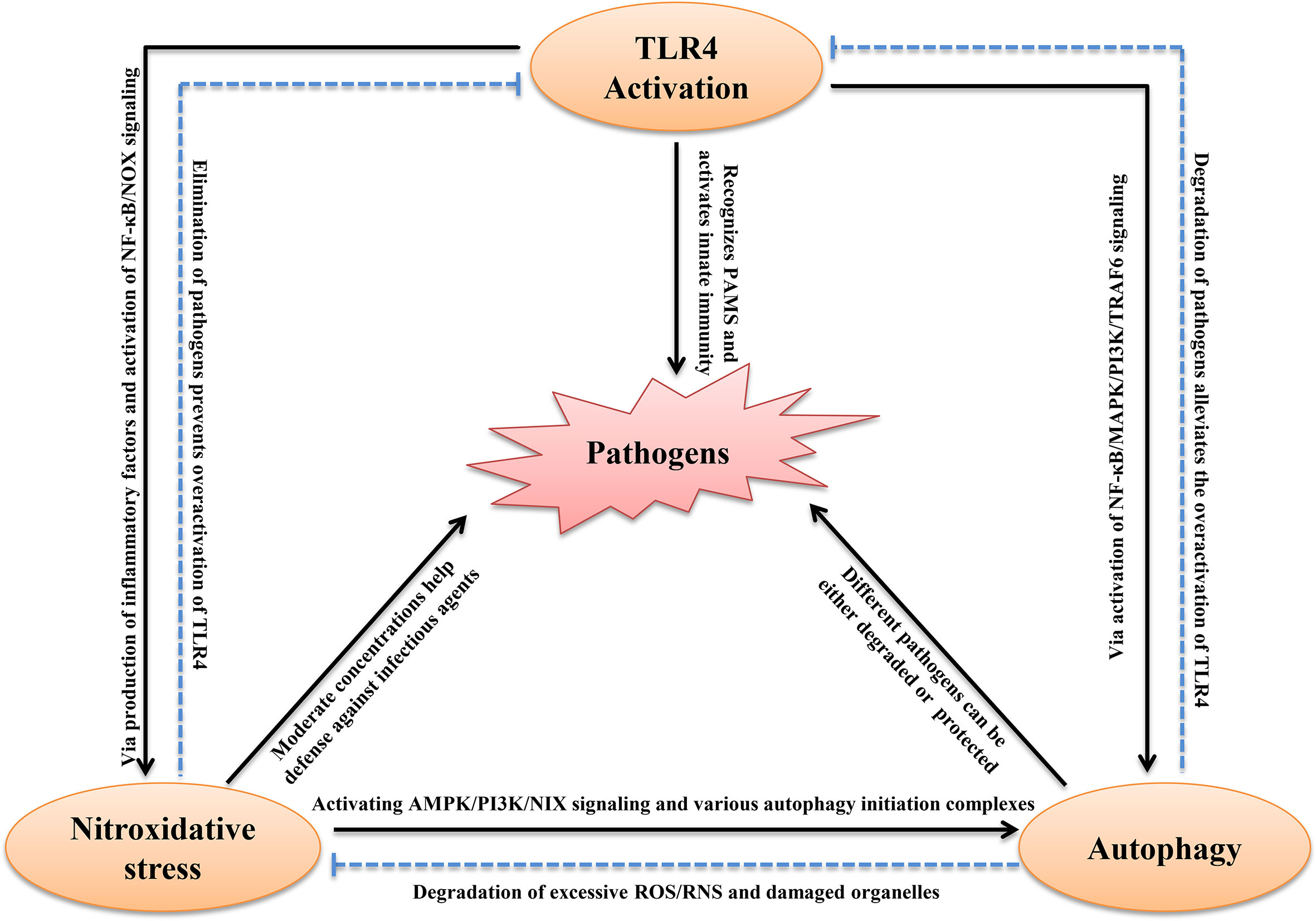
Figure 2 Schematic diagram of the interplay between TLR4, autophagy and nitroxidative stress in infectious Diseases. See text for explanation.
Autophagy Regulates Reactive Intermediates Production and TLR4 Activity
A recent study reported autophagy also regulated nitroxidative stress. Autophagy often helps remove damaged organelles and excess metabolic intermediates to maintain redox homeostasis in all types of cells (Kroemer et al., 2010). Autophagic dysfunction results in mitochondrial damage and the accumulation of oxidative intermediates, which promote the production of proinflammatory cytokines that can lead to ROS-mediated cell death (Larabi et al., 2020). Deletions of autophagy-related genes, such as ATG5 and ATG16 promoted high cellular ROS levels (Asano et al., 2017; Saxena et al., 2018). Moreover, nitroxidation intermediates have dual roles in the regulation of autophagy. NO activates autophagy via mTOR activity. Furthermore, NO accumulation triggered nitrosative stress, which promoted ATM/LKB1/AMPK/TSC2 signaling cascades that initiated autophagy via the inhibition of mTORC1 (Tripathi et al., 2013). NOS, another nitroxidation intermediate, interacted with PINK1 to activate selective autophagy by triggering the translocation of Parkin (Han et al., 2015). To date, no studies have shown that autophagy directly eliminates RNS. However, evidence has shown that autophagy removes substances that induce RNS production. For example, autophagy eliminated damaged mitochondria and inflammatory cytokines to reduce RNS accumulation (Shi et al., 2012; Kaminskyy and Zhivotovsky, 2014). Therefore, autophagy can be considered a non-canonical antioxidant system that helps to degrade excessive ROS/RNS. The intracellular redox state can affect the fate of cells, and the timely removal of oxidizing molecules is beneficial to the maintenance of homeostasis. Following the removal of pathogenic microorganisms by autophagy and nitroxidative stress, the activation of TLR4 is alleviated. During infection by pathogenic microorganisms, this physiological homeostasis helps to eliminate pathogens or helps pathogens survive. Conversely, if this homeostasis is disrupted, the host will be seriously challenged. The above studies have identified extensive crosstalk between TLR4, autophagy and nitroxidative stress (Figure 2).
Concluding Remarks
As a pattern recognition receptor, TLR4 is involved in mediating pathogen-inducing inflammation. Moderate inflammatory responses help fight against pathogenic infections, but the excessive activation of inflammatory responses can trigger cytokine storm, which causes the immune system to attack the host leading to multiple organ failure and even death. Autophagy and nitroxidative stress are involved in the regulation of inflammation, and TLR4, autophagy, and nitroxidative stress have vital roles during pathogenic microorganism-induced host immune responses. Following the pathways of autophagy and nitroxidative stress and the signaling transduction of TLR4 has indicated a strong connection between them. Studying the detailed regulatory relationships between TLR4, autophagy and nitroxidative stress might help enhance our understanding of the interactions between pathogenic microorganisms and hosts. The activation of TLR4 always contributes to autophagy and the production of reactive intermediates including ROS/RNS. mTOR and TRAF6 are key factors that connect TLR4 signaling, autophagy and nitroxidative stress. Furthermore, promoting mTOR and TRAF6-dependent autophagy and nitroxidative stress via upstream TLR4-MyD88-NF-κB/MAPK and TLR4-TRAF6 signaling pathways, and downstream NOX, AMPK signaling and inflammatory cytokines regulates the elimination of pathogens and maintenance of body health. Furthermore, nitroxidative stress can induce autophagy through the activation of NF-κB, AMPK, and Nrf2 pathways. Then, autophagy helps to degrade excessive ROS/RNS to balance the intracellular redox state.
An important question is how to harness the links between TLR4, autophagy, and nitroxidative stress to control pathogenic microorganism infections. Many natural substances and small molecular compounds were reported to control autophagy and oxidation by regulating TLR4 activity (Jamali-Raeufy et al., 2021b). Of note, TLR4 activity, autophagy and oxidative stress all affect the survival of pathogenic microorganisms. According to the above ideas, we should identify targets from the signal pathways or key node proteins involved in TLR4/autophagy/nitroxidative stress simultaneously. Among them, mTOR, TRAF, NF-κB and their upstream and downstream molecules are worth investigating. In addition, we should choose different treatment strategies according to the types of pathogenic microorganisms and the characteristics of the infected organs. Finally, there are some outstanding questions: What are the differences between different TLR4 downstream signaling-mediated autophagy and nitroxidative stress pathways? Are TLR activation, autophagy and nitroxidative stress caused by infection and non-infectious diseases different? What are the specific molecular mechanisms and processes of the autophagic degradation of ROS/RNS? What are the detailed mechanisms of the activation of autophagy by endogenously produced RNS? Does excessive autophagy help to eliminate the ROS/RNS? If so, what is the relationship between excessive autophagy-mediated cell death and low ROS/RNS levels? These questions need to be answered in future studies.
Author Contributions
SW, KZ, and SD conceived the manuscript. SW, KZ, and QH wrote the manuscript. SW, KZ, QH and YY prepared the Figure. SW and QH revised the manuscript. Funding acquisition was by SW and JL. All authors contributed to the article and approved the submitted version.
Funding
The authors thank the following funding sources: National Natural Science Foundation of China (32002153, 32002298), Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science (R2019YJ-YB2004, R2019YJ-YB2005), Science and Technology Planning Project of Guangzhou (202102020177, 202102020385).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank J. Ludovic Croxford, PhD, from Liwen Bianji for editing the language of a draft of this manuscript.
References
Akira, S., Uematsu, S., Takeuchi, O. (2006). Pathogen Recognition and Innate Immunity. Cell 124 (4), 783–801. doi: 10.1016/j.cell.2006.02.015
Asano, J., Sato, T., Ichinose, S., Kajita, M., Onai, N., Shimizu, S., et al. (2017). Intrinsic Autophagy Is Required for the Maintenance of Intestinal Stem Cells and for Irradiation-Induced Intestinal Regeneration. Cell Rep. 20 (5), 1050–1060. doi: 10.1016/j.celrep.2017.07.019
Bernard, M., Yang, B., Migneault, F., Turgeon, J., Dieude, M., Olivier, M. A., et al. (2020). Autophagy Drives Fibroblast Senescence Through MTORC2 Regulation. Autophagy 16 (11), 2004–2016. doi: 10.1080/15548627.2020.1713640
Bhattacharya, M., Sharma, A. R., Mallick, B., Sharma, G., Lee, S. S., Chakraborty, C. (2020). Immunoinformatics Approach to Understand Molecular Interaction Between Multi-Epitopic Regions of SARS-Cov-2 Spike-Protein With TLR4/MD-2 Complex. Infect. Genet. Evol. 85:104587. doi: 10.1016/j.meegid.2020.104587
Cekic, C., Casella, C. R., Sag, D., Antignano, F., Kolb, J., Suttles, J., et al. (2011). Myd88-Dependent SHIP1 Regulates Proinflammatory Signaling Pathways in Dendritic Cells After Monophosphoryl Lipid a Stimulation of TLR4. J. Immunol. 186 (7), 3858–3865. doi: 10.4049/jimmunol.1001034
Chassin, C., Kocur, M., Pott, J., Duerr, C. U., Gutle, D., Lotz, M., et al. (2010). Mir-146a Mediates Protective Innate Immune Tolerance in the Neonate Intestine. Cell Host Microbe 8 (4), 358–368. doi: 10.1016/j.chom.2010.09.005
Chen, Y., He, J., Tian, M., Zhang, S. Y., Guo, M. R., Kasimu, R., et al. (2014). UNC51-Like Kinase 1, Autophagic Regulator and Cancer Therapeutic Target. Cell Prolif. 47 (6), 494–505. doi: 10.1111/cpr.12145
Chen, G., Wu, D., Guo, W., Cao, Y., Huang, D., Wang, H., et al. (2020). Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J. Clin. Invest. 130 (5), 2620–2629. doi: 10.1172/JCI137244
Chen, X. F., Wu, J., Zhang, Y. D., Zhang, C. X., Chen, X. T., Sun, J. H., et al. (2018). Role of Zc3h12a in Enhanced IL-6 Production by Newborn Mononuclear Cells in Response to Lipopolysaccharide. Pediatr. Neonatol. 59 (3), 288–295. doi: 10.1016/j.pedneo.2017.09.006
Chen, S., Yuan, J., Yao, S., Jin, Y., Chen, G., Tian, W., et al. (2015). Lipopolysaccharides may Aggravate Apoptosis Through Accumulation of Autophagosomes in Alveolar Macrophages of Human Silicosis. Autophagy 11 (12), 2346–2357. doi: 10.1080/15548627.2015.1109765
Choudhury, A., Mukherjee, S. (2020). In Silico Studies on the Comparative Characterization of the Interactions of SARS-Cov-2 Spike Glycoprotein With ACE-2 Receptor Homologs and Human Tlrs. J. Med. Virol. 92 (10), 2105–2113. doi: 10.1002/jmv.25987
Cianfanelli, V., Cecconi, F. (2015). AMBRA1: When Autophagy Meets Cell Proliferation. Autophagy 11 (9), 1705–1707. doi: 10.1080/15548627.2015.1053681
Dai, J., Chen, H., Chai, Y. (2019). Advanced Glycation End Products (Ages) Induce Apoptosis of Fibroblasts by Activation of NLRP3 Inflammasome via Reactive Oxygen Species (ROS) Signaling Pathway. Med. Sci. Monit. 25, 7499–7508. doi: 10.12659/MSM.915806
Dajon, M., Iribarren, K., Cremer, I. (2017). Toll-Like Receptor Stimulation in Cancer: A Pro- and Anti-Tumor Double-Edged Sword. Immunobiology 222 (1), 89–100. doi: 10.1016/j.imbio.2016.06.009
Das, A., Kim, S. H., Arifuzzaman, S., Yoon, T., Chai, J. C., Lee, Y. S., et al. (2016). Transcriptome Sequencing Reveals That LPS-Triggered Transcriptional Responses in Established Microglia BV2 Cell Lines are Poorly Representative of Primary Microglia. J. Neuroinflamm. 13 (1), 182. doi: 10.1186/s12974-016-0644-1
Das, S., Pandey, K., Kumar, A., Sardar, A. H., Purkait, B., Kumar, M., et al. (2012). TGF-Beta1 Re-Programs TLR4 Signaling in L. Donovani Infection: Enhancement of SHP-1 and Ubiquitin-Editing Enzyme A20. Immunol. Cell Biol. 90 (6), 640–654. doi: 10.1038/icb.2011.80
Deng, J. S., Jiang, W. P., Chen, C. C., Lee, L. Y., Li, P. Y., Huang, W. C., et al. (2020). Cordyceps Cicadae Mycelia Ameliorate Cisplatin-Induced Acute Kidney Injury by Suppressing the TLR4/NF-Kappab/MAPK and Activating the HO-1/Nrf2 and Sirt-1/AMPK Pathways in Mice. Oxid. Med. Cell Longev. 2020, 7912763. doi: 10.1155/2020/7912763
Deng, S., Wu, Q., Yu, K., Zhang, Y., Yao, Y., Li, W., et al. (2012). Changes in the Relative Inflammatory Responses in Sheep Cells Overexpressing of Toll-Like Receptor 4 When Stimulated With LPS. PLoS One 7 (10), e47118. doi: 10.1371/journal.pone.0047118
Deng, S., Yu, K., Zhang, B., Yao, Y., Wang, Z., Zhang, J., et al. (2015). Toll-Like Receptor 4 Promotes NO Synthesis by Upregulating GCHI Expression Under Oxidative Stress Conditions in Sheep Monocytes/Macrophages. Oxid. Med. Cell Longev 2015, 359315. doi: 10.1155/2015/359315
Di Meo, S., Venditti, P. (2020). Evolution of the Knowledge of Free Radicals and Other Oxidants. Oxid. Med. Cell Longev. 2020, 9829176. doi: 10.1155/2020/9829176
Drexler, A. L., Pietri, J. E., Pakpour, N., Hauck, E., Wang, B., Glennon, E. K., et al. (2014). Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and Plasmodium Falciparum Resistance in Anopheles Stephensi. PLoS Pathog. 10 (6), e1004231. doi: 10.1371/journal.ppat.1004231
Ermolaeva, M. A., Michallet, M. C., Papadopoulou, N., Utermohlen, O., Kranidioti, K., Kollias, G., et al. (2008). Function of TRADD in Tumor Necrosis Factor Receptor 1 Signaling and in TRIF-Dependent Inflammatory Responses. Nat. Immunol. 9 (9), 1037–1046. doi: 10.1038/ni.1638
Fei, Y. Y., Bhat, J. A., Gai, J. Y., Zhao, T. J. (2020). Global Transcriptome Profiling of Enterobacter Strain NRS-1 in Response to Hydrogen Peroxide Stress Treatment. Appl. Biochem. Biotechnol. 191 (4), 1638–1652. doi: 10.1007/s12010-020-03313-x
Filomeni, G., De Zio, D., Cecconi, F. (2015). Oxidative Stress and Autophagy: The Clash Between Damage and Metabolic Needs. Cell Death Differ. 22 (3), 377–388. doi: 10.1038/cdd.2014.150
Fratelli, M., Goodwin, L. O., Orom, U. A., Lombardi, S., Tonelli, R., Mengozzi, M., et al. (2005). Gene Expression Profiling Reveals a Signaling Role of Glutathione in Redox Regulation. Proc. Natl. Acad. Sci. U. S. A. 102 (39), 13998–14003. doi: 10.1073/pnas.0504398102
Fullgrabe, J., Klionsky, D. J., Joseph, B. (2014). The Return of the Nucleus: Transcriptional and Epigenetic Control of Autophagy. Nat. Rev. Mol. Cell Biol. 15 (1), 65–74. doi: 10.1038/nrm3716
Halliwell, B. (1996). Antioxidants in Human Health and Disease. Annu. Rev. Nutr. 16, 33–50. doi: 10.1146/annurev.nu.16.070196.000341
Han, J. Y., Kang, M. J., Kim, K. H., Han, P. L., Kim, H. S., Ha, J. Y., et al. (2015). Nitric Oxide Induction of Parkin Translocation in PTEN-Induced Putative Kinase 1 (PINK1) Deficiency: Functional Role of Neuronal Nitric Oxide Synthase During Mitophagy. J. Biol. Chem. 290 (16), 10325–10335. doi: 10.1074/jbc.M114.624767
Han, S. H., Korm, S., Han, Y. G., Choi, S. Y., Kim, S. H., Chung, H. J., et al. (2019). GCA Links TRAF6-ULK1-Dependent Autophagy Activation in Resistant Chronic Myeloid Leukemia. Autophagy 15 (12), 2076–2090. doi: 10.1080/15548627.2019.1596492
Harrison, A. G., Lin, T., Wang, P. (2020). Mechanisms of SARS-Cov-2 Transmission and Pathogenesis. Trends Immunol. 41 (12), 1100–1115. doi: 10.1016/j.it.2020.10.004
He, L., He, T., Farrar, S., Ji, L., Liu, T., Ma, X. (2017). Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell Physiol. Biochem. 44 (2), 532–553. doi: 10.1159/000485089
Hernansanz-Agustin, P., Choya-Foces, C., Carregal-Romero, S., Ramos, E., Oliva, T., Villa-Pina, T., et al. (2020). Na(+) Controls Hypoxic Signalling by the Mitochondrial Respiratory Chain. Nature 586 (7828), 287–291. doi: 10.1038/s41586-020-2551-y
Hinchy, E. C., Gruszczyk, A. V., Willows, R., Navaratnam, N., Hall, A. R., Bates, G., et al. (2018). Mitochondria-Derived ROS Activate AMP-Activated Protein Kinase (AMPK) Indirectly. J. Biol. Chem. 293 (44), 17208–17217. doi: 10.1074/jbc.RA118.002579
Huang, C. Y., Deng, J. S., Huang, W. C., Jiang, W. P., Huang, G. J. (2020). Attenuation of Lipopolysaccharide-Induced Acute Lung Injury by Hispolon in Mice, Through Regulating the TLR4/PI3K/Akt/Mtor and Keap1/Nrf2/HO-1 Pathways, and Suppressing Oxidative Stress-Mediated ER Stress-Induced Apoptosis and Autophagy Nutrients 12 (6), 1742. doi: 10.3390/nu12061742
Hurley, P. J., Bunz, F. (2007). ATM and ATR: Components of an Integrated Circuit. Cell Cycle 6 (4), 414–417. doi: 10.4161/cc.6.4.3886
Hurley, J. H., Young, L. N. (2017). Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 86, 225–244. doi: 10.1146/annurev-biochem-061516-044820
Islam, M. T. (2017). Oxidative Stress and Mitochondrial Dysfunction-Linked Neurodegenerative Disorders. Neurol. Res. 39 (1), 73–82. doi: 10.1080/01616412.2016.1251711
Jamali-Raeufy, N., Alizadeh, F., Mehrabi, Z., Mehrabi, S., Goudarzi, M. (2021a). Acetyl-L-Carnitine Confers Neuroprotection Against Lipopolysaccharide (LPS) -Induced Neuroinflammation by Targeting TLR4/Nfkappab, Autophagy, Inflammation and Oxidative Stress. Metab. Brain Dis. 36 (6), 1391–1401. doi: 10.1007/s11011-021-00715-6
Jamali-Raeufy, N., Alizadeh, F., Mehrabi, Z., Mehrabi, S., Goudarzi, M. (2021b). Correction to: Acetyl-L-Carnitine Confers Neuroprotection Against Lipopolysaccharide (LPS) -Induced Neuroinflammation by Targeting TLR4/Nfkappab, Autophagy, Inflammation and Oxidative Stress. Metab. Brain Dis. 36 (6), 1403. doi: 10.1007/s11011-021-00721-8
Janeway, C. A., Jr. (2013). Pillars Article: Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harb. Symp. Quant. Biol. 54 1989, 1–13. doi: 10.1101/SQB.1989.054.01.003
Kaminskyy, V. O., Zhivotovsky, B. (2014). Free Radicals in Cross Talk Between Autophagy and Apoptosis. Antioxid Redox Signal 21 (1), 86–102. doi: 10.1089/ars.2013.5746
Kandadi, M. R., Frankel, A. E., Ren, J. (2012). Toll-Like Receptor 4 Knockout Protects Against Anthrax Lethal Toxin-Induced Cardiac Contractile Dysfunction: Role of Autophagy. Br. J. Pharmacol. 167 (3), 612–626. doi: 10.1111/j.1476-5381.2012.02040.x
Kapuy, O., Papp, D., Vellai, T., Banhegyi, G., Korcsmaros, T. (2018). Systems-Level Feedbacks of NRF2 Controlling Autophagy Upon Oxidative Stress Response. Antioxidants (Basel) 7 (3), 39. doi: 10.3390/antiox7030039
Kawai, T., Akira, S. (2011). Toll-Like Receptors and Their Crosstalk With Other Innate Receptors in Infection and Immunity. Immunity 34 (5), 637–650. doi: 10.1016/j.immuni.2011.05.006
Kawasaki, T., Kawai, T. (2014). Toll-Like Receptor Signaling Pathways. Front. Immunol. 5, 461. doi: 10.3389/fimmu.2014.00461
Kim, T. S., Jin, Y. B., Kim, Y. S., Kim, S., Kim, J. K., Lee, H. M., et al. (2019). SIRT3 Promotes Antimycobacterial Defenses by Coordinating Mitochondrial and Autophagic Functions. Autophagy 15 (8), 1356–1375. doi: 10.1080/15548627.2019.1582743
Kim, J., Lee, H. W., Rhee, D. K., Paton, J. C., Pyo, S. (2017). Pneumolysin-Induced Autophagy Contributes to Inhibition of Osteoblast Differentiation Through Downregulation of Sp1 in Human Osteosarcoma Cells. Biochim. Biophys. Acta Gen. Subj. 1861 (11 Pt A), 2663–2673. doi: 10.1016/j.bbagen.2017.07.008
Kim, E. Y., Shin, H. Y., Kim, J. Y., Kim, D. G., Choi, Y. M., Kwon, H. K., et al. (2010). ATF3 Plays a Key Role in Kdo2-Lipid a-Induced TLR4-Dependent Gene Expression via NF-Kappab Activation. PLoS One 5 (12), e14181. doi: 10.1371/journal.pone.0014181
Koren, I., Reem, E., Kimchi, A. (2010). Autophagy Gets a Brake: DAP1, a Novel Mtor Substrate, is Activated to Suppress the Autophagic Process. Autophagy 6 (8), 1179–1180. doi: 10.4161/auto.6.8.13338
Ko, S. H., Rho, D. J., Jeon, J. I., Kim, Y. J., Woo, H. A., Kim, N., et al. (2016). Crude Preparations of Helicobacter Pylori Outer Membrane Vesicles Induce Upregulation of Heme Oxygenase-1 via Activating Akt-Nrf2 and Mtor-Ikappab Kinase-NF-Kappab Pathways in Dendritic Cells. Infect. Immun. 84 (8), 2162–2174. doi: 10.1128/IAI.00190-16
Kroemer, G., Marino, G., Levine, B. (2010). Autophagy and the Integrated Stress Response. Mol. Cell 40 (2), 280–293. doi: 10.1016/j.molcel.2010.09.023
Larabi, A., Barnich, N., Nguyen, H. T. T. (2020). New Insights Into the Interplay Between Autophagy, Gut Microbiota and Inflammatory Responses in IBD. Autophagy 16 (1), 38–51. doi: 10.1080/15548627.2019.1635384
Lavandero, S., Chiong, M., Rothermel, B. A., Hill, J. A. (2015). Autophagy in Cardiovascular Biology. J. Clin. Invest. 125 (1), 55–64. doi: 10.1172/JCI73943
Lee, N. R., Ban, J., Lee, N. J., Yi, C. M., Choi, J. Y., Kim, H., et al. (2018). Activation of RIG-I-Mediated Antiviral Signaling Triggers Autophagy Through the MAVS-TRAF6-Beclin-1 Signaling Axis. Front. Immunol. 9, 2096. doi: 10.3389/fimmu.2018.02096
Lee, J. S., Park, S., Jeong, H. W., Ahn, J. Y., Choi, S. J., Lee, H., et al. (2020). Immunophenotyping of COVID-19 and Influenza Highlights the Role of Type I Interferons in Development of Severe COVID-19. Sci. Immunol. 5 (49), eabd1554. doi: 10.1126/sciimmunol.abd1554
Lee, J. W., Park, S., Takahashi, Y., Wang, H. G. (2010). The Association of AMPK With ULK1 Regulates Autophagy. PLoS One 5 (11), e15394. doi: 10.1371/journal.pone.0015394
Levine, B., Kroemer, G. (2019). Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176 (1-2), 11–42. doi: 10.1016/j.cell.2018.09.048
Liang, Y., Zeng, J., Luo, B., Li, W., He, Y., Zhao, W., et al. (2020). TET2 Promotes IL-1beta Expression in J774.1 Cell Through TLR4/MAPK Signaling Pathway With Demethylation of TAB2 Promoter. Mol. Immunol. 126, 136–142. doi: 10.1016/j.molimm.2020.08.003
Li, Y., Deng, S. L., Lian, Z. X., Yu, K. (2019). Roles of Toll-Like Receptors in Nitroxidative Stress in Mammals. Cells 8 (6), 576. doi: 10.3390/cells8060576
Linares, J. F., Duran, A., Yajima, T., Pasparakis, M., Moscat, J., Diaz-Meco, M. T. (2013). K63 Polyubiquitination and Activation of Mtor by the P62-TRAF6 Complex in Nutrient-Activated Cells. Mol. Cell 51 (3), 283–296. doi: 10.1016/j.molcel.2013.06.020
Lipinski, M. M., Hoffman, G., Ng, A., Zhou, W., Py, B. F., Hsu, E., et al. (2010). A Genome-Wide Sirna Screen Reveals Multiple Mtorc1 Independent Signaling Pathways Regulating Autophagy Under Normal Nutritional Conditions. Dev. Cell 18 (6), 1041–1052. doi: 10.1016/j.devcel.2010.05.005
Liu, J., Han, C., Xie, B., Wu, Y., Liu, S., Chen, K., et al. (2014b). Rhbdd3 Controls Autoimmunity by Suppressing the Production of IL-6 by Dendritic Cells via K27-Linked Ubiquitination of the Regulator NEMO. Nat. Immunol. 15 (7), 612–622. doi: 10.1038/ni.2898
Liu, W., Jiang, Y., Sun, J., Geng, S., Pan, Z., Prinz, R. A., et al. (2018). Activation of TGF-Beta-Activated Kinase 1 (TAK1) Restricts Salmonella Typhimurium Growth by Inducing AMPK Activation and Autophagy. Cell Death Dis. 9 (5), 570. doi: 10.1038/s41419-018-0612-z
Liu, J., Qian, C., Cao, X. (2016a). Post-Translational Modification Control of Innate Immunity. Immunity 45 (1), 15–30. doi: 10.1016/j.immuni.2016.06.020
Liu, J. N., Suh, D. H., Trinh, H. K., Chwae, Y. J., Park, H. S., Shin, Y. S. (2016b). The Role of Autophagy in Allergic Inflammation: A New Target for Severe Asthma. Exp. Mol. Med. 48 (7), e243. doi: 10.1038/emm.2016.38
Liu, G. Q., Wu, Z. Y., Yang, H. S., Zheng, J. L., Lin, Z. (2003). The Inhibition Effect of Interleukin-1 Receptor Antagonist on Interleukin-1 Alpha-Induced Intercellular Adhesion Molecule-1 Expression on Orbital Fibroblasts and Adhesion of Peripheral Blood Mononuclear Cells. Zhonghua Yan Ke Za Zhi 39 (9), 528–532. doi: 10.3760/cma.j.issn.0412-4081.2003.09.106
Liu, H., Zhang, J., Zhang, S., Yang, F., Thacker, P. A., Zhang, G., et al. (2014a). Oral Administration of Lactobacillus Fermentum I5007 Favors Intestinal Development and Alters the Intestinal Microbiota in Formula-Fed Piglets. J. Agric. Food Chem. 62 (4), 860–866. doi: 10.1021/jf403288r
Lu, L. H., Chao, C. H., Yeh, T. M. (2019). Inhibition of Autophagy Protects Against Sepsis by Concurrently Attenuating the Cytokine Storm and Vascular Leakage. J. Infect. 78 (3), 178–186. doi: 10.1016/j.jinf.2018.12.003
Luo, Z., Xu, X., Sho, T., Zhang, J., Xu, W., Yao, J., et al. (2019). ROS-Induced Autophagy Regulates Porcine Trophectoderm Cell Apoptosis, Proliferation, and Differentiation. Am. J. Physiol. Cell Physiol. 316 (2), C198–C209. doi: 10.1152/ajpcell.00256.2018
Lu, Y. C., Yeh, W. C., Ohashi, P. S. (2008). LPS/TLR4 Signal Transduction Pathway. Cytokine 42 (2), 145–151. doi: 10.1016/j.cyto.2008.01.006
Lu, J., Zavorotinskaya, T., Dai, Y., Niu, X. H., Castillo, J., Sim, J., et al. (2013). Pim2 is Required for Maintaining Multiple Myeloma Cell Growth Through Modulating TSC2 Phosphorylation. Blood 122 (9), 1610–1620. doi: 10.1182/blood-2013-01-481457
Lv, G., Zhu, H., Zhou, F., Lin, Z., Lin, G., Li, C. (2014). AMP-Activated Protein Kinase Activation Protects Gastric Epithelial Cells From Helicobacter Pylori-Induced Apoptosis. Biochem. Biophys. Res. Commun. 453 (1), 13–18. doi: 10.1016/j.bbrc.2014.09.028
Mahalingaiah, P. K., Singh, K. P. (2014). Chronic Oxidative Stress Increases Growth and Tumorigenic Potential of MCF-7 Breast Cancer Cells. PLoS One 9 (1), e87371. doi: 10.1371/journal.pone.0087371
Mansell, A., Smith, R., Doyle, S. L., Gray, P., Fenner, J. E., Crack, P. J., et al. (2006). Suppressor of Cytokine Signaling 1 Negatively Regulates Toll-Like Receptor Signaling by Mediating Mal Degradation. Nat. Immunol. 7 (2), 148–155. doi: 10.1038/ni1299
Min, Y., Kim, M. J., Lee, S., Chun, E., Lee, K. Y. (2018). Inhibition of TRAF6 Ubiquitin-Ligase Activity by PRDX1 Leads to Inhibition of NFKB Activation and Autophagy Activation. Autophagy 14 (8), 1347–1358. doi: 10.1080/15548627.2018.1474995
Mistry, J. J., Marlein, C. R., Moore, J. A., Hellmich, C., Wojtowicz, E. E., Smith, J. G. W., et al. (2019). ROS-Mediated PI3K Activation Drives Mitochondrial Transfer From Stromal Cells to Hematopoietic Stem Cells in Response to Infection. Proc. Natl. Acad. Sci. U. S. A. 116 (49), 24610–24619. doi: 10.1073/pnas.1913278116
Molteni, C. G., Principi, N., Esposito, S. (2014). Reactive Oxygen and Nitrogen Species During Viral Infections. Free Radic. Res. 48 (10), 1163–1169. doi: 10.3109/10715762.2014.945443
Monlish, D. A., Bhatt, S. T., Schuettpelz, L. G. (2016). The Role of Toll-Like Receptors in Hematopoietic Malignancies. Front. Immunol. 7, 390. doi: 10.3389/fimmu.2016.00390
Mrityunjaya, M., Pavithra, V., Neelam, R., Janhavi, P., Halami, P. M., Ravindra, P. V. (2020). Immune-Boosting, Antioxidant and Anti-Inflammatory Food Supplements Targeting Pathogenesis of COVID-19. Front. Immunol. 11, 570122. doi: 10.3389/fimmu.2020.570122
Mu, H. H., Hasebe, A., Van Schelt, A., Cole, B. C. (2011). Novel Interactions of a Microbial Superantigen With TLR2 and TLR4 Differentially Regulate IL-17 and Th17-Associated Cytokines. Cell Microbiol. 13 (3), 374–387. doi: 10.1111/j.1462-5822.2010.01540.x
Nairz, M., Schleicher, U., Schroll, A., Sonnweber, T., Theurl, I., Ludwiczek, S., et al. (2013). Nitric Oxide-Mediated Regulation of Ferroportin-1 Controls Macrophage Iron Homeostasis and Immune Function in Salmonella Infection. J. Exp. Med. 210 (5), 855–873. doi: 10.1084/jem.20121946
Nakatogawa, H., Ohbayashi, S., Sakoh-Nakatogawa, M., Kakuta, S., Suzuki, S. W., Kirisako, H., et al. (2012). The Autophagy-Related Protein Kinase Atg1 Interacts With the Ubiquitin-Like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation. J. Biol. Chem. 287 (34), 28503–28507. doi: 10.1074/jbc.C112.387514
Nazio, F., Strappazzon, F., Antonioli, M., Bielli, P., Cianfanelli, V., Bordi, M., et al. (2013). Mtor Inhibits Autophagy by Controlling ULK1 Ubiquitylation, Self-Association and Function Through AMBRA1 and TRAF6. Nat. Cell Biol. 15 (4), 406–416. doi: 10.1038/ncb2708
Nozawa, T., Aikawa, C., Minowa-Nozawa, A., Nakagawa, I. (2017). The Intracellular Microbial Sensor NLRP4 Directs Rho-Actin Signaling to Facilitate Group a Streptococcus-Containing Autophagosome-Like Vacuole Formation. Autophagy 13 (11), 1841–1854. doi: 10.1080/15548627.2017.1358343
O’Neill, L. A., Bowie, A. G. (2007). The Family of Five: TIR-Domain-Containing Adaptors in Toll-Like Receptor Signalling. Nat. Rev. Immunol. 7 (5), 353–364. doi: 10.1038/nri2079
O’Neill, L. A., Dunne, A., Edjeback, M., Gray, P., Jefferies, C., Wietek, C. (2003). Mal and Myd88: Adapter Proteins Involved in Signal Transduction by Toll-Like Receptors. J. Endotoxin Res. 9 (1), 55–59. doi: 10.1179/096805103125001351
Pagano, A. F., Py, G., Bernardi, H., Candau, R. B., Sanchez, A. M. (2014). Autophagy and Protein Turnover Signaling in Slow-Twitch Muscle During Exercise. Med. Sci. Sports Exerc. 46 (7), 1314–1325. doi: 10.1249/MSS.0000000000000237
Papp, L. V., Lu, J., Holmgren, A., Khanna, K. K. (2007). From Selenium to Selenoproteins: Synthesis, Identity, and Their Role in Human Health. Antioxid Redox Signal 9 (7), 775–806. doi: 10.1089/ars.2007.1528
Park, H. S., Jung, H. Y., Park, E. Y., Kim, J., Lee, W. J., Bae, Y. S. (2004). Cutting Edge: Direct Interaction of TLR4 With NAD(P)H Oxidase 4 Isozyme is Essential for Lipopolysaccharide-Induced Production of Reactive Oxygen Species and Activation of NF-Kappa B. J. Immunol. 173 (6), 3589–3593. doi: 10.4049/jimmunol.173.6.3589
Rhen, M. (2019). Salmonella and Reactive Oxygen Species: A Love-Hate Relationship. J. Innate Immun. 11 (3), 216–226. doi: 10.1159/000496370
Sasaki, M., Ikeda, H., Sato, Y., Nakanuma, Y. (2008). Proinflammatory Cytokine-Induced Cellular Senescence of Biliary Epithelial Cells is Mediated via Oxidative Stress and Activation of ATM Pathway: A Culture Study. Free Radic. Res. 42 (7), 625–632. doi: 10.1080/10715760802244768
Saxena, A., Lopes, F., McKay, D. M. (2018). Reduced Intestinal Epithelial Mitochondrial Function Enhances In Vitro Interleukin-8 Production in Response to Commensal Escherichia Coli. Inflamm. Res. 67 (10), 829–837. doi: 10.1007/s00011-018-1172-5
Scherz-Shouval, R., Shvets, E., Elazar, Z. (2007). Oxidation as a Post-Translational Modification That Regulates Autophagy. Autophagy 3 (4), 371–373. doi: 10.4161/auto.4214
Schieber, M., Chandel, N. S. (2014). ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 24 (10), R453–R462. doi: 10.1016/j.cub.2014.03.034
Settembre, C., De Cegli, R., Mansueto, G., Saha, P. K., Vetrini, F., Visvikis, O., et al. (2013). TFEB Controls Cellular Lipid Metabolism Through a Starvation-Induced Autoregulatory Loop. Nat. Cell Biol. 15 (6), 647–658. doi: 10.1038/ncb2718
Shariq, M., Quadir, N., Sharma, N., Singh, J., Sheikh, J. A., Khubaib, M., et al. (2021). Mycobacterium Tuberculosis Ripa Dampens TLR4-Mediated Host Protective Response Using a Multi-Pronged Approach Involving Autophagy, Apoptosis, Metabolic Repurposing, and Immune Modulation. Front. Immunol. 12, 636644. doi: 10.3389/fimmu.2021.636644
Sharma, V., Makhdoomi, M., Singh, L., Kumar, P., Khan, N., Singh, S., et al. (2021). Trehalose Limits Opportunistic Mycobacterial Survival During HIV Co-Infection by Reversing HIV-Mediated Autophagy Block. Autophagy 17 (2), 476–495. doi: 10.1080/15548627.2020.1725374
Sheedy, F. J., Palsson-McDermott, E., Hennessy, E. J., Martin, C., O’Leary, J. J., Ruan, Q., et al. (2010). Negative Regulation of TLR4 via Targeting of the Proinflammatory Tumor Suppressor PDCD4 by the Microrna MiR-21. Nat. Immunol. 11 (2), 141–147. doi: 10.1038/ni.1828
Shi, S., Blumenthal, A., Hickey, C. M., Gandotra, S., Levy, D., Ehrt, S. (2005). Expression of Many Immunologically Important Genes in Mycobacterium Tuberculosis-Infected Macrophages is Independent of Both TLR2 and TLR4 But Dependent on IFN-Alphabeta Receptor and STAT1. J. Immunol. 175 (5), 3318–3328. doi: 10.4049/jimmunol.175.5.3318
Shi, C. S., Kehrl, J. H. (2008). Myd88 and Trif Target Beclin 1 to Trigger Autophagy in Macrophages. J. Biol. Chem. 283 (48), 33175–33182. doi: 10.1074/jbc.M804478200
Shin, H. J., Kim, H., Oh, S., Lee, J. G., Kee, M., Ko, H. J., et al. (2016). AMPK-SKP2-CARM1 Signalling Cascade in Transcriptional Regulation of Autophagy. Nature 534 (7608), 553–557. doi: 10.1038/nature18014
Shi, C. S., Shenderov, K., Huang, N. N., Kabat, J., Abu-Asab, M., Fitzgerald, K. A., et al. (2012). Activation of Autophagy by Inflammatory Signals Limits IL-1beta Production by Targeting Ubiquitinated Inflammasomes for Destruction. Nat. Immunol. 13 (3), 255–263. doi: 10.1038/ni.2215
Si, M., Wang, J., Xiao, X., Guan, J., Zhang, Y., Ding, W., et al. (2015). Ohr Protects Corynebacterium Glutamicum Against Organic Hydroperoxide Induced Oxidative Stress. PLoS One 10 (6), e0131634. doi: 10.1371/journal.pone.0131634
Smallwood, M. J., Nissim, A., Knight, A. R., Whiteman, M., Haigh, R., Winyard, P. G. (2018). Oxidative Stress in Autoimmune Rheumatic Diseases. Free Radic. Biol. Med. 125, 3–14. doi: 10.1016/j.freeradbiomed.2018.05.086
Sohrabi, Y., Lagache, S. M. M., Schnack, L., Godfrey, R., Kahles, F., Bruemmer, D., et al. (2018). Mtor-Dependent Oxidative Stress Regulates Oxldl-Induced Trained Innate Immunity in Human Monocytes. Front. Immunol. 9, 3155. doi: 10.3389/fimmu.2018.03155
Song, C., Mitter, S. K., Qi, X., Beli, E., Rao, H. V., Ding, J., et al. (2017). Oxidative Stress-Mediated Nfkappab Phosphorylation Upregulates P62/SQSTM1 and Promotes Retinal Pigmented Epithelial Cell Survival Through Increased Autophagy. PLoS One 12 (2), e0171940. doi: 10.1371/journal.pone.0171940
Stamler, J. S., Singel, D. J., Loscalzo, J. (1992). Biochemistry of Nitric Oxide and its Redox-Activated Forms. Science 258 5090, 1898–1902. doi: 10.1126/science.1281928
Starr, T., Child, R., Wehrly, T. D., Hansen, B., Hwang, S., Lopez-Otin, C., et al. (2012). Selective Subversion of Autophagy Complexes Facilitates Completion of the Brucella Intracellular Cycle. Cell Host Microbe 11 (1), 33–45. doi: 10.1016/j.chom.2011.12.002
Su, P. H., Hsu, Y. W., Huang, R. L., Weng, Y. C., Wang, H. C., Chen, Y. C., et al. (2017). Methylomics of Nitroxidative Stress on Precancerous Cells Reveals DNA Methylation Alteration at the Transition From In Situ to Invasive Cervical Cancer. Oncotarget 8 (39), 65281–65291. doi: 10.18632/oncotarget.18370
Tiruppathi, C., Soni, D., Wang, D. M., Xue, J., Singh, V., Thippegowda, P. B., et al. (2014). The Transcription Factor DREAM Represses the Deubiquitinase A20 and Mediates Inflammation. Nat. Immunol. 15 (3), 239–247. doi: 10.1038/ni.2823
Tripathi, D. N., Chowdhury, R., Trudel, L. J., Tee, A. R., Slack, R. S., Walker, C. L., et al. (2013). Reactive Nitrogen Species Regulate Autophagy Through ATM-AMPK-TSC2-Mediated Suppression of Mtorc1. Proc. Natl. Acad. Sci. U.S.A. 110 (32), E2950–E2957. doi: 10.1073/pnas.1307736110
Umezawa, K., Akaike, T., Fujii, S., Suga, M., Setoguchi, K., Ozawa, A., et al. (1997). Induction of Nitric Oxide Synthesis and Xanthine Oxidase and Their Roles in the Antimicrobial Mechanism Against Salmonella Typhimurium Infection in Mice. Infect. Immun. 65 (7), 2932–2940. doi: 10.1128/iai.65.7.2932-2940.1997
Valko, M., Leibfritz, D., Moncol, J., Cronin, M. T., Mazur, M., Telser, J. (2007). Free Radicals and Antioxidants in Normal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol. 39 (1), 44–84. doi: 10.1016/j.biocel.2006.07.001
Valko, M., Rhodes, C. J., Moncol, J., Izakovic, M., Mazur, M. (2006). Free Radicals, Metals and Antioxidants in Oxidative Stress-Induced Cancer. Chem. Biol. Interact. 160 (1), 1–40. doi: 10.1016/j.cbi.2005.12.009
Vasselon, T., Detmers, P. A. (2002). Toll Receptors: A Central Element in Innate Immune Responses. Infect. Immun. 70 (3), 1033–1041. doi: 10.1128/IAI.70.3.1033-1041.2002
Wang, Y., Chen, T., Han, C., He, D., Liu, H., An, H., et al. (2007). Lysosome-Associated Small Rab Gtpase Rab7b Negatively Regulates TLR4 Signaling in Macrophages by Promoting Lysosomal Degradation of TLR4. Blood 110 (3), 962–971. doi: 10.1182/blood-2007-01-066027
Wang, S., Ge, W., Harns, C., Meng, X., Zhang, Y., Ren, J. (2018b). Ablation of Toll-Like Receptor 4 Attenuates Aging-Induced Myocardial Remodeling and Contractile Dysfunction Through Ncori-HDAC1-Mediated Regulation of Autophagy. J. Mol. Cell Cardiol. 119, 40–50. doi: 10.1016/j.yjmcc.2018.04.009
Wang, S., Song, X., Zhang, K., Deng, S., Jiao, P., Qi, M., et al. (2020a). Overexpression of Toll-Like Receptor 4 Affects Autophagy, Oxidative Stress, and Inflammatory Responses in Monocytes of Transgenic Sheep. Front. Cell Dev. Biol. 8, 248. doi: 10.3389/fcell.2020.00248
Wang, C., Wang, H., Zhang, D., Luo, W., Liu, R., Xu, D., et al. (2018a). Phosphorylation of ULK1 Affects Autophagosome Fusion and Links Chaperone-Mediated Autophagy to Macroautophagy. Nat. Commun. 9 (1), 3492. doi: 10.1038/s41467-018-05449-1
Wang, Y. H., Wu, J. J., Lei, H. Y. (2009). The Autophagic Induction in Helicobacter Pylori-Infected Macrophage. Exp. Biol. Med. (Maywood) 234 (2), 171–180. doi: 10.3181/0808-RM-252
Wang, Y. T., Zaitsev, K., Lu, Q., Li, S., Schaiff, W. T., Kim, K. W., et al. (2020b). Select Autophagy Genes Maintain Quiescence of Tissue-Resident Macrophages and Increase Susceptibility to Listeria Monocytogenes. Nat. Microbiol. 5 (2), 272–281. doi: 10.1038/s41564-019-0633-0
Wang, S., Zhu, X., Xiong, L., Zhang, Y., Ren, J. (2016). Toll-Like Receptor 4 Knockout Alleviates Paraquat-Induced Cardiomyocyte Contractile Dysfunction Through an Autophagy-Dependent Mechanism. Toxicol. Lett. 257, 11–22. doi: 10.1016/j.toxlet.2016.05.024
Weigert, A., von Knethen, A., Fuhrmann, D., Dehne, N., Brune, B. (2018). Redox-Signals and Macrophage Biology. Mol. Aspects Med. 63, 70–87. doi: 10.1016/j.mam.2018.01.003
West, A. P., Brodsky, I. E., Rahner, C., Woo, D. K., Erdjument-Bromage, H., Tempst, P., et al. (2011). TLR Signalling Augments Macrophage Bactericidal Activity Through Mitochondrial ROS. Nature 472 (7344), 476–480. doi: 10.1038/nature09973
Wi, S. M., Moon, G., Kim, J., Kim, S. T., Shim, J. H., Chun, E., et al. (2014). TAK1-ECSIT-TRAF6 Complex Plays a Key Role in the TLR4 Signal to Activate NF-Kappab. J. Biol. Chem. 289 (51), 35205–35214. doi: 10.1074/jbc.M114.597187
Wiseman, H., Halliwell, B. (1996). Damage to DNA by Reactive Oxygen and Nitrogen Species: Role in Inflammatory Disease and Progression to Cancer. Biochem. J. 313 (Pt 1), 17–29. doi: 10.1042/bj3130017
Xia, M., Liu, J., Wu, X., Liu, S., Li, G., Han, C., et al. (2013). Histone Methyltransferase Ash1l Suppresses Interleukin-6 Production and Inflammatory Autoimmune Diseases by Inducing the Ubiquitin-Editing Enzyme A20. Immunity 39 (3), 470–481. doi: 10.1016/j.immuni.2013.08.016
Xu, Y., Shen, J., Ran, Z. (2020). Emerging Views of Mitophagy in Immunity and Autoimmune Diseases. Autophagy 16 (1), 3–17. doi: 10.1080/15548627.2019.1603547
Yamashita, M., Chattopadhyay, S., Fensterl, V., Saikia, P., Wetzel, J. L., Sen, G. C. (2012). Epidermal Growth Factor Receptor is Essential for Toll-Like Receptor 3 Signaling. Sci. Signal 5 (233), ra50. doi: 10.1126/scisignal.2002581
Yao, L., Kan, E. M., Kaur, C., Dheen, S. T., Hao, A., Lu, J., et al. (2013). Notch-1 Signaling Regulates Microglia Activation via NF-Kappab Pathway After Hypoxic Exposure In Vivo and In Vitro. PLoS One 8 (11), e78439. doi: 10.1371/journal.pone.0078439
Yao, Z., Zhang, Q., Li, X., Zhao, D., Liu, Y., Zhao, K., et al. (2014). Death Domain-Associated Protein 6 (Daxx) Selectively Represses IL-6 Transcription Through Histone Deacetylase 1 (HDAC1)-Mediated Histone Deacetylation in Macrophages. J. Biol. Chem. 289 (13), 9372–9379. doi: 10.1074/jbc.M113.533992
Ye, Y., Tan, S., Zhou, X., Li, X., Jundt, M. C., Lichter, N., et al. (2015). Inhibition of P-Ikappabalpha Ubiquitylation by Autophagy-Related Gene 7 to Regulate Inflammatory Responses to Bacterial Infection. J. Infect. Dis. 212 (11), 1816–1826. doi: 10.1093/infdis/jiv301
Zhao, H., Chen, H., Xiaoyin, M., Yang, G., Hu, Y., Xie, K., et al. (2019). Autophagy Activation Improves Lung Injury and Inflammation in Sepsis. Inflammation 42 (2), 426–439. doi: 10.1007/s10753-018-00952-5
Zhao, Y., Kuang, M., Li, J., Zhu, L., Jia, Z., Guo, X., et al. (2021). SARS-Cov-2 Spike Protein Interacts With and Activates TLR41. Cell Res. 31 (7), 818–820. doi: 10.1038/s41422-021-00495-9
Zhao, H., Zhang, M., Zhou, F., Cao, W., Bi, L., Xie, Y., et al. (2016). Cinnamaldehyde Ameliorates LPS-Induced Cardiac Dysfunction via TLR4-NOX4 Pathway: The Regulation of Autophagy and ROS Production. J. Mol. Cell Cardiol. 101, 11–24. doi: 10.1016/j.yjmcc.2016.10.017
Zhong, Z., Umemura, A., Sanchez-Lopez, E., Liang, S., Shalapour, S., Wong, J., et al. (2016). NF-Kappab Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 164 (5), 896–910. doi: 10.1016/j.cell.2015.12.057
Zhou, M., Xu, W., Wang, J., Yan, J., Shi, Y., Zhang, C., et al. (2018). Boosting Mtor-Dependent Autophagy via Upstream TLR4-Myd88-MAPK Signalling and Downstream NF-Kappab Pathway Quenches Intestinal Inflammation and Oxidative Stress Injury. EBioMedicine 35, 345–360. doi: 10.1016/j.ebiom.2018.08.035
Zhu, Y., Yin, Q., Wei, D., Yang, Z., Du, Y., Ma, Y. (2019). Autophagy in Male Reproduction. Syst. Biol. Reprod. Med. 65 (4), 265–272. doi: 10.1080/19396368.2019.1606361
Keywords: TLR4, autophagy, nitroxidative stress, interaction, homeostasis
Citation: Zhang K, Huang Q, Deng S, Yang Y, Li J and Wang S (2021) Mechanisms of TLR4-Mediated Autophagy and Nitroxidative Stress. Front. Cell. Infect. Microbiol. 11:766590. doi: 10.3389/fcimb.2021.766590
Received: 29 August 2021; Accepted: 04 October 2021;
Published: 22 October 2021.
Edited by:
Hua Niu, Affiliated Hospital of Guilin Medical University, ChinaCopyright © 2021 Zhang, Huang, Deng, Yang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sutian Wang, wstlyt@126.com; Jianhao Li, jianhao63@sina.com
†These authors have contributed equally to this work
 Kunli Zhang
Kunli Zhang Qiuyan Huang
Qiuyan Huang Shoulong Deng
Shoulong Deng Yecheng Yang
Yecheng Yang Jianhao Li
Jianhao Li Sutian Wang
Sutian Wang