- 1State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory for Aquatic Economic Animals and Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 4Marine Fisheries Development Center of Guangdong Province, Huizhou, China
Beta-hydroxysteroid dehydrogenases (β-HSDs) are a group of steroidogenic enzymes that are involved in steroid biosynthesis and metabolism, and play a crucial role in mammalian physiology and development, including sex determination and differentiation. In the present study, a genome-wide analysis identified the numbers of β-hsd genes in orange-spotted grouper (Epinephelus coioides) (19), human (Homo sapiens) (22), mouse (Mus musculus) (24), chicken (Gallus gallus) (16), xenopus (Xenopus tropicalis) (24), coelacanth (Latimeria chalumnae) (17), spotted gar (Lepisosteus oculatus) (14), zebrafish (Danio rerio) (19), fugu (Takifugu rubripes) (19), tilapia (Oreochromis niloticus) (19), medaka (Oryzias latipes) (19), stickleback (Gasterosteus aculeatus) (17) and common carp (Cyprinus carpio) (27) samples. A comparative analysis revealed that the number of β-hsd genes in teleost fish was no greater than in tetrapods due to gene loss followed by a teleost-specific whole-genome duplication event. Based on transcriptome data from grouper brain and gonad samples during sex reversal, six β-hsd genes had relatively high expression levels in the brain, indicating that these genes may be required for neurogenesis or the maintenance of specific biological processes in the brain. In the gonad, two and eight β-hsd genes were up- and downregulated, respectively, indicating their important roles in sex reversal. Our results demonstrated that β-hsd genes may be involved in the sex reversal of grouper by regulating the synthesis and metabolism of sex steroid hormones.
Introduction
In almost all vertebrates, sexual reproduction requires both females and males to maintain survival and variation. Therefore, sex determination and differentiation are important processes for the continuation of species (Kobayashi et al., 2013). The orange-spotted grouper (Epinephelus coioides) is a protogynous hermaphrodite species. The gonads of most groupers develop into ovaries during the first sex differentiation; however, the ovaries of some individuals then change to testis after female sexual maturation is complete in a process known as sex reversal (Liu and de Mitcheson, 2009). Gonadal changes in sex-changing fish are accompanied by changes in plasma levels of sex steroids (Godwin, 2010), and the balance between estrogen and androgen controls the direction of gonadal development during sex reversal (Liu et al., 2017).
All classes of steroid hormones are synthesized from the common precursor cholesterol. The synthesis of sex steroid hormones is carried out under the catalysis of a series of enzymes, and the genes encoding these enzymes include hsd3b1, hsd11b2, hsd17b1, hsd17b3, and hsd20b2 of the beta-hydroxysteroid dehydrogenases (β-HSDs) superfamily, and cyp11a1, cyp11b, cyp17a1, cyp19a1a, and cyp21a1 of the cytochrome P450 (cP450) superfamily (Tokarz et al., 2015). Hsd3b1 is responsible for the oxidation and isomerization of Δ5-3β-hydroxysteroid precursors in order to form Δ4-3-ketosteroid (Nakamoto et al., 2012). Hsd11b2 catalyzes the conversion of cortisol to cortisone (Fuller et al., 2012), and also plays a crucial role in the conversion of 11β-OH-androstenedione to 11-ketotestosterone (11-KT) (Tokarz et al., 2015), which affects the production of the most important androgen in teleost fish. Furthermore Hsd17b1 converts inactive estrone to active estradiol (Mindnich et al., 2004), Hsd17b3 is an essential enzyme for the synthesis of 11-KT (Mindnich et al., 2005) and Hsd20b2 converts 17alpha-hydroxyprogesterone to 17alpha,20beta-DP, which has been identified as the maturation-inducing hormone of several teleosts (Senthilkumaran et al., 2002).
HSDs catalyze the conversion of a hydroxy group to a keto group or vice versa on a steroid ring in a specific position and orientation (Yang et al., 2005). Different types of β-HSDs are found in various tissues including brain and gonad (He et al., 2018), and most belong to the short chain dehydrogenase/reductase (SDR) family, except for HSD17B5, which belongs to the structural family of aldo-keto reductases (AKRs) (Day et al., 2008). To the best of our knowledge, systematic identification of the SDR and HSD gene families has only been performed in plants (Moummou et al., 2012) and bacteria (Kisiela et al., 2012), respectively. In addition, functional studies of β-hsd genes have mainly utilized human, mouse and zebrafish model organisms, while little is known about their expression and function in other species.
Orange-spotted grouper is an important marine fish cultured in China and Southeast Asian countries. Recently, sequencing and assembly of the grouper genome, and RNA-sequencing of the brain and gonad of grouper during sex reversal were completed (unpublished data), making it a good model for genome-wide identification and gonadal expression profiling of β-hsd genes.
Here we report the genome-wide identification of β-hsd genes in grouper, human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), xenopus (Xenopus tropicalis), coelacanth (Latimeria chalumnae), spotted gar (Lepisosteus oculatus), zebrafish (Danio rerio), fugu (Takifugu rubripes), tilapia (Oreochromis niloticus), medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), and common carp (Cyprinus carpio). Furthermore, RNA-seq data was used to analyze the expression of β-hsd genes during the sex reversal of grouper. Our study may provide a greater understanding of the evolution and possible functions of β-hsd genes in teleosts.
Materials and Methods
Sample Preparation and Gonadal Classification
All animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Sun Yat-sen University. Grouper were cultivated in the Marine Fisheries Development Center of Guangdong Province. In April 2015, 100 groupers weighing approximately 5 kg were sacrificed for tissue sampling after being anesthetized by MS222. For each fish, the gonad and brain were taken and stored at −80°C for RNA extraction, and a piece of gonadal tissue was fixed in Bouin’s fluid for histological examination.
Histological examination was performed as described previously (Xiao et al., 2018). Each gonad was assigned to one of five periods based on the gonadal morphology previously reported (Liu and de Mitcheson, 2009): ovarian-phase-2 (Ov2), ovarian-phase-3 (Ov3), ovarian-phase-4 (Ov4), bisexual-phase (Bi) and testis (Te). Ov2 gonad was predominated by primary-growth stage oocytes, Ov3 gonad was accompanied by the development of cortical-alveolus stage oocytes, and the appearance of vitellogenic stage oocytes indicated that the ovary had matured and belonged to Ov4. During the sex reversal of grouper, spermatogenic cysts and primary-growth stage oocytes are present in the bisexual-phase gonad simultaneously. The oocyte completely disappears and the gonad is filled with male germ cells, indicating that the sex reversal has been completed and the gonad has changed to testis.
Genome-Wide Identification of β-hsd Genes
The genome sequences and predicted protein sequences of human1, mouse2, chicken3, xenopus4, coelacanth5, spotted gar6, zebrafish7, fugu8, tilapia9, medaka10, and stickleback11 were downloaded from Ensembl, and the sequences of the common carp were downloaded from NCBI12. The genomic sequence of grouper has been recently sequenced, with 1.03 GB of genomic data assembled and 23043 encoded genes predicted, of which 23,013 genes could be annotated (PRJEB28248 in the NCBI Sequence Read Archive site).
To identify candidate β-hsd genes in these thirteen species, we collected the gene and protein names of the β-hsd gene family from the literature (Payne et al., 1997; Dai et al., 2003; Mindnich et al., 2004; Seckl, 2004; Hoffmann and Maser, 2007; Meier et al., 2009; Mindnich and Adamski, 2009; Sreenivasulu and Senthilkumaran, 2009; Rotinen et al., 2010; Kisiela et al., 2012; Saloniemi et al., 2012; Tokarz et al., 2012; Lin et al., 2013; He et al., 2018). Secondly, the amino acid sequences of human and zebrafish β-hsd genes were collected from Ensembl and GenBank by searching for their gene and protein names. Next, specific hidden Markov models (HMMs) of human and zebrafish β-HSDs were developed by hmmbuild command in HMMER (HMMER3.1B213) and then used as the query sequences to search the protein databases of all the studied species with hmmsearch command in HMMER (E-value = 0.01). Because all the β-hsd genes belong to the SDR family, except for hsd17b5, which belongs to the AKR superfamily, there was concern that the HMMs-based search would miss hsd17b5. Therefore, the amino acid sequence of human HSD17B5 was used as query sequence to search the protein databases of other species by local BLASTP with a cut off E-value of 1e-5 (BLAST 2.2.26 release from NCBI by anonymous FTP14) (Altschul et al., 1997). All the suspected sequences were aligned with the NCBI non-redundant protein sequence database by online BLASTP program to obtain reliable sequences of β-hsd genes.
To explore the structural diversity of the β-HSD family, the 19 grouper β-HSD protein sequences were submitted to the online CD-Search in CDD program15 (Marchler-Bauer et al., 2017) to search for functional domains (E-value = 0.01). In addition, the online tool GSDS 2.016 (Hu et al., 2015) was used to display the gene structure and functional domains of grouper β-hsd genes.
Phylogenetic Analysis
Multiple alignment of the amino acid sequences of all β-HSDs from six species including grouper, human, mouse, zebrafish, tilapia, and stickleback was performed using ClustalX-2.1 program (Jeanmougin et al., 1998). The maximum likelihood tree and neighbor-joining phylogenetic trees were constructed using the MEGA7 program (Kumar et al., 2016).
Transcriptome-Based Analysis of Expression Profiling of the β-hsd Genes
Brain and gonad tissue from two fish in each stage were selected for transcriptome sequencing. RNA extraction and detection, library construction, sequencing, gene expression calculation and differential gene expression analyses were performed as previously described (Xiao et al., 2018). These RNA-seq data (PRJNA564153 in the NCBI Sequence Read Archive site) were used to analyze the β-hsd gene expression profiles during sex reversal. The expression of β-hsd genes was shown using R packages (i386 3.4.0). The RPKM values were transformed into Z scores before drawing heatmap. Z scores were plotted according to Z = (x - μ)/σ, where x is the log2 transformed gene expression measurement and μ and σ are the mean and standard deviations of expression of the gene.
Gene Expression Profiling by Quantitative Real-Time PCR (qRT-PCR)
To examine the reliability of RNA-seq results, 10 β-hsd genes were selected for validation using qRT-PCR. Because there were only four bisexual-phase fish among the 100 sacrificed individuals, four fish were used for qRT-PCR in each period. Total RNA was extracted and reverse-transcribed using TRIzolTM reagent (Invitrogen, United States) and a Transcriptor First Strand cDNA Synthesis Kit (Roche, United States), respectively. Specific primers (Supplementary Table S3) were designed by Primer Premier 6 software. β-actin was chosen to be the housekeeping gene for its stable expression in brain and gonad tissue during sex reversal. The qRT-PCR reactions based on SYBR (LightCycler 480 SYBR Green I Master, Roche, United States) were performed with a LightCycler 480 system (Roche, United States). Relative expression levels were calculated using the 2–△ △ Ct method (Livak and Schmittgen, 2001). Analysis and visualization of quantitative results was performed by GraphPad Prism 6.0, and one-way ANOVA statistical analysis was used in the analysis.
Results
Identification of β-hsd Genes
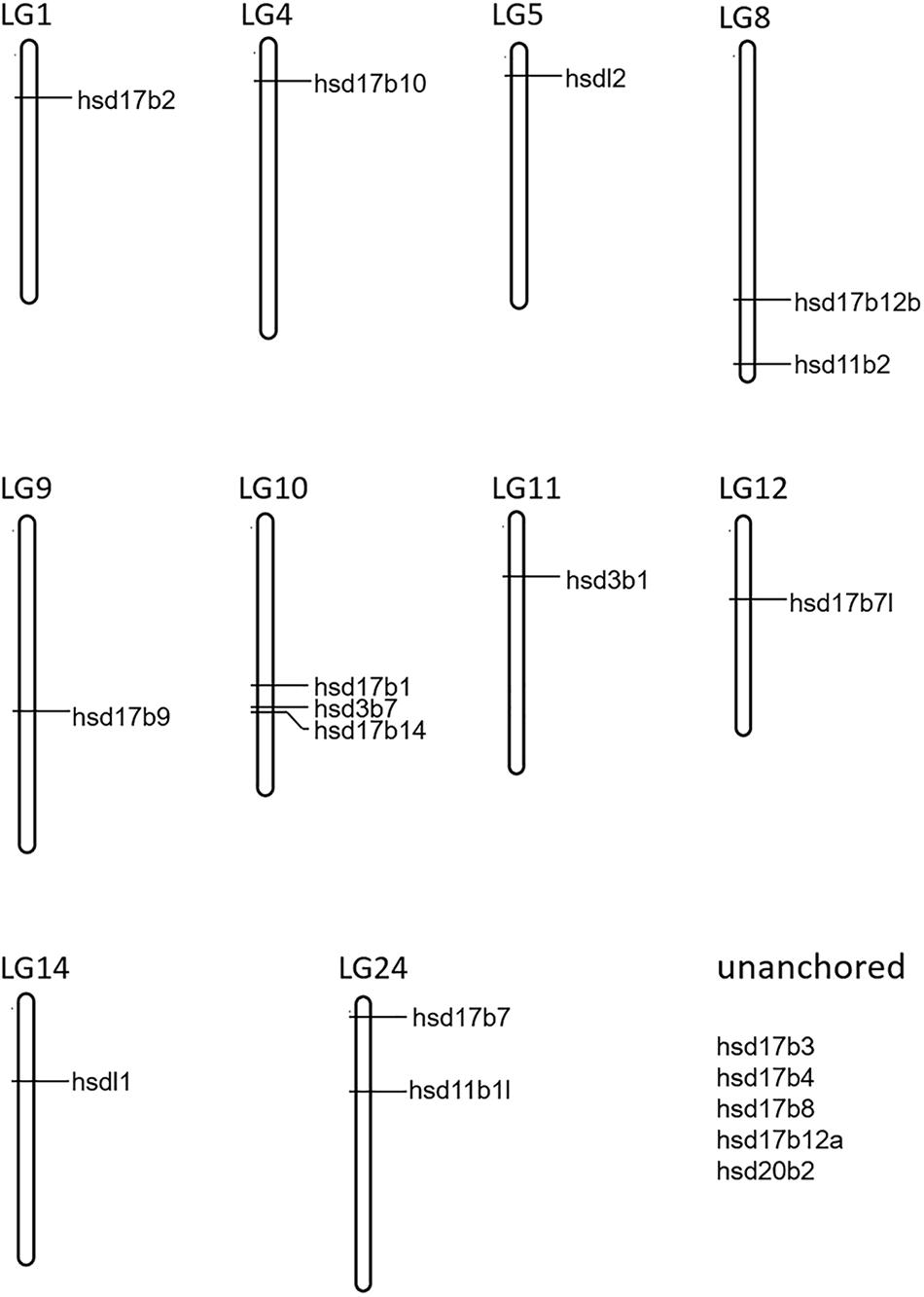
A total of 22 and 19 β-hsd genes were collected from the human and zebrafish genome, respectively, which were used to construct HMM models. A set of 19 putative members of the β-hsd genes have been identified from the orange-spotted grouper genome (Table 1). Among the 19 β-hsd genes, a total of 14 were distributed across 10 linkage groups (LGs) in the grouper genome (Figure 1). In addition, we also identified 24, 16, 24, 17, 14, 19, 19, 19, 17, and 27 β-hsd genes in mouse, chicken, xenopus, coelacanth, spotted gar, fugu, tilapia, medaka, stickleback, and common carp, respectively (Table 1). The gene and protein IDs of the β-hsd genes of all species are listed in the attached table (Supplementary Table S1).
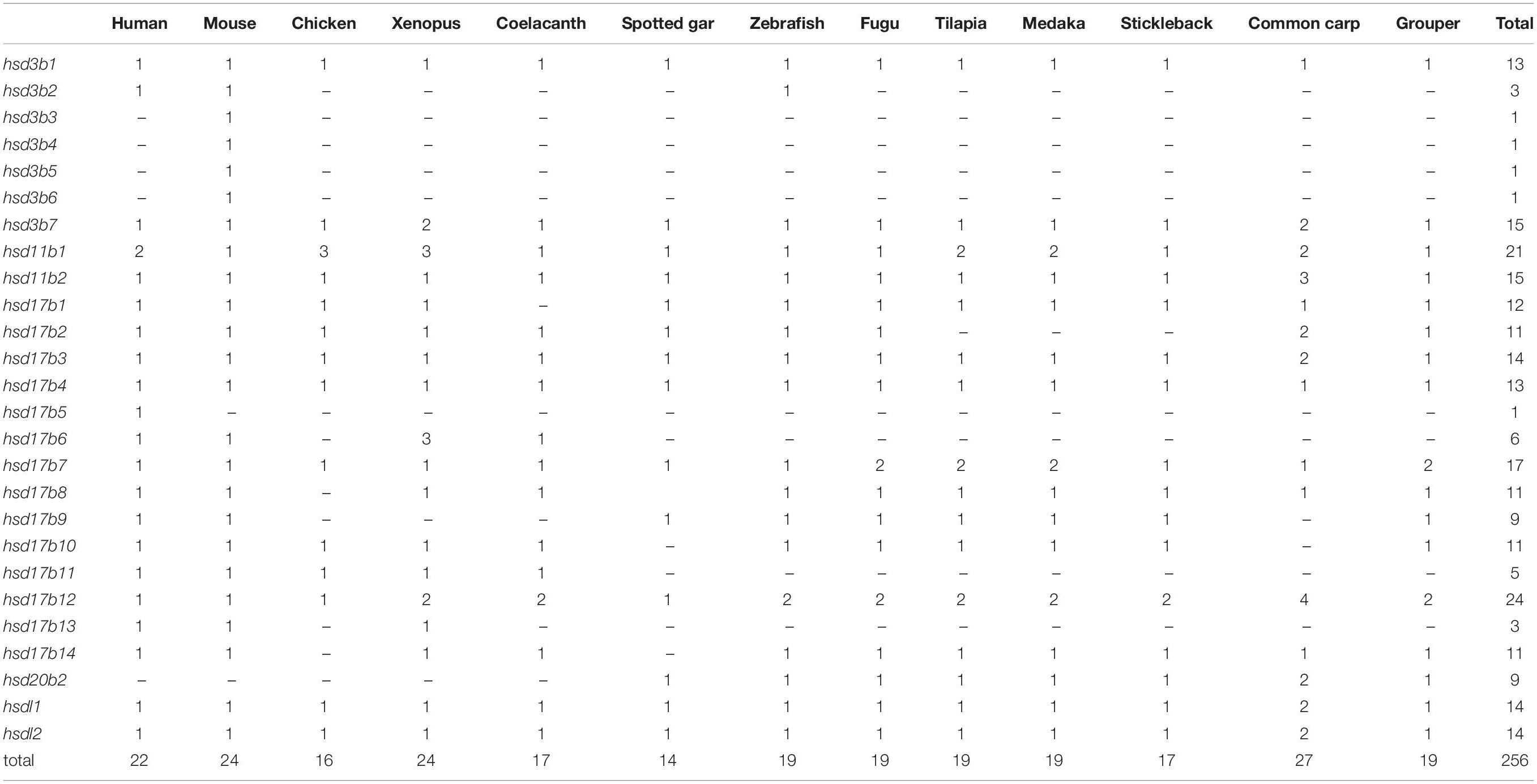
Table 1. Number variation of β-hsd genes in the orange-spotted grouper and the other surveyed animals.
Gene Structure and Conserved Domains
The exon-intron structure of the grouper β-hsd genes was further characterized, and showed that the number of introns in each β-hsd gene varied from 2–16 (Figure 2B). Hsd3b1 was the gene with the lowest number of introns, containing only two, while hsd17b4 had the greatest amount at 16. In addition, a phylogenetic tree was constructed using the grouper β-HSD protein sequences to determine whether the exon-intron structure is consistent with the phylogenetic tree (Figure 2A). As expected, several genes with similar exon/intron structures were clustered together on the phylogenetic tree, such as hsd17b7/hsd17b7l, hsd20b2/hsd17b12a/hsd17b12b. However, due to large number of exon/intron structure types of the grouper β-hsd genes, only a few genes shared similar structures.
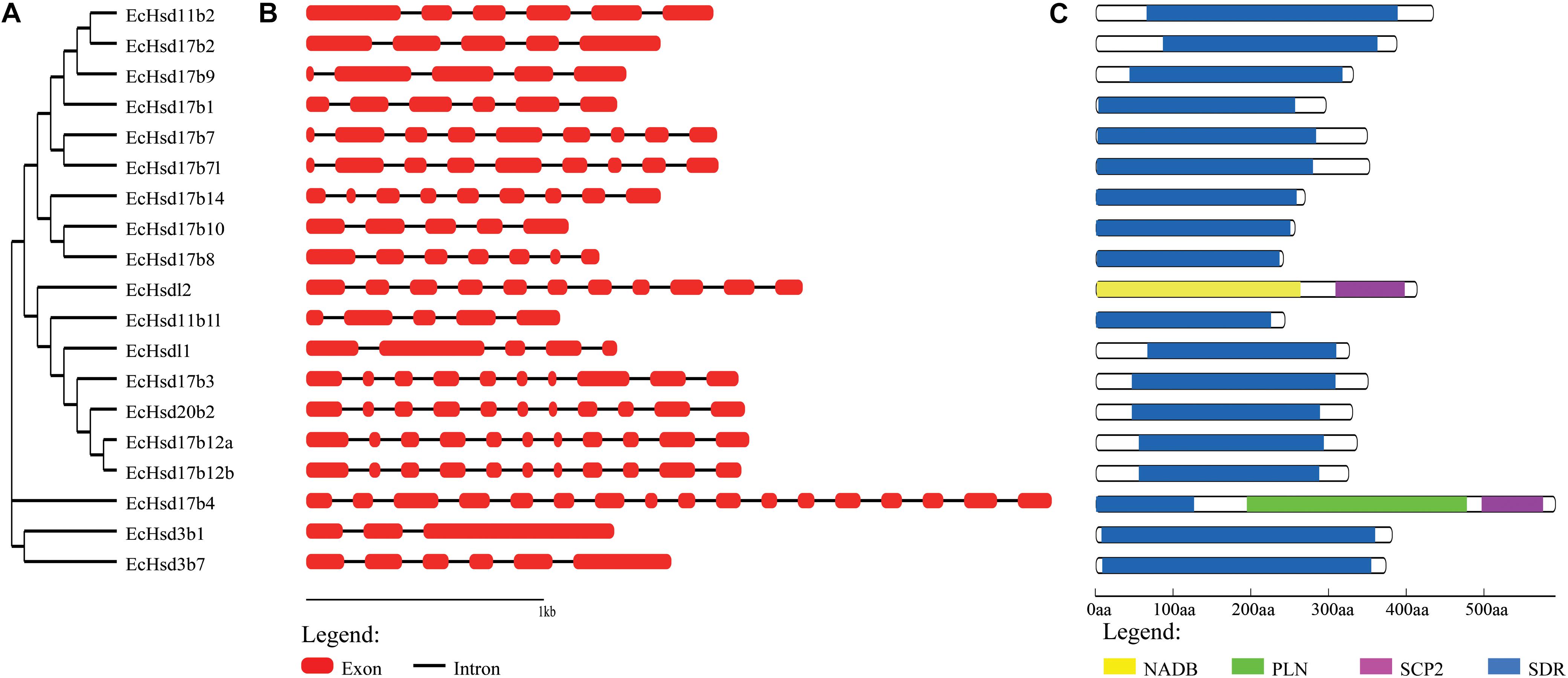
Figure 2. Phylogenetic analysis, gene structure and conserved domains of β-hsd genes in orange-spotted grouper. (A) The phylogenetic tree of all β-Hsd in grouper was constructed using Neighbor-Joining method. (B) The exon/intron organization of β-hsd genes of grouper. The red boxes represent exons and black lines indicate introns. (C) The conserved protein domains in the β-Hsd were identified using CD-Search program. Each domain is indicated with a specific color. Yellow: NADB_Rossmann superfamily, green: PLN02864 superfamily, purple: SCP2 superfamily, blue: SDR superfamily.
The functional domains of grouper β-HSD were predicted by the CD-Search (Marchler-Bauer et al., 2017) based on their protein sequences (Figure 2C). As shown in Figure 2C, 18 of the 19 grouper β-HSDs possessed an SDR superfamily domain, except Hsdl2, indicating a high level of conservation of β-hsd genes belonging to the SDR superfamily. In addition to the SDR superfamily domain, Hsd17b4 also contained two other domains: PLN02864 superfamily domain and SCP2 superfamily domain. Notably, Hsdl2 was a member of the SDR superfamily, but no SDR superfamily domain existed in grouper Hsdl2, according to the CDD prediction. Therefore, we also predicted the functional domain of Hsdl2 protein sequences of other species by CD-Search (Supplementary Figure S1). As shown in Supplementary Figure S1, six Hsdl2 protein sequences contained the SDR superfamily domain and did not contain the NADB_Rossmann superfamily domain, while the other eight Hsdl2 protein sequences, including grouper Hsdl2, were the opposite.
Phylogenetic Analysis
To clarify the evolutionary relationships among β-hsd genes, a phylogenetic tree was constructed with amino acid sequences of 96 β-HSDs collected from grouper, human, zebrafish, stickleback, and tilapia. Both maximum likelihood (ML, Figure 3) and Neighboring-Joining method (NJ, Supplementary Figure S2) strategies were devoted to construct phylogenetic trees based on the alignment of their amino acid sequences. The trees produced by these two methods differed only in a small number of branches, indicating that the evolutionary tree is credible.
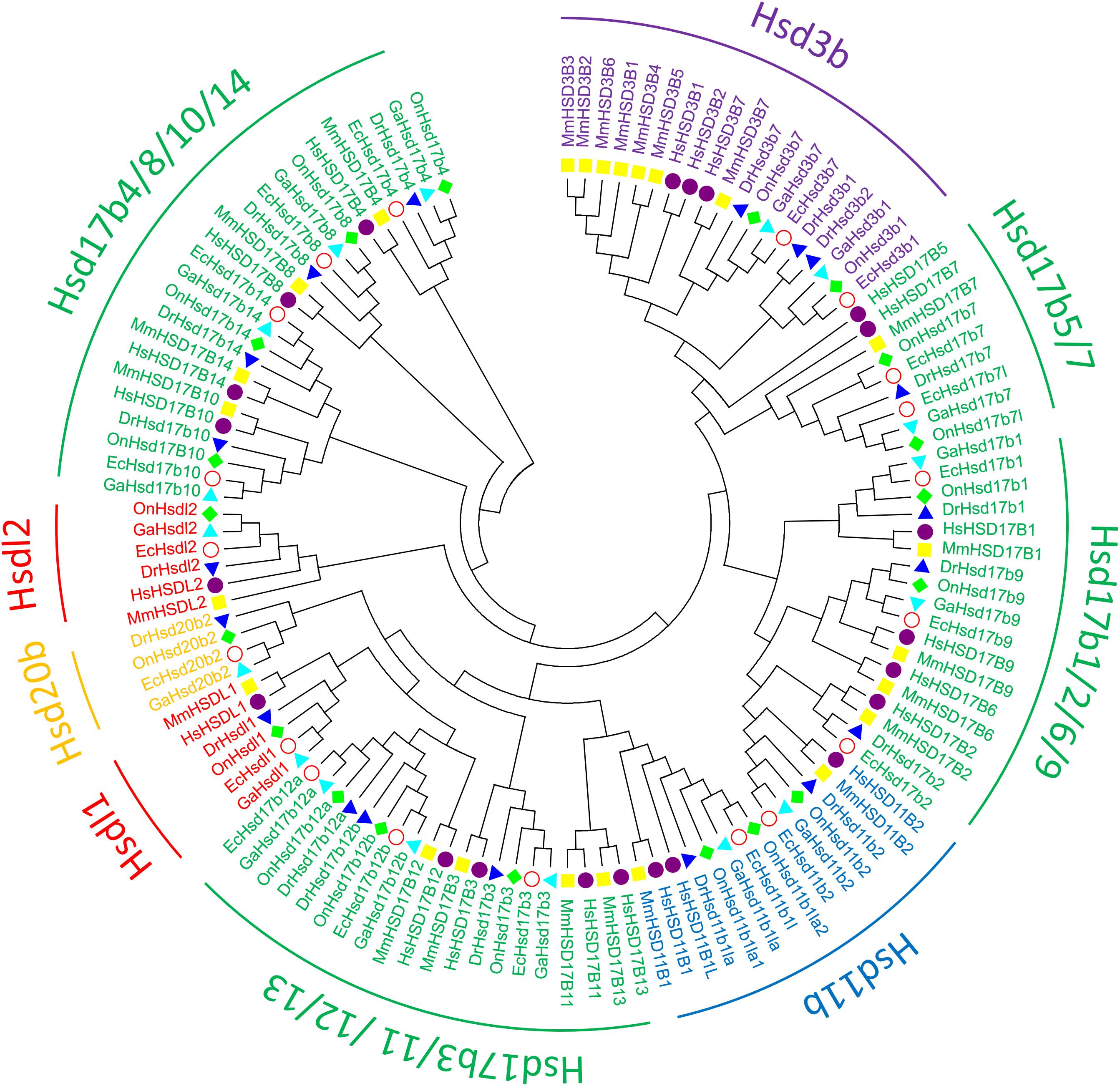
Figure 3. Phylogenetic relationships of β-Hsd proteins from grouper and other five species. The phylogenetic tree was constructed using MEGA7 by Maximum Likelihood method with the amino acid sequences of β-Hsd from grouper (Epinephelus coioides, Ec), (Homo sapiens, Hs), mouse (Mus musculus, Mm), zebrafish (Danio rerio, Dr), tilapia (Oreochromis niloticus, On), and stickleback (Gasterosteus aculeatus, Ga).
Expression of β-hsd Genes During Sex Reversal of Grouper
We used transcriptome data of grouper brains and gonads at five developmental stages, namely Ov2(ovarian-phase-2), Ov3(ovarian-phase-3), Ov4(ovarian-phase-4), Bi(bisexual-phase) and Te(testis), to profile the expression of β-hsd genes during sex reversal of grouper. By transcriptomic analysis, 18 of the 19 β-hsd genes were detected in the brains and gonads during sex reversal, except for hsd17b2 (Supplementary Table S2). According to the criteria that a gene is considered to be expressed if it exhibits an expression level with RPKM value ≥2, we found that except for hsd11b1l, hsd17b9, and hsd20b2, the other 15 β-hsd genes were expressed in the brains or gonads in at least one development stage (Figure 4 and Supplementary Table S2). Among these expressed β-hsd genes, eleven members (hsd3b1, hsd17b1, hsd17b4, hsd17b7l, hsd17b8, hsd17b10, hsdl17b12a, hsd17b12b, hsd17b14, hsdl1, and hsdl2) and six members (hsd11b2, hsd17b3, hsd17b10, hsd17b12b, hsdl1, and hsdl2) presented high expression (RPKM value ≥10) in gonads and brain in at least one developmental stage, respectively. As shown in Figure 4, the expression levels of eight β-hsd genes (hsd3b7, hsd17b1, hsd17b7l, hsd17b10, hsd17b12a, hsd17b12b, hsdl1, and hsdl2) were reduced, while two β-hsd genes (hsd3b1 and hsd17b14) were up-regulated in the gonad during sex reversal.
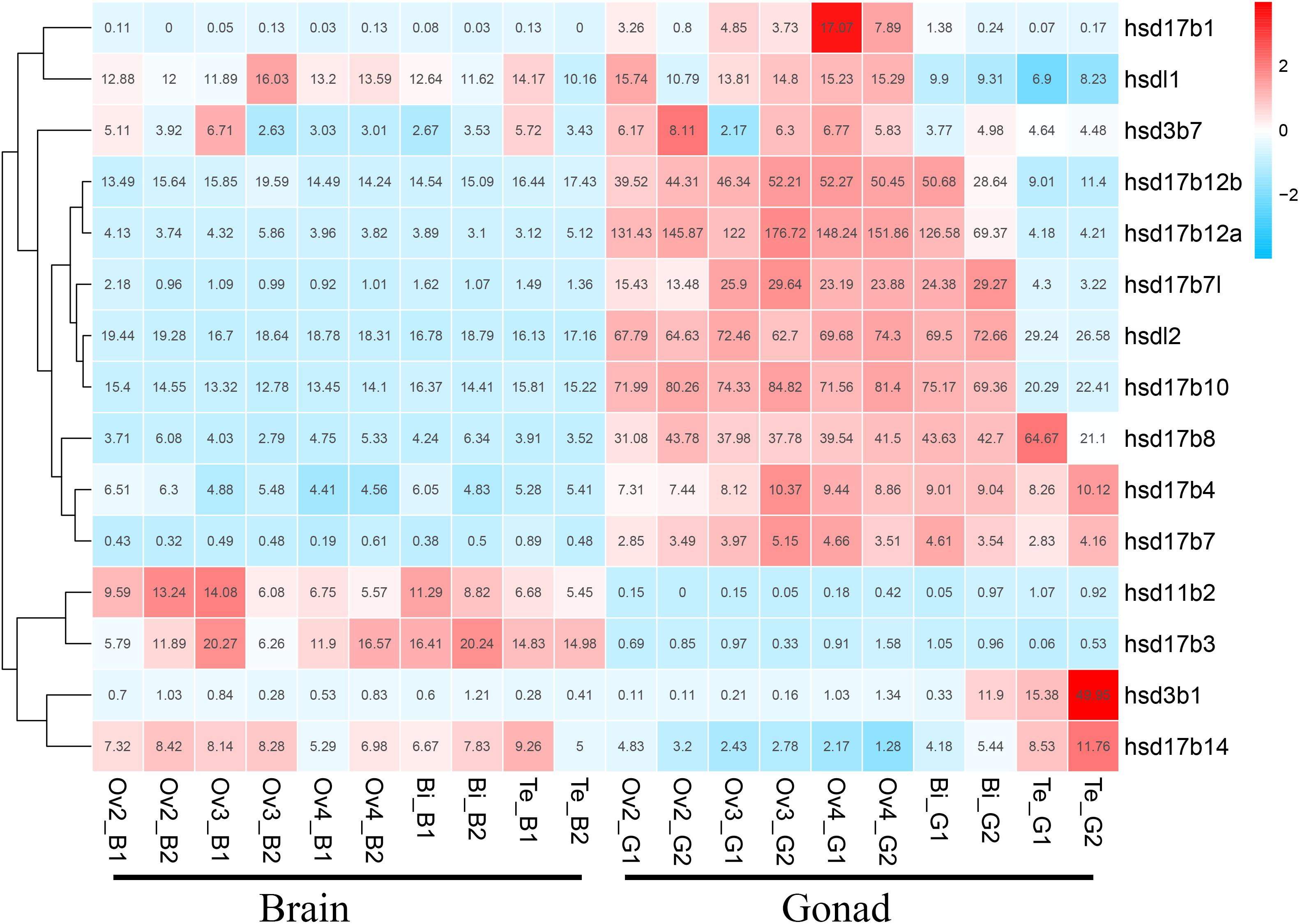
Figure 4. Clustering of the expression profiles of 19 grouper β-hsd genes during sex reversal. Genes were clustered according to phylogenetic relationships in expression profiles. The RPKM values were transformed into Z scores. Z scores were plotted according to Z = (x - μ)/σ, where x is the log 2 transformed gene expression measurement and μ and σ are the mean and standard deviations of expression of the gene. Red and blue colors indicate high and low relative expression levels after homogenization, respectively. The RPKM value for each gene and each stage was provided in the figure.
To further verify the expression levels of β-hsd genes during sex reversal of grouper, qRT-PCR validation experiments were conducted using gene-specific primers (Supplementary Table S3). The four genes involved in the synthesis of sex steroid hormones (hsd3b1, hsd11b2, hsd17b3, and hsd20b2) and eight genes with RPKM value ≥20 (a total of ten genes after de-duplication) were selected for qRT-PCR validation. Expression patterns during sex reversal of these 10 genes were shown to be consistent with transcriptome data (Figures 4, 5 and Supplementary Table S2).
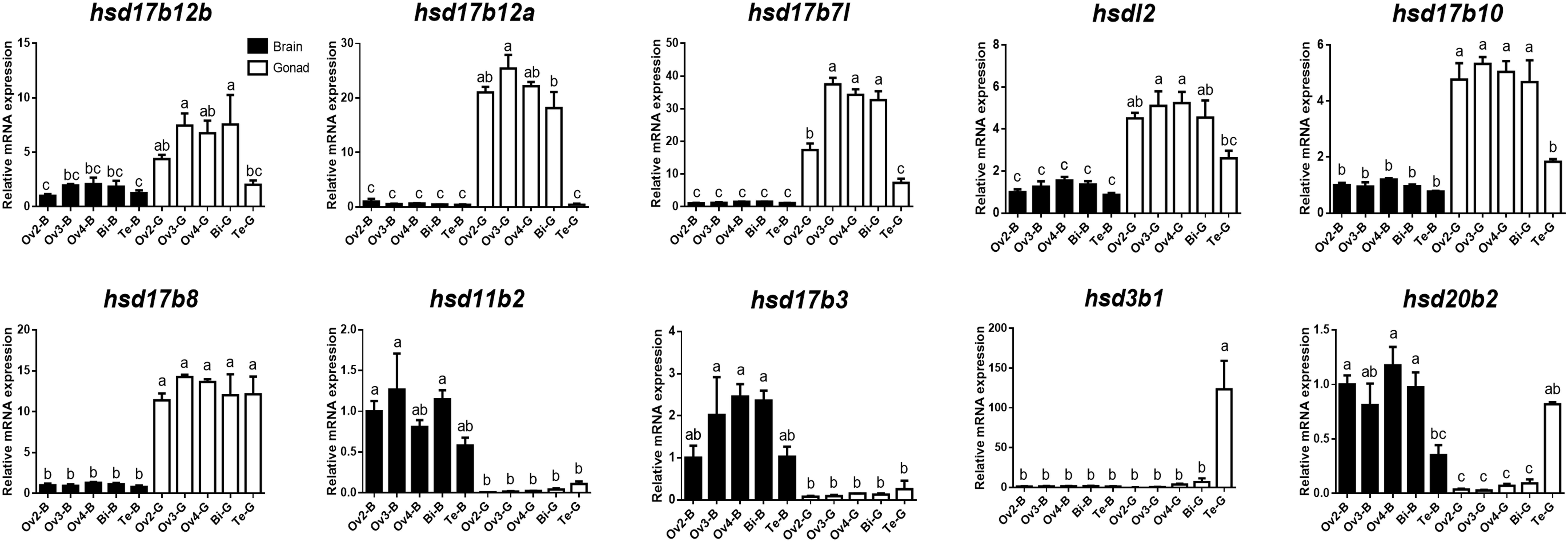
Figure 5. Quantitative RT-PCR examination of 10 β-hsd genes expressions in the brain and gonad during sex reversal of grouper. Values represent the relative mRNA expression in relation to internal control (β-actin gene). Data were expressed as the mean ± SEM for four replicates. In each panel, different lowercase letters indicate significant differences (p < 0.05).
Discussion
HSDs are a group of steroidogenic enzymes that are involved in steroid biosynthesis and metabolism (Ye et al., 2014). To the best of our knowledge, HSD gene repertoires have previously been described in bacteria (Kisiela et al., 2012), plants (Moummou et al., 2012), zebrafish (Mindnich et al., 2004; Mindnich and Adamski, 2009) and several mammals (Payne et al., 1997; Baker, 2001; Belyaeva and Kedishvili, 2006; Rotinen et al., 2010), but no studies have summarized all the β-hsd genes. Little is known about the corresponding grouper genes; however, the availability of the complete grouper genome sequence has made it possible to identify all the β-hsd genes in this species for the first time. In the present study, we systematically idntified 19 putative β-hsd genes in the E. coioides genome, and subsequently characterized the genes in terms of position in the genome, exon-intron structure and conserved domains. In addition, we also isolated β-hsd genes in samples of human, mouse, chicken, xenopus, coelacanth, spotted gar, zebrafish, fugu, tilapia, medaka, stickleback, and common carp. Finally, an analysis of grouper β-hsd gene expression levels during sex reversal indicated that these genes may play important roles in this significant physiological process. Our study represented the first comprehensive investigation of the fish β-hsd gene family, and the resulting data will undoubtedly be useful for future analyses, including further research into the role of β-hsd genes and sex steroids in sex reversal of grouper.
Comparative analysis revealed several evolutionary phenomena of the β-hsd genes during the separation of fish species from other animals. First, unlike some other gene families which have undergone an expansion in the teleost fish, the number of β-hsd genes in teleost fish was no greater than in tetrapods. Studies have reported after two major radiations of jawed vertebrate, teleosts shared another round of whole-genome duplication events (the 3R WGD) (Kasahara et al., 2007), and common carp underwent an additional whole-genome duplication event (the 4R WGD) (Xu et al., 2014). For the nuclear receptor (NR) superfamily, 48, 75, and 137 members have been reported in human, tilapia and common carp, respectively, suggesting that NRs may have expanded along with multiple duplication events (Cheng et al., 2015). The sox gene family has also undergone a continuous expansion in the teleost fish following their whole genome duplication (Wei et al., 2016). However, for the β-hsd genes, expansion after the 3R WGD and the 4R WGD was not as obvious as in the NR gene family and sox gene family. For example, hsd3b1 and hsd17b4 had only one copy in all species, and hsd17b3, hsdl1, and hsdl2 had two copies in common carp which had undergone the 4R WGD, and only one copy in other species. Such a phenomenon is considered to be a result of gene loss followed by the WGD. The duplication-degeneration-complementation (DDC) model proposed by Lynch and Conery (2000) on the fate of duplicated genes suggested that the common fate of a duplicated gene was lost from the genome owing to non-functionalization unless one of the duplicate genes acquired a new function, or the functions of the ancestral gene are sub-divided between the sister duplicate genes. Gene loss was thought to be a mechanism to maintain a balance for duplicated genes post genome duplication events. Gene loss is evident in the β-hsd gene family, but not so obvious in the NR and sox gene families, which may be due to different selective pressures or different evolutionary rates among different gene catalogs. Secondly, several genes were identified only in a particular species. For instance, among all the species we studied, Hsd3b3, Hsd3b4, Hsd3b5, and Hsd3b6 were specifically identified in the mouse genome. The six mouse Hsd3b genes (MnHsd3b1, MnHsd3b2 and four mouse-specific Hsd3b genes) were located in a small region (98499071-98859794) of chromosome 3 and clustered together in the phylogenetic tree, seemingly suggesting that the four mouse-specific Hsd3b genes were more likely a product of mouse-specific gene duplication rather than gene loss in other species. In addition, hsd20b2 was not identified in tetrapod genomes, apparently indicating that hsd20b2 may be lost in the tetrapod genome but retained in the teleost genome following their divergence.
The orange-spotted grouper is a typical protogynous hermaphrodite species. Previous studies have shown that groupers in the natural environment generally develop into female individuals during the first sex differentiation (Liu and de Mitcheson, 2009). Then the sex reversal from female to male occur in a small part of mature/functional females, while the males retain the same sex throughout their life span (Bhandari et al., 2003). Sex steroids play a very important role in sex reversal of groupers. Low serum E2 levels and increased 11-KT levels are probably the events mediating protogynous sex change in grouper (Bhandari et al., 2003). In addition, exogenous androgens could artificially induce the sex reversal of groupers from female to male (Chen et al., 2011; Oh et al., 2013; Murata et al., 2014). However, little is known about the expression profiles of sex steroid synthesis and metabolism genes in the natural sex reversal process. HSDs participate in the synthesis and metabolism of sex steroid hormones by catalyzing the conversion of a hydroxy group to a keto group or vice versa on a steroid ring in a specific position and orientation (Yang et al., 2005). Studies on the expression profiles of hsd genes at different stages of grouper sex reversal may provide us a better understanding on the roles of hsd genes in sex reversal.
The transcriptome data indicated that six (hsd11b2, hsd17b3, hsd17b10, hsd17b12b, hsdl1, and hsdl2) and eleven (hsd3b1, hsd17b1, hsd17b4, hsd17b7l, hsd17b8, hsd17b10, hsdl17b12a, hsd17b12b, hsd17b14, hsdl1, and hsdl2) β-HSD genes had relatively high expression levels (RPKM >10) in the brain and gonad during sex reversal of grouper, respectively. HSDs are indispensable neuro-steroidogenic enzymes, and the neuro-steroids synthesized in neurons and glia can act on various receptors to regulate neuroexcitability while also playing important roles in brain development, neuroprotection and neurogenesis as well as cognition and memory (He et al., 2018). For example, human HSD17B10 is found in various brain regions and is essential for the maintenance of neurosteroid homeostasis (He et al., 2018); and Hsd11b2 is involved in the negative feedback regulation of cortisol post stress in the brain of zebrafish (Alderman and Vijayan, 2012). In grouper, transcriptome data has shown that the expression levels of six β-hsd genes highly expressed in the brain did not change significantly during sexual reversal, seemingly indicating that these genes may be required for neurogenesis or the maintenance of specific biological processes in the brain, but not for sex reversal events.
In the gonad, two (hsd3b1 and hsd17b14) and eight (hsd3b7, hsd17b1, hsd17b7l, hsd17b10, hsd17b12a, hsd17b12b, hsdl1, and hsdl2) β-HSD genes were up- and downregulated during sex reversal, respectively. Hsd3b1 catalyzes the second step of steroidogenesis and is required for the synthesis of all steroids including 11-KT (Hsu et al., 2009). In human testis specimens, HSD17B14 protein has shown immunoreactivity in most of the seminiferous epithelium as well as in peritubular areas harboring Leydig cells (Sivik et al., 2012). Transfection of HSD17B14 in human breast cancer cells significantly decreased the levels of estradiol (Jansson et al., 2006), and further studies have suggested a role for HSD17B14 in the local inactivation of steroid (Lukacik et al., 2007). Therefore, we hypothesized that the increased expression levels of hsd3b1 during the sex reversal of grouper may promote the synthesis of 11-KT, while the increased expression level of hsd17b14 may lead to the inactivation of estradiol in this study.
HSD17B1 is mainly expressed in the ovary (Mindnich and Adamski, 2009), and catalyzes lower estrogenic active estrone (E1) to highly active estradiol (E2) (Hakkarainen et al., 2015; Jarvensivu et al., 2015). HSD17B12 has been implicated in the conversion of estrone to estradiol as well as in the synthesis of arachidonic acid (AA), and plays important roles in ovarian function and female fertility (Kemilainen et al., 2016). Accordingly, we propose that the high expression levels of hsd17b1, hsd17b12a and hsd17b 12b in the ovary may be due to its involvement in the maintenance of the ovary, and the degeneration of ovary leads to the downregulation of these genes during the sex reversal grouper. The 3-beta-hydroxy-Delta(5)-C(27)-steroid oxidoreductase, which is encoded by HSD3B7 gene, is a membrane-bound enzyme of the endoplasmic reticulum that catalyzes an early step in the synthesis of bile acids from cholesterol, and mutation of the HSD3B7 gene causes neonatal cholestasis (Cheng et al., 2003). Hsd17b7l was identified only in grouper, fugu, tilapia and medaka in this study, and its function has not been studied yet. HSD17B10 catalyzes the oxidation of neuroactive steroids and degradation of isoleucine in the nervous system (Yang et al., 2014). HSDL1 and HSDL2 is highly expressed in human testis and ovary tissue (Huang et al., 2001; Dai et al., 2003). However, the function of Hsd3b7, Hsd17b7l, Hsd17b10, Hsdl1, and Hsdl2 in the gonad is not well understood. Therefore, further studies are required to reveal their functions in the gonad and their roles in the sex reversal of grouper.
Conclusion
Theβ-hsd genes play important roles in the biosynthesis and metabolism of steroids. In the present study, a genome-wide analysis identified numbers of the β-hsd genes in the human (22), mouse (24), chicken (16), xenopus (24), coelacanth (17), spotted gar (14), zebrafish (19), fugu (19), tilapia (19), medaka (19), stickleback (17), grouper (19), and common carp (27) samples. A comparative analysis revealed that the number of β-hsd genes in teleost fish was no greater than in tetrapods due to gene loss followed by the teleost-specific whole-genome duplication event. Transcriptome-based expression profiling uncovered the expressions of the β-hsd genes during the sex reversal of grouper. The exact roles of these differentially expressed β-hsd genes during sex reversal need to be precisely characterized in the future.
Ethics Statement
All animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Sun Yat-sen University.
Author Contributions
LX performed the experiments and wrote the manuscript. YG and DW analyzed the data. MZ and XH contributed the reagents and materials. YZ, SL, and HL provided guidance on the whole manuscript. All authors reviewed and approved the final submission.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31802274, 31802266, 31972769, and 31902346), Guangdong Provincial Natural Science Foundation (2018A030313890 and 2018B030311026), Guangdong Provincial Science and Technology Program (2017B090904022), Guangdong Provincial Special Fund For Modern Agriculture Industry Technology Innovation Teams (2019KJ143), GDAS Special Project of Science and Technology Development (No. 2019GDASYL-0103065), Science and Technology Planning Project of Huizhou (20170214023102296), Special Fund of State Key Laboratory of Developmental Biology of Freshwater (2018KF001), and open fund of Guangdong South China Sea Key Laboratory of Aquaculture for Aquatic Economic Animals (KFKT2019ZD07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00161/full#supplementary-material
Footnotes
- ^ http://asia.ensembl.org/Homo_sapiens/Info/Index
- ^ http://asia.ensembl.org/Mus_musculus/Info/Index
- ^ http://asia.ensembl.org/Gallus_gallus/Info/Index
- ^ http://asia.ensembl.org/Xenopus_tropicalis/Info/Index
- ^ http://asia.ensembl.org/Latimeria_chalumnae/Info/Index
- ^ http://asia.ensembl.org/Lepisosteus_oculatus/Info/Index
- ^ http://asia.ensembl.org/Danio_rerio/Info/Index
- ^ http://asia.ensembl.org/Takifugu_rubripes/Info/Index
- ^ http://asia.ensembl.org/Oreochromis_niloticus/Info/Index
- ^ http://asia.ensembl.org/Oryzias_latipes_hni/Info/Index
- ^ http://asia.ensembl.org/Gasterosteus_aculeatus/Info/Index
- ^ ftp://ftp.ncbi.nlm.nih.gov/genomes/Cyprinus_carpio/
- ^ http://www.hmmer.org/
- ^ ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://gsds.cbi.pku.edu.cn/
References
Alderman, S. L., and Vijayan, M. M. (2012). 11beta-Hydroxysteroid dehydrogenase type 2 in zebrafish brain: a functional role in hypothalamus-pituitary-interrenal axis regulation. J. Endocrinol. 215, 393–402. doi: 10.1530/JOE-12-0379
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Baker, M. E. (2001). Evolution of 17beta-hydroxysteroid dehydrogenases and their role in androgen, estrogen and retinoid action. Mol. Cell. Endocrinol. 171, 211–215. doi: 10.1016/s0303-7207(00)00414-7
Belyaeva, O. V., and Kedishvili, N. Y. (2006). Comparative genomic and phylogenetic analysis of short-chain dehydrogenases/reductases with dual retinol/sterol substrate specificity. Genomics 88, 820–830. doi: 10.1016/j.ygeno.2006.06.004
Bhandari, R. K., Komuro, H., Nakamura, S., Higa, M., and Nakamura, M. (2003). Gonadal restructuring and correlative steroid hormone profiles during natural sex change in protogynous honeycomb grouper (Epinephelus merra). Zool. Sci. 20, 1399–1404. doi: 10.2108/zsj.20.1399
Chen, H., Zhang, Y., Li, S., Lin, M., Shi, Y., Sang, Q., et al. (2011). Molecular cloning, characterization and expression profiles of three estrogen receptors in protogynous hermaphroditic orange-spotted grouper (Epinephelus coioides). Gen. Comp. Endocrinol. 172, 371–381. doi: 10.1016/j.ygcen.2011.03.025
Cheng, J. B., Jacquemin, E., Gerhardt, M., Nazer, H., Cresteil, D., Heubi, J. E., et al. (2003). Molecular genetics of 3-beta-hydroxy-Delta5-C27-steroid oxidoreductase deficiency in 16 patients with loss of bile acid synthesis and liver disease. J. Clin. Endocrinol. Metab. 88, 1833–1841. doi: 10.1210/jc.2002-021580
Cheng, Y., Tao, W., Chen, J., Sun, L., Zhou, L., Song, Q., et al. (2015). Genome-wide identification, evolution and expression analysis of nuclear receptor superfamily in Nile tilapia, Oreochromis niloticus. Gene 569, 141–152. doi: 10.1016/j.gene.2015.05.057
Dai, J., Xie, Y., Wu, Q., Wang, L., Yin, G., Ye, X., et al. (2003). Molecular cloning and characterization of a novel human hydroxysteroid dehydrogenase-like 2 (HSDL2) cDNA from fetal brain. Biochem. Genet. 41, 165–174.
Day, J. M., Tutill, H. J., Purohit, A., and Reed, M. J. (2008). Design and validation of specific inhibitors of 17beta-hydroxysteroid dehydrogenases for therapeutic application in breast and prostate cancer, and in endometriosis. Endocr. Relat. Cancer 15, 665–692. doi: 10.1677/ERC-08-0042
Fuller, P. J., Yao, Y., Yang, J., and Young, M. J. (2012). Mechanisms of ligand specificity of the mineralocorticoid receptor. J. Endocrinol. 213, 15–24. doi: 10.1530/JOE-11-0372
Godwin, J. (2010). Neuroendocrinology of sexual plasticity in teleost fishes. Front. Neuroendocrinol. 31:203–216. doi: 10.1016/j.yfrne.2010.02.002
Hakkarainen, J., Jokela, H., Pakarinen, P., Heikela, H., Katkanaho, L., Vandenput, L., et al. (2015). Hydroxysteroid (17beta)-dehydrogenase 1-deficient female mice present with normal puberty onset but are severely subfertile due to a defect in luteinization and progesterone production. FASEB J. 29, 3806–3816. doi: 10.1096/fj.14-269035
He, X. Y., Dobkin, C., and Yang, S. Y. (2018). 17beta-Hydroxysteroid dehydrogenases and neurosteroid metabolism in the central nervous system. Mol. Cell. Endocrinol. 489, 92–97. doi: 10.1016/j.mce.2018.10.002
Hoffmann, F., and Maser, E. (2007). Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab. Rev. 39, 87–144. doi: 10.1080/03602530600969440
Hsu, H. J., Lin, J. C., and Chung, B. C. (2009). Zebrafish cyp11a1 and hsd3b genes: structure, expression and steroidogenic development during embryogenesis. Mol. Cell. Endocrinol. 312, 31–34. doi: 10.1016/j.mce.2009.07.030
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, Y., Tang, R., Dai, J., Gu, S., Zhao, W., Cheng, C., et al. (2001). A novel human hydroxysteroid dehydrogenase like 1 gene (HSDL1) is highly expressed in reproductive tissues. Mol. Biol. Rep. 28, 185–191.
Jansson, A. K., Gunnarsson, C., Cohen, M., Sivik, T., and Stal, O. (2006). 17beta-hydroxysteroid dehydrogenase 14 affects estradiol levels in breast cancer cells and is a prognostic marker in estrogen receptor-positive breast cancer. Cancer Res. 66, 11471–11477. doi: 10.1158/0008-5472.can-06-1448
Jarvensivu, P., Saloniemi-Heinonen, T., Awosanya, M., Koskimies, P., Saarinen, N., and Poutanen, M. (2015). HSD17B1 expression enhances estrogen signaling stimulated by the low active estrone, evidenced by an estrogen responsive element-driven reporter gene in vivo. Chem. Biol. Interact 234, 126–134. doi: 10.1016/j.cbi.2015.01.008
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405.
Kasahara, M., Naruse, K., Sasaki, S., Nakatani, Y., Qu, W., Ahsan, B., et al. (2007). The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719.
Kemilainen, H., Adam, M., Maki-Jouppila, J., Damdimopoulou, P., Damdimopoulos, A. E., Kere, J., et al. (2016). The hydroxysteroid (17beta) dehydrogenase family gene HSD17B12 is involved in the prostaglandin synthesis pathway, the ovarian function, and regulation of fertility. Endocrinology 157, 3719–3730. doi: 10.1210/en.2016-1252
Kisiela, M., Skarka, A., Ebert, B., and Maser, E. (2012). Hydroxysteroid dehydrogenases (HSDs) in bacteria – a bioinformatic perspective. J. Steroid Biochem. Mol. Biol. 129, 31–46. doi: 10.1016/j.jsbmb.2011.08.002
Kobayashi, Y., Nagahama, Y., and Nakamura, M. (2013). Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 7, 115–125. doi: 10.1159/000342009
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lin, S. X., Poirier, D., and Adamski, J. (2013). A challenge for medicinal chemistry by the 17beta-hydroxysteroid dehydrogenase superfamily: an integrated biological function and inhibition study. Curr. Top. Med. Chem. 13, 1164–1171. doi: 10.2174/15680266113139990004
Liu, H., Todd, E. V., Lokman, P. M., Lamm, M. S., Godwin, J. R., and Gemmell, N. J. (2017). Sexual plasticity: a fishy tale. Mol. Reprod. Dev. 84, 171–194. doi: 10.1002/mrd.22691
Liu, M., and de Mitcheson, Y. S. (2009). Gonad development during sexual differentiation in hatchery-produced orange-spotted grouper (Epinephelus coioides) and humpback grouper (Cromileptes altivelis) (Pisces: Serranidae. Epinephelinae). Aquaculture 287, 191–202. doi: 10.1016/j.aquaculture.2008.10.027
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lukacik, P., Keller, B., Bunkoczi, G., Kavanagh, K. L., Lee, W. H., Adamski, J., et al. (2007). Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem. J. 402, 419–427. doi: 10.1042/bj20061319
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Meier, M., Tokarz, J., Haller, F., Mindnich, R., and Adamski, J. (2009). Human and zebrafish hydroxysteroid dehydrogenase like 1 (HSDL1) proteins are inactive enzymes but conserved among species. Chem. Biol. Interact 178, 197–205. doi: 10.1016/j.cbi.2008.10.036
Mindnich, R., and Adamski, J. (2009). Zebrafish 17beta-hydroxysteroid dehydrogenases: an evolutionary perspective. Mol. Cell. Endocrinol. 301, 20–26. doi: 10.1016/j.mce.2008.12.002
Mindnich, R., Deluca, D., and Adamski, J. (2004). Identification and characterization of 17 beta-hydroxysteroid dehydrogenases in the zebrafish Danio rerio. Mol. Cell. Endocrinol. 215, 19–30. doi: 10.1016/j.mce.2003.11.010
Mindnich, R., Haller, F., Halbach, F., Moeller, G., Hrabe, D. A. M., and Adamski, J. (2005). Androgen metabolism via 17beta-hydroxysteroid dehydrogenase type 3 in mammalian and non-mammalian vertebrates: comparison of the human and the zebrafish enzyme. J. Mol. Endocrinol. 35, 305–316. doi: 10.1677/jme.1.01853
Moummou, H., Kallberg, Y., Tonfack, L. B., Persson, B., and van der Rest, B. (2012). The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 12:219. doi: 10.1186/1471-2229-12-219
Murata, R., Kobayashi, Y., Karimata, H., Kishimoto, K., Kimura, M., and Nakamura, M. (2014). Transient sex change in the immature Malabar grouper, Epinephelus malabaricus, androgen treatment. Biol. Reprod. 91:25. doi: 10.1095/biolreprod.113.115378
Nakamoto, M., Fukasawa, M., Tanaka, S., Shimamori, K., Suzuki, A., Matsuda, M., et al. (2012). Expression of 3beta-hydroxysteroid dehydrogenase (hsd3b), star and ad4bp/sf-1 during gonadal development in medaka (Oryzias latipes). Gen. Comp. Endocrinol. 176, 222–230. doi: 10.1016/j.ygcen.2012.01.019
Oh, S. R., Kang, H. C., Lee, C. H., Hur, S. W., and Lee, Y. D. (2013). Sex reversal and masculinization according to growth in longtooth grouper Epinephelus bruneus. Dev. Reprod. 17, 79–85. doi: 10.12717/DR.2013.17.2.079
Payne, A. H., Abbaszade, I. G., Clarke, T. R., Bain, P. A., and Park, C. H. (1997). The multiple murine 3 beta-hydroxysteroid dehydrogenase isoforms: structure, function, and tissue- and developmentally specific expression. Steroids 62, 169–175. doi: 10.1016/s0039-128x(96)00177-8
Rotinen, M., Villar, J., Celay, J., and Encio, I. (2010). Type 10 17beta-hydroxysteroid dehydrogenase expression is regulated by C/EBPbeta in HepG2 cells. J. Steroid. Biochem. Mol. Biol. 122, 164–171. doi: 10.1016/j.jsbmb.2010.07.003
Saloniemi, T., Jokela, H., Strauss, L., Pakarinen, P., and Poutanen, M. (2012). The diversity of sex steroid action: novel functions of hydroxysteroid (17beta) dehydrogenases as revealed by genetically modified mouse models. J. Endocrinol. 212, 27–40. doi: 10.1530/JOE-11-0315
Seckl, J. R. (2004). 11beta-hydroxysteroid dehydrogenases: changing glucocorticoid action. Curr. Opin. Pharmacol. 4, 597–602. doi: 10.1016/j.coph.2004.09.001
Senthilkumaran, B., Sudhakumari, C. C., Chang, X. T., Kobayashi, T., Oba, Y., Guan, G., et al. (2002). Ovarian carbonyl reductase-like 20beta-hydroxysteroid dehydrogenase shows distinct surge in messenger RNA expression during natural and gonadotropin-induced meiotic maturation in nile tilapia. Biol. Reprod. 67, 1080–1086. doi: 10.1095/biolreprod67.4.1080
Sivik, T., Vikingsson, S., Green, H., and Jansson, A. (2012). Expression patterns of 17beta-hydroxysteroid dehydrogenase 14 in human tissues. Horm. Metab. Res. 44, 949–956. doi: 10.1055/s-0032-1321815
Sreenivasulu, G., and Senthilkumaran, B. (2009). New evidences for the involvement of 20beta-hydroxysteroid dehydrogenase in final oocyte maturation of air-breathing catfish. Gen. Comp. Endocrinol. 163, 259–269. doi: 10.1016/j.ygcen.2009.04.022
Tokarz, J., Mindnich, R., Norton, W., Moller, G., Hrabe, D. A. M., and Adamski, J. (2012). Discovery of a novel enzyme mediating glucocorticoid catabolism in fish: 20beta-hydroxysteroid dehydrogenase type 2. Mol. Cell. Endocrinol. 349, 202–213. doi: 10.1016/j.mce.2011.10.022
Tokarz, J., Moller, G., Hrabe, D. A. M., and Adamski, J. (2015). Steroids in teleost fishes: A functional point of view. Steroids 103, 123–144. doi: 10.1016/j.steroids.2015.06.011
Wei, L., Yang, C., Tao, W., and Wang, D. (2016). Genome-wide identification and transcriptome-based expression profiling of the Sox gene family in the Nile Tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 17:270. doi: 10.3390/ijms17030270
Xiao, L., Wang, D., Guo, Y., Tang, Z., Liu, Q., Li, S., et al. (2018). Comparative transcriptome analysis of diploid and triploid hybrid groupers (Epinephelus coioides♀ x E. lanceolatus♂ Genomics 111, 251–259. doi: 10.1016/j.ygeno.2018.11.010
Xu, P., Zhang, X., Wang, X., Li, J., Liu, G., Kuang, Y., et al. (2014). Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219.
Yang, S. Y., He, X. Y., Isaacs, C., Dobkin, C., Miller, D., and Philipp, M. (2014). Roles of 17beta-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J. Steroid. Biochem. Mol. Biol. 143, 460–472. doi: 10.1016/j.jsbmb.2014.07.001
Yang, S. Y., He, X. Y., and Schulz, H. (2005). 3-Hydroxyacyl-CoA dehydrogenase and short chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. FEBS J. 272, 4874–4883. doi: 10.1111/j.1742-4658.2005.04911.x
Keywords: β-HSD, orange-spotted grouper (Epinephelus coioides), genome-wide analysis, gene expression, sex reversal
Citation: Xiao L, Guo Y, Wang D, Zhao M, Hou X, Li S, Lin H and Zhang Y (2020) Beta-Hydroxysteroid Dehydrogenase Genes in Orange-Spotted Grouper (Epinephelus coioides): Genome-Wide Identification and Expression Analysis During Sex Reversal. Front. Genet. 11:161. doi: 10.3389/fgene.2020.00161
Received: 07 January 2019; Accepted: 11 February 2020;
Published: 04 March 2020.
Edited by:
Yi Zhao, Beijing University of Chinese Medicine, ChinaReviewed by:
Biju Issac, Leidos Biomedical Research, Inc., United StatesXiaopeng Zhu, Carnegie Mellon University, United States
Copyright © 2020 Xiao, Guo, Wang, Zhao, Hou, Li, Lin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuisheng Li, lshuish@mail.sysu.edu.cn; Yong Zhang, lsszy@mail.sysu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Ling Xiao1,2†
Ling Xiao1,2† Dengdong Wang
Dengdong Wang Shuisheng Li
Shuisheng Li Haoran Lin
Haoran Lin Yong Zhang
Yong Zhang