Barrier to Gene Flow of Grey Mangrove Avicennia marina Populations in the Malay Peninsula as Revealed From Nuclear Microsatellites and Chloroplast Haplotypes
- 1Ecology and Biodiversity Research Group, Biology Department, Vrije Universiteit Brussel (VUB), Brussels, Belgium
- 2Mangrove Research Unit, Institute of Oceanography and Environment, Universiti Malaysia Terengganu (UMT), Kuala Terengganu, Malaysia
- 3Mangrove Specialist Group (MSG), Species Survival Commission (SSC), International Union for the Conservation of Nature (IUCN), Gland, Switzerland
- 4Department of Genetics and Plant Breeding, Sylhet Agricultural University, Sylhet, Bangladesh
- 5Systems Ecology and Resource Management Research Unit (SERM), Department of Organism Biology, Université Libre de Bruxelles, Brussels, Belgium
Contemporary mangrove forest areas took shape historically and their genetic connectivity depends on sea-faring propagules, subsequent settlement, and persistence in suitable environments. Mangrove species world-wide may experience genetic breaks caused by major land barriers or opposing ocean currents influencing their population genetic structure. For Malay Peninsula, several aquatic species showed strong genetic differentiation between East and West coast regions due to the Sunda shelf flooding since the Last Glacial Maximum. In this study genetic diversity and structure of Avicennia marina populations in Malay Peninsula were assessed using nuclear microsatellite markers and chloroplast sequences. Even though all populations showed identical morphological features of A. marina, three evolutionary significant units were obtained with nuclear and cytoplasmic markers. Avicennia marina along a 586 km stretch of the West coast differed strongly from populations along an 80 km stretch of the East coast featuring chloroplast capture of Avicennia alba in an introgressive A. marina. Over and above this expected East-West division, an intra-regional subdivision was detected among A. marina populations in the narrowest region of the Strait of Malacca. The latter genetic break was supported by an AMOVA, STRUCTURE, and BARRIER analysis whereas RST > FST indicated an evolutionary signal of long-lasting divergence. Two different haplotypes along the Western coast showed phylogeographic relationship with either a northern or a putative southern lineage, thereby assuming two Avicennia sources facing each other during Holocene occupation with prolonged separation in the Strait of Malacca. MIGRATE-n model testing supported a northward unidirectional stepping-stone migration route, although with an unclear directionality at the genetic break position, most likely due to weak oceanic currents. Low levels of genetic diversity and southward connectivity was detected for East coast Avicennia populations. We compared the fine-scale spatial genetic structure (FSGS) of Avicennia populations along the exposed coast in the East vs. the sheltered coast in the West. A majority of transects from both coastlines revealed no within-site kinship-based FSGS, although the remoteness of the open sea is important for Avicennia patches to maintain a neighborhood. The results provide new insights for mangrove researchers and managers for future in-depth ecological-genetic-based species conservation efforts in Malay Peninsula.
Introduction
Coastal communities living adjacent to the mangrove ecosystems in tropical, subtropical and warm temperate regions of the world are receiving manifold ecological and socio-economic benefits since the ancient times (Walters et al., 2008; Spalding et al., 2010; Lee et al., 2014; Seddon et al., 2020; Dahdouh-Guebas et al., 2021). The mangrove species occupying different environmental gradients in a habitat serve collectively for land building, coastal protection, water quality improvement, phytoremediation, carbon sequestration, breeding or nursery ground to various aquatic and terrestrial fauna (Donato et al., 2011; Cohen et al., 2013; Analuddin et al., 2017; del Valle et al., 2020; Osland et al., 2020; Kathiresan et al., 2021). Due to the increased public awareness and extensive mangrove plantation schemes in recent years, the (average) annual loss of global mangrove cover declined to 2–4% and opened the doors for conservation optimism (Dahdouh-Guebas et al., 2020; Friess et al., 2020). Contemporary mangrove forest areas took shape historically and their genetic connectivity depends on sea-faring propagules, subsequent establishment, and persistence in suitable environments (Van der Stocken et al., 2019a). Mangrove seed/propagules experience both short to long-distance dispersal by tidal action (hydrochory) and grow either close to the mother tree or in other suitable locations. In this context, the physical land barriers and water current or circulation patterns were found to be crucial in bringing changes to the species' abundance and distribution of plants (Van der Stocken et al., 2019b) and animals (Fairuz-Fozi et al., 2021).
Several researchers have observed the land-barrier effect of Malay Peninsula and found genetically different animal populations (e.g., Tachypleus gigas, T. tridentatus, Carcinoscorpius rotundicauda, Barbonymus schwanenfeldii, Varuna litterata) between East and West coasts (Kamarudin and Esa, 2009; Yang et al., 2009; Ismail et al., 2011; Adibah et al., 2015; Liew et al., 2015; Suppapan et al., 2017; Fairuz-Fozi et al., 2021). In case of mangrove, species such as Avicennia alba Blume, Bruguiera gymnorrhiza (L.) Lamk., Ceriops decandra (Griff.) Ding Hou, Lumnitzera racemosa Willd. and Sonneratia alba J. Smith also revealed such genetic separation (Su et al., 2006; Liao et al., 2007; Huang et al., 2008; Wee et al., 2020). However, Wee et al. (2020) elucidated that the Malay Peninsula is acting like a filter, rather than a barrier, to the gene flow of mangroves by considering dispersal potential of its seeds or propagules. This East-West divide is therefore an increasingly documented phenomenon explained from the Sunda shelf flooding during the Holocene and likewise re-population of East and West Malay coastlines from different source populations since the Last Glacial Maximum. During the global de-glaciation period, the flooding of the Sunda Shelf facilitated mangrove species to undergo range shift toward the Malay Peninsula East and West coast (Wee et al., 2014, 2015). Peninsular Malaysia has ca. 110,952 ha of mangrove cover (18% of the country's mangrove extent), of which nearly 17,570 ha (3%) are distributed along the East coast and 93,382 ha (15%) on the West coast (Hamdan and Misman, 2020). The less mangrove cover on the East coast is due to strong waves and current from the South China Sea, especially during the northeast monsoon, whereas the West coast facing the Strait of Malacca and sheltered by Indonesia receives a weaker current (Akhir et al., 2015; Zainol et al., 2021). Such contrasting ocean dynamics may also result in a different spatial genetic structure of mangroves, both at regional and local scales (Triest, 2008).
Mangrove propagules do not have a dormant stage and propagule dispersal is affected by factors such as buoyancy, propagule viability and timely establishment (Rabinowitz, 1978). Thus, the persistence of the population depends solely on the formation, release, distribution and establishment of propagules as clonal growth and vegetative dispersal is absent in mangroves. Once established, and before reaching sexual maturity, propagules and seedlings are subject to predation (Dahdouh-Guebas et al., 2011), environmental factors affecting early growth (Krauss et al., 2008) and anthropogenic pressure (Cannicci et al., 2008). Tidal influence, ocean currents and wind action predict possible distribution patterns and colonization of mangrove species over long oceanic distances (Clarke, 1993; Van der Stocken et al., 2015, 2019a). Many studies illustrated a long-range connection of mangrove trees in relation to ocean currents and direction, especially along same coastlines (Mori et al., 2015a; De Ryck et al., 2016; Ngeve et al., 2017; Hodel et al., 2018; Van der Stocken et al., 2019b). Among others, a stepping-stone model of migration between estuaries was portrayed for Avicennia species (Do et al., 2019; Wee et al., 2020; Triest et al., 2021a,b,c). This contributes to the importance of coastal connectivity through dispersal of propagules, thus gene flow, which is the only natural cohesive force between longer-term estuaries for a species to maintain its evolutionary units. However, as explained previously, the barriers to genetic connectivity may come from land masses and different migration histories (Triest, 2008; Hodel et al., 2018; Wee et al., 2020; Triest et al., 2021b), opposing ocean currents (Mori et al., 2015b; Ngeve et al., 2017) or very large rivers (Triest et al., 2018). In general, the expectation that migration routes followed major ocean and coastal currents is largely confirmed by genetic diversity and population-level genetic structure approaches.
However, connectivity patterns of mangrove settlement between estuaries of the same coastline and different habitats thereof are more complicated. Estuarine landscapes are highly diverse and unique in their complexity such that the establishment of mangrove propagules is dependent on suitable habitats due to sedimentation patterns of coastal and major river systems, channels or creeks and sandbar dunes (Triest and Van der Stocken, 2021). Spontaneous processes of propagule dispersal and mangrove vegetation formation are expected to result in different neighborhood sizes of individual trees in exposed seaward vs. sheltered landward populations (Triest and Van der Stocken, 2021), distance from open sea (Triest et al., 2021a), river flow intensity (Ngeve et al., 2017; Chablé Iuit et al., 2020), channel structures (Triest et al., 2020) or degree of fragmentation (Hasan et al., 2018; Bryan-Brown et al., 2020). Persistence of Avicennia trees is influenced by the coastal landform (Triest and Van der Stocken, 2021). The positioning of mangroves along the coast can show gradients of oceanic influences, from highly exposed coastal areas—projecting into open seas—to far inland estuaries along rivers (Ngeve et al., 2017; Chablé Iuit et al., 2020), or even no tidal influence at all (Triest et al., 2021a). Dispersal and settlement that have occurred over the past few and overlapping generations along a gradient of different tidal and ocean currents left traces in the locally recorded amount of genetic diversity (Triest et al., 2021a) and very often in their neighborhood associated fine-scale spatial genetic structure (FSGS) (Triest et al., 2020; Triest and Van der Stocken, 2021). Sufficient prior knowledge of polymorphic genetic markers is available for the geographically widespread gray mangrove Avicennia marina (Forsk.) Vierh. and related species (Triest, 2008) to allow resolution within an estuary and even of individual trees at the local fine-scale level (Hasan et al., 2018; Do et al., 2019; Triest et al., 2020, 2021a,c; Triest and Van der Stocken, 2021). Microsatellite loci of Avicennia species partially cross-amplify or show unique alleles at taxon level, so cases of hybridization are detectable (Mori et al., 2015a). Hybridization is not uncommon in mangroves (Ragavan et al., 2017) and admixtures or genetic introgression may represent cryptic evolutionary units or even be subject to “cryptic ecological degradation” (Koedam and Dahdouh-Guebas, 2008), a concept of conservation relevance.
The Malay Peninsula is expected to be a land barrier hindering gene flow between the East and West coast (Duke et al., 2002). Nevertheless, the difference in genetic structure across mangrove species suggest that the Malay Peninsula act as a “filter” rather than a strict geographical barrier, depending on the different dispersal capacities of mangrove species and on the ocean currents (Wee et al., 2020). Avicennia spp. having a low long distance dispersal potential are more likely to be restricted than other mangrove representatives. In this study, we verify for an East-West genetic break of A. marina in Malay Peninsula and more profoundly, at regional scale, focused on the connectivity pattern along each coastline. We hypothesize that connectivity between locations would be more pronounced along the East than to the West and that directionality of historically accumulated gene flow would follow main contemporary ocean currents. We specifically aim to (1) analyze the genetic diversity and structure of A. marina populations on both East and West coasts; (2) estimate the likelihood of different migration models between populations along a same coastline; and (3) compare the extent of a FSGS of A. marina at different distances from the open sea or tidal influence. To achieve this, we considered a combination of nuclear microsatellites and a chloroplast intron sequence (trnH-psbA). Understanding the processes and traces of spontaneous dispersal, establishment and persistence of mangrove areas in a regional context is extremely useful for future research, conservation and management priority settings.
Materials and Methods
Study Area
A total of 13 sampling sites were selected from nine mangrove locations in Peninsular Malaysia (Figure 1). Whereas, six sites from three locations namely, Tumpat (E1-A and E1-B), Tok Bali (E2-A and E2-B), and Setiu Wetlands (E3-A and E3-B) are located along an 80 km stretch of the East coast, seven sites from six locations namely, Penang (W1), Matang (W2-A and W2-B), Kuala Selangor (W3), Malacca (W4), Sungai Sebatu (W5), and Pontian (Johor) (SW) were along an 586 km stretch of the West coast (Figure 1; Supplementary Figure 1). On the East, mangroves in Tumpat were patchy, separated by intermittent water canals and largely exposed to the South China Sea (Satyanarayana et al., 2010), unlike they are river adjoining and sheltered from open sea by a sandbar at Tok Bali and Setiu Wetlands. For the West, all locations are directly exposed to the Strait of Malacca and only Matang mangroves were patchy and separated by major river channels. The pioneer A. marina is widely distributed in all locations of the present study. In fact, this species is common throughout its range and found across the Indo-Pacific (Tomlinson, 2016).
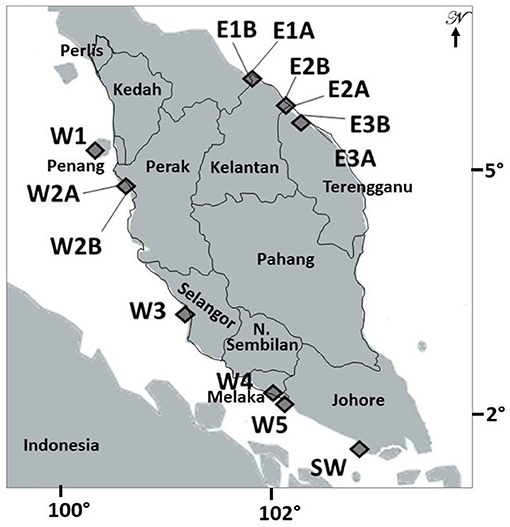
Figure 1. Overview of the studied populations of Avicennia marina along the West coast (W1 to W5 and SW) and of introgressed Avicennia marina on the East coast (E1 to E3) of the Malay Peninsula. Population codes are denoted as in Table 1 and in Supplementary Figure 1 with images of all locations.
Sample Collection
Fieldwork was conducted during the period of May-July 2017 and a total number of 486 samples were collected (179 from East coast and 307 from West coast). Young healthy leaves of A. marina, not damaged by insects or diseases, were handpicked from the tip of the branches of adult as well as juvenile vegetation. We considered ≥2.5 cm stem diameter (D130) for the adult trees and saplings with three leaf pairs or less for the juveniles (Brokaw and Thompson, 2000; Goessens et al., 2014). Mature leaves were not considered due to the high content of polysaccharides, polyphenols and tannins (Ibrahim, 2011), which may disturb the amplification of DNA during the polymerase chain reaction. The sample collection was done through a transect approach for pure stands and random methods for mixed distant patches. The GPS (Garmin 45, USA) location of each sampled tree or juvenile was recorded to estimate the length of the transect line. The sampled leaves were collected into numbered paper envelopes, allowed to air or sun dry until no moisture traces, and further preserved with silica gel for transportation and handling within 1 month.
While observing transects at a given distance to the open sea (Table 1; Supplementary Figure 1), the interval between each sampled individual along the transect was also measured. The average number of sampled trees was 37 per transect though ranged exceptionally from 16 to 60 for a few sites (Table 2). Shortest distance intervals between the neighboring trees, mostly within 10 m, were considered. The gaps between mangrove patches were included into the total distance as fine-scaled analysis focuses on pairs of individual trees within a distance class below 100 m, regardless of the patch location.
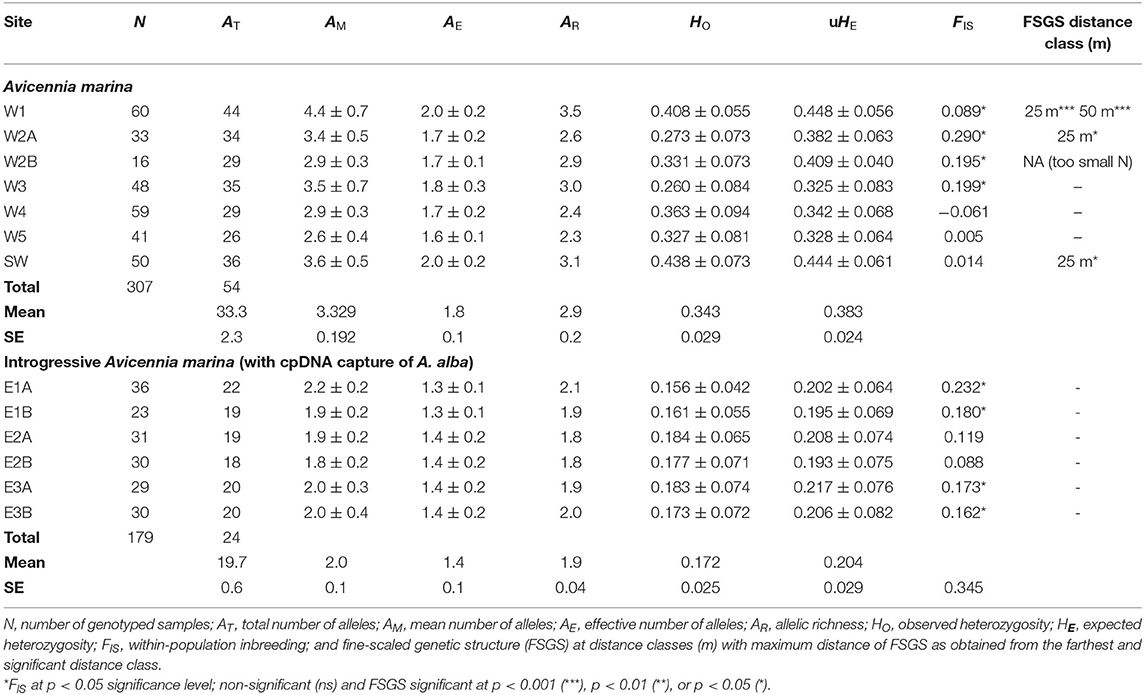
Table 2. Population genetic variables of Avicennia marina transects in mangrove areas along Malacca Strait on the West Malay Peninsula and of introgressed Avicennia marina (with captured cpDNA of A. alba) on the East Malay Peninsula.
For comparative reasons we additionally considered samples of Avicennia species from outside the study area (Supplementary Table 2), namely A. marina from China, Vietnam, Bangladesh, Kenya and South Africa; A. alba, from Vietnam and The Philippines; A. rumphiana Hallier f. from The Philippines and Avicennia officinalis L. from Bangladesh that were available in the BRVU (part of BR herbarium) collection at the Vrije Universiteit Brussel.
DNA Extraction, Microsatellite Genotyping, and Chloroplast DNA Sequencing
Genomic DNA was extracted from approximately 20 mg of each dried leaf tissue using the E.Z.N.A. SP plant DNA Mini kit (Omega bio-tek, Norcross, GA, USA). The concentration of individual samples ranged from 10 to 200 ng/μl. The multiplexed PCR reactions consisted of 13 microsatellite markers: Avma1, Avma02, Avma6, Avma8, Avma10, Avma14, Avma17 (Geng et al., 2007); Am3, Am81, (Maguire et al., 2000a); Aa22, Aa23, Aa67 (Teixeira et al., 2003); and AMK6 (Triest et al., 2020). Primers were fluorescence-labeled with four different dye-labels (6FAM/VIC/NED/PET), and a mixture of 0.2 μM of each primer. About 6.25 μl master mix (Qiagen Multiplex PCR kit), 1.25 μl primer mix, 2.5 μl H2O and 2.5 μl of genomic DNA were used for multiplex PCR reactions. PCR was performed in a thermal cycler (Bio-Rad MyCycler) under the conditions of initial denaturation at 95°C for 15 min followed by 35 cycles of 30 s denaturation at 95°C, 90 s annealing at 57°C, 80 s elongation at 72°C and a final extension of 30 min at 60°C. All PCR products were separated on an ABI3730XL sequencer (Macrogen, Seoul, Korea) and allele sizes were determined with GeneMarker v.2.60 (SoftGenetics LLC, State College, USA).
We used a non-coding region (trnH-psbA) with primer pairs aF-aR and cF-cR (Sang et al., 1997). PCR amplification was carried out in 25 μL of reaction mixture containing 2.5 μL of genomic DNA, 2.5 μL 10 × PCR buffer, 0.2 mM of each dNTP, 1.6 mM MgCl2, 200 nM of the forward and reverse primer, 80 μg mL−1 bovine serum albumin (BSA) and 1 U Taq DNA polymerase (Promega GoTaq DNA Polymerase; Promega, Madison, USA). PCR reactions were performed in a thermal cycler (Bio-Rad MyCycler) and started with 2 min at 94°C, followed by 30 cycles of 45 s at 92°C, 1 min at 50°C for the reactions with trnH-psbA primers, 2 min at 72°C, and a final extension at 72°C for 5 min. All PCR products of trnH-psbA were sequenced on an ABI3730XL sequencer (Macrogen, Seoul, Korea).
Chloroplast Sequence Data Analysis
With an assumption of potential different gene pools, we tested the samples for chloroplast identity. The trnH-psbA sequences of the chloroplast genome in 3 to 7 samples from each site (43 in total) were identified and compared with Avicennia species from Asian (A. marina, A. alba, A. rumphiana and Avicennia officinalis) and African (A. marina) regions (Supplementary Table 2). Sequences of trnH-psbA were aligned (492 bp) and analyzed in Geneious Prime v.2019.2.1 software using MAFFT alignment and MrBayes Maximum Likelihood phylogenetic analysis (GTR substitution model with gamma rate variation; 4 heated chains of 1,100,000 chain length and 100,000 burn-in; Avicennia officinalis from Bangladesh was taken as outgroup).
Microsatellite Data Analysis
Prior to population and individual sample-based analysis, we tested data for genotypic disequilibrium, potential null alleles and overall resolution of the selected microsatellite markers. A linkage test between all pairs of loci (1,000 permutations) identified no genotypic disequilibrium at the 0.05 level (FSTAT v.2.9.3) (Goudet, 2001). MICRO-CHECKER indicated no scoring errors or large allele dropouts (Van Oosterhout et al., 2004). However, null alleles were present in all populations of the East coast for two loci (Avma14 and Avma2) and in all populations of the West coast (Avma14 and Aa22). We decided to omit these loci for profound data analysis of each regional group separately and thus considered only 11 out of 13 loci for analysis at either East or at West coast level (Supplementary Table 1).
The probability of identity (PI), namely whether two individuals could share an identical multilocus genotype by chance, gave a cumulative probability of identity for all polymorphic microsatellite loci in each site of 6.0 × 10−3 on average, thereby providing ample resolution (done with GenAlEx v.6.5, Peakall and Smouse, 2012). Basic population genetic variables were measured separately for each coastline and site: total number of alleles (AT), mean number of alleles (AM), effective number of alleles (AE), allelic richness (AR), observed heterozygosity (HO), unbiased expected heterozygosity (uHE), population inbreeding coefficient (FIS)—with 1,000 permutations test—using FSTAT and GenAlEx. Pairwise genotypic differentiation at individual and population level was used to produce a PCoA in GenAlEx.
On basis of co-dominant alleles (F-statistics) and microsatellite repeat lengths (R-statistics), the genetic structure among sites (FST, RST), inbreeding within sites (FIS, RIS), overall inbreeding (FIT, RIT), and a pairwise genotypic differentiation matrix (FST) of each species were calculated via AMOVA-FST and AMOVA- RST at 999 random permutations (GenAlEx). This allowed further to test for an evolutionary signal (when RST > FST) and to roughly estimate overall connectivity levels as Nm = FST/(1-4FST) under the assumption of an island migration model, very likely to be violated. Therefore, specific hypotheses to estimate gene flow were tested with Migrate-n (Beerli, 2006; Beerli and Palczewski, 2010) from the mutation-scaled population sizes (Theta) and immigration rates (M). The Brownian model was tested locus by locus along with the product of all distributions of all loci. Uni- and bidirectional historical migration/expansion models were tested. Uniform prior distribution settings (min, max, delta) were as follows for Theta = 0.0, 20.0, 2 and for M = 0.0, 20, 2. The number of recorded steps was 106 at a sampling frequency of 103 after an initial burn-in. The effective number of immigrants per generation (Nem) was calculated as [Theta × M]/4. Specific hypotheses testing on directionality were considered in panmixia, source-sink and stepping-stone models for the migration between mangrove locations situated along the same coastline. More precisely, we considered six A. marina populations of the West coast to test the hypothesis of a northward or southward coastal current direction in the Strait of Malacca. Similarly, we considered three Avicennia populations from the East coast to test the hypothesis of prevailing southward coastal current direction of the South China Sea. The Brownian motion mutation model was adopted for randomly generated subsamples of 20 individuals in a transect, following the abovementioned settings, computing two replicate chains (with different seed), and using the Bezier thermodynamic integration (Beerli and Palczewski, 2010) for calculation of the Bayes factors from marginal likelihoods giving the model probabilities.
A Bayesian clustering analysis at individual level for three data sets was carried out in STRUCTURE v.2.3.4 (Pritchard et al., 2000) using an admixture model with correlated allele frequencies. We considered all 13 Avicennia populations (13 polymorphic microsatellite loci) and in further detail only the Eastern (11 polymorphic loci, excluding Avma14 and Avma2) and Western (11 polymorphic loci, excluding Avma14 and Aa22) populations separately. The model ran 10 iterations for each K value from 1 to 13; the burn-in period was 50,000 with 500,000 Markov chain Monte Carlo (MCMC) repeats. The optimal K was inferred with the ΔK statistic (Evanno et al., 2005) and LnPK using Structure Harvester (Earl and von Holdt, 2012) calculated with StructureSelector (Li and Liu, 2018).
The BARRIER v.2.2 software (Manni et al., 2004) was used to detect the location of sharp genetic changes between neighboring populations on basis of 10 pairwise FST matrices of every microsatellite locus, allowing a maximum of one barrier per matrix. We opted to calculate from superposition of raw data from FST matrices at locus level. The thickness of barrier lines thus will be based on the additivity of matrices accounting for the independent microsatellite loci that we consider as a preferred informative and valid method over bootstrapping a single mean FST matrix. A more detailed test of the West coast populations using pairwise FST and RST was performed at four distance classes of 127, 205, 388, 586 km (automatically generated when considering the option of a similar amount of pairwise comparisons) and 1,000 permutations with SPAGeDi 1.5a (Hardy and Vekemans, 2002), thereby allowing to test for an evolutionary signal (when RST > FST) specifically within each distance class. These distance classes represent threshold values instead of a full regression of a Mantel test. Linear-regression and log-regression over the full distance range were calculated with b-slope significance level (one-sided test; 1,000 permutations) for both FST and RST approaches.
The overall FIJ kinship coefficient (Loiselle et al., 1995) for all pairs of individuals of within-site comparisons was tested for equal distance intervals in four classes (up to 25, 50, 75, and 100 m) by SPAGeDi 1.5a (Hardy and Vekemans, 2002). The FSGS i.e., the spatial autocorrelation of individual populations, were all tested for significance with 1,000 permutations. We computed the log-slope (-b) of linear regressions between pairwise kinship coefficients and geographical distance over restricted distance with 1,000 permutations. The potential influence of the open sea on transect FSGS was verified by plotting FIJ kinship values of the first distance class (25 m) against the km distance from starting point of transect to open sea. We also checked the possible effect of resolution to detect FSGS, caused by the lower total number of alleles by plotting kinship values over 25 m distance against the total number of alleles within each population.
Results
Chloroplast Sequences and Haplotype Differentiation
The sequences of trnH-psbA from 43 samples resulted in three distinct haplotypes along Malay Peninsula (Figure 2; Supplementary Table 2). All populations from the East coast revealed a particular A. alba haplotype (451 bp) with a sequence similarly to A. alba from Vietnam and closely related to A. alba from The Philippines. This taxon, from the East Malay Peninsula, has identical morphological features resembling A. marina and thus it could be hypothesized as an introgressive A. marina with a captured chloroplast of A. alba following an earlier hybridization event. In contrast, populations on the West coast harbored two different trnH-psbA variants of A. marina that occurred geographically well-separated. Populations from the northern W1 to W3 sites contained a single haplotype (485 bp) that appeared to be more related to A. marina from Bangladesh than to those from Vietnam and China, whereas populations from the southern W4, W5, and SW sites had a unique haplotype (487 bp) clustered separately and closer to African A. marina than to the Asian samples. The two chloroplast sequences of A. marina along the West coast differed consistently in three mononucleotide repeats (T7 or T8; C7 or C10; T8 or T6), one transversion (A/C), and one transition (A/G). The captured A. alba haplotypes along the East coast showed 21 substitutions, three indels and two mononucleotide repeats when compared to A. marina. We could exclude the presence of A. rumphiana (458 bp) or A. officinalis (445 bp) haplotypes in the studied samples.
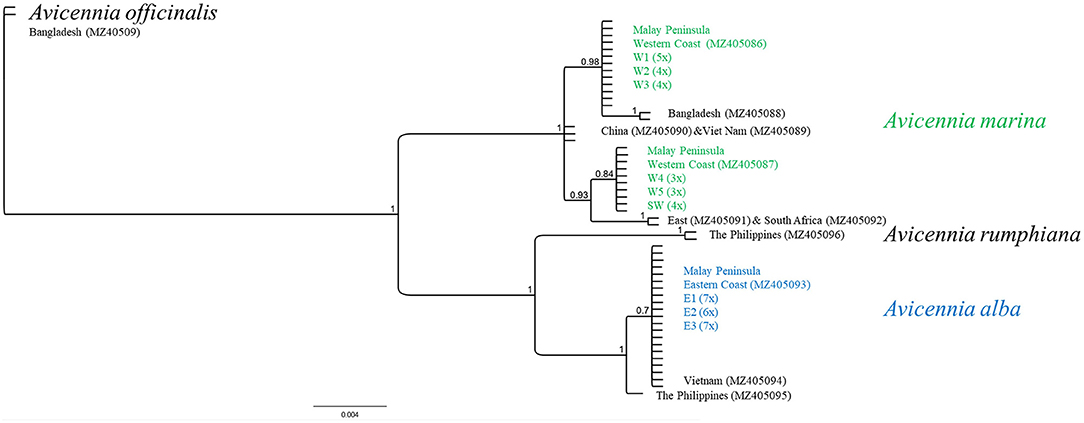
Figure 2. Maximum Likelihood tree of trnH-psbA intron with chloroplast identity of samples from East and West Malay Peninsula when compared to Avicennia marina from other countries and to other Avicennia species. Note the captured chloroplast of A. alba on the East Malay Peninsula and the two different haplotypes of A. marina along the West Malay Peninsula showing a break between W3 and W4.
Genetic Diversity of Nuclear Microsatellites
There are 54 alleles (on average 33 and 26-44 per transect) in 11 loci from the seven sampling sites along the West coast of Malay Peninsula (586 km stretch), with a mean (AM) number of 2.6–4.4, effective (AE) number of 1.6–2.0 and an adjusted richness (AR) of 2.3–3.5 (Table 2). The overall observed heterozygosity (HO = 0.343) was slightly lower than the expected heterozygosity (uHE = 0.383). The within-population inbreeding (mean FIS = 0.082) ranging from −0.061 to 0.290 was significant (p < 0.05) for four sites (Table 2).
AMOVA- FST results from the West coast revealed 37% genetic variation among A. marina populations, 5% among individuals and 58% within individuals, giving an estimate of FST = 0.369, FIS = 0.082 and FIT = 0.421 (Table 3). AMOVA- RST gave higher estimates of genetic variance among the populations than FST with RST = 0.572 (55%), hence an evolutionary signal was indicated by larger differentiation due to allele repeat length of the microsatellites (mutational steps) instead of allele identity only (Table 3). Considering the cpDNA differentiation between the northern (W1, W2, W3) and the southern (W4, W5, SW) sampling sites, a pairwise genetic differentiation of the nuclear microsatellites was highest for all populations between the northern (W1, W2, W3) and the southern (W4, W5, SW) sampling sites (FST = 0.356–0.541), whereas differences between the locations were more distinct for northern sites (FST = 0.120–0.301) than to the southern sites (FST = 0.010–0.071) (Supplementary Table 3). This indicates a strong connectivity among the southern A. marina populations with a very low or no connectivity with the northern ones. The PCoA fully separated these two geographical groups at the population as well as at the individual levels (Supplementary Figure 2).
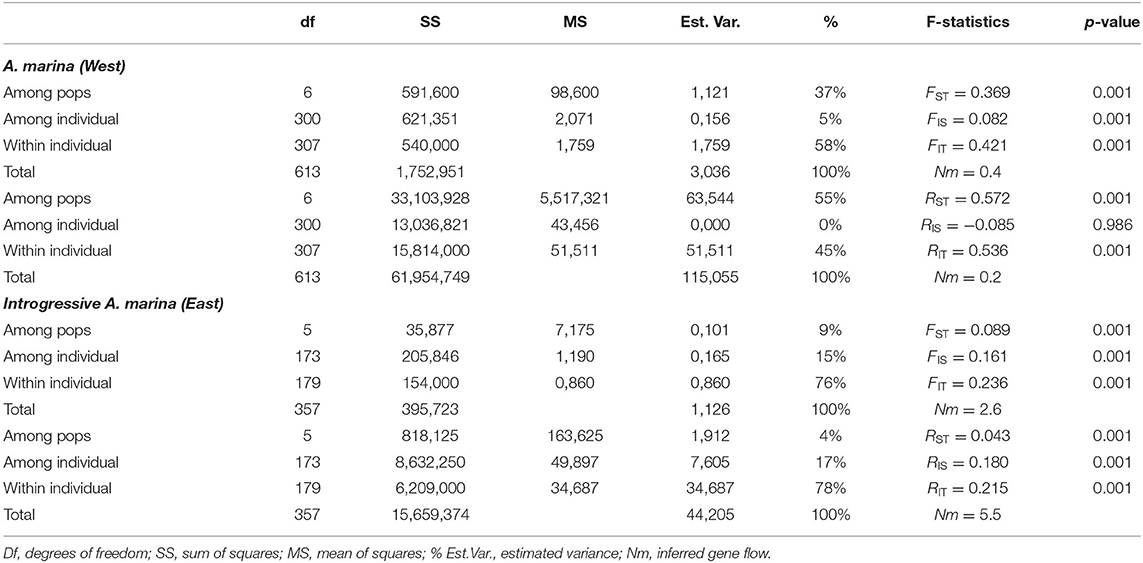
Table 3. Summary of AMOVA with F-statistics and R-statistics of Avicennia marina (Western populations) and introgressive Avicennia marina (Eastern populations with captured cpDNA of Avicennia alba) mangrove fragments along coasts of the Malay Peninsula.
On the other hand, six sampling sites along the East coast of Malay Peninsula (80 km stretch) indicated 24 alleles (on average 20 and 18–22 per transect) in 11 loci, with a mean number (AM) of 1.8–2.2, effective number (AE) of 1.3–1.4 and an adjusted richness (AR) of 1.8–2.1 (Table 2). The observed heterozygosity (HO = 0.172) was slightly lower than the expected heterozygosity (uHE = 0.204). The within population inbreeding (mean FIS = 0.161) ranging from −0.088 to 0.232 was significant (p < 0.05) for four sites (Table 2).
AMOVA- FST results explained 9% genetic variation among the introgressive A. marina populations on the East coast, 15% among individuals and 76% within individuals, giving an estimate of FST = 0.089, FIS = 0.161, and FIT = 0.236 (Table 3). AMOVA- RST provided much lower estimates of genetic variance among these populations (4%) than FST, hence differences in allele repeat length of the microsatellites were non-explanatory (Table 3). Pairwise genetic differentiation was highest between transects E1 and E2 (FST = 0.151–0.207), whereas FST values between locations E2 and E3 remained low though significant (0.037–0.089) (Supplementary Table 4). All within-site comparisons of transects were non-significant (Supplementary Table 4). The PCoA showed a single cluster at individual level, but clustered transects separately at site level (Supplementary Figure 2).
Genetic Structure, Coastal Connectivity, and Gene Flow Models
The Bayesian clustering analysis with 486 A. marina samples indicated well-separated gene pools and assigned every individual to a single gene pool without any admixture (Figure 3). Delta K was high for K = 2 and K = 3. The latter outcome of K = 3 was supported by each iteration (low standard deviation), approached the LnP(K) plateau and fully corresponds to the previously defined haplotype groups. Also, the STRUCTURE analysis of each coastline revealed K = 1 for the Eastern (N = 179) and K = 2 (N = 307) for the Western populations (namely comprising a northern vs. a southern cluster on the West coast), hence we considered an overall structure of K = 3 clusters. The BARRIER analysis showed two major genetic breaks, one between East and West coasts of Malay Peninsula and another between the populations of W3 and W4 in the Strait of Malacca on the West coast (Figure 4). Implicit spatial analysis of the populations along the West coast using four distance classes revealed a significant pairwise lower FST and RST in the first distance class up to 127 km and a marginal significant b-log regression over the full distance of 586 km (one-sided test: FST r2 = 0.47, p = 0.01; RST r2 = 0.40, p = 0.02) (Supplementary Table 5).
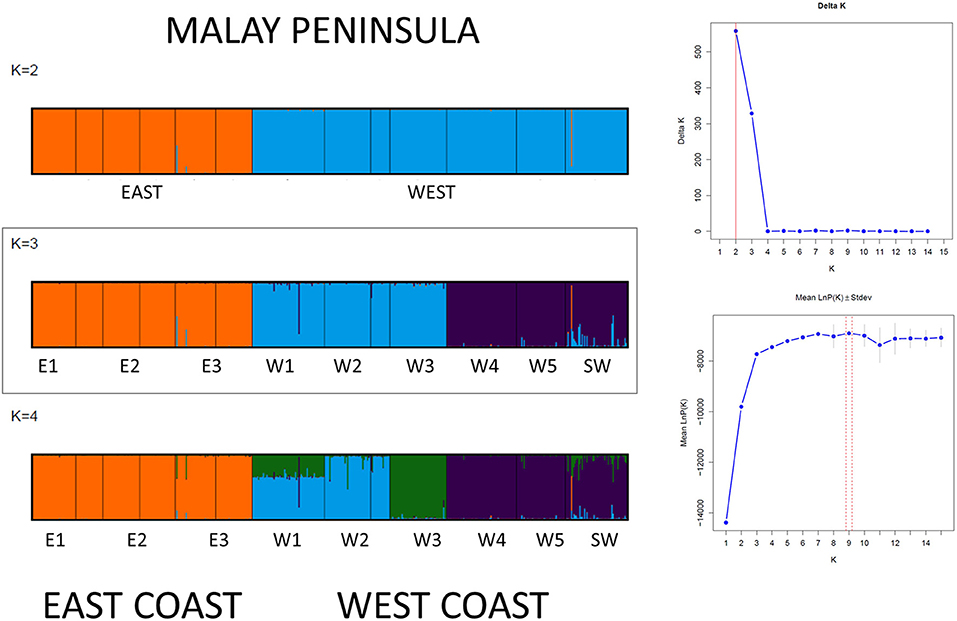
Figure 3. STRUCTURE results of Avicennia marina populations showing a break between W3 and W4 along the West coast and for introgressive Avicennia marina populations reflecting a single gene pool on the East coastline of the Malay Peninsula.
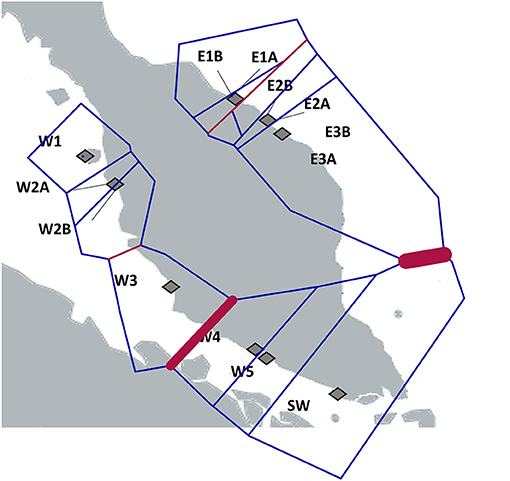
Figure 4. Genetic barriers (red lines) computed from FST distance matrices of ten independent microsatellite loci using BARRIER analysis. The thickness of the red line reflects the importance of the barriers. Note the barrier between W3 and W4 in the Strait of Malacca in addition to a previously well-documented East-West barrier.
Specific testing with migrate-n on the directionality for A. marina across six mangrove locations of the West coast indicated that panmixia or bidirectional stepping-stone models (from South to North as well as from North to South) are less likely than a customized unidirectional South to North migration as well as a local bidirectionality between W3 and W4 sites (Table 4; Figure 5). Highest estimated gene flow values were observed from W3 toward W2 (Nem = 1.4) and from W5 toward W4 (Nem = 1.1). The lowest gene flow estimate for the bidirectional scenario of W4–W3 (Nem = 0.5) and an absence of gene flow for the reciprocal (Nem = 0.01) were all in agreement with the genetic break evidenced from BARRIER and STRUCTURE analyses. Migration of the introgressive A. marina across three mangrove locations on the East coast were supported by a unidirectional stepping-stone model (with gene flow estimates of Nem = 0.3–0.5), following the main ocean current directionality of the South China Sea (Table 4; Figure 5).
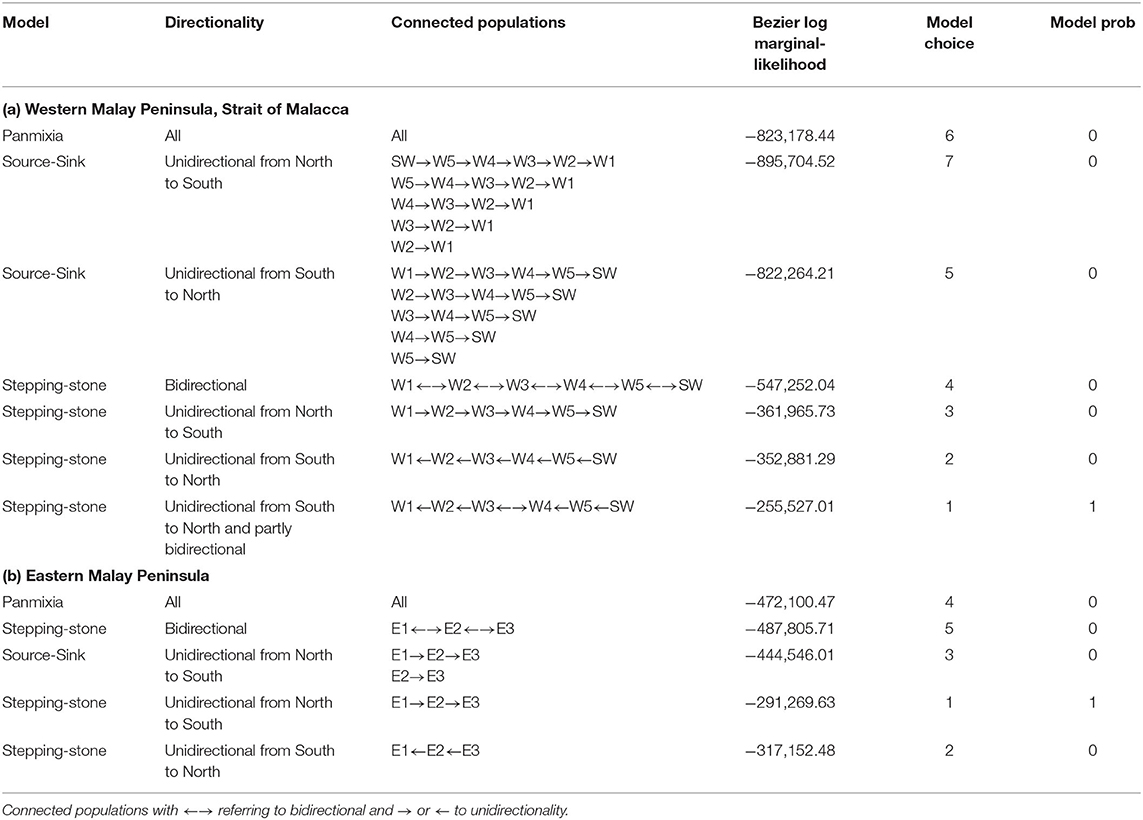
Table 4. Comparison of migration models on the directionality of (a) Avicennia marina populations for West Malay Peninsula sites and hypothesis of Northward Coastal current direction in the narrowest and shallow part of the Strait of Malacca; and (b) Introgressive Avicennia marina (with captured cpDNA of Avicennia alba) populations for East Malay Peninsula sites and hypothesis of prevailing Southward Coastal current direction in the South China Sea; with Nem included for each scenario.
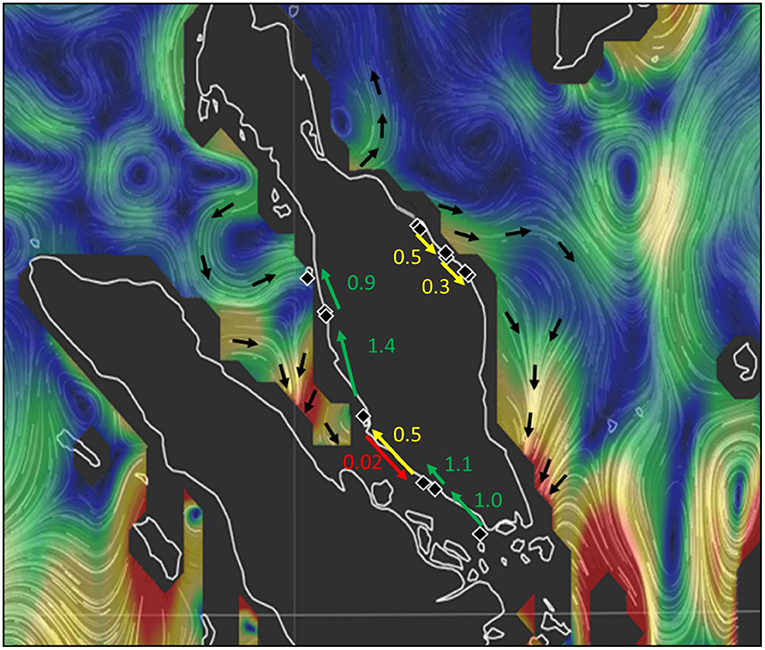
Figure 5. Migration model with directions and gene flow estimates between populations (black rhombs). Strength of principal ocean currents of the study area are indicated in a gradient from blue and green (strong) to yellow and red (weak). Oceanic currents around Peninsula Malaysia in July 2017. Data from ESR (2009), OSCAR third degree resolution ocean surface currents. Ver. 1. PO.DAAC, CA, USA. Dataset accessed (2018-07-28) at http://dx.doi.org/10.5067/OSCAR-03D01.
Fine-Scale Genetic Structure
The overall FIJ kinship coefficient for within-site comparisons revealed positive values for only three transects with the shorter distance classes of 25 or 50 m (Table 2), while the other transects lacked a fine-scale structure. The (FIJ)1 value of the first distance class (< 25 m) indicated a potential relationship with the distance to the open sea, because of an absence of FSGS (zero values) for mangroves along the West coast when situated closer to the coast than about 250 m (Supplementary Figure 3). However, absence of a detectable and significant FSGS could also be explained from lower allelic diversity and smaller sample size in transects on the East coast (Supplementary Figure 3).
Discussion
Evolutionary Significant Units and Hybridization
Avicennia marina, with a least concern (LC) status of the IUCN, has limited focus for conservation and management in many parts of the world including Malaysia. In Peninsular Malaysia, Rhizophora apiculata Blume and R. mucronata Lam. are more popular than Avicennia spp. due to their abundant seed bank, highest growth rate and commercial importance (Goessens et al., 2014; Tangah et al., 2020). However, the forestry measures such as planting, sustainable use, legal protection of areas, etc., are still applicable to A. marina in all countries wherever is needed (IUCN, 2021). Inbreeding and genetic diversity that are common to mangrove populations were also observed for A. marina (Maguire et al., 2000b; Dodd et al., 2002; Yan et al., 2016). Therefore, continued monitoring and in-depth research are necessary to provide further scientific insights on this population diversity, especially from the areas like Malay Peninsula where land blockage effect is protuberant (Azman et al., 2020; Wee et al., 2020; Fairuz-Fozi et al., 2021).
Though all sampled populations showed identical morphological features of A. marina, its actual abundance and distribution seems different along the East and West coasts of Malay Peninsula. Three evolutionary significant units could be confirmed from both nuclear and cytoplasmic markers. Avicennia marina along the 586 km stretch of West coast differentiated strongly from an introgressive A. marina featuring a captured chloroplast of A. alba along the 80 km stretch of East coast. Over and above this expected East-West division, an intra-regional subdivision was detected among A. marina populations in the narrowest region of the Strait of Malacca. This genetic break was clearly supported by the AMOVA, STRUCTURE, and BARRIER analyses whereas RST > FST indicates an evolutionary signal of this long-lasting divergence. Two different haplotypes along the West coast showed phylogeographic relationship with either a northern or a putative southern lineage, thereby assuming two Avicennia sources facing each other during the Holocene occupation with a prolonged separation in the Strait of Malacca.
The Malay Peninsula is expected to be a land barrier hindering gene flow between the East and West coast (Duke et al., 2002). The East-West genetic divergence across the Malay Peninsula has already been highlighted for several mangrove species such as S. alba (Wee et al., 2017; Yang et al., 2017) and A. alba (Wee et al., 2020), and to a lesser extent for B. gymnorhiza (Minobe et al., 2010; Wee et al., 2020). However, it has been shown that Malay Peninsula does not act the same depending on the species or genera studied. Indeed, Rhizophora mucronata and R. apiculata did not show the same population structures across Malay Peninsula: while R. apiculata showed some divergence between the two coasts, this was not the case for R. mucronata which showed higher dispersal potential (Inomata et al., 2009; Wee et al., 2020). The difference in genetic structure across species suggest that the Malay Peninsula act as a “filter” rather than a strict geographical barrier, depending on dispersal capacities of mangrove species and ocean currents (Wee et al., 2020). Avicennia spp. having a low long distance dispersal potential are more likely to be restricted to within their oceanic basin (Maguire et al., 2000b), as supported by our results and thus potentially indicating correlation between dispersal potential and genetic admixture after the LGM for Avicennia species.
Along the East coast an introgressive A. marina featuring a captured chloroplast of A. alba could be demonstrated because we could compare with trnH-psbA of pure A. alba samples from different locations in Vietnam and different islands in The Philippines. We mentioned one from each country in this article because the sequences were similar for the Bayes ML and the focus here was on Malay Peninsula diversity. Therefore, the cpDNA of the East coast samples belongs to A. alba and is not a misidentification of trnH-psbA. Since for the East coast samples, their nuclear genome largely amplifies the A. marina microsatellite loci and their cpDNA fully matches A. alba, then the chloroplast must be derived from an A. alba egg-cell in the case of maternal inheritance. Although pollination can go either way, the recipient mother plant in this case was definitely A. alba. Hybridization is not uncommon in mangroves. According to Ragavan et al. (2017), the natural hybrids in mangroves are predominantly found in seven genera namely, Acrostichum, Avicennia, Bruguiera, Ceriops, Lumnitzera, Rhizophora, and Sonneratia. In the case of Avicennia, two hybrids - one from Thailand with the parental species of A. marina and A. rumphiana and another from Brazil with the parental species of A. germinans (L.) Stearn and A. bicolor Standley were reported (Huang et al., 2014; Mori et al., 2015b). For Peninsular Malaysia, both East and West coasts are known to have the mangrove hybrids like Bruguiera x hainesii C. G. Rogers, Bruguiera x rhynchopetala (W. C. Ko) X.-J. Ge and N. C. Duke, Rhizophora x annamalayana Kathir., and Sonneratia x hainanensis W. C. Ko, E. Y. Chen and W. Y. Chen (Wan Juliana et al., 2014; Jamilah et al., 2015; Razali et al., 2016). In addition, presence of Rhizophora x lamarckii Montr., Rhizophora x mohanii P. Ragavan, Sonneratia x gulngai N. C. Duke, in the Malaysian mangroves was informed by Ragavan et al. (2017). We assume that field reports on possible cases of Avicennia hybridization were not available, because the leaf features of those trees that were found to be introgessed hybrids in our study resembled A. marina morphologically. Therefore, it is necessary to supplement nuclear DNA markers with chloroplast sequences in biogeographical regions where more than one Avicennia species occurs.
Connectivity Along Same Coastline
Migrate-n model testing supported northward unidirectional stepping-stone along the West coast, though directionality at the genetic break point (narrowest region of the Strait of Malacca) was unclear most likely due to weaker oceanic currents. Long-distance dispersal of A. marina propagules can be attributed to its buoyancy and viability characteristics as well as the hydrodynamics i.e., tidal inundation (Breitfuss et al., 2003), current speed and direction (Steinke and Ward, 2003). Wind may play a limited role in the dispersal of Avicennia propagules as they show low surface water contact and less influence by the drag force (Van der Stocken et al., 2015). Successful establishment and growth of the propagules after short to long-distance dispersal patterns (Rabinowitz, 1978; Clarke, 1993) becomes a responsible source for historically accumulated gene flow between populations (Duke et al., 1998). In this post-establishment phase, propagule predators are known to play a key role in the Americas, Africa and Asia (Smith, 1987; McGuinness, 1997; Dahdouh-Guebas et al., 1998; Langston et al., 2017). Once the crypto-viviparous propagules of A. marina released into seawater, they are expected to undergo different phases like floating with unshed pericarp, floating with shed pericarp, sunken with unshed pericarp, sunken with shed pericarp, etc., over a period of time (B. Satyanarayana, pers. comm.). If propagules do not reach to a suitable substratum immediately, then they may remain viable in seawater for several days to weeks (Steinke, 1986; Clarke and Myerscough, 1991; Clarke, 1993; Clarke et al., 2001) and would be able to disperse between geographically disjunct estuaries. Extended floating periods of several months have been observed for other Avicennia species, e.g., A. germinans (Rabinowitz, 1978; Alleman and Hester, 2011).
Our estimation of genetic connectivity among A. marina populations along the West coast of Malay Peninsula through comparison of migration models supported a putative unidirectional dispersal route that was congruent with prevailing ocean currents (Rizal et al., 2010, 2012; Haditiar et al., 2020). This further emphasizes the relevance of coastal connectivity to mangrove persistence (Van der Stocken et al., 2019a,b) as well as the importance of apparently discrete estuaries. The connectivity between adjacent mangroves was supported by the stepping-stone migration model. The dispersal of Avicennia propagules is likely restricted to a few tens of kilometers (e.g., Clarke, 1993; Duke et al., 1998; Melville and Burchett, 2002). Recently, Binks et al. (2018) found a long-distance dispersal up to 100 km, which is comparable to the 80 km stretch between northern and southern populations (E1–E3) of the East Malay Peninsula to demonstrate their genetic connectivity in a stepping-stone manner. The present findings also add to the emerging evidence of Avicennia species in general following an adjacent migration, such as the unidirectional way obtained for A. alba on the West coast (Wee et al., 2020), A. marina in eastern Africa (Triest et al., 2021c) or along Leyte island in The Philippines (Triest et al., 2021a) and bidirectional ways for A. germinans along each of the Caribbean and Pacific coasts of central America (Ochoa-Zavala et al., 2019). Although repeated bottlenecks or founder effects of the pioneering Avicennia species may have caused the differentiation of populations (Maguire et al., 2000a; Arnaud-Haond et al., 2006), we noticed only limited evidence of such conditions in the present study region.
The weaker currents slow down propagule dispersal and increase the risk of their entrapment among mangrove roots or settlement on unsuitable substratum such as a sand bar or beach. Low or absent tidal currents in the mangrove patches are responsible for leaving a trace of elevated kinship values. Such FSGS traces were also estimated within the spatial extents of a few meters to several hundreds of meters (Mori et al., 2015b; Do et al., 2019; Chablé Iuit et al., 2020; Triest et al., 2020; Triest and Van der Stocken, 2021). When the populations were sheltered (Triest and Van der Stocken, 2021) or severely fragmented and confined to artificial dikes (Hasan et al., 2018), these kinship values became enhanced. For instance, R. mangle showed a FSGS up to 90 m in different estuarine conditions of the Caribbean mangroves (Yucatan, Mexico), and even further up to 240 m when along a river (Chablé Iuit et al., 2020). In a high rainfall area of the Cameroon Estuary Complex, R. racemosa showed no or only limited autocorrelation within 25 m due to strong hydrodynamic situations (Ngeve et al., 2017). Nonetheless, one may not forget that the FSGS and mangrove populations diversity are also determined by the cumulative effect of insect, wind and bird pollination (Hermansen et al., 2014; Wee et al., 2015) and not solely by propagule dispersal patterns. The flowers of Avicennia are visited by numerous pollinators that largely comprise insects (i.e., honeybees – one of the frequent visitors), bats and birds (Clarke and Myerscough, 1991) that shows the possibility of cross-pollination as well as mating of siblings. The elevated levels of inbreeding in many sites should be explained also from a lack of pollen flow and from non-random mating, which requires a different design of study.
Conservation Relevance
Avicennia marina is largely distributed at the river mouths as a pioneer group of vegetation in Peninsular Malaysia (WIM, 2015). Besides the less human aided propagations for Avicennia spp., they are still considered as protective forest along the coastlines (Roslan and Nik Mohd Shah, 2014). Hence, these populations are mostly of natural origin and strongly support the genetic diversity observations in the present study. The clear-cut distinction of A. marina and introgressive A. marina populations on the East and the West coasts aid for species-level conservation and benefit researchers for future ecological genetic studies, without taxonomic errors. We call for the precautionary principle to not just treat the introgressive A. marina populations that captured a chloroplast of A. alba as Avicennia marina stands sensu stricto. The ecological justification for this resides in the concept of ‘cryptic ecological degradation', defined as 'functional ecological degradation that involves a qualitative decline of typical, stenotopic, vulnerable, valuable and functional species that is masked by a quantitative increase of less typical, eurytopic, disturbance-resistant, less valuable and less functional species. In a more general context, it is a qualitative ecological and socio-economic degradation of one ecosystem component that is masked by an easily detectable quantitative status quo or even increase of another component (Dahdouh-Guebas et al., 2005a,b; Dahdouh-Guebas et al., 2021). Cryptic ecological degradation has been reported in plant ecology (Koedam and Dahdouh-Guebas, 2008) and animal ecology (Bartolini et al., 2011). In the context of the present study the genetic introgression may imply yet another form of the concept of cryptic ecological degradation.
The present study also explains local FSGS with the farthest distance to the open sea in sheltered mangroves, although its absence in some areas could come from lack of distinctive power. The latter can be relevant for gene pools such as found in the introgressive A. marina on the East coast with an intrinsically low allelic and genetic diversity. Micro-evolutionary processes that involved introgressive hybridization with chloroplast capture of another species might leads to severe bottlenecks and pose constraints on the nuclear genome, allowing only a limited amount of variation to be transmitted for future generations. Low levels of diversity and a southward connectivity were also detected for Avicennia populations on this East coast. Irrespective of the morphological entity of A. marina in Malay Peninsula, there are three evolutionary significant units characterized by nuclear and chloroplast markers that merit conservation attention. The high likelihood of historical unidirectional stepping-stone migration between adjacent mangroves and the persistence of sharp genetic breaks, both add to the importance of considering coastal connectivity and conservation strategies of designing marine parks that should aim to preserve the natural cohesive forces on longer term.
Data Availability Statement
The datasets presented in this study can be found in online repositories. Genbank accession numbers of microsatellite loci and of trnH-psbA sequences are provided in Supplementary Tables 1, 2 of this article.
Author Contributions
LT: conceptualization (lead), data curation (lead), formal analysis (lead), funding acquisition (lead), investigation (lead), methodology (lead), resources (lead), visualization (supporting), writing-original draft (lead), and writing-review and editing (lead). BS: conceptualization (lead), data curation (supporting), formal analysis (supporting), funding acquisition (lead), investigation (lead), methodology (supporting), resources (lead), visualization (supporting), writing-original draft (lead), and writing-review and editing (supporting). OD and KS: conceptualization (supporting), data curation (supporting), formal analysis (supporting), funding acquisition (supporting), investigation (supporting), methodology (supporting), resources (supporting), visualization (supporting), writing-original draft (supporting), and writing-review and editing (supporting). TS: conceptualization (supporting), data curation (lead), formal analysis (supporting), funding acquisition (supporting), investigation (lead), methodology (lead), resources (supporting), visualization (supporting), writing-original draft (supporting), and writing-review and editing (supporting). FD-G: conceptualization (supporting), data curation (supporting), formal analysis (supporting), funding acquisition (lead), investigation (supporting), methodology (supporting), resources (supporting), visualization (supporting), writing-original draft (supporting), and writing-review and editing (supporting). All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Vrije Universiteit Brussel (VUB; grant number BAS42), and by the European Commission-funded Erasmus Mundus Masters Course in Tropical Biodiversity and Ecosystems (TROPIMUNDO). Also, the Higher Institution Centre of Excellence (HICoE) grant (vote #66928) awarded to the INOS, UMT has extended partial support to complete the fieldwork.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors sincerely thank EMMC TROPIMUNDO for facilitating scholarship holder's research. Authors are grateful to the Forestry Department for their kind permission to conduct this research. Mr. Mohammad Nazrin Ishak (INOS) extended full support to organize the fieldwork most successfully and collect the samples from each place.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2021.727819/full#supplementary-material
References
Adibah, A. B., Ng, W. L., and Tan, S. G. (2015). The Malay Peninsula as a barrier to gene flow in an Asian horseshoe crab species, Carcinoscorpius rotundicauda Latreille. Biochem. Syst. Ecol. 60, 204–210. doi: 10.1016/j.bse.2015.04.026
Akhir, M. F., Daryabor, F., Husain, M. L., and Tangang, F. (2015). Evidence of upwelling along Peninsular Malaysia during southwest monsoon. Open J. Mar. Sc. 5, 273–279. doi: 10.4236/ojms.2015.53022
Alleman, L. K., and Hester, M. W. (2011). Reproductive ecology of black mangrove (Avicennia germinans) along the Louisiana coast: propagule production cycles, dispersal limitations, and establishment elevations. Estuar. Coast. 34, 1068–1077. doi: 10.1007/s12237-011-9404-8
Analuddin, K., Sharma, S., Septiana, A., Sahidin, I., Rianse, U., and Nadaoka, K. (2017). Heavy metal bioaccumulation in mangrove ecosystem at the coral triangle ecoregion, Southeast Sulawesi, Indonesia. Mar. Pollut. Bull. 125, 472–480. doi: 10.1016/j.marpolbul.2017.07.065
Arnaud-Haond, S., Teixeira, S., Massa, S. L., Billot, C., Saenger, P., Coupland, G., et al. (2006). Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 15, 3515–3525. doi: 10.1111/j.1365-294X.2006.02997.x
Azman, A., Ng, K.-K.-S., Ng, C.-H., Lee, C.-T., Tnah, L.-H., Zakaria, N.-F., et al. (2020). Low genetic diversity indicating the threatened status of Rhizophora apiculata (Rhizophoraceae) in Malaysia: declined evolution meets habitat destruction. Nature 10:19112. doi: 10.1038/s41598-020-76092-4
Bartolini, F., Cimò, F., Dahdouh-Guebas, F., Fusi, M., Penha Lopes, G., and Cannicci, S. (2011). The effect of sewage discharge on the ecosystem engineering activities of two East African fiddler crab species: consequences for mangrove ecosystem functioning. Mar. Environm. Res. 71, 53–61. doi: 10.1016/j.marenvres.2010.10.002
Beerli, P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345. doi: 10.1093/bioinformatics/bti803
Beerli, P., and Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326. doi: 10.1534/genetics.109.112532
Binks, R. M., Byrne, M., McMahon, K., Pitt, G., Murray, K., and Evans, R. D. (2018). Habitat discontinuities from strong barriers to gene flow among mangrove populations, despite the capacity for long-distance dispersal. Divers. Distrib. 25, 298–309. doi: 10.1111/ddi.12851
Breitfuss, M. J., Connolly, R. M., and Dale, P. E. R. (2003). Mangrove distribution and mosquito control: transport of Avicennia marina propagules by mosquito-control runnels in southeast Queensland saltmarshes. Estuar. Coast. Shelf Sci 56, 573–579. doi: 10.1016/S0272-7714(02)00207-X
Brokaw, N., and Thompson, J. (2000). The H for DBH. For. Ecol. Manag. 129, 89–91. doi: 10.1016/S0378-1127(99)00141-3
Bryan-Brown, D. N., Connolly, R. M., Richards, D. R., Adame, F., Friess, D. A., and Brown, C. J. (2020). Global trends in mangrove forest fragmentation. Sci. Rep. 10, 7117. doi: 10.1038/s41598-020-63880-1
Cannicci, S., Burrows, D., Fratini, S., Lee, S.Y., Smith, T.J. III., Offenberg, J., et al. (2008). Faunal impact on vegetation structure and ecosystem function in mangrove forests: a review. Aquat. Bot. 89, 186–200. doi: 10.1016/j.aquabot.2008.01.009
Chablé Iuit, L. R., Machkour-M'Rabet, S., Espinoza-Ávalos, J., Hernández-Arana, H. A., López-Adame, H., and Hénaut, Y. (2020). Genetic structure and connectivity of the Red mangrove at different geographic scales through a complex transverse hydrological system from freshwater to marine ecosystems. Diversity 12:48. doi: 10.3390/d12020048
Clarke, P. J. (1993). Dispersal of Gray Mangrove (Avicennia marina) propagules in Southeastern Australia. Aquat. Bot. 43, 195–204. doi: 10.1016/0304-3770(93)90021-N
Clarke, P. J., Kerrigan, R. A., and Westphal, C. J. (2001). Dispersal patterns and early growth in 14 tropical mangroves: do early life history traits correlate with patterns of adult distribution. J. Ecol. 89, 648–659. doi: 10.1046/j.0022-0477.2001.00584.x
Clarke, P. J., and Myerscough, P. J. (1991). Buoyancy of Avicennia marina propagules in South-Eastern Australia. Aust. J. Bot. 39, 77–83. doi: 10.1071/BT9910077
Cohen, R., Kaino, J., Okello, J. A., Bosire, J. O., Kairo, J. G., Huxham, M., et al. (2013). Propagating uncertainty to estimates of above-ground biomass for Kenyan mangroves: a scaling procedure from tree to landscape level. Forest Ecol. Manage. 310, 968–982. doi: 10.1016/j.foreco.2013.09.047
Dahdouh-Guebas, F., Ajonina, G. N., Amir, A. A., Andradi-Brown, D. A., Aziz, I., Balke, T., et al. (2020). Public perceptions of mangrove forests matter for their conservation. Front. Mar. Sci. 7:603651. doi: 10.3389/fmars.2020.603651
Dahdouh-Guebas, F., Hettiarachchi, S., Sooriyarachchi, S., Lo Seen, D., Batelaan, O., Jayatissa, L. P., et al. (2005a). Transitions in ancient inland freshwater resource management in Sri Lanka affect biota and human populations in and around coastal lagoons. Curr. Biol. 15, 579–586. doi: 10.1016/j.cub.2005.01.053
Dahdouh-Guebas, F., Hug'e, J., Abuchahla, G. M. O., Cannicci, S., Jayatissa, L. P., Kairo, J. G., et al. (2021). Reconciling nature, people and policy in the mangrove social-ecological system through the adaptive cycle heuristic. Estuar. Coast. Shelf Sc. 248:106942. doi: 10.1016/j.ecss.2020.106942
Dahdouh-Guebas, F., Jayatissa, L. P., Di Nitto, D., Bosire, J. O., Lo Seen, D., and Koedam, N. (2005b). How effective were mangroves as a defence against the recent tsunami? Curr. Biol. 15, 443–447. doi: 10.1016/j.cub.2005.06.008
Dahdouh-Guebas, F., Koedam, N., Satyanarayana, B., and Cannicci, S. (2011). Human hydrographical changes interact with propagule predation behaviour in Sri Lankan mangrove forests. J. Experim. Mar. Biol. and Ecol. 399, 188–200. doi: 10.1016/j.jembe.2010.11.012
Dahdouh-Guebas, F., Verneirt, M., Tack, J. F., Van Speybroeck, D., and Koedam, N. (1998). Propagule predators in Kenyan mangroves and their possible effect on regeneration. Mar. Freshw. Res. 49, 345–650. doi: 10.1071/MF97108
De Ryck, D. J. R., Koedam, N., Van der Stocken, T., van der Ven, R. M., Adams, J., and Triest, L. (2016). Dispersal limitation of the mangrove Avicennia marina at its South African range limit in strong contrast to connectivity in its core East African region. Mar. Ecol. Prog. Ser. 545, 123–134. doi: 10.3354/meps11581
del Valle, A., Eriksson, M., Ishizawa, O. A., and Miranda, J. J. (2020). Mangroves protect coastal economic activity from hurricanes. PNAS 117, 265–270. doi: 10.1073/pnas.1911617116
Do, B. T. N., Koedam, N., and Triest, L. (2019). Avicennia marina maintains genetic structure whereas Rhizophora stylosa connects mangroves in a flooded, former inner sea (Vietnam). Estuar. Coast. Shelf Sci. 222, 195–204. doi: 10.1016/j.ecss.2019.04.005
Dodd, R. S., Afzal-Rafii, Z., Kashani, N., and Budrick, J. (2002). Land barriers and open oceans: effects on gene diversity and population structure in Avicennia germinans L. (Avicenniaceae). Mol. Ecol. 11, 1327–1338. doi: 10.1046/j.1365-294X.2002.01525.x
Donato, D. C., Kauffman, J. B., Murdiyarso, D., Kurnianto, S., Stidham, M., and Kanninen, M. (2011). Mangroves among the most carbon-rich forests in the tropics. Nat. Geosci. 4, 293–297. doi: 10.1038/ngeo1123
Duke, N. C., Benzie, J. A. H., Goodall, J. A., and Ballment, E. R. (1998). Genetic structure and evolution of species in the mangrove genus Avicennia (Avicenniaceae) in the Indo-West Pacific. Evolution 52, 1612–1626. doi: 10.1111/j.1558-5646.1998.tb02242.x
Duke, N. C., Lo, E. Y. Y., and Sun, M. (2002). Global distribution and genetic discontinuities of mangrove -emerging patterns in the evolution of Rhizophora. Trees 16, 65–79. doi: 10.1007/s00468-001-0141-7
Earl, D. M., and von Holdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Fairuz-Fozi, N., Triest, L., Ashikin Mat Zauki, A., Kaben, A. M., Nelson, B. R., Chatterji, A., et al. (2021). Mangrove horseshoe crab (Carcinoscorpius rotundicauda Latreille, 1802) populations reveal genetic break in Strait of Malacca, with connectivity along southern coasts of Peninsular Malaysia. Aquat. Cons. Mar. Freshw. Ecosyst. 31, 1559–1569. doi: 10.1002/aqc.3552
Friess, D. A., Yando, E. S., Abuchahla, G. M. O., Adams, J. B., Cannicci, S., Canty, S. W. J., et al. (2020). Mangroves give cause for conservation optimism, for now. Curr. Biol. 30, PR153–R154. doi: 10.1016/j.cub.2019.12.054
Geng, Q. F., Lian, C. L., Tao, M., Li, Q., and Hogetsus, T. (2007). Isolation and characterization of 10 new compound microsatellite markers for a mangrove tree species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Mol. Ecol. Notes 7, 1208–1210. doi: 10.1111/j.1471-8286.2007.01834.x
Goessens, A., Satyanarayana, B., Van der Stocken, T., Quispe Zuniga, M., Mohd-Lokman, H., Sulong, I., et al. (2014). Is Matang mangrove forest in Malaysia sustainably rejuvenating after more than a century of conservation and harvesting management? PLoS ONE 9:e105069. doi: 10.1371/journal.pone.0105069
Goudet, J. (2001). FSTAT version 2.9.3: a program to estimate and test gene diversities and fixation indices (update from version 1.2, Goudet 1995): a computer program to calculate F-statistic. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Haditiar, Y., Putri, M. R., Ismail, N., Muchlisin, Z. A., Ikhwan, M., and Rizal, S. (2020). Numerical study of tides in the Malacca Strait with a 3-D model. Heliyon 6:e04828. doi: 10.1016/j.heliyon.2020.e04828
Hamdan, O., and Misman, M. A. (2020). “Extents and distribution of mangroves in Malaysia,” in Status of Mangroves in Malaysia, Chapter 1, eds O. Hamdan, M. H. Tariq, P. Ismail (Malaysia: Forest Research Institute Malaysia (FRIM)), Special publication no. 50 (ISBN # 978-967-2149-81-1), 2–41.
Hardy, O. J., and Vekemans, X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. doi: 10.1046/j.1471-8286.2002.00305.x
Hasan, S., Triest, L., Afrose, S., and De Ryck, D. (2018). Migrant pool model of dispersal explains strong connectivity of Avicennia officinalis within Sundarban mangrove areas: effect of fragmentation and replantation. Estuar. Coast. Shelf Sci. 214, 38–47. doi: 10.1016/j.ecss.2018.09.007
Hermansen, T. D., Britton, D. R., Ayre, D. J., and Minchonton, T. E. (2014). Identifying the real pollinators? Exotic honeybees are the dominant flower visitors and only effective pollinators of Avicennia marina in Australian temperate mangroves. Estuar. Coast. 37, 621–635. doi: 10.1007/s12237-013-9711-3
Hodel, R. G. J., Knowles, L. L., McDaniel, S. F., Payton, A. C., Dunaway, J. F., Soltis, P. S., et al. (2018). Terrestrial species adapted to sea dispersal: differences in propagule dispersal of two Caribbean mangroves. Mol. Ecol. 27, 4612–4626. doi: 10.1111/mec.14894
Huang, L., Li, X., Huang, V., Shi, S., and Zhou, R. (2014). Molecular evidence for natural hybridization in the mangrove genus Avicennia. Pakistan J. Bot. 46, 1577–1584.
Huang, Y., Tan, F., Su, G., Deng, S., He, H., and Shi, S. (2008). Population genetic structure of three tree species in the mangrove genus Ceriops (Rhizophoraceae) from the Indo West Pacific. Genetica 133, 47–56. doi: 10.1007/s10709-007-9182-1
Ibrahim, R. I. H. (2011). A modified CTAB protocol for DNA extraction from young flower petals of some medicinal plant species. Geneconserve 10, 165–182.
Inomata, N., Wang, W.-R., Changtragoon, S., and Szmidt, A. E. (2009). Levels and patterns of DNA variation in two sympatric mangrove species, Rhizophora apiculata and R. mucronata from Thailand. Genes Genet. Syst. 84, 277–286. doi: 10.1266/ggs.84.277
Ismail, N., Taib, M., Shamsuddin, A.A., and Shazani, S. (2011). Genetic variability of wild horseshoe crab, Tachypleus gigas (Müller) in Tanjung Dawai, Kedah and Cherating, Pahang of Peninsular Malaysia. Eur. J. Sci. Res. 60, 592–601.
IUCN (2021). The IUCN Red List of Threatened Species. International Union for Conservation of Nature. Available online at: https://www.iucnredlist.org/species/178828/7619457#conservation-actions (accessed 12 June 2021).
Jamilah, M. S., Faridah, M., and Shahrudin, R. (2015). “Setiu: more than a wetland,” in Setiu Wetlands: Species, Ecosystems and Livelihood, eds F. Mohamad, J. M. Salim, J. M. Jani, and R. Shahrudin (Malaysia: Universiti Malaysia Terengganu), 87–100.
Kamarudin, K. R., and Esa, Y. (2009). Phylogeny and phylogeography of Barbonymus schwanenfeldii (Cyprinidae) from Malaysia inferred using partial cytochrome b mtDNA gene. J. Trop. Biol. Conserv. 5, 1–13.
Kathiresan, K., Rajendran, N., Balakrishnan, B., Thiruganasambandam, R., and Narayanasamy, R. (2021). Carbon sequestration and storage in planted mangrove stands of Avicennia marina. Region. Stud. Mar. Sc. 43:101701. doi: 10.1016/j.rsma.2021.101701
Koedam, N., and Dahdouh-Guebas, F. (2008). Ecological quality changes precede changes in quantity in mangrove forests. Science.
Krauss, K. W., Lovelock, C. E., McKee, K. L., Lopez-Hoffman, L., Ewe, S. M., and Sousa, W. P. (2008). Environmental drivers in mangrove establishment and early development: a review. Aquat. Bot. 89, 105–127. doi: 10.1016/j.aquabot.2007.12.014
Langston, A. K., Kaplan, D. A., and Angelini, C. (2017). Predation restricts black mangrove (Avicennia germinans) colonization at its northern range limit along Florida's Gulf Coast. Hydrobiologia 803, 317–331. doi: 10.1007/s10750-017-3197-0
Lee, S. Y., Primavera, J. H., Dahdouh-Guebas, F., McKee, K., Bosire, J. O., Cannicci, S., et al. (2014). Ecological role and services of tropical mangrove ecosystems: a reassessment. Glob. Ecol. Biogeogr. 23, 726–743. doi: 10.1111/geb.12155
Li, Y. L., and Liu, J. X. (2018). StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 18, 176–177. doi: 10.1111/1755-0998.12719
Liao, P. C., Havanond, S., and Huang, S. (2007). Phylogeography of Ceriops tagal (Rhizophoraceae) in Southeast Asia: The land barrier of the Malay Peninsula has caused population differentiation between the Indian Ocean and South China Sea. Cons. Genet. 8, 89–98. doi: 10.1007/s10592-006-9151-8
Liew, P. L., Ng, W. L., and Tan, S. G. (2015). Levels and patterns of genetic variation in an Asian horseshoe crab species, Tachypleus gigas Müller, from the Malay Peninsula. Mar. Biol. Res. 11, 879–886. doi: 10.1080/17451000.2015.1024135
Loiselle, B., Sork, V. L., Nason, J., and Graham, C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82, 1420–1425. doi: 10.1002/j.1537-2197.1995.tb12679.x
Maguire, T. L., Edwards, K. J., Saenger, P., and Henry, R. (2000a). Characterisation and analysis of microsatellite loci in a mangrove species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Theor. Appl. Genet. 101, 279–285. doi: 10.1007/s001220051480
Maguire, T. L., Saenger, P., Baverstock, P., and Henry, R. (2000b). Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.)Vierh. (Avicenniaceae). Mol. Ecol. 9, 1853–1862. doi: 10.1046/j.1365-294x.2000.01089.x
Manni, F., Guérard, E., and Heyer, E. (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier?s algorithm. Hum. Biol. 76, 173–190. doi: 10.1353/hub.2004.0034
McGuinness, K. A. (1997). Seed predation in a tropical mangrove forest: a test of the dominance-predation model in northern Australia. J. Trop. Ecol. 13, 293–302. doi: 10.1017/S0266467400010464
Melville, F., and Burchett, M. (2002). Genetic variation in Avicennia marina in three estuaries of Sydney (Australia) and implications for rehabilitation and management. Mar. Pollut. Bull. 44, 469–479. doi: 10.1016/S0025-326X(01)00259-4
Minobe, S., Fukui, S., Saiki, R., Kajita, T., Changtragoon, S., Shukor, N. A. A., et al. (2010). Highly differentiated population structure of a Mangrove species, Bruguiera gymnorhiza (Rhizophoraceae) revealed by one nuclear GapCp and one chloroplast intergenic spacer trnF-trnL. Conserv. Genet. 11, 301–310. doi: 10.1007/s10592-009-9806-3
Mori, G. M., Zucchi, M. I., Sampaio, I., and Souza, A. P. (2015a). Species distribution and introgressive hybridization of two Avicennia species from the Western Hemisphere unveiled by phylogeographic patterns. BMC Evol. Biol. 15, 1–15. doi: 10.1186/s12862-015-0343-z
Mori, G. M., Zucchi, M. I., and Souza, A. P. (2015b). Multiple-geographic-scale genetic structure of two mangrove tree species: the roles of mating system, hybridization, limited dispersal and extrinsic factors. PLoS ONE 10:e0118710. doi: 10.1371/journal.pone.0118710
Ngeve, M., Van der Stocken, T., Menemenlis, D., Koedam, N., and Triest, L. (2017). Hidden founders? Strong bottlenecks and fine-scale genetic structure in mangrove populations of the Cameroon Estuary complex. Hydrobiologia 803, 189–207. doi: 10.1007/s10750-017-3369-y
Ochoa-Zavala, M., Jaramillo-Correa, J. P., Piñero, D., Nettel-Hernanz, A., and Núñez-Farfán, J. (2019). Contrasting colonization patterns of black mangrove (Avicennia germinans (L.) L.) gene pools along the Mexican coasts. J. Biogeogr. 46, 884–898. doi: 10.1111/jbi.13536
Osland, M. J., Feher, L. C., Spivak, A. C., Nestlerode, J. A., Almario, A. E., Cormier, N., et al. (2020). Rapid peat development beneath created, maturing mangrove forests: ecosystem changes across 25-year chronosequence. Ecol. Applic. 30, e02085. doi: 10.1002/eap.2085
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard, J. K., Stephens, M., and Donelly, P. S. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Rabinowitz, D. (1978). Dispersal properties of mangrove propagules. Biotropica 10, 47–57. doi: 10.2307/2388105
Ragavan, P., Zhou, R., Ng, W. L., Rana, T. S., Mageswaran, T., Mohan, P. M., et al. (2017). Natural hybridization in mangroves – an overview. Bot. J. Linnean Soc. 185, 208–224. doi: 10.1093/botlinnean/box053
Razali, M. S., Zulkifli, M. K. F., and Wan Ahmad, W. J. (2016). “Distribution and rarity of mangrove and coastal plants in developing indicators of hotspots in Setiu Wetlands,” in Proceedings of the Setiu Wasteland Scientific Expedition Seminar “Connecting Science to Society” (Malaysia: Universiti Malaysia Terengganu), 14–23.
Rizal, S., Damm, P., Wahid, M. A., Sundermann, J., Ilhamsyah, Y., and Iskandar, T. M. (2012). General circulation in the Malacca Strait and Andaman Sea: a numerical model study. Am. J. Environm. Sc. 8, 479–488. doi: 10.3844/ajessp.2012.479.488
Rizal, S., Setiawan, I., Iskandar, T., Ilhamsyah, Y., Wahid, M. A., and Musman, M. (2010). Currents simulation in the Malacca Straits by using three-dimensional numerical model. Sains Malaysiana 39, 519–524.
Roslan, A., and Nik Mohd Shah, N. M. (2014). A Working Plan for the Matang Mangrove Forest Reserve, Perak: The First 10- Year Period (2010–2019) of the Third Rotation (6th Revision). Malaysia: State Forestry Department of Perak, 229p.
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi: 10.2307/2446155
Satyanarayana, B., Idris, I. F., Mohamad, K. A., Mohd-Lokman, H., Shazili, N. A. M., and Dahdouh-Guebas, F. (2010). Mangrove species distributions and abundances in relation to local environmental settings: a case-study at Tumpat, Kelantan Delta, East coast of Peninsular Malaysia. Bot. Mar. 53, 79–88. doi: 10.1515/BOT.2010.006
Seddon, N., Chausson, A., Berry, P., Girardin, C. A. J., and Smith, Turner, B. (2020). Understanding the value and limits of nature-based solutions to climate change and other global challenges. Philos. Trans. R. Soc. B Biol. Sci. 375:20190120. doi: 10.1098/rstb.2019.0120
Smith, T. J III. (1987). Seed predation in relation to tree dominance and distribution in mangrove forests. Ecology 68, 266–273. doi: 10.2307/1939257
Steinke, T. D. (1986). A preliminary study of buoyancy behavior in Avicennia marina propagules. S. Afr. J. Bot. 52, 559–565. doi: 10.1016/S0254-6299(16)31492-2
Steinke, T. D., and Ward, C. J. (2003). Use of plastic drift cards as as indicators of possible dispersal of propagules of the Mangrove Avicennia Marina by ocean currents. Afr. J. Mar. Sci. 25, 169–176. doi: 10.2989/18142320309504007
Su, G. H., Huang, Y. L., Tan, F. X., Ni, X. W., Tang, T., and Shi, S. H. (2006). Genetic variation in Lumnitzera racemosa, a mangrove species from the Indo-West Pacific. Aquat. Bot. 84, 341–346. doi: 10.1016/j.aquabot.2006.01.001
Suppapan, J., Pechsiri, J. O., Thong, S., Vanichanon, A., Sangthong, P., and Supmee, V. (2017). Population genetic analysis of oceanic paddle crab (Varuna litterata) in Thailand. Sains Malaysiana 46, 2251–2261. doi: 10.17576/jsm-2017-4612-01
Tangah, J., Chung, A. Y. C., Nilus, R., Abdullah, S. Z. S., Abu, S., and Lohuji, P. L. (2020). “Management system of mangroves in Sabah: issues and challenges,” in Status of Mangroves in Malaysia, Chapter 9, eds O. Hamdan, M. H. Tariq, P. Ismail (Malaysia: Forest Research Institute Malaysia (FRIM)), Special publication no. 50, (ISBN # 978-967-2149-81-1), 142–155.
Teixeira, S., Arnaud-Haond, S., Duarte, C., and Serrao, E. (2003). Polymorphic microsatellite DNA markers in the mangrove tree Avicennia alba. Mol. Ecol. Notes 3, 544–546. doi: 10.1046/j.1471-8286.2003.00505.x
Triest, L. (2008). Molecular ecology and biogeography of mangrove trees towards conceptual insights on gene flow and barriers: a review. Aquat. Bot. 89, 138–154. doi: 10.1016/j.aquabot.2007.12.013
Triest, L., Del Socorro, A., Gado, V. J., Mazo, A. M., and Sierens, T. (2021a). Avicennia genetic diversity and fine-scaled structure influenced by coastal proximity of mangrove fragments. Front. Mar. Sci. 8:643982. doi: 10.3389/fmars.2021.643982
Triest, L., Hasan, S., Mitro, P., De Ryck, D., and Van der Stocken, T. (2018). Geographical distance and large rivers shape genetic structure of Avicennia officinalis in the highly dynamic sundarbans mangrove forest and Ganges Delta Region. Estuaries Coast. 41, 908–920. doi: 10.1007/s12237-017-0309-z
Triest, L., Van der Stocken, T, Sierens, T., Deus, E. K., Mangora, M. M., and Koedam, N. (2021c). Connectivity of Avicennia marina populations within a proposed marine transboundary conservation area between Kenya and Tanzania. Biol. Cons. 256:109040. doi: 10.1016/j.biocon.2021.109040
Triest, L., and Van der Stocken, T. (2021). Coastal landform constrains dispersal in mangroves. Front. Mar. Sci. 8:617855. doi: 10.3389/fmars.2021.617855
Triest, L., Van der Stocken, T., Akinyi, A. A., Sierens, T., Kairo, J., and Koedam, N. (2020). Channel network structure determines genetic connectivity of landward–seaward Avicennia marina populations in a tropical bay. Ecol Evol. 10, 12059–12075. doi: 10.1002/ece3.6829
Triest, L., Van der Stocken, T., De Ryck, D., Kochzius, M., Lorent, S., Ngeve, M., et al. (2021b). Expansion of the mangrove species Rhizophora mucronata in the Western Indian Ocean launched contrasting genetic patterns. Sci. Rep. 11:4987. doi: 10.1038/s41598-021-84304-8
Van der Stocken, T., Carroll, D., Menemenlis, D., Simard, M., and Koedam, N. (2019b). Global-scale dispersal and connectivity in mangroves. Proc. Natl. Acad. Sci. U. S. A. 116, 915–922. doi: 10.1073/pnas.1812470116
Van der Stocken, T., Vanschoenwinkel, B., De Ryck, D. J. R., Bouma, T. J., Dahdouh-Guebas, F., and Koedam, N. (2015). Interaction between water and wind as a driver of passive dispersal in mangroves. PLoS ONE 10:e0121593. doi: 10.1371/journal.pone.0121593
Van der Stocken, T., Wee, A. K. S., De Ryck, D. J. R., Vanschoenwinkel, B., Friess, D. A., Dahdouh-Guebas, F., et al. (2019a). A general framework for propagule dispersal in mangroves. Biol. Rev. 94, 1547–1575. doi: 10.1111/brv.12514
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Walters, B. B., Rönnbäck, P., Kovacs, J. M., Crona, B., Hussain, S. A., Badola, R., et al. (2008). Ethnobiology, socio-economics and management of mangrove forests: a review. Aquat. Bot. 89, 220–236. doi: 10.1016/j.aquabot.2008.02.009
Wan Juliana, W. A., Razali, M. S., Ngadi, E., Zulkifli, F., Razak, A., Mohd-Arrabe, A. B., et al. (2014). Langkawi Geopark is home to rare mangrove flora. Malaysian Naturalist 67, 50–53.
Wee, A. K. S., Low, S. Y., and Webb, E. L. (2015). Pollen limitation affects reproductive outcome in the bird-pollinated mangrove Bruguiera gymnorrhiza (Lam.) in a highly urbanized environment. Aquat. Bot. 120, 240–243. doi: 10.1016/j.aquabot.2014.09.001
Wee, A. K. S., Noreen, A. M. E., Ono, J., Takayama, K., Kumar, P. P., Tan, H. T. W., et al. (2020). Genetic structure across a biogeographical barrier reflect dispersal potential of four Southeast Asian mangrove plant species. J. Biogr. 47, 1258–1271. doi: 10.1111/jbi.13813
Wee, A. K. S., Takayama, K., Asakawa, T., Thompson, B., Onrizal, S. S., Tung, N. X., et al. (2014). Oceanic currents, not land masses, maintain the genetic structure of the mangrove Rhizophora mucronata Lam. (Rhizophoraceae) in Southeast Asia. J. Biogeogr. 41, 954–964. doi: 10.1111/jbi.12263
Wee, A. K. S., Teo, J. X. H., Chua, J. L., Takayama, K., Asakawa, T., Meenakshisundaram, S. H., et al. (2017). Vicariance and oceanic barriers drive contemporary genetic structure of widespread mangrove species Sonneratia alba J. Sm in the Indo-West Pacific. Forests 8:483. doi: 10.3390/f8120483
Yan, Y.-B., Duke, N. C., and Sun, M. (2016). Comparative analysis of the pattern of population genetic diversity in three Indo-West Pacific Rhizophora mangrove species. Front. Plant Sci. 7:1434. doi: 10.3389/fpls.2016.01434
Yang, M.-C., Chen, C.-P., Hsieh, H.-L., Huang, H., and Chen, C. A. (2009). “Phylogeography, demographic history, and reserves network of horseshoe crab, Tachypleus tridentatus, in the South and East China seaboards,” in Biology and Conservation of Horseshoe Crabs, eds J. Tanacredi, M. Botton, and D. Smith (Boston, MA: Springer), 163–181.
Yang, Y., Li, J., Yang, S., Li, X., Fang, L. U., Zhong, C., et al. (2017). Effects of Pleistocene sea-level fluctuations on mangrove population dynamics: a lesson from Sonneratia alba. BMC Evolut. Biol. 17:22. doi: 10.1186/s12862-016-0849-z
Keywords: Avicennia, genetic structure, connectivity, microsatellites, trnH-psbA
Citation: Triest L, Satyanarayana B, Delange O, Sarker KK, Sierens T and Dahdouh-Guebas F (2021) Barrier to Gene Flow of Grey Mangrove Avicennia marina Populations in the Malay Peninsula as Revealed From Nuclear Microsatellites and Chloroplast Haplotypes. Front. Conserv. Sci. 2:727819. doi: 10.3389/fcosc.2021.727819
Received: 19 June 2021; Accepted: 27 August 2021;
Published: 23 September 2021.
Edited by:
Yoshiaki Tsuda, University of Tsukuba, JapanReviewed by:
Wei Lun Ng, Xiamen University Malaysia, MalaysiaDamien Daniel Hinsinger, INRA Centre Versailles-Grignon, France
Copyright © 2021 Triest, Satyanarayana, Delange, Sarker, Sierens and Dahdouh-Guebas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludwig Triest, ltriest@vub.be
 Ludwig Triest
Ludwig Triest Behara Satyanarayana
Behara Satyanarayana Olga Delange
Olga Delange Kishore Kumar Sarker
Kishore Kumar Sarker Tim Sierens
Tim Sierens Farid Dahdouh-Guebas
Farid Dahdouh-Guebas