- Gene Silencing Section, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Understanding the functional effects of the wide-range of aberrant genetic characteristics associated with the human chromosome 8q24 region in cancer remains daunting due to the complexity of the locus. The most logical target for study remains the MYC proto-oncogene, a prominent resident of 8q24 that was first identified more than a quarter of a century ago. However, many of the amplifications, translocation breakpoints, and viral integration sites associated with 8q24 are often found throughout regions surrounding large expanses of the MYC locus that include other transcripts. In addition, chr.8q24 is host to a number of single nucleotide polymorphisms associated with cancer risk. Yet, the lack of a direct correlation between cancer risk alleles and MYC expression has also raised the possibility that MYC is not always the target of these genetic associations. The 8q24 region has been described as a “gene desert” because of the paucity of functionally annotated genes located within this region. Here we review the evidence for the role of other loci within the 8q24 region, most of which are non-coding transcripts, either in concert with MYC or independent of MYC, as possible candidate gene targets in malignancy.
Introduction
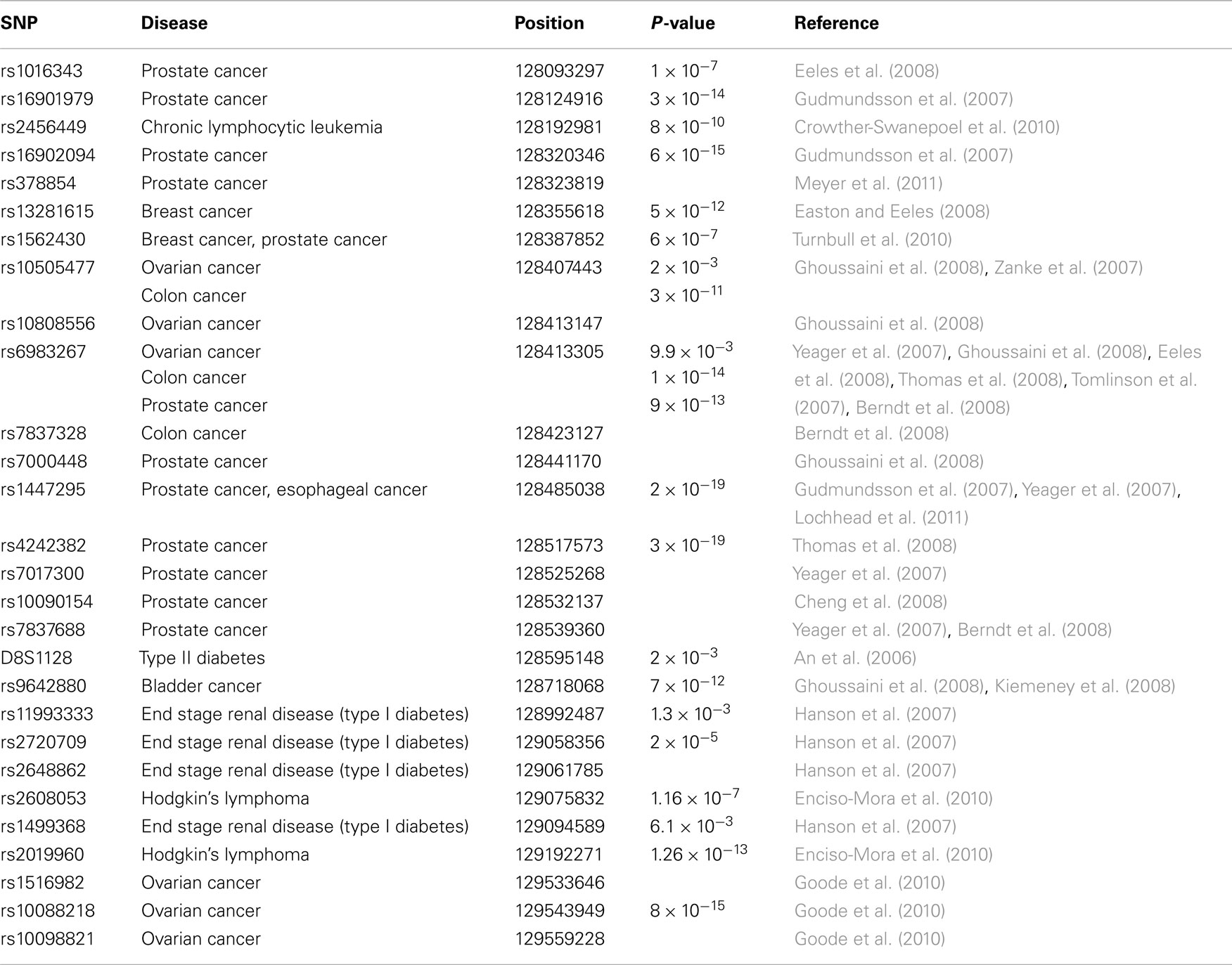
Genome wide association studies (GWAS) have identified a large number of single nucleotide polymorphisms (SNPs) in a segment of about 2 Mb mapping to human chromosome 8q24 (chr.8q24) that are linked to susceptibility for different diseases including cancers of the prostate, breast, esophagus, head and neck, ovarian, colon, and pancreas (Easton and Eeles, 2008; Grisanzio and Freedman, 2010; Figure 1). One gene found within this chr.8q24 region, MYC, stands out as the most likely candidate to be functionally linked to these cancer risk-associated SNPs given its well defined role as an oncogenic transcription factor (Meyer et al., 2006). In addition, to GWAS linkage, there are several other cancer-associated features of this region of chr.8q24 (128.0–130.0 Mb) that serve to further establish the significance of 8q24 in driving tumor development that also complicates its’ study as well. While MYC is recognized to be the most frequently amplified protein-coding gene across all cancer types (Beroukhim et al., 2010), the fact that increased copies of MYC are often accompanied by co-amplification of an adjacent non-coding locus, PVT1 is not fully appreciated nor understood (Asker et al., 1988; Shtivelman and Bishop, 1989; Bakkus et al., 1990; Minarovits et al., 1990; Figure 1). Furthermore, chromosomal translocations that appear to target MYC are often found in Burkitt’s lymphoma (BL) and other non-Hodgkin’s lymphoma types, but a subset of these lymphomas (15–20%) exhibiting breakpoints as far as 300–400 kb downstream of MYC on 8q24, is a feature that complicates the hypothesis that singular MYC is always the intended target.
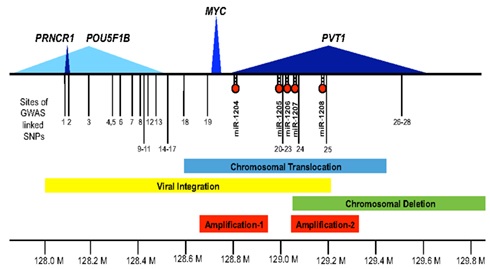
Figure 1. The 8q24 gene desert. Shown along the 1.8-Mb region of human chromosome 8q24 (128.0 Mb thru 129.8 Mb) are the extent of the protein encoding MYC transcript (blue), the lincRNA PVT1 (dark blue), the putative lincRNA PRNCR1 (dark blue), the less defined POU5F1P1 transcript (light blue), and the microRNAs, miR-1204 through miR-1208. Also depicted are the locations of GWAS defined SNPs (1–28) associated with disease susceptibility (see Table 1 for details). The bars denote focused regions of amplification (shown in red), chromosomal translocation (blue), viral integration (yellow), or deletion (green) that have been observed in different tumor types (see text for details).
In addition to MYC, the transcripts, POU5F1B and PVT1, reside within the 128.0- to 130.0-Mb region of chr.8q24 most often associated with GWAS, translocation, or integration events (Figure 1). The POU5F1B gene was long considered as a pseudogene (POU5F1P1), however, a recent report now proposes that it could encode a weak transcriptional activator (Kastler et al., 2010). The PVT1 locus expresses several alternatively spliced non-coding transcripts (Shtivelman et al., 1989; Huppi et al., 1990; Shtivelman and Bishop, 1990) and PVT1 is the host to a cluster of microRNAs (miR-1204 ∼ 1208; Huppi et al., 2008). Nevertheless, no clear functional role for any of these transcripts has been identified and the paucity of coding region genes within this chromosomal region has lead to its description as the “8q24 gene desert” (Ghoussaini et al., 2008; Wasserman et al., 2010). In this review we discuss the growing evidence that the 8q24 gene desert in fact, has a complex pattern of transcription that, in addition to MYC, should be considered when assaying for disease candidate loci.
SNPs Across the 8q24 Region are Associated with Disease-Risk
Several years ago, our laboratory and others (Rothberg and Otto, 1995; Siwarski et al., 2001) identified SNPs within the coding region of MYC (S11N, CAA-33, and S288K). For one of the SNPs (S288K), we were able to follow the inheritance of the rare non-synonymous variant allele in a small family pedigree and compare expression of MYC between the two alleles. Our report highlighted the possibility of reduced expression from two rare variant MYC alleles (S288K and CAA-33) and thus, demonstrated for the first time, differential expression levels of germline MYC alleles (Siwarski et al., 2001). At the time, we could not distinguish whether reduced MYC expression was due to the differential binding of transcription factors (such as YY1) to the actual SNP sequence or whether associated 5′ or 3′ base changes accompanied the presence of these alleles. The limitations of this study were the small size of the cohort, coupled with the inability to find the rare variant outside of this family. However, with development of high throughput, population-based SNP analysis, GWAS are empowered with the ability to avoid false positive discovery through study of large, well-documented case control and cohort groups that avoid bias as a result of for example, survivorship and sample integrity.
One of the first cancer susceptibility GWAS to be published linked prostate cancer susceptibility to SNP variants in the region of chr.8q24, far upstream of MYC in the Icelandic population (Amundadottir et al., 2006). Subsequent reports from the study of Caucasian men in Sweden and the US further confirmed the linkage to prostate adenocarcinoma and narrowed the region of risk to two alleles, the −8 allele at the marker DG8S737 and the A allele of rs1447295 (Amundadottir et al., 2006). The incidence of the DG8S737 marker was also increased in patients at risk for prostate cancer in an independent study of African Americans but this study also suggested that differences in the overall frequency of the DG8S737 allele in Caucasians versus African Americans might bias the link to prostate cancer (Freedman et al., 2006). As a result of the many additional studies of SNPs from the region of chr.8q24 (Gudmundsson et al., 2007; Haiman et al., 2007; Yeager et al., 2007; Eeles et al., 2008; Ghoussaini et al., 2008; Thomas et al., 2008), several sub-regions of susceptibility can be assigned to prostate cancer risk in a region of ∼2.7 Mb of DNA extending from a position centromeric to POU5F1B to include a region of several hundred kb of DNA proximal of MYC (Figure 1; Table 1).
While many of the chr.8q24 associated SNPs were originally identified as susceptibility markers for prostate cancer, additional SNPs in the region of chr.8q24 have now been associated with many other cancers (Figure 1; Table 1). For example, two SNPs (i.e., rs13281615 and rs1562430) are linked to breast cancer (Easton and Eeles, 2008), and the SNP rs9642880 has been uniquely associated with bladder cancer (Ghoussaini et al., 2008). Some SNPs including rs10505477 (positioned at 128.40 Mb), rs1562430 (position 128.38 Mb), and rs6983267 (position 128.41 Mb) are found to be associated with increased susceptibility in multiple cancers whereas some SNP variants appear to be unique susceptibility markers to a singular cancer type. An example of the unique cancer based SNP is rs2456449, which to date, has only been associated with susceptibility to B-cell chronic lymphocytic leukemia (Crowther-Swanepoel et al., 2010). In addition to the SNP variants located to the proximal (5′) side of MYC, a few susceptibility variants have been identified 3′ of MYC in the PVT1 region.
A number of SNP variants for ovarian cancer have been identified on both the 5′ (Ghoussaini et al., 2008) and 3′ side of MYC (Goode et al., 2010), respectively. Although this might implicate MYC as the central target in ovarian cancer, some doubt as to the strength of these GWAS studies has been raised, as the positioning of the risk alleles appears to be report specific (White et al., 2010; Braem et al., 2011). The fact that ovarian cancer is a heterogeneous disease may also be a factor leading to the lack of reproducibility in assigning ovarian cancer risk. Nevertheless, independent findings such as, frequent amplification in ovarian cancer (see below) still suggests that the 8q24 locus may be an important region in the progression of ovarian cancer.
The presence of multiple loci associated with susceptibility to different cancer specificities raises another provocative question as to whether the clustering of many 8q24 SNP variants could be pleiotropic. For instance, is the region of 8q24 unique in the genome in harboring many cancer risk loci or will additional regions of multiple cancer risk eventually be identified. A possible clue comes from a study of disease susceptibility in end stage renal disease (ESRD). A series of SNP variants located within the PVT1 region have been identified as candidate loci for ESRD in type 1-diabetes (Hanson et al., 2007). Further characterization of the PVT1 gene in diabetic kidney disease studies suggest a possible molecular mechanism for the non-coding PVT1 gene in extracellular matrix (ECM) accumulation (Alvarez and DiStefano, 2011). More importantly, it relies on a function of PVT1 independent of MYC, in the progression of ESRD and it underscores the possibility that the 8q24 locus is multifaceted and not simply dominated by a single regulatory element or gene. Interestingly, diabetes and cancer are known to share common risk factors although for prostate cancer in particular, the risk is inversely correlated with diabetes (Giovannucci et al., 2010).
The challenge of population-based cancer susceptibility in GWAS is the establishment of a connection between the position of SNP variants and functionality. This is critical because most cancer-associated SNP variants have been found to be synonymous, mapping to regions of intervening DNA, often very distant from candidate protein-coding genes. This has been especially apparent in the chr.8q24 region where MYC resides as the most likely candidate gene target yet establishing a functional correlation with MYC expression has been surprisingly inconsistent.
MYC
The MYC gene (128748315–128753680) is comprised of three exons; exon 1 is non-coding, exons 2 and 3 are protein-coding (for review see Pelengaris and Khan, 2003). The translation start signal (TSS) in exon 2 generates a protein of 439 amino acids (64 kDa). Alternative translational initiation start sites are capable of generating a larger protein, p67 and a shorter one, MYC. Transcription of the MYC gene initiates from one of two promoters, P1 or P, and there are several well-characterized elements that bind the MYC promoter including CT, FUSE, TFIIH, and FIR. When chromosomal translocations displace P1 and P2 with the immunoglobulin enhancers (as observed in BL), de-regulated MYC expression persists from a series of cryptic promoters within the intron.
MYC is one of the most studied oncogenes stemming from its association with a large number of diseases and indeed, a link to MYC expression for alleles associated with particular SNPs would be a first candidate gene of choice. In concert with this hypothesis, Sole et al. (2008) found consistent up-regulated expression of MYC in normal prostate samples with regard to at least one risk locus (rs1447295). However, another group (Pomerantz et al., 2009a) failed to find any correlation between MYC expression and a risk allele (including rs1447295) in their normal prostate samples. Possible reasons for the inconsistencies could be differences in tissue purity or integrity of RNA, but a third study now suggests that the ideal window to examine the affects of risk allele and expression of MYC may be earlier in the formation of the tumor than what is usually tested (Wasserman et al., 2010). MYC expression in association with disease-risk alleles has also been examined in colorectal tissues and once again, three independent studies have failed to establish any correlation between MYC expression and the risk alleles (Zanke et al., 2007; Pomerantz et al., 2009b; Tuupanen et al., 2009). In a more detailed study of a single risk allele rs6983267 in colorectal samples, Wright et al. (2010) dissected MYC expression into specific chromosomes and found MYC expression to be allele specific. As noted above, SNPs both upstream and downstream of MYC have been linked to ovarian cancer suggesting MYC is the target gene in this case. However, studies of risk-associated alleles in ovarian cancer have failed to show correlation with MYC expression (Goode et al., 2010). Thus, the issue of association between risk alleles and MYC expression remains inconsistent.
The co-localization of epigenetic landmarks with SNP variants could also be a source of transacting regulation of nearby genes such as MYC. In fact, chromatin immunoprecipitation studies have found acetylation and histone markers consistent with regulatory activity for one SNP variant (rs6983267) in colorectal cancer and reporter assays have shown allele specific enhancement of MYC expression in colorectal cancer lines (Pomerantz et al., 2009b; Wright et al., 2010). Several other SNP variants in prostate and breast cancer show similar properties in a tissue specific fashion (Ahmadiyeh et al., 2010). Further, the rs6983267 risk allele has been shown to bind avidly to the TCF7L2 transcription factor compared to the non-risk allele (Sotelo et al., 2010). The question is, do these enhancers interact with MYC or with other genes in the chr.8q24 region or with transacting elements elsewhere in the genome? Several studies have now demonstrated direct binding of the rs6983267 risk region with promoter regions of MYC in colorectal cancer cell lines (Pomerantz et al., 2009b; Wright et al., 2010) and prostate cancer cell lines (Sotelo et al., 2010). However, the results in the prostate study are conflicting in that the greatest enhancer activity was found not in association with the risk locus rs3983267 but with a neighboring region of DNA (Sotelo et al., 2010). Furthermore, enhancer activity associated with rs3983267 was seen only in the presence of both TCF7L2 and CTNNB1 and reporter assays showed enhanced expression with the non-risk allele compared to the risk allele. Nevertheless, Wasserman et al. (2010) are able to show that the risk allele does indeed bind more avidly than the non-risk allele in vivo (prostate and coagulating glands). With the inconsistency of these results, it is apparent that the lack of correlation between MYC expression and the presence of the risk allele in prostate cancer would indicate that either the window of accessibility is earlier in the development of the tumor or the gene target is not MYC. Consistent with this argument are the studies of Cole and colleagues (Wright et al., 2010) who show that the rs3983267 enhancer has significant binding activity to both MYC and also the neighboring PVT1 promoters. While this could be explained as co-regulated expression of both MYC and PVT1, it is also possible that the intended target in some of these malignancies is something other than MYC. For example, in an examination of expression in prostate cancer specimens, significant increased expression was observed with at least one PVT1 transcript variant (Pomerantz et al., 2009b).
PVT1
The “plasmacytoma variant translocation” or PVT1 locus was originally defined as a cluster of chromosomal translocation or viral integration breakpoints located several hundred kilobases (kb) downstream of MYC in B-cell or T cell lymphomas (Erikson et al., 1983; Webb et al., 1984). The term “variant” referred to the frequency of chromosomal breakpoints in BL or mouse plasmacytoma found in the PVT1 region (10–20%) compared to those breakpoints found near MYC (80–90%). Although the breakpoints fuse a segment of PVT1 with immunoglobulin light chain gene, it was originally thought that interruption of PVT1 simply lead to de-regulated expression of nearby MYC. However, lack of consistent up-regulation of MYC in a number of lymphomas, lead to the search and discovery of PVT1 transcripts in both human and mouse (Shtivelman et al., 1989; Huppi et al., 1990). The human PVT1 transcript starts with either exon 1a (128806779) or exon 1b (128808208) ∼40–42 kb 3′ of the MYC transcript and with extensive alternative splicing, the PVT1 transcript can extend 1.1 Mb distal to MYC to create transcripts of between 2.7 and 3.3 kb in length (Figure 2). Among the human PVT1 transcripts identified, expression of exon 1a far out-weighs the utilization of exon 1b. Most importantly, exons 1a and 1b are mutually exclusive in that they have not been found spliced together in cDNAs. In fact, it is possible that exon 1b is only expressed in malignant cell lines containing amplified PVT1 (Guan et al., 2007). Although the most commonly used exons are 1a, 2, 3b, 4b, 7, 8, and 9, it should be emphasized that alternative splicing in different tissues makes it difficult to assign a single reference consensus sequence (Figure 2). One of the most remarkable features of the PVT1 locus since the discovery of cDNA clones in the early 1990s is the conservation of transcriptional activity in this region across synteny. PVT1 transcripts have been cloned from rat (Koehne et al., 1989; Tsichlis et al., 1989), mouse (Huppi et al., 1990), and human (Shtivelman et al., 1989) and yet no consistent protein encoding open reading frames could be established despite the utilization of as many as 8–10 alternative exons. Despite the consistent localization of PVT1 transcripts close to MYC in several different species, the inconsistency of the location of exons in these species coupled with a complete lack of sequence conservation across species suggested early on to investigators, including ourselves, that PVT1 was not translated as a protein product from the normal transcript. Thus PVT1 became an early example of a non-coding RNA (for a current review of non-coding RNA see Esteller, 2011), similar to the loci H19 (Brannan et al., 1990) and XIST (Brockdorff et al., 1992; Brown et al., 1992). Even though suggestions were made that PVT1 could act as a regulatory RNA, a lack of defined function left PVT1 transcription as a curiosity (Huppi and Siwarski, 1994).
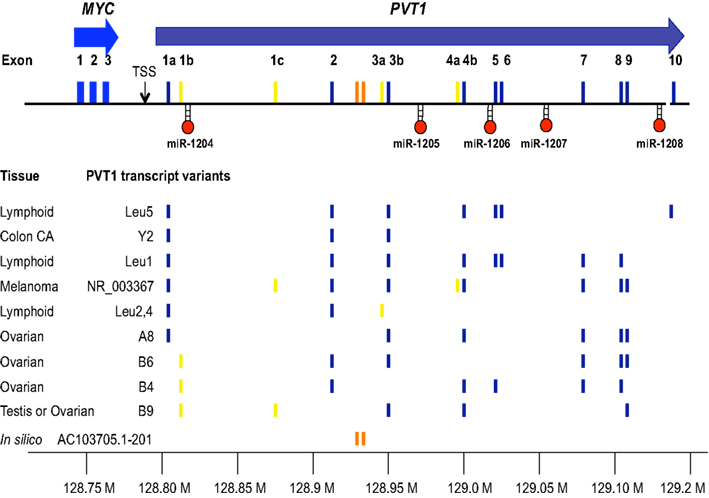
Figure 2. Transcripts from the 8q24 region downstream of MYC. MYC (blue) is used as the anchoring position for depicting the series of alternative spliced transcripts from PVT1 and the location of miRNAs (miR-1204 through miR-1208). The positions of the exons are located with respect to the chromosome 8 address (128.80–129.20 Mb) and are colored with respect to whether they are frequently used (dark blue), infrequently used (yellow), or part of a putative lincRNA (orange). Shown are the PVT1 exon composition observed in cDNA clones from the tissues indicated [Leu1, Leu2, 4, Leu5 (Huppi, unpublished), A8, B6, B4, B9 (Guan et al., 2007) Y2, NR_003367 (Shtivelman et al., 1989), AC103705.1-201 (Ensembl annotation lincRNA pipeline)]. Only unique variants have been listed to abbreviate the list. The reader is encouraged to review the complete list online (UCSC database).
Our improved understanding of the importance of long RNAs (lincRNAs) and the finding of multiple disease-risk SNPs close to the PVT1 have prompted renewed interest and investigation into this locus. It was a search for methylated chromatin (H3K4me3 and H3K36me3) that led to the prediction, and ultimately, the identification of three mouse PVT1 cDNAs (Pvt1-201, Pvt1-202, Pvt1-203) as potential lincRNAs (Guttman et al., 2009). As a human lincRNA counterpart in the PVT1 region has been predicted from the Ensembl lincRNA annotation pipeline (AC103705.1-201 in Figure 2) there is a potential for lincRNA based PVT1 function, but this still needs to be validated in vivo. Recent evidence has also noted PVT1 as among the top fold decrease in expression during the transition from induced pluripotent stem cells (iPSCs) to neurons (Lin et al., 2011). Additional evidence for a functional role for PVT1 has also come from the study of the effect of silencing of PVT1 transcripts via RNAi. Specifically, Gray and colleagues were able to induce apoptosis in ovarian or breast cell lines following silencing of PVT1 in cell lines with amplified chr.8q24 but they found no apoptosis in non-amplified cell lines (Guan et al., 2007). Why 8q24 amplified cell lines as opposed to non-amplified cell lines are uniquely susceptible to silencing of PVT1 is puzzling and will need further clarification. Evidence that PVT1 was acting independently of MYC in these experiments came from the fact that inhibition of MYC alone failed to induce apoptosis. Further, a recent publication (Meyer et al., 2011) has identified a risk variant rs378854 that is located within a DNase hypersensitive site and within a repetitive region ∼0.5 Mb upstream of the PVT1 promoter (also upstream of MYC). This region had not been detected previously due to the need to extract repetitive sequences from DNase analysis. This risk allele (rs378854) appears to be prostate tissue specific. Binding of the transcription factor YY1 is observed on the risk variant and further studies with chromatin conformation capture (3C) have shown the ability of this region to interact long range with MYC, PVT1 (but not FAM84B a gene located centromeric of POU5F1B). This study also correlated reduced expression of PVT1 (in the absence of a change in MYC expression) with the loss of YY1 binding to the risk allele. Another hint at a possible functional role for PVT1 has been found in a transposon-based genetic screen of Gemcitabine sensitivity in pancreatic cells (You et al., 2011). Although this study needs to be validated, transposon-based inactivation of the PVT1 transcript alone (independent of MYC) resulted in increased sensitivity to Gemcitabine in the pancreatic cell line ASPC-1 whereas over-expression of PVT1 led to increased cellular proliferation even in the presence of Gemcitabine.
The discovery of miRNAs several years ago also encouraged us to re-examine the PVT1 locus for the presence of RNAs that may have been too small to be detected in the original cDNA cloning studies. Our detailed study of the refined region of 400 kb surrounding the PVT1 region lead to the discovery of a cluster of miRNAs (miR-1204 ∼ 1208) residing within the PVT1 locus but not overlapping with any PVT1 exons (Huppi et al., 2008). One miRNA, miR-1204 resides adjacent to the little used exon 1b of PVT1 and includes a very short overlapping segment with the exon. From 5′ RACE experiments, it is clear that miR-1204 and PVT1 share promoters and probably share regulatory elements for the most part as expression of both PVT1 and miR-1204 often appear to respond together (Barsotti et al., 2011). Each of the other miRNAs, miR-1205 ∼ 1208, appear to be differentially regulated from PVT1 and miR-1204 (Huppi et al., 2008). Interestingly, in the study of prostate cancer specimens and the risk allele rs378854, Ponder and colleagues observe evidence of a possible correlation between expression of miR-1208 and prostate cancer (Meyer et al., 2011), although this finding will need further study. In another recent study of human diploid fibroblasts, miR-1204 was noted as having the greatest increase in expression levels among miRNAs in senescent versus early passage fibroblasts (Marasa et al., 2010) and miR-1204 was also noted as showing increased expression during the iPSCs neuronal differentiation (Lin et al., 2011). Although preliminary, these findings implicate miR-1204 in the subset of miRNAs associated with suppression of tumor growth. Our recent findings have shown that PVT1 and miR-1204 are both responsive to Daunorubicin treatment in a number of cell lines including colon, fibroblast, and lymphoid (Barsotti et al., 2011). As the presence of a p53 binding site in the promoter region of PVT1 and miR-1204 is the basis for the Daunorubicin mediated response to DNA damage in several different tissue types, these results implicate a pro-survival role for PVT1 consistent with the findings of You et al. (2011) in the Gemcitabine experiments in pancreatic cells. In contrast, activation of miR-1204 by binding of p53 appears to be anti-proliferative suggesting a very interesting series of opposing actions of PVT1 versus miR-1204. Further studies on the apparent opposing actions of PVT1 and miR-1204 are being carried out.
Studies in type 1-diabetes ESRD (Millis et al., 2007; Alvarez and DiStefano, 2011) have ruled MYC out as the candidate gene whereas certain variants of the PVT1 transcript were highly expressed in renal cells and specifically up-regulated in hyperglycemic conditions. Furthermore, silencing of PVT1 appeared to reduce the expression of specific genes, including FN1, COL4, TGFB1, and PAI-1 that are involved in the composition of the ECM affecting glomerular filtration (Alvarez and DiStefano, 2011). Perhaps for the first time, a specific function might be assigned to the non-coding PVT1 transcript independent of MYC, but this finding will need to be confirmed.
POU5F1B and PRNC1
A series of transcripts that map centromeric to the GWAS SNP variants, 1.2 Mb proximal to MYC and just distal to FAM84B on chr.8q24 are associated with POU5F1B (128427857–128429455). POU5F1B was originally considered to be a pseudogene (POU5F1P1) of OCT4, an important transcription factor involved in stem cell pluripotency and reprogramming. Extensive 5′ RACE experiments have identified several alternatively spliced transcripts (Kastler et al., 2010), differing in their TSS that all seem to splice to a single main exon carrying the ORF. The transcript variant with the most distant TSS actually starts within the intron of FAM84B generating a transcript that extends over a region of 860 kb. There is evidence that transcription of POU5F1B is increased in many cases of prostate cancer, whereas some common prostate cell lines (i.e., DU-145, PC-3) do not appear to express the transcript. Kastler et al. (2010) have used an antibody to Oct4 assuming that Oct4 is not expressed in prostate tissue to demonstrate that a protein is indeed detectable in prostatic tissue. However, the signal for this protein is weak and direct demonstration of a protein specific to POU5F1B needs to be performed to validate these results. In addition, a long non-coding RNA (PRNCR1) of 13 kb was recently isolated from one of the recognized susceptibility regions just distal to the SNP variant rs1456315 (Chung et al., 2011). Up-regulated expression of PRNCR1 has been noted in a number of microdissected prostate cancer samples and prostate intraepithelial neoplasia compared to normal adjacent tissue (Chung et al., 2011). Further validation of this transcript will be needed to determine whether it plays a role in prostate cancer or other cancers.
Cancer-Associated Genomic Alterations and the 8q24 Region
In addition to the clustering of cancer-associated SNPs that map to chr.8q24, the 8q24 region has recently been identified in a large-scale study across human cancers as the most frequently amplified region (14%; Beroukhim et al., 2010). Detailed studies of individual cancer type studies have also revealed extensive evidence of gene amplification involving the chr.8q24 region, often with reference to increased MYC copy number and/or increased MYC expression. Although most reports focus primarily on MYC as the target of these amplification events, more detailed analyses with additional probes often finds adjacent regions of 8q24 within these amplicons. MYC amplification, for example, is considered to be a prognostic marker of early stage lung adenocarcinoma (ADC) as MYC amplification correlates with poor prognosis (Iwakawa et al., 2011). Specifically five sub-regions of amplification were defined in the chr.8q24 region including a MYC containing sub-region three in 10.8% of primary lung ADCs (n = 65) and 25% of lung ADC cell lines (n = 40). While PVT1 and miR-1208 mapped to what the authors refer to as amplification sub-region four, no other sub-regions (1, 2, or 5) corresponded to any candidate transcripts. In a sequencing study of the lung ADC cell line, NCI-H2171, a fusion gene comprised of CHD7 and PVT1 was identified (Campbell et al., 2008). Subsequent fusions between CHD7 and exon 1 of PVT1 were also identified in LU-135 (Pleasance et al., 2010). In another large study of lung ADC (n = 371), amplification identified by “Genomic Identification of Significant Targets in Cancer” (GISTIC) analysis focused on the region between 129.18 and 129.34 Mb (q = 9.06 × 10−13) that refers to MYC as the affected proto-oncogene (Weir et al., 2007). The more precise localization of this amplification is actually the region of distal PVT1 and miR-1208 (Figure 1 – Amplification-2).
Genomic Identification of Significant Targets in Cancer analysis in a study of 52 ovarian tumors found 59% with a gain of MYC and PVT1 but only significant over-expression of PVT1 relative to normal samples (Haverty et al., 2009). Consistent with these results are the combined studies of ovarian and breast cancer by Gray and colleagues that show PVT1 and MYC contribute independently to pathogenesis with a strong correlation to overall chr.8q24 amplification (Guan et al., 2007). RNAi-mediated silencing in ovarian and breast cell lines showed reduced proliferation in 8q24 amplified cell lines for either MYC or PVT1, whereas silencing of just PVT1 increased apoptosis and only in cell lines with amplification of 8q24. However, as mentioned above, no mechanistic explanation for the specificity of this observation in cell lines with amplified 8q24 has emerged as yet. High-resolution comparative genomic arrays of chromosome 8q have also been utilized in a study of gastroesophageal junction ADCs that include esophagus and cardia cancers (van Duin et al., 2007). Three commonly overexpressed regions of genomic gain were identified on chr.8q24 with the most distinctive encompassing MYC at 128–132 Mb. The other regions that were identified are between 124 and 127 Mb (referred to as Region 1) that included FAM84B and a region far downstream between 142 and 146 Mb (referred to as Region 4). In studies of expression of gastric esophageal junction tumors versus normal adjacent tissue, van Duin et al. (2007) saw a significant increase in expression of MYC, but not FAM84B. Many examples of amplified chr.8q24, particularly those from older studies have exclusively compared expression of MYC without the benefit of knowing additional transcripts may exist. In this case, a large bulk of the literature contains evidence of chr.8q24 amplification in combination with MYC over-expression (Borg et al., 1992; Mangano et al., 1998; Kim et al., 2006). One of the first studies of PVT1 transcription by Shtivelman and Bishop (1989), intuitively suggested that PVT1 or at least the first exon of PVT1 may co-amplify with MYC and the expression of both MYC and PVT1 transcripts may be coordinately up-regulated. These findings arose from studies of amplified chr.8q24 in colon, SCLC, and neuroepithelioma cell lines. Additional studies also found co-amplification of the first exon of PVT1 and MYC in 2/26 cases of multiple myeloma (Bakkus et al., 1990). With the high frequency of co-amplification that seems to be focused on the immediate region surrounding MYC, PVT1, exons 1a and 1b, and miR-1204 in many different tumors, we have designated this region 1 of amplification in 8q24 (Figure 1 – Amplification-1).
Chromosomal Translocation
The 8q region is among the earliest cytogenetic examples of a human chromosomal translocation (Zech et al., 1976). The T(8:14) translocation and subsequent variant translocations of T(8:22) and T(2:8) became the hallmark lesion in BL, a form of non-Hodgkin’s lymphoma prevalent in sub-Saharan regions of Africa. Interestingly, it was the similar mouse B-cell disease, the plasmacytoma that suggested the first molecular recognition of MYC as the targeted fusion gene in the chr.8q24 region (Shen-Ong et al., 1982). Although other forms of non-Hodgkin’s lymphoma such as a subset of diffuse large B-cell lymphoma, follicular lymphoma, or mantle cell lymphoma may also present with MYC translocations, the predominance of the chr.8q24-based translocation in BL has become the hallmark lesion for WHO designation as BL. While the fusion in most (frequency of 80%) of these diseases is between the immunoglobulin (Ig) heavy chain and MYC, variant translocations (at a frequency of 20%) also implicating the chr.8q24 region but with a different fusion partner (Ig light chain genes) are also recognizable BL specific lesions. The target of essentially all variant translocations to the Ig light chain genes is PVT1. Some T cell leukemias also carry T(8;14) translocations but the fusion partner is one of the TCR loci (TCR alpha or delta juxtaposed to either MYC or PVT1. In a very interesting study describing the cytogenetic events associated with a rare disease of hematodermic neoplasm, the authors have identified a series of changes including deletion of the region 3′ of PVT1 (Jardin et al., 2009). What they report is an arrest of plasmacytoid dendritic cells at the G1/S transition possibly connected to the loss of several miRNAs (miR-1206, miR-1207, and miR-1208) from the Chr.8q24 cluster (Jardin et al., 2009).
Viral Integration
Infection with human papilloma virus (HPV) is believed to be important in the pathogenesis of anogenital carcinomas and other epithelial carcinomas (zur Hausen, 2000). It is also believed the HPV will persist in an epifocal state in early dysplasia with the presence of E6 and E7 proteins capable of interfering with cellular control mechanisms. Eventual integration of HPV into the genome is found in advanced and high risk carcinomas that might suggest the additional recruitment of cellular oncogenes in the transformation process. However, the essential randomness of HPV integration throughout the entire genome makes the argument of a specific target tenuous at best. The most common site of HPV integration in genital neoplasia (10.7% or 26/243) is the Chr.8q24 region with two sub-clusters of integration sites around 500 and 60 kb upstream of MYC (Durst et al., 1987; Popescu et al., 1987; Peter et al., 2006). Originally, it was thought that only HPV18 integrated into the 8q24 locus (Ferber et al., 2003), but more recent studies have documented HPV16 and HPV45 to be integrated into the 8q24 region as well (Peter et al., 2006; Kraus et al., 2008). In one of the most often-characterized cervical cancer cell lines, HeLa, integration of HPV18 into the 8q24 locus was identified with accompanying increased MYC expression. It now appears that the basis for the over-expression of MYC in HeLa and other ADCs with 8q24 integrated HPV sequences is co-amplification of MYC and adjacent sequences including PVT1a, miR-1204 with the integrated viral sequences of HPV (Herrick et al., 2005; Peter et al., 2006). An important histological distinction has been noted in that ADCs frequently exhibit an 8q24 HPV integration whereas squamous cell carcinomas contain other random chromosomal integration events (Lombard et al., 1998; Peter et al., 2006). Thus, it may not be the type of HPV found integrated at 8q24 that is so important in the development of cervical CA, but the location and the accompanying amplification events.
Summary
It is well known that the oasis is a small but significant component of the desert ecosystem. By analogy, it may be that the presence of not just the MYC oncogene, but also the adjacent loci of PVT1, PRNCR1, the miRNA cluster of miR-1204 ∼ 1208 or POU5F1P1 represent a more comprehensive transcriptional oasis in the 8q24 gene desert. In most studies involving the 8q24 locus, the MYC proto-oncogene is the focus of attention as neighboring non-coding transcripts have struggled for the establishment of functionality. However, it is often but not always the case that increased MYC expression is accompanied by increased expression of the neighboring transcripts, principally PVT1. The reason for this could be epigenetic (chromatin accessibility) or the suggestion that at least in one case, MYC binds to the promoter of PVT1, thereby activating PVT1 and miR-1204 transcription (Carramusa et al., 2007). Another means of co-activating MYC, PVT1, and miR-1204 derives from the observed enhancer binding activity of the rs3983267 risk region to the promoters of PVT1, MYC, and miR-1204 (Wright et al., 2010). In at least one study of the pathophysiology of ovarian and breast cancer, it has been shown that MYC and PVT1 act independently and at least for PVT1, this action is mediated through apoptosis (Guan et al., 2007). Furthermore, this action may be associated specifically with amplification of the 8q24 region. Induction of apoptosis by over-expression of miR-1204 and perhaps the other miRNAs in the miR-1204 ∼ 1208 cluster as well suggests an anti-proliferative role for the miRNAs independent of MYC (Barsotti et al., 2011). Obviously, it will also be important to elucidate the specific nature of interactions between the miR-1204 ∼ 1208 cluster and the non-coding PVT1 host transcript as well. An interesting genetic analog to the PVT1 region is human chromosome 13q14 that contains two miRNAs, miR15a, miR-16-1, within the intron 4 of the host non-coding transcript DLEU2 (Migliazza et al., 2001). Although the specific role for DLEU2 is not understood, it appears that deletion of the entire region is required in the development of an aggressive phenotype of chronic lymphocytic leukemia suggesting a combined role for miRNAs and a non-coding transcript (Klein et al., 2010). In this paper, we have attempted to raise the awareness of other transcripts besides MYC in the 8q24 region to emphasize possible roles they may play as well in what appears to one of the most cited regions (8q24) in the human genome.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by the Intramural Research Program (Center for Cancer Research, NCI) of the NIH. The authors thank Dr. Ashish Lal for helpful comments and suggestions in review of the manuscript prior to publication.
References
Ahmadiyeh, N., Pomerantz, M. M., Grisanzio, C., Herman, P., Jia, L., Almendro, V., He, H. H., Brown, M., Liu, X. S., Davis, M., Caswell, J. L., Beckwith, C. A., Hills, A., MacConaill, L., Coetzee, G. A., Regan, M. M., and Freedman, M. L. (2010). 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. U.S.A. 107, 9742–9746.
Alvarez, M. L., and DiStefano, J. K. (2011). Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 6, e18671. doi: 10.1371/journal.pone.0018671
Amundadottir, L. T., Sulem, P., Gudmundsson, J., Helgason, A., Baker, A., Agnarsson, B. A., Sigurdsson, A., Benediktsdottir, K. R., Cazier, J. B., Sainz, J., Jakobsdottir, M., Kostic, J., Magnusdottir, D. N., Ghosh, S., Agnarsson, K., Birgisdottir, B., Le Roux, L., Olafsdottir, A., Blondal, T., Andresdottir, M., Gretarsdottir, O. S., Bergthorsson, J. T., Gudbjartsson, D., Gylfason, A., Thorleifsson, G., Manolescu, A., Kristjansson, K., Geirsson, G., Isaksson, H., Douglas, J., Johansson, J. E., Bälter, K., Wiklund, F., Montie, J. E., Yu, X., Suarez, B. K., Ober, C., Cooney, K. A., Gronberg, H., Catalona, W. J., Einarsson, G. V., Barkardottir, R. B., Gulcher, J. R., Kong, A., Thorsteinsdottir, U., and Stefansson, K. L. (2006). A common variant associated with prostate cancer in European and African populations. Nat. Genet. 38, 652–658.
An, P., Freedman, B. I., Rich, S. S., Mandel, S. A., Arnett, D. K., Myers, R. H., Chen, Y. D., Hunt, S. C., and Rao, D. C. (2006). Quantitative trait loci on chromosome 8q24 for pancreatic beta-cell function and 7q11 for insulin sensitivity in obese nondiabetic white and black families: evidence from genome-wide linkage scans in the NHLBI Hypertension Genetic Epidemiology Network (HyperGEN) Study. Diabetes 55, 551–558.
Asker, C., Mareni, C., Coviello, D., Ingvarsson, S., Sessarego, M., Origone, P., Klein, G., and Sumeigi, J. (1988). Amplification of c-myc and pvt-1 homologous sequences in acute nonlymphatic leukemia. Leuk. Res. 12, 523–527.
Bakkus, M. H., Brakel-van Peer, K. M., Michiels, J. J., van’t Veer, M. B., and Benner, R. (1990). Amplification of the c-myc and the pvt-like region in human multiple myeloma. Oncogene 5, 1359–1364.
Barsotti, A., Beckerman, R., Laptenko, O., Huppi, K., Caplen, N. J., and Prives, C. (2011). P53-dependent induction of PVT1 and MIR-1204. J. Biol. Chem. 287, 2509–2519.
Berndt, S. I., Potter, J. D., Hazra, A., Yeager, M., Thomas, G., Makar, K. W., Welch, R., Cross, A. J., Huang, W. Y., Schoen, R. E., Giovannucci, E., Chan, A. T., Chanock, S. J., Peters, U., Hunter, D. J., and Hayes, R. B. (2008). Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum. Mol. Genet. 17, 2665–2672.
Beroukhim, R., Mermel, C. H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J., Barretina, J., Boehm, J. S., Dobson, J., Urashima, M., Mc Henry, K. T., Pinchback, R. M., Ligon, A. H., Cho, Y. J., Haery, L., Greulich, H., Reich, M., Winckler, W., Lawrence, M. S., Weir, B. A., Tanaka, K. E., Chiang, D. Y., Bass, A. J., Loo, A., Hoffman, C., Prensner, J., Liefeld, T., Gao, Q., Yecies, D., Signoretti, S., Maher, E., Kaye, F. J., Sasaki, H., Tepper, J. E., Fletcher, J. A., Tabernero, J., Baselga, J., Tsao, M. S., Demichelis, F., Rubin, M. A., Janne, P. A., Daly, M. J., Nucera, C., Levine, R. L., Ebert, B. L., Gabriel, S., Rustgi, A. K., Antonescu, C. R., Ladanyi, M., Letai, A., Garraway, L. A., Loda, M., Beer, D. G., True, L. D., Okamoto, A., Pomeroy, S. L., Singer, S., Golub, T. R., Lander, E. S., Getz, G., Sellers, W. R., and Meyerson, M. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905.
Borg, A., Baldetorp, B., Ferno, M., Olsson, H., and Sigurdsson, H. (1992). c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int. J. Cancer 51, 687–691.
Braem, M. G., Schouten, L. J., Peeters, P. H., den Brandt, P. A., and Onland-Moret, N. C. (2011). Genetic susceptibility to sporadic ovarian cancer: a systematic review. Biochim. Biophys. Acta 1816, 132–146.
Brannan, C. I., Dees, E. C., Ingram, R. S., and Tilghman, S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36.
Brockdorff, N., Ashworth, A., Kay, G. F., McCabe, V. M., Norris, D. P., Cooper, P. J., Swift, S., and Rastan, S. (1992). The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526.
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., and Willard, H. F. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542.
Campbell, P. J., Stephens, P. J., Pleasance, E. D., O’Meara, S., Li, H., Santarius, T., Stebbings, A., Leroy, C., Edkins, S., Hardy, C., Teague, J. W., Menzies, A., Goodhead, I., Turner, D. J., Clee, C. M., Quail, M. A., Cox, A., Brown, C., Durbin, R., Hurles, M. E., Edwards, P. A. W., Bignell, G. R., Stratton, M. R., and Futrea, P. A. (2008). Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729.
Carramusa, L., Contino, F., Ferro, A., Minafra, L., Perconti, G., Giallongo, A., and Feo, S. (2007). The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J. Cell. Physiol. 213, 511–518.
Cheng, I., Plummer, S. J., Jorgenson, E., Liu, X., Rybicki, B. A., Casey, G., and Witte, J. S. (2008). 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur. J. Hum. Genet. 16, 496–505.
Chung, S., Nakagawa, H., Uemura, M., Piao, L., Ashikawa, K., Hosono, N., Takata, R., Akamatsu, S., Kawaguchi, T., Morizono, T., Tsunoda, T., Daigo, Y., Matsuda, K., Kamatani, N., Nakamura, Y., and Kubo, M. (2011). Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 102, 245–252.
Crowther-Swanepoel, D., Broderick, P., Di Bernardo, M. C., Dobbins, S. E., Torres, M., Mansouri, M., Ruiz-Ponte, C., Enjuanes, A., Rosenquist, R., Carracedo, A., Jurlander, J., Campo, E., Juliusson, G., Montserrat, E., Smedby, K. E., Dyer, M. J., Matutes, E., Dearden, C., Sunter, N. J., Hall, A. G., Mainou-Fowler, T., Jackson, G. H., Summerfield, G., Harris, R. J., Pettitt, A. R., Allsup, D. J., Bailey, J. R., Pratt, G., Pepper, C., Fegan, C., Parker, A., Oscier, D., Allan, J. M., Catovsky, D., Houlston, R. S., and Institute of Cancer Research, Sutton, Surrey, UK. (2010). Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42, 132–136.
Durst, M., Croce, C. M., Gissmann, L., Schwarz, E., and Huebner, K. (1987). Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. U.S.A. 84, 1070–1074.
Easton, D. F., and Eeles, R. A. (2008). Genome-wide association studies in cancer. Hum. Mol. Genet. 17, R109–R115.
Eeles, R. A., Kote-Jarai, Z., Giles, G. G., Olama, A. A., Guy, M., Jugurnauth, S. K., Mulholland, S., Leongamornlert, D. A., Edwards, S. M., Morrison, J., Field, H. I., Southey, M. C., Severi, G., Donovan, J. L., Hamdy, F. C., Dearnaley, D. P., Muir, K. R., Smith, C., Bagnato, M., Ardern-Jones, A. T., Hall, A. L., O’Brien, L. T., Gehr-Swain, B. N., Wilkinson, R. A., Cox, A., Lewis, S., Brown, P. M., Jhavar, S. G., Tymrakiewicz, M., Lophatananon, A., Bryant, S. L., UK Genetic Prostate Cancer Study Collaborators, British Association of Urological Surgeons’ Section of Oncology, UK ProtecT Study Collaborators, Horwich, A., Huddart, R. A., Khoo, V. S., Parker, C. C., Woodhouse, C. J., Thompson, A., Christmas, T., Ogden, C., Fisher, C., Jamieson, C., Cooper, C. S., English, D. R., Hopper, J. L., Neal, D. E., and Easton, D. F. (2008). Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 40, 316–321.
Enciso-Mora, V., Broderick, P., Ma, Y., Jarrett, R. F., Hjalgrim, H., Hemminki, K., Van Den Berg, A., Olver, B., Lloyd, A., Dobbins, S. E., Lightfoot, T., Van Leeuwen, F. E., Försti, A., Diepstra, A., Broeks, A., Vijayakrishnan, J., Shield, L., Lake, A., Montgomery, D., Roman, E., Engert, A., Von Strandmann, E. P., Reiners, K. S., Nolte, I. M., Smedby, K. E., Adami, H.-O., Russell, N. S., Glimelius, B., Hamilton-Dutoit, S., De Bruin, M., Ryder, L. P., Molin, D., Sorensen, K. M., Chang, E. T., Taylor, M., Cooke, R., Hofstra, R., Westers, H., Van Wezel, T., Van Eijk, R., Ashworth, A., Rostgaard, K., Melbye, M., Swerdlow, A. J., and Houlston, R. S. (2010). A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat. Genet. 42, 1126–1130.
Erikson, J., Nishikura, K., ar-Rushdi, A., Finan, J., Emanuel, B., Lenoir, G., Nowell, P. C., and Croce, C. M. (1983). Translocation of an immunoglobulin kappa locus to a region 3’ of an unrearranged c-myc oncogene enhances c-myc transcription. Proc. Natl. Acad. Sci. U.S.A. 80, 7581–7585.
Ferber, M. J., Thorland, E. C., Brink, A. A., Rapp, A. K., Phillips, L. A., McGovern, R., Gostout, B. S., Cheung, T. H., Chung, T. k. H., Fu, W. Y., and Smith, D. I. (2003). Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene 22, 7233–7242.
Freedman, M. L., Haiman, C. A., Patterson, N., McDonald, G. J., Tandon, A., Waliszewska, A., Penney, K., Steen, R. G., Ardlie, K., John, E. M., Oakley-Girvan, I., Whittemore, A. S., Cooney, K. A., Ingles, S. A., Altshuler, D., Henderson, B. E., and Reich, D. (2006). Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. U.S.A. 103, 14068–14073.
Ghoussaini, M., Song, H., Koessler, T., Al Olama, A. A., Kote-Jarai, Z., Driver, K. E., Pooley, K. A., Ramus, S. J., Kjaer, S. K., Hogdall, E., DiCioccio, R. A., Whittemore, A. S., Gayther, S. A., Giles, G. G., Guy, M., Edwards, S. M., Morrison, J., Donovan, J. L., Hamdy, F. C., Dearnaley, D. P., Ardern-Jones, A. T., Hall, A. L., O’Brien, L. T., Gehr-Swain, B. N., Wilkinson, R. A., Brown, P. M., Hopper, J. L., Neal, D. E., Pharoah, P. D., Ponder, B. A., Eeles, R. A., Easton, D. F., Dunning, A. M., and UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, UK ProtecT Study Collaborators. (2008). Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl. Cancer Inst. 100, 962–966.
Giovannucci, E., Harlan, D. M., Archer, M. C., Bergenstal, R. M., Gapstur, S. M., Habel, L. A., Pollak, M., Regensteiner, J. G., and Yee, D. (2010). Diabetes and cancer: a consensus report. Diabetes and cancer: a consensus report. CA Cancer J. Clin. 60, 207–221.
Goode, E. L., Chenevix-Trench, G., Song, H., Ramus, S. J., Notaridou, M., Lawrenson, K., Widschwendter, M., Vierkant, R. A., Larson, M. C., Kjaer, S. K., Birrer, M. J., Berchuck, A., Schildkraut, J., Tomlinson, I., Kiemeney, L. A., Cook, L. S., Gronwald, J., Garcia-Closas, M., Gore, M. E., Campbell, I., Whittemore, A. S., Sutphen, R., Phelan, C., Anton-Culver, H., Pearce, C. L., Lambrechts, D., Rossing, M. A., Chang-Claude, J., Moysich, K. B., Goodman, M. T., Dörk, T., Nevanlinna, H., Ness, R. B., Rafnar, T., Hogdall, C., Hogdall, E., Fridley, B. L., Cunningham, J. M., Sieh, W., McGuire, V., Godwin, A. K., Cramer, D. W., Hernandez, D., Levine, D., Lu, K., Iversen, E. S., Palmieri, R. T., Houlston, R., van Altena, A. M., Aben, K. K., Massuger, L. F., Brooks-Wilson, A., Kelemen, L. E., Le, N. D., Jakubowska, A., Lubinski, J., Medrek, K., Stafford, A., Easton, D. F., Tyrer, J., Bolton, K. L., Harrington, P., Eccles, D., Chen, A., Molina, A. N., Davila, B. N., Arango, H., Tsai, Y. Y., Chen, Z., Risch, H. A., McLaughlin, J., Narod, S. A., Ziogas, A., Brewster, W., Gentry-Maharaj, A., Menon, U., Wu, A. H., Stram, D. O., Pike, M. C., Wellcome Trust Case-Control Consortium, Beesley, J., Webb, P. M., Australian Cancer Study (Ovarian Cancer), Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium (OCAC), Chen, X., Ekici, A. B., Thiel, F. C., Beckmann, M. W., Yang, H., Wentzensen, N., Lissowska, J., Fasching, P. A., Despierre, E., Amant, F., Vergote, I., Doherty, J., Hein, R., Wang-Gohrke, S., Lurie, G., Carney, M. E., Thompson, P. J., Runnebaum, I., Hillemanns, P., Dürst, M., Antonenkova, N., Bogdanova, N., Leminen, A., Butzow, R., Heikkinen, T., Stefansson, K., Sulem, P., Besenbacher, S., Sellers, T. A., Gayther, S. A., Pharoah, P. D., and Ovarian Cancer Association Consortium (OCAC). (2010). A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 42, 874–879.
Grisanzio, C., and Freedman, M. L. (2010). Chromosome 8q24-associated cancers and MYC. Genes Cancer 1, 555–559.
Guan, Y., Kuo, W. L., Stilwell, J. L., Takano, H., Lapuk, A. V., Fridlyand, J., Mao, J. H., Yu, M., Miller, M. A., Santos, J. L., Kalloger, S. E., Carlson, J. W., Ginzinger, D. G., Celniker, S. E., Mills, G. B., Huntsman, D. G., and Gray, J. W. (2007). Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin. Cancer Res. 13, 5745–5755.
Gudmundsson, J., Sulem, P., Manolescu, A., Amundadottir, L. T., Gudbjartsson, D., Helgason, A., Rafnar, T., Bergthorsson, J. T., Agnarsson, B. A., Baker, A., Sigurdsson, A., Benediktsdottir, K. R., Jakobsdottir, M., Xu, J., Blondal, T., Kostic, J., Sun, J., Ghosh, S., Stacey, S. N., Mouy, M., Saemundsdottir, J., Backman, V. M., Kristjansson, K., Tres, A., Partin, A. W., Albers-Akkers, M. T., Godino-Ivan Marcos, J., Walsh, P. C., Swinkels, D. W., Navarrete, S., Isaacs, S. D., Aben, K. K., Graif, T., Cashy, J., Ruiz-Echarri, M., Wiley, K. E., Suarez, B. K., Witjes, J. A., Frigge, M., Ober, C., Jonsson, E., Einarsson, G. V., Mayordomo, J. I., Kiemeney, L. A., Isaacs, W. B., Catalona, W. J., Barkardottir, R. B., Gulcher, J. R., Thorsteinsdottir, U., Kong, A., and Stefansson, K. (2007). Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 39, 631–637.
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., Huarte, M., Zuk, O., Carey, B. W., Cassady, J. P., Cabili, M. N., Jaenisch, R., Mikkelsen, T. S., Jacks, T., Hacohen, N., Bernstein, B. E., Kellis, M., Regev, A., Rinn, J. L., and Lander, E. S. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227.
Haiman, C. A., Patterson, N., Freedman, M. L., Myers, S. R., Pike, M. C., Waliszewska, A., Neubauer, J., Tandon, A., Schirmer, C., McDonald, G. J., Greenway, S. C., Stram, D. O., Le Marchand, L., Kolonel, L. N., Frasco, M., Wong, D., Pooler, L. C., Ardlie, K., Oakley-Girvan, I., Whittemore, A. S., Cooney, K. A., John, E. M., Ingles, S. A., Altshuler, D., Henderson, B. E., and Reich, D. (2007). Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 39, 638–644.
Hanson, R. L., Craig, D. W., Millis, M. P., Yeatts, K. A., Kobes, S., Pearson, J. V., Lee, A. M., Knowler, W. C., Nelson, R. G., and Wolford, J. K. (2007). Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56, 975–983.
Haverty, P. M., Hon, L. S., Kaminker, J. S., Chant, J., and Zhang, Z. (2009). High-resolution analysis of copy number alterations and associated expression changes in ovarian tumors. BMC Med. Genomics 2, 21. doi: 10.1186/1755-8794-2-21
Herrick, J., Conti, C., Teissier, S., Thierry, F., Couturier, J., Sastre-Garau, X., Favre, M., Orth, G., and Bensimon, A. (2005). Genomic organization of amplified MYC genes suggests distinct mechanisms of amplification in tumorigenesis. Cancer Res. 65, 1174–1179.
Huppi, K., and Siwarski, D. (1994). Chimeric transcripts with an open reading frame are generated as a result of translocation to the Pvt-1 region in mouse B-cell tumors. Int. J. Cancer 59, 848–851.
Huppi, K., Siwarski, D., Skurla, R., Klinman, D., and Mushinski, J. F. (1990). Pvt-1 transcripts are found in normal tissues and are altered by reciprocal (6;15) translocations in mouse plasmacytomas. Proc. Natl. Acad. Sci. U.S.A. 87, 6964–6968.
Huppi, K., Volfovsky, N., Runfola, T., Jones, T. L., Mackiewicz, M., Martin, S. E., Mushinski, J. F., Stephens, R., and Caplen, N. J. (2008). The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res. 6, 212–221.
Iwakawa, R., Kohno, T., Kato, M., Shiraishi, K., Tsuta, K., Noguchi, M., Ogawa, S., and Yokota, J. (2011). MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin. Cancer Res. 17, 1481–1489.
Jardin, F., Callanan, M., Penther, D., Ruminy, P., Troussard, X., Kerckaert, J. P., Figeac, M., Parmentier, F., Rainville, V., Vaida, I., Bertrand, P., Duval, A. B., Picquenot, J. M., Chaperot, L., Marolleau, J. P., Plumas, J., Tilly, H., and Bastard, C. (2009). Recurrent genomic aberrations combined with deletions of various tumour suppressor genes may deregulate the G1/S transition in CD4+CD56+ haematodermic neoplasms and contribute to the aggressiveness of the disease. Leukemia 23, 698–707.
Kastler, S., Honold, L., Luedeke, M., Kuefer, R., Moller, P., Hoegel, J., Vogel, W., Maier, C., and Assum, G. (2010). POU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinoma. Prostate 70, 666–674.
Kiemeney, L. A., Thorlacius, S., Sulem, P., Geller, F., Aben, K. K., Stacey, S. N., Gudmundsson, J., Jakobsdottir, M., Bergthorsson, J. T., Sigurdsson, A., Blondal, T., Witjes, J. A., Vermeulen, S. H., Hulsbergen-van de Kaa, C. A., Swinkels, D. W., Ploeg, M., Cornel, E. B., Vergunst, H., Thorgeirsson, T. E., Gudbjartsson, D., Gudjonsson, S. A., Thorleifsson, G., Kristinsson, K. T., Mouy, M., Snorradottir, S., Placidi, D., Campagna, M., Arici, C., Koppova, K., Gurzau, E., Rudnai, P., Kellen, E., Polidoro, S., Guarrera, S., Sacerdote, C., Sanchez, M., Saez, B., Valdivia, G., Ryk, C., de Verdier, P., Lindblom, A., Golka, K., Bishop, D. T., Knowles, M. A., Nikulasson, S., Petursdottir, V., Jonsson, E., Geirsson, G., Kristjansson, B., Mayordomo, J. I., Steineck, G., Porru, S., Buntinx, F., Zeegers, M. P., Fletcher, T., Kumar, R., Matullo, G., Vineis, P., Kiltie, A. E., Gulcher, J. R., Thorsteinsdottir, U., Kong, A., Rafnar, T., and Stefansson, K. (2008). Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 40, 1307–1312.
Kim, Y. H., Girard, L., Giacomini, C. P., Wang, P., Hernandez-Boussard, T., Tibshirani, R., Minna, J. D., and Pollack, J. R. (2006). Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 25, 130–138.
Klein, U., Lia, M., Crespo, M., Siegel, R., Shen, Q., Mo, T., Ambesi-Impiombato, A., Califano, A., Migliazza, A., Bhagat, G., and Dalla-Favera, R. (2010). The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40.
Koehne, C. F., Lazo, P. A., Alves, K., Lee, J. S., Tsichlis, P. N., and O’Donnell, P. V. (1989). The Mlvi-1 locus involved in the induction of rat T-cell lymphomas and the pvt-1/Mis-1 locus are identical. J. Virol. 63, 2366–2369.
Kraus, I., Driesch, C., Vinokurova, S., Hovig, E., Schneider, A., von Knebel Doeberitz, M., and Dürst, M. (2008). The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 68, 2514–2522.
Lin, M., Pedrosa, E., Shah, A., Hrabovsky, A., Maqbool, S., Zheng, D., and Lachman, H. M. (2011). RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE 6, e23356. doi: 10.1371/journal.pone.0023356
Lochhead, P., Ng, M. T., Hold, G. L., Rabkin, C. S., Vaughan, T. L., Gammon, M. D., Risch, H. A., Lissowska, J., Mukhopadhya, I., Chow, W. H., and El-Omar, E. M. (2011). Possible association between a genetic polymorphism at 8q24 and risk of upper gastrointestinal cancer. Eur. J. Cancer Prev. 20, 54–57.
Lombard, I., Vincent-Salomon, A., Validire, P., Zafrani, B., de la Rochefordiere, A., Clough, K., Favre, M., Pouillart, P., and Sastre-Garau, X. (1998). Human papillomavirus genotype as a major determinant of the course of cervical cancer. J. Clin. Oncol. 16, 2613–2619.
Mangano, R., Piddini, E., Carramusa, L., Duhig, T., Feo, S., and Fried, M. (1998). Chimeric amplicons containing the c-myc gene in HL60 cells. Oncogene 17, 2771–2777.
Marasa, B. S., Srikantan, S., Martindale, J. L., Kim, M. M., Lee, E. K., Gorospe, M., and Abdelmohsen, K. (2010). MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2, 333–343.
Meyer, K. B., Maia, A. T., O’Reilly, M., Ghoussaini, M., Prathalingam, R., Porter-Gill, P., Ambs, S., Prokunina-Olsson, L., Carroll, J., and Ponder, B. A. (2011). A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 7, e1002165. doi: 10.1371/journal.pgen.1002165
Meyer, N., Kim, S. S., and Penn, L. Z. (2006). The Oscar-worthy role of Myc in apoptosis. Semin. Cancer Biol. 16, 275–287.
Migliazza, A., Bosch, F., Komatsu, H., Cayanis, E., Martinotti, S., Toniato, E., Guccione, E., Qu, X., Chien, M., Murty, V. V., Gaidano, G., Inghirami, G., Zhang, P., Fischer, S., Kalachikov, S. M., Russo, J., Edelman, I., Efstratiadis, A., and Dalla-Favera, R. (2001). Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood 97, 2098–2104.
Millis, M. P., Bowen, D., Kingsley, C., Watanabe, R. M., and Wolford, J. K. (2007). Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 56, 3027–3032.
Minarovits, J., Steinitz, M., Boldog, F., Imreh, S., Wirschubsky, Z., Ingvarsson, S., Hedenskog, M., Minarovits-Kormuta, S., and Klein, G. (1990). Differences in c-myc and pvt-1 amplification in SEWA sarcoma sublines selected for adherent or non-adherent growth. Int. J. Cancer 45, 514–520.
Peter, M., Rosty, C., Couturier, J., Radvanyi, F., Teshima, H., and Sastre-Garau, X. (2006). MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene 25, 5985–5993.
Pleasance, E. D., Stephens, P. J., O’Meara, S., McBride, D. J., Meynert, A., Jones, D., Lin, M. L., Beare, D., Lau, K. W., Greenman, C., Varela, I., Nik-Zainal, S., Davies, H. R., Ordoñez, G. R., Mudie, L. J., Latimer, C., Edkins, S., Stebbings, L., Chen, L., Jia, M., Leroy, C., Marshall, J., Menzies, A., Butler, A., Teague, J. W., Mangion, J., Sun, Y. A., McLaughlin, S. F., Peckham, H. E., Tsung, E. F., Costa, G. L., Lee, C. C., Minna, J. D., Gazdar, A., Birney, E., Rhodes, M. D., McKernan, K. J., Stratton, M. R., Futreal, P. A., and Campbell, P. J. (2010). A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190.
Pomerantz, M. M., Beckwith, C. A., Regan, M. M., Wyman, S. K., Petrovics, G., Chen, Y., Hawksworth, D. J., Schumacher, F. R., Mucci, L., Penney, K. L., Stampfer, M. J., Chan, J. A., Ardlie, K. G., Fritz, B. R., Parkin, R. K., Lin, D. W., Dyke, M., Herman, P., Lee, S., Oh, W. K., Kantoff, P. W., Tewari, M., McLeod, D. G., Srivastava, S., and Freedman, M. L. (2009a). Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 69, 5568–5574.
Pomerantz, M. M., Ahmadiyeh, N., Jia, L., Herman, P., Verzi, M. P., Doddapaneni, H., Beckwith, C. A., Chan, J. A., Hills, A., Davis, M., Yao, K., Kehoe, S. M., Lenz, H.-J., Haiman, C. A., Yan, C., Henderson, B. E., Frenkel, B., Barretina, J., Bass, A., Tabernero, J., Baselga, J., Regan, M. M., Manak, J. R., Shivdasani, R., Coetzee, G. A., and Freedman, M. L. (2009b). The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 41, 882–884.
Popescu, N. C., DiPaolo, J. A., and Amsbaugh, S. C. (1987). Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet. Cell Genet. 44, 58–62.
Rothberg, P. G., and Otto, Y. M. (1995). A polymorphic variant of human c-Myc: Asn11→Ser. Mamm. Genome 6, 209–211.
Shen-Ong, G. L., Keath, E. J., Piccoli, S. P., and Cole, M. D. (1982). Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell 31, 443–452.
Shtivelman, E., and Bishop, J. M. (1989). The PVT gene frequently amplifies with MYC in tumor cells. Mol. Cell. Biol. 9, 1148–1154.
Shtivelman, E., and Bishop, J. M. (1990). Effects of translocations on transcription from PVT. Mol. Cell. Biol. 10, 1835–1839.
Shtivelman, E., Henglein, B., Groitl, P., Lipp, M., and Bishop, J. M. (1989). Identification of a human transcription unit affected by the variant chromosomal translocations 2;8 and 8;22 of Burkitt lymphoma. Proc. Natl. Acad. Sci. U.S.A. 86, 3257–3260.
Siwarski, D., Kim, J., Diaw, L., and Huppi, K. (2001). Identification of a transcriptionally compromised allele of c-MYC in a North American family. J. Med. Genet. 38, 47–49.
Sole, X., Hernandez, P., de Heredia, M. L., Armengol, L., Rodriguez-Santiago, B., Gómez, L., Maxwell, C. A., Aguiló, F., Condom, E., Abril, J., Pérez-Jurado, L., Estivill, X., Nunes, V., Capellá, G., Gruber, S. B., Moreno, V., and Pujana, M. A. (2008). Genetic and genomic analysis modeling of germline c-MYC overexpression and cancer susceptibility. BMC Genomics 9, 12. doi: 10.1186/1471-2164-9-12
Sotelo, J., Esposito, D., Duhagon, M. A., Banfield, K., Mehalko, J., Liao, H., Stephens, R. M., Harris, T. J., Munroe, D. J., and Wu, X. (2010). Long-range enhancers on 8q24 regulate c-Myc. Proc. Natl. Acad. Sci. U.S.A. 107, 3001–3005.
Thomas, G., Jacobs, K. B., Yeager, M., Kraft, P., Wacholder, S., Orr, N., Yu, K., Chatterjee, N., Welch, R., Hutchinson, A., Crenshaw, A., Cancel-Tassin, G., Staats, B. J., Wang, Z., Gonzalez-Bosquet, J., Fang, J., Deng, X., Berndt, S. I., Calle, E. E., Feigelson, H. S., Thun, M. J., Rodriguez, C., Albanes, D., Virtamo, J., Weinstein, S., Schumacher, F. R., Giovannucci, E., Willett, W. C., Cussenot, O., Valeri, A., Andriole, G. L., Crawford, E. D., Tucker, M., Gerhard, D. S., Fraumeni, J. F., Hoover, R., Hayes, R. B., Hunter, D. J., and Chanock, S. J. (2008). Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 40, 310–315.
Tomlinson, I., Webb, E., Carvajal-Carmona, L., Broderick, P., Kemp, Z., Spain, S., Penegar, S., Chandler, I., Gorman, M., Wood, W., Barclay, E., Lubbe, S., Martin, L., Sellick, G., Jaeger, E., Hubner, R., Wild, R., Rowan, A., Fielding, S., Howarth, K., Silver, A., Atkin, W., Muir, K., Logan, R., Kerr, D., Johnstone, E., Sieber, O., Gray, R., Thomas, H., Peto, J., Cazier, J.-B., and Houlston, R. (2007). A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 39, 984–988.
Tsichlis, P. N., Shepherd, B. M., and Bear, S. E. (1989). Activation of the Mlvi-1/mis1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 86, 5487–5491.
Turnbull, C., Ahmed, S., Morrison, J., Pernet, D., Renwick, A., Maranian, M., Seal, S., Ghoussaini, M., Hines, S., Healey, C. S., Hughes, D., Warren-Perry, M., Tapper, W., Eccles, D., Evans, D. G., The Breast Cancer Susceptibility Collaboration (UK), Hooning, M., Schutte, M., van den Ouweland, A., Houlston, R., Ross, G., Langford, C., Pharoah, P. D. P., Stratton, M. R., Dunning, A. M., Nazneen, Rahman, N., and Easton, D. F. (2010). Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507.
Tuupanen, S., Turunen, M., Lehtonen, R., Hallikas, O., Vanharanta, S., Kivioja, T., Björklund, M., Wei, G., Yan, J., Niittymäki, I., Mecklin, J. P., Järvinen, H., Ristimäki, A., Di-Bernardo, M., East, P., Carvajal-Carmona, L., Houlston, R. S., Tomlinson, I., Palin, K., Ukkonen, E., Karhu, A., Taipale, J., and Aaltonen, L. A. (2009). The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 41, 885–890.
van Duin, M., van Marion, R., Vissers, K. J., Hop, W. C., Dinjens, W. N., Tilanus, H. W., Siersema, P. D., and van Dekken, H. (2007). High-resolution array comparative genomic hybridization of chromosome 8q: evaluation of putative progression markers for gastroesophageal junction adenocarcinomas. Cytogenet. Genome Res. 118, 130–137.
Wasserman, N. F., Aneas, I., and Nobrega, M. A. (2010). An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 20, 1191–1197.
Webb, E., Adams, J. M., and Cory, S. (1984). Variant (6; 15) translocation in a murine plasmacytoma occurs near an immunoglobulin kappa gene but far from the myc oncogene. Nature 312, 777–779.
Weir, B. A., Woo, M. S., Getz, G., Perner, S., Ding, L., Beroukhim, R., Lin, W. M., Province, M. A., Kraja, A., Johnson, L. A., Shah, K., Sato, M., Thomas, R. K., Barletta, J. A., Borecki, I. B., Broderick, S., Chang, A. C., Chiang, D. Y., Chirieac, L. R., Cho, J., Fujii, Y., Gazdar, A. F., Giordano, T., Greulich, H., Hanna, M., Johnson, B. E., Kris, M. G., Lash, A., Lin, L., Lindeman, N., Mardis, E. R., McPherson, J. D., Minna, J. D., Morgan, M. B., Nadel, M., Orringer, M. B., Osborne, J. R., Ozenberger, B., Ramos, A. H., Robinson, J., Roth, J. A., Rusch, V., Sasaki, H., Shepherd, F., Sougnez, C., Spitz, M. R., Tsao, M. S., Twomey, D., Verhaak, R. G., Weinstock, G. M., Wheeler, D. A., Winckler, W., Yoshizawa, A., Yu, S., Zakowski, M. F., Zhang, Q., Beer, D. G., Wistuba, I. I., Watson, M. A., Garraway, L. A., Ladanyi, M., Travis, W. D., Pao, W., Rubin, M. A., Gabriel, S. B., Gibbs, R. A., Varmus, H. E., Wilson, R. K., Lander, E. S., and Meyerson, M. (2007). Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898.
White, K. L., Sellers, T. A., Fridley, B. L., Vierkant, R. A., Phelan, C. M., Tsai, Y. Y., Kalli, K. R., Berchuck, A., Iversen, E. S., Hartmann, L. C., Liebow, M., Armasu, S., Fredericksen, Z., Larson, M. C., Duggan, D., Couch, F. J., Schildkraut, J. M., Cunningham, J. M., and Goode, E. L. (2010). Variation at 8q24 and 9p24 and risk of epithelial ovarian cancer. Twin Res. Hum. Genet. 13, 43–56.
Wright, J. B., Brown, S. J., and Cole, M. D. (2010). Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol. 30, 1411–1420.
Yeager, M., Orr, N., Hayes, R. B., Jacobs, K. B., Kraft, P., Wacholder, S., Minichiello, M. J., Fearnhead, P., Yu, K., Chatterjee, N., Wang, Z., Welch, R., Staats, B. J., Calle, E. E., Feigelson, H. S., Thun, M. J., Rodriguez, C., Albanes, D., Virtamo, J., Weinstein, S., Schumacher, F. R., Giovannucci, E., Willett, W. C., Cancel-Tassin, G., Cussenot, O., Valeri, A., Andriole, G. L., Gelmann, E. P., Tucker, M., Gerhard, D. S., Fraumeni, J. F. Jr., Hoover, R., Hunter, D. J., Chanock, S. J., and Thomas, G. (2007). Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649.
You, L., Chang, D., Du, H. Z., and Zhao, Y. P. (2011). Genome-wide screen identifies PVT1 as a regulator of gemcitabine sensitivity in human pancreatic cancer cells. Biochem. Biophys. Res. Commun. 407, 1–6.
Zanke, B. W., Greenwood, C. M., Rangrej, J., Kustra, R., Tenesa, A., Farrington, S. M., Prendergast, J., Olschwang, S., Chiang, T., Crowdy, E., Ferretti, V., Laflamme, P., Sundararajan, S., Roumy, S., Olivier, J. F., Robidoux, F., Sladek, R., Montpetit, A., Campbell, P., Bezieau, S., O’Shea, A. M., Zogopoulos, G., Cotterchio, M., Newcomb, P., McLaughlin, J., Younghusband, B., Green, R., Green, J., Porteous, M. E., Campbell, H., Blanche, H., Sahbatou, M., Tubacher, E., Bonaiti-Pellié, C., Buecher, B., Riboli, E., Kury, S., Chanock, S. J., Potter, J., Thomas, G., Gallinger, S., Hudson, T. J., and Dunlop, M. G. (2007). Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 39, 989–994.
Zech, L., Haglund, U., Nilsson, K., and Klein, G. (1976). Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int. J. Cancer 17, 47–56.
Keywords: MYC, PVT1, miR-1204, 8q24
Citation: Huppi K, Pitt JJ, Wahlberg BM and Caplen NJ (2012) The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front. Gene. 3:69. doi: 10.3389/fgene.2012.00069
Received: 18 January 2012; Accepted: 10 April 2012;
Published online: 30 April 2012.
Edited by:
Mike Eccles, University of Otago, New ZealandCopyright: © 2012 Huppi, Pitt, Wahlberg and Caplen. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Konrad Huppi, Gene Silencing Section, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Building 37, Room 6128, Bethesda, MD 20892, USA. e-mail: huppi@helix.nih.gov
†Present address: Jason J. Pitt, University of Chicago, Chicago, IL, USA;
Brady M. Wahlberg, Kirksville College of Osteopathic Medicine, Kirksville, MO, USA.