- 1Department of Health Sciences, Carleton University, Ottawa, ON, Canada
- 2Department of Complex Continuing Care, Saint Vincent Hospital, Ottawa, ON, Canada
- 3Centre for Infection, Immunity and Inflammation, University of Ottawa, Ottawa, ON, Canada
Delayed wound healing can cause significant issues for immobile and ageing individuals as well as those living with co-morbid conditions such as diabetes, cardiovascular disease, and cancer. These delays increase a patient’s risk for infection and, in severe cases, can result in the formation of chronic, non-healing ulcers (e.g., diabetic foot ulcers, surgical site infections, pressure ulcers and venous leg ulcers). Chronic wounds are very difficult and expensive to treat and there is an urgent need to develop more effective therapeutics that restore healing processes. Sustained innate immune activation and inflammation are common features observed across most chronic wound types. However, the factors driving this activation remain incompletely understood. Emerging evidence suggests that the composition and structure of the wound microbiome may play a central role in driving this dysregulated activation but the cellular and molecular mechanisms underlying these processes require further investigation. In this review, we will discuss the current literature on: 1) how bacterial populations and biofilms contribute to chronic wound formation, 2) the role of bacteria and biofilms in driving dysfunctional innate immune responses in chronic wounds, and 3) therapeutics currently available (or underdevelopment) that target bacteria-innate immune interactions to improve healing. We will also discuss potential issues in studying the complexity of immune-biofilm interactions in chronic wounds and explore future areas of investigation for the field.
Introduction
Wounds are a broad category of injuries that include everything from minor cuts and scrapes to surgical incisions and serious trauma. In healthy individuals, minor wounds heal quickly without complication. However, larger wounds take more time to heal, increasing risk of infection. Delayed wound healing is a significant issue among ageing individuals, those with immobility and those with chronic diseases such as diabetes, vascular disease, and cancer (1–3). In the most severe cases, these delays can result in the formation of non-healing or chronic ulcers, which can cause significant pain, prolonged hospitalization, loss of function, and may eventually lead to amputations and/or the development of sepsis (1, 4). Wound care (acute and chronic) is labor-intensive and represents a substantial economic burden on healthcare systems, costing billions of dollars annually in North America (5). As the global population continues to age and experience increasing rates of co-morbid chronic diseases such as diabetes, there is an urgent need to understand the pathophysiology of delayed healing or the formation of non-healing wounds to develop more effective therapies that can repair tissue damage and restore health (3).
There is an increasing interest to understand how dysregulated host-pathogen interactions affect healing processes. For instance, colonization of the wound bed with low levels of bacteria does not affect healing (6). However, local infection with high levels of replicating bacteria plays a significant role in delayed healing and in non-healing ulcer formation (6, 7). Chronic wounds show a considerable diversity in the bacterial species at the site of injury, but it is unclear how these differences contribute to chronicity. Further, these bacteria form robust biofilms, which embed the bacterial cells in a self-produced polymeric matrix, protecting them from host immune responses and antibiotics. This structure provides numerous advantages to the community such as metabolic cooperation, passive resistance, and horizontal gene transfer (8). It has also been shown to impair the tissue repair processes and promote a low-grade inflammatory response (7, 9). In this review we will discuss the current understanding of how interactions between bacterial biofilms and innate immune cells drive damaging inflammatory processes that contribute to delayed healing in chronic wounds. We will also discuss how we can target these interactions to develop novel therapeutics for individuals with difficult to treat chronic wounds.
Contribution of Bacterial Biofilms to Chronic Wounds
Overview of Bacterial Biofilms
Bacteria exist as single, planktonic cells or as multicellular communities and aggregates with or without surface attachment, called biofilms (10–12). Biofilms possess distinct characteristics compared to planktonic cells, including increased antibiotic tolerance, changes in gene expression, and altered host interactions (13). Bacteria within these structures are embedded in a self-produced extracellular polymeric substance (EPS) composed of extracellular DNA, proteins, exopolysaccharides and water. In addition to the microbial components, the EPS can also include host substances such as proteins, DNA, immunoglobulins, and blood components (14, 15).
Biofilms are complex and diverse structures that can be composed of single species or can be polymicrobial (15). They are up to 1,000 times more tolerant to antimicrobial agents and disinfectants than planktonic cells (16). Further, the immune system is often inefficient in combating biofilm-related infections (17, 18). Several factors contribute to the increased robustness of biofilms including low growth rates, high cell density, the presence of persister cells, nutrient and oxygen gradients, horizontal gene transfer, efflux pumps, and high rates of mutation (16, 19). The presence of the EPS matrix is also considered a physical barrier against antimicrobial agents and the host immune responses since it reduces the diffusion of drugs, antibodies and immune cells into the biofilm.
Biofilm-growing bacteria have been shown to colonize medical devices (e.g., contact lenses, cardiovascular valves, implants, ortho-dental prosthetics, urinary and central vascular catheters) and a variety of host tissues, causing many chronic infections, including osteomyelitis, vaginosis, lung infections in cystic fibrosis patients, ventilator-associated pneumonia, device-related infections, and chronic wound infections (15, 20). It is estimated that bacteria in biofilms cause up to 80% of all human infections (21) and are involved in more than 60% of all chronic wound infections (15). These biofilms are composed of bacterial species found in the normal flora of the skin, the gut and oral mucosa as well as in external environments (22).
Human Skin Microbiome
The human body is naturally colonized by thousands of different microbial species that collectively form a complex ecosystem called the human microbiome (23). More than 1,000 different bacterial species can be found on the human body, and it is estimated that there are up to 150 times more microbial genes than human genes within the human body (24, 25). These microorganisms selectively colonize different parts of the body, such as the skin, gastrointestinal tract, conjunctiva, oral cavity, vagina, uterus, and lungs (26). The human microbiome plays essential roles in health, including protection against invading pathogens, metabolism of molecules, nutrient acquisition, control of cellular proliferation and differentiation, and development of the immune system (25, 27, 28). However, the composition and diversity of the microbiome can be altered by several factors (e.g., diet, use of antibiotics, mode of birth, and age) and it can become associated with opportunistic pathogens and specific isolates that cause severe infections (24, 28). For example, certain isolates of Staphylococcus aureus can be either commensal, opportunistic or pathogenic.
The highest number of microbes are found in the colon (1014 bacteria), followed by the skin (1012 bacteria) (26). The skin microbiome is highly adapted to the skin’s physiological environment, such as the absence of several nutrients, an acidic pH and temperature. The bacterial microbiota found in each skin region depends on the microbe’s ability to thrive in these conditions. For example, bacteria from the Staphylococci genus use urea from the sweat as a nitrogen source whereas Cutibacterium acnes (formerly Propionibacterium acnes) produce fatty acids by metabolizing triglycerides present in sebum (27, 29). The skin is naturally considered a physical barrier against external stressors, and the natural skin microbiota protects the body by competing with pathogenic microbes and impairing their development, catabolizes natural skin products, such as lipids, and modulates the immune system (29). If the skin is disrupted, or there is an imbalance between pathogenic and the natural microbiota and infections can occur (27, 29).
Overall, most microbes from the skin microbiota belong to four different phyla: Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria (29). While certain species are found in multiple microenvironments (e.g., C. acnes, Staphylococcus epidermidis, and Staphylococcus capitis), the composition of the skin microbiome differs significantly according to the environmental conditions and location. Figure 1 highlights some of the most common bacterial species found in dry, sebaceous and moist environments. Dry areas represent the most diverse microbial environments. In these regions, streptococcus species are very common but actinobacteria, proteobacteria, firmicutes, and bacteroidetes are also detected including Corynebacterium tuberculostearicum, Staphylococcus sp., Veillonella parvula and Micrococcus luteus (27, 30). In moist and sebaceous sites corynebacterium are highly prevalent. However, these microenvironments are also populated with other common species including Staphylococcus hominis, Enhydrobacter aerosaccus, Streptococcus mitis and Micrococcus lutues (27, 29, 30).
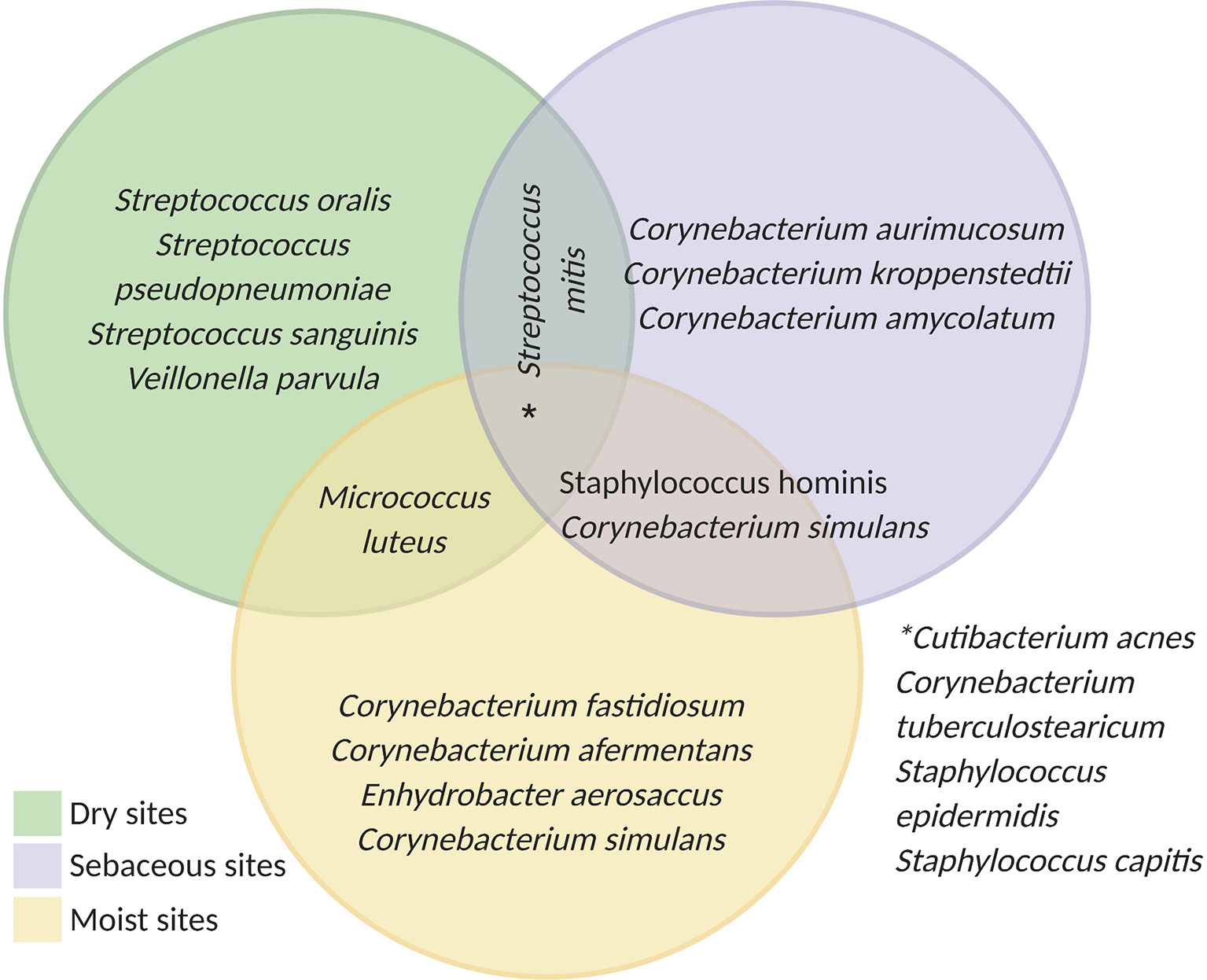
Figure 1 Schematic of skin microbiota according to the physiological sites: dry (green): buttock, volar forearm, hypothenar palm; moist (yellow): plantar heel, popliteal fossa, toe web space, axillary vault, and nare; sebaceous (purple): back, occiput, retroauricular crease, and glabella. Developed using data from (27).
It is important to acknowledge, that while bacteria are the most prevalent microorganisms found on the skin, fungal species also contribute to the microbiota (27). Malassezia is among the most common fungal genera accounting for 53-80% of the fungal population (29). Findley et al. found that Malassezia is found on several sites including retroauricular crease, nare, palm, back, and volar forearm. Other fungal species were also detected, with the plantar heel showing the greatest diversity with approximately 80 genera, including Malassezia, Aspergillus, Cryptococcus, Rhodoturulla, Epicoccum (31).
From Contamination to Infection
The human skin microbiota can influence the wound healing process and affect the severity of infections. In chronic wounds, microbial contamination and colonization can evolve to an infection, a process that involves different microorganisms (32). Contamination refers to the presence of non-proliferating bacteria originating from the natural microbiota or the environment. All wounds are contaminated by microorganisms; however, if they encounter favorable conditions that support survival, the wounds will become colonized. Colonization is characterized by the presence of multiplying microorganisms on the surface of the wound (33–35). In these initial processes, the host immune defenses are not triggered, and there are no overt clinical signs of infection. This is likely because the wound is colonized by Gram-positive bacteria, especially those belonging to the Staphylococci genus, which do not elicit strong immune responses (32, 36).
In the later stages, Gram-negative bacteria, mainly rod-shaped bacteria such as Pseudomonas sp., Escherichia coli, Klebsiella pneumoniae, and Enterobacter spp. contaminate the wound and become the predominant species in this microenvironment. These species typically originate from the urogenital tract of the patient or from the nearby environment. Colonization of may also be affected by any antibiotic use (local or systemic). This acute colonization is an intermediate stage before deeper tissue infections and only elicits a localized immune reaction (32, 37). In this stage, the bioburden increases, and the presence of these microbes can delay the healing process. Finally, the last step is infection, in which the microorganisms invade the deep tissues, the body cannot control the levels of microbes, and an intense host response is induced (32, 36). This stage is also characterized by the rapid consumption of oxygen by aerobic bacteria, which favors anaerobic microbes (32, 33).
Bioburden
Many aspects of wound bioburden correlate with healing outcomes. These include microbial load, microbial diversity, and presence of pathogenic organisms (38). Microbial load is commonly used to diagnosis a wound infection when clinical manifestations are absent (38). While the skin microbiota contains approximately 1 billion bacteria/cm2 of tissue (39, 40), a microbial load higher than 105 colony-forming units (CFU) per gram of tissue is generally considered the standard reference for diagnosing infection (41). However, this value can vary according to the type of infection and type of wound evaluated. In diabetic foot ulcer (DFU), the microbial load may be higher than in venous leg ulcer (VLU) patients (38, 41).
Microbial Diversity
Microbial composition and diversity are other factors associated with delayed healing. The most common bacteria found in the wound bed are aerobes such as Staphylococci ssp., Corynebacterium spp, Pseudomonas spp. as well as anaerobes such as Anaerococcus spp and Finegoldia spp (42–45). However, a higher diversity of microbial species in the wound bed can be associated with impaired healing outcomes (38). In addition to overall diversity, the presence of specific pathogenic organisms or even certain isolates can also contribute to chronic wound formation. Some strains of S. aureus and group A streptococci, have been shown to cause severe infections and trigger an intense inflammatory response, affecting the healing process (32). Moreover, high levels of S. aureus and P. aeruginosa can affect healing outcomes because these organisms are often resistant to different classes of antibiotics, they form biofilms with antiphagocytic activities and they produce several virulence factors, including secretion of toxins and enzymes, which drive further tissue damage (34, 46). Further studies are required to understand how single species vs. poly-microbial biofilms may contribute to these processes.
Methodologies for Characterization
Several factors must be taken into consideration when studying the skin and wound microbiota. Among the most important, are the sample collection technique and the method used to characterize the microbial population (i.e., culture-based or molecular methods) (47). At the level of sample collection, wound swabs and tissue biopsies (or debridement tissue) are commonly used for microbiota analysis. Swabbing the wound surface is often preferred because it is non-invasive, and it can be performed multiple times for longitudinal studies. However, dry swabs generally collect a low biomass and do not capture the diversity of species found in deeper tissues (48). Nakatsuji et al. found the bacterial diversity on the surface was not the same as that in the sub-epidermis, which included high levels of proteobacteria (49). Further, Travis et al. found there was minimal correlation between the number and types of bacteria found in swabs compared to tissue biopsies. They also found tissue biopsies contained an overall greater diversity of bacterial species but that the frequency of potential pathogens was higher in wound swabs (50). While biopsies represent the gold standard to capture the true diversity of bacteria found in skin and wound samples, it is invasive and requires a skilled clinician.
The two techniques most widely used to characterize bacteria in chronic wound infections are: A) culture-based methods, in which the microbial culture is collected from the affected tissue and bacteria are grown on selective or nonselective media; and B) molecular-based methods, in which the bacterial 16S rRNA is sequenced and microbes are identified based on databases (41, 51–53). While the culturing of bacteria is primarily used in the hospital setting, this method is usually limited to growing certain strains of bacteria and does not allow for the detection of fastidious and slow-growing bacteria, viable but nonculturable bacteria (VBNC) or dormant bacteria (12). The use of molecular methods, such as RNA sequencing, has increased in recent years but is still predominantly used for research. In comparison to culturing methods, these approaches allow for the identification of a larger diversity bacterial species in the wound. It can also be used to elucidate microbial activities, behaviors, strategies, and processes during infections (12). However, RNA sequencing is associated with a substantial demand for financial, time and bioinformatic support. Moreover, it cannot distinguish between living, dead or dormant bacteria and might overlook minority species (52). To complement these approaches, microscopy can be used to capture the complexity and the organization of bacterial populations in the wound environment. It can be used to visualize and characterize individual cells within a population/structure (e.g., biofilm) and provide important insights into their interactions and structural organization (48, 54). Regardless of the assay used, several other factors impact the diversity (or our ability of evaluate the diversity) of the wound microbiota including patient demographics, personal hygiene, grade of wound severity, patient’s immune status, and ongoing or previous use of antimicrobial therapies (12, 52).
Bacterial Diversity Across Chronic Wound Types
Chronic wounds or non-healing wounds are commonly defined as wounds that have failed to proceed through the normal phases of wound healing in an orderly and timely manner (55). The most common types of these wounds are diabetic foot ulcers, pressure ulcers, venous leg ulcers, abscesses and surgical site infections. While bacteria contribute to the pathophysiology of delayed healing and chronic wound formation, the molecular and cellular mechanisms underlying these processes remain incompletely understood. To date, most studies have been cross-sectional nature (Table 1). Few have used longitudinal approaches to evaluate how bacterial communities change in different chronic wounds over the course of infection. Moreover, fewer studies have investigated correlations between the microbiota and specific wound outcomes (53).
Diabetic Foot Ulcers
To date, most studies analyzing microbial communities in chronic wounds have focused on infections from patients with diabetic foot ulcers. These investigations have been recently reviewed in significant detail and are beyond the scope of the current review (52, 56–59). Briefly, the most frequently identified genera in DFU include Staphylococcus, Corynebacterium, Pseudomonas, Streptococcus, Stenotrophomonas, Enterobacter, Escherichia, Enterococcus, Serratia, Acinetobacter, Peptoniphilus, Anaerococcus and Finegoldia (42–45, 50, 60–67). The most common pathogens found were the Gram-positive bacteria S. aureus, S. epidermidis, Enterococcus faecalis and Streptococcus agalactiae and the Gram-negative bacteria P. aeruginosa, Stenotrophomonas maltophilia, E. coli and Acinetobacter baumannii (43–45, 62).
Other Types of Chronic Wounds
The bacterial communities of other types of wounds, including venous leg ulcers, pressure ulcers, surgical site infections, and abscesses, have not been fully evaluated. Previous studies have shown that like DFU, commonly identified genera include Staphylococcus, Corynebacterium, Streptococcus, Enterococcus, Pseudomonas, Stenotrophomonas, Enterobacteriaceae, Acinetobacter and Finegoldia (Table 2, and (43, 60, 78–82). To date, Wolcott et al. have performed one of the largest studies collecting samples from 916 venous leg ulcers, 767 decubitus ulcers, and 370 samples from nonhealing surgical wounds and evaluating bacterial diversity using 16s rDNA sequencing. They found that the most frequent bacterial species found across all wound types were S. aureus, S. epidermidis, E. faecalis, P. aeruginosa, S. maltophilia and Finegoldia magna. They also observed a high prevalence of anaerobic bacteria in wound samples, including Finegoldia spp. and Anaerococcus spp (present in 24% and 25% of wounds, respectively) and variety of commensal bacteria including Staphylococci and Corynebacteria across wound types (43). Interestingly, unlike DFU, these wounds contained Cutibacterium acnes and other Cutibacterium species suggesting an important role for the location of the wounds in spectrum of microbial colonization (43, 83, 84). Further, most of these infections were polymicrobial biofilms consisting of many different species. Only 7% of wound samples were found to be mono-species biofilms with P. aeruginosa and S. epidermidis being the most common bacteria growing in single-species biofilms in wounds (43). More in vivo studies with larger samples sizes are urgently needed to fully understand the importance of microbial diversity, biofilms and the wound microbiome in chronic wounds infections and to elucidate the impact of aerobic, anaerobic, pathogenic and commensal bacteria in driving chronic immune activation and inflammation in situations of delayed wound healing.
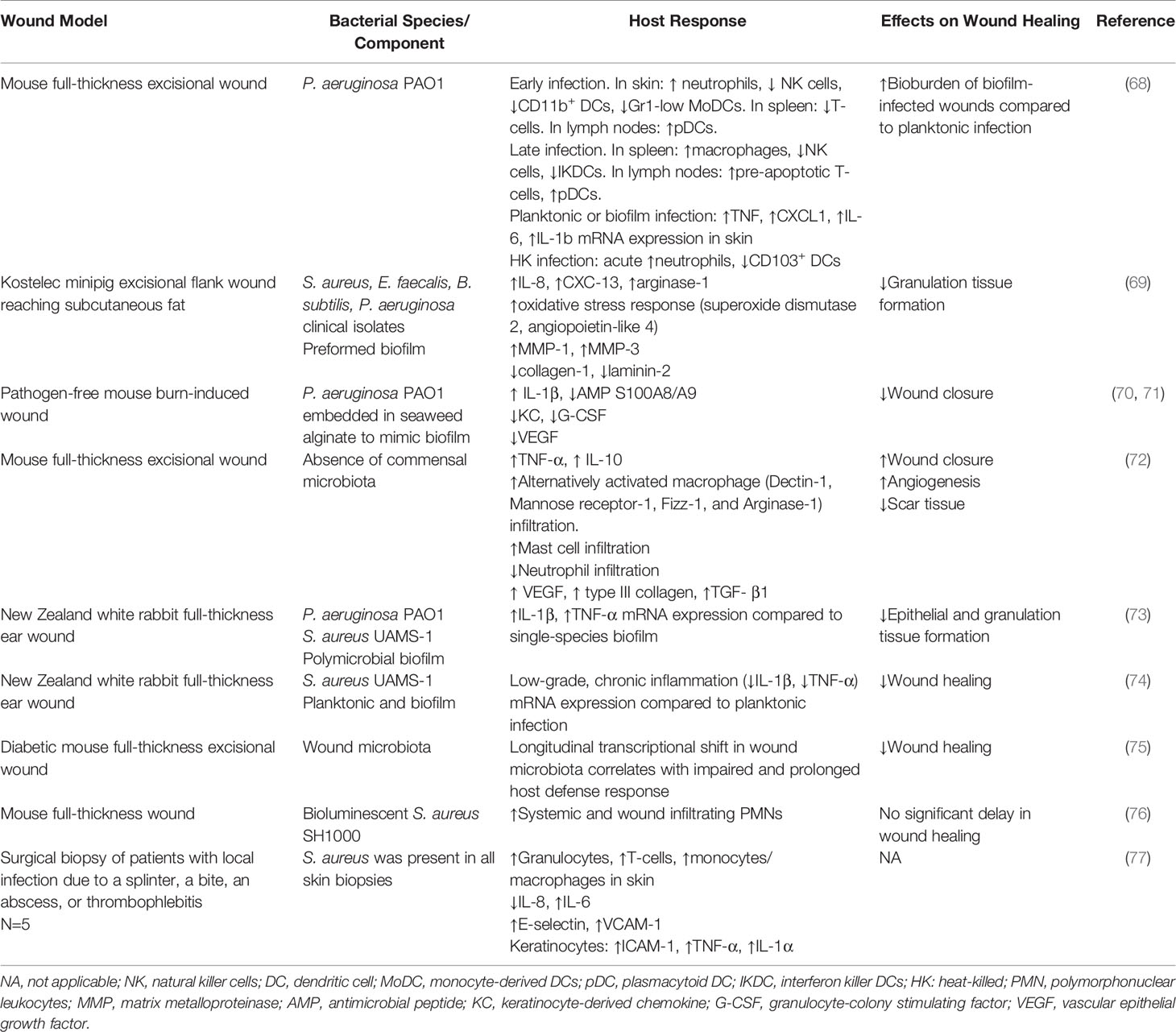
Table 2 Summary of studies characterizing host immune responses to bacteria and bacterial biofilms in wound models.
Contribution of Chronic Innate Immune Activation and Inflammation to Chronic Wounds
Overview of Wound Healing
Wound healing has been extensively described elsewhere, and for that reason, we will only briefly summarize the process here (Figure 2) (34, 85–92). Normal healing (Figure 2A, described as Steps 1-4) is comprised of four coordinated phases: hemostasis, inflammation, proliferation, and tissue remodeling (34). Hemostasis includes platelet aggregation and activation, which initiates the coagulation cascade and the formation of a transient fibrin scaffold (Step 1) (86, 91, 92). During this process, platelet degranulation releases damage-associated molecular patterns (DAMPs), cytokines, chemokines, and growth factors, which accumulate within the scaffold to generate a chemotactic gradient for immune cell infiltration (Step 2) (86, 88, 90). This infiltration is required to clear dead or damaged cells, cellular debris, and any pathogens that colonize the wound bed. It also prepares the wound for the healing phases. The innate immune system plays a critical role in modulating these processes as well as the transition from inflammation to proliferation phases, which includes a transition of inflammatory macrophages (M1) into anti-inflammatory wound healing cells (M2; Step 3) (86, 87). This anti-inflammatory transition activates keratinocytes and fibroblasts in the wound bed where they proliferate and contribute to healing processes (91). Keratinocytes are essential for wound re-epithelialization (86). Fibroblasts deposit collagen to form the extracellular matrix (ECM) or granulation tissue, which replaces the temporary fibrin scaffold (90, 92). During the proliferation stage, angiogenesis also restores tissue vascularity (86). Finally, in the remodeling phase, fibroblasts replace granulation tissue with scar tissue and contraction occurs, resulting in wound closure (Step 4) (34, 86, 88).
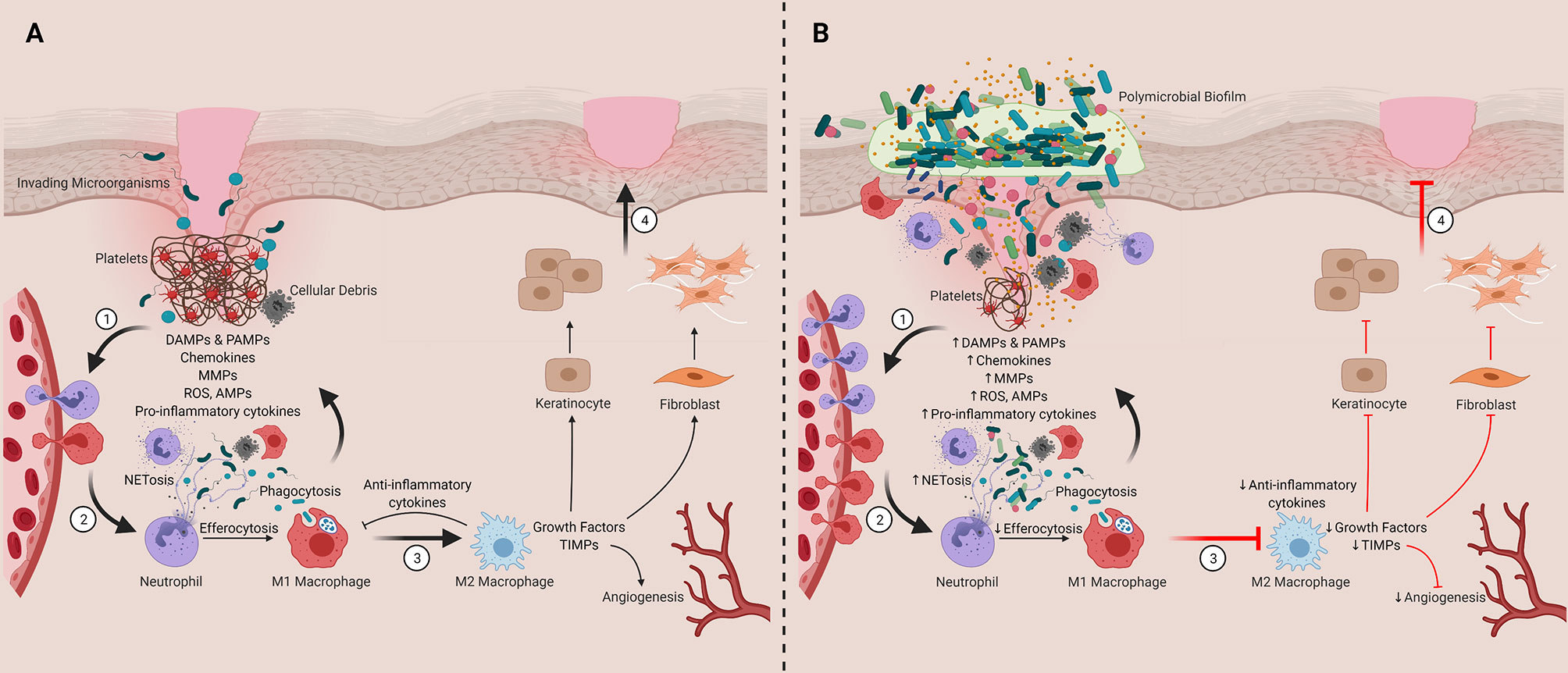
Figure 2 Contribution of innate immune cells and inflammation to timely and delayed wound healing. (A) Representation of the four phases of wound healing ([1] Hemostasis, [2] Inflammation, [3] Proliferation and [4] Tissue Remodeling). (B) Chronic wounds are stalled in the inflammatory stage. We hypothesize that this inflammation is sustained by chronic activation of the innate immune system, which is driven their interactions and responses to polymicrobial biofilms found in and on the wound bed. DAMPs, damage-associate molecular patterns; PAMPs, pathogen-associated molecular patterns; MMPs, matrix metalloproteinases; ROS, reactive oxygen species; AMPs, antimicrobial peptides; TIMPs, tissue inhibitor of metalloproteinases. Created with BioRender.com.
Role of the Innate Immune System in the Inflammatory Phase of Healing
Although the inflammatory phase involves a wide range of immune cells, such as mast cells, dermal dendritic cells (Langerhans cells), and T lymphocytes, innate immune cells such as neutrophils and macrophages play a central role in regulating healing process (85, 91–93). For the purposes of this review, we will focus on these two cell types.
Neutrophils
Neutrophils play a central role in healing damaged tissues and resolving infections. Early after the initial tissue damage, DAMPs, pathogen-associated molecular patterns (PAMPs), leukotriene B4 (LTB4), and CXCL8 family chemokines induce and augment neutrophil chemotaxis via CXCR2 into the wound bed (94–97). As the first cells recruited to the site of injury, neutrophils perform several diverse functions. First, they amplify inflammatory processes by releasing cytokines such as IL-1β, TNF-α, and IL-6 (85, 98) and function to prevent microbial invasion by phagocytosing microorganisms and releasing reactive oxygen species (ROS), proteases, and antimicrobial peptides (99, 100). Further, they undergo NETosis, where they form neutrophil extracellular traps (NETs) made of decondensed chromatin, histones, granular enzymes, antimicrobial peptides and proteases to immobilize and destroy exogenous pathogens (91, 100). They also release serine proteases, such as cathepsin G and elastase, and matrix metalloproteases (MMP), such as MMP2, 8, and 9 that degrade the ECM, facilitating tissue debridement that makes room for additional immune cell influx (92, 99). Neutrophils also contribute to tissue remodeling and repair. Theilgaard-Mönch et al. demonstrated that infiltrating neutrophils activate and induce the transcritption of genes involved in angiogenesis, keratinocyte adhesion, and migration and proliferation of keratinocytes and fibroblasts at the site of injury (98).
Macrophages
After damage has occurred, resident macrophages become activated by DAMPs and PAMPs in the local microenvironment, initiating the inflammatory response required to initiate the healing process (101, 102). Pro-inflammatory chemokines and cytokines such as CCL2, IFN-γ, and TNF-α recruit neutrophils and then circulating monocytes into the wound (102, 103). In mouse models, the expression of chemokine receptors CCR2 and CX3R1 has been shown to be critical for mediating monocyte recruitment to damaged tissues (104, 105). These infiltrating monocytes differentiate into macrophages, and along with resident macrophages, potentiate the inflammatory and healing processes.
Macrophages display incredible phenotypic plasticity, existing on a spectrum of inflammatory and anti-inflammatory activation states (106, 107). During the inflammatory phase of wound healing, macrophages display a proinflammatory or a classically activated M1-like phenotype, producing inflammatory mediators such as TNF-α, IL-6, IL-1β, IL-12, IL-23 to facilitate additional leukocyte recruitment and activation (34, 88, 89, 92). M1 cells also work cooperatively with neutrophils to phagocytose damaged cells and clear the wound of bacteria and tissue debris by producing ROS-containing phagosomes (92). Like neutrophils, they also produce MMPs to degrade the wound extracellular matrix to allow for further immune cell infiltration (92, 102). This digested matrix feeds back as DAMPs to amplify inflammatory signaling (108).
Following the clearance of debris and invading microorganisms, macrophages adopt an anti-inflammatory or an alternatively activated M2-like phenotype to support tissue remodeling and repair (106, 107). In vitro studies have shown that IL-4 and IL-13 drives M2 activation and wound healing functions (107). However, in vivo, the mechanisms driving this M1 to M2 phenotypic switch may be more complicated and may be regulated by IL-10, glucocorticoids, prostaglandins, miRNAs, and adenosine and peroxisome proliferator-activated receptor (PPAR-γ) signaling in the wound microenvironment (86–88, 109). At the end of the inflammatory phase, macrophage phagocytosis of apoptotic neutrophils (i.e. efferocytosis) may also aid in the transition to the repair phases of wound healing (110).
To initiate tissue remodeling and repair, M2 cells produce IL-10, IL-12, and TGF-β to suppress inflammatory effects of TNF-α and IL-1β (90). During the proliferation phase they also release growth factors, such as vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and fibroblast growth factor-2 (FGF-2), which promote angiogenesis and keratinocyte and fibroblast proliferation (86, 88, 89, 91, 111). Finally, during the repair and remodeling phase, macrophages stimulate fibroblasts to mature into myofibroblasts that deposit collagen into the wound bed to replace the temporary fibrin scaffold with granulation tissue (86, 92, 112). They also regulate MMPs and tissue inhibitors of metalloproteinases (TIMPs) production to allow for both ECM formation and degradation of excess cellular components to restore homeostasis (92, 113). This M1 to M2 phenotypic switch makes macrophages a central cellular player in regulating the transition from inflammation to proliferation during wound healing (86).
Dysregulated Inflammatory Processes in Chronic Wounds
In chronic wounds, the orderly process of healing is thought to be stalled in the inflammatory phase and is characterized by persistent activation of the innate immune response (Figure 2B, described as Steps 1-4) (55, 87). This sustained inflammation drives additional immune cell infiltration and activation (Step 1), which amplifies MMP, collagenase, and elastase activity and suppresses TIMPs (114–119). This imbalance in proteolytic activity leads to excessive degradation of growth factors, anti-inflammatory cytokines, and ECM components, hindering progression through the phases of healing (120–122). This inflammation also drives excessive NETosis and reactive oxygen production (ROS), which contributes to further tissue damage and impaired healing (Step 2) (123–127). Impaired M1-M2 phenotypic switching has also been implicated in chronic wound formation (Step 3). In venous ulcers, iron overload has been linked to sustained proinflammatory M1 macrophage activation (128, 129). Further, studies in diabetic mice have demonstrated that dysfunctional efferocytosis of apoptotic neutrophils by pro-inflammatory macrophages results in apoptotic cell burden that causes sustained inflammation, preventing macrophages from transition into their anti-inflammatory state (130). In further support of these observations, depletion of M2 macrophages in mice with surgical wounds has been shown to increase neutrophil and M1 macrophage infiltration, which prolonged the inflammatory phase and decreased collagen deposition in wounds (131). Collectively these findings suggest sustained innate immune activation plays a central role driving chronic wound formation. Critically, it is unclear what exactly is driving this chronic innate immune activation. Further, it is unclear if the mechanisms driving this innate immune dysfunction including dysregulated M1-M2 switching differs across tissue microenvironments (e.g., wound types), particularly as emerging data suggests differences between antimicrobial and inflammatory responses across wound types (132)
Role of Bacteria in Driving Innate Immune Activation and Inflammation in Chronic Wounds
Emerging evidence suggests the wound microbiome and the formation of bacterial biofilms may contribute to delayed wound healing (45, 133–138). However, the causal relationship between microbiome composition, biofilm formation, dysregulated innate immune activation and persistent inflammation in chronic wounds remains poorly understood (12, 139). It is unclear what comes first – if bacteria and biofilm formation drive innate immune dysfunction, or if innate immune dysfunction makes the wound microenvironment more susceptible to biofilm formation. Understanding the role of bacteria-innate immune interactions in driving persistent inflammation and impaired healing in chronic wounds may offer new opportunities to restore healing processes (140). Previous reviews have explored key findings from in vitro studies (141–143). Here, we will focus on in vivo evidence from animal models and patient samples. We will summarize the existing knowledge on the role of bacteria-innate immune interactions in driving persistent inflammation in chronic wounds and explore novel treatments currently under development to target these interactions.
General Host Response to Bacteria or Bacterial Components in Wounds
While in vitro studies are important for identifying potential cellular and molecular mechanisms underlying innate dysfunction in chronic wounds, these models cannot capture the dynamic and complex nature of the immune response in vivo. Animal models and patient samples are better positioned to capture this complexity and can be used to evaluate localized as well as systemic immune responses.
Several studies have shown that wound bacterial infections can alter local and systemic immune responses (Table 2). Sweere et al. found that mice with chronic P. aeruginosa (PAO1) wound infections showed time-dependent changes in wound, lymph node, and spleen immune cell populations. In this model, early stages were associated with increased skin neutrophil infiltration and a reduction in the number of skin natural killer (NK) cells, CD11b+ dendritic cells (DCs), and Gr1-low monocyte-derived DCs (MoDCs) (68). Similarly, Kim et al. found wound colonization by S. aureus increased systemic polymorphonuclear leukocytes (PMN) by twofold and dramatically increased PMN recruitment into the wound bed (76). Systemic immune responses were more pronounced in later stages of wound infection and included increased lymph node plasmacytoid DCs (pDCs), increased splenic macrophages and lymph node pre-apoptotic T-cells and a reduction in NK cells and interferon killer DCs (IKDCs). Based on these findings, authors concluded that adaptive immune responses might not play a significant role in driving the inflammatory response against P. aeruginosa (68). In human skin biopsies, Van der Laan et al. found diverse injuries result in different local inflammatory responses (77). Compared to aseptic conditions, wounds infected with S. aureus contained increased granulocytes, T-cells, and monocytes/macrophages in the dermal layer and dermis (77). However, further studies are required to understand changes to systemic responses in humans with chronic wounds.
Animals models have also been used to evaluate differences in immune responses to planktonic bacteria vs. biofilm infections in wounds. Sweere et al. found that P. aeruginosa infection was associated with increased TNF, CXCL1, IL6, IL1B mRNA expression in mouse skin but that these responses did not differ between planktonic bacteria vs. biofilms, despite the higher bioburden associated with the biofilm structure (68). However, Trostrup et al. found that P. aeruginosa biofilms inhibit wound healing by suppressing VEGF, antimicrobial peptide production (S100A8/A9) and neutrophil effector cytokine production (70, 71). In dermal punch wounds in rabbit ears, Gurjala et al. found that S. aureus biofilms trigger lower levels of inflammation in the wound bed compared to planktonic cells. However, they found that the persistent nature of the immune response to biofilms was shown to impair epithelial migration and granulation over time (74). Interestingly, using a similar rabbit model, Seth et al. found that polymicrobial biofilms containing P. aeruginosa (PAO1) and S. aureus (UAMS-1) triggered substantially higher inflammatory responses compared to single species structures (73). This elevated inflammatory response impaired wound epithelialization and granulation tissue formation. These authors also found that biofilm-deficient mutant strains of S. aureus were associated with reduced cytokine mRNA expression suggesting that the biofilm structure may, at least in part, contribute to the inflammatory response (73). In a porcine model, polymicrobial biofilms containing S. aureus, E. faecalis, Bacillus subtilis, P. aeruginosa were shown to prolong inflammation, increase necrosis, delay granulation, and impair the development of the extracellular matrix. Gene expression analysis revealed an upregulation of inflammatory mediators such as IL8, CXCL13, and arginase-1 (ARG1), as well as genes associated with oxidative stress response including superoxide dismutase 2 (SOD2) and angiopoietin-like 4 (ANGPTL4) (69).
Several studies have highlighted that the skin commensal microbiota play important functions in the regulation of wound healing and in the innate immune defense against infection (144–146). In fact, using longitudinal transcriptional profiling, Grice et al. found a shift in the wound microbiota of diabetic mice, and found this shift correlated with impaired healing and a prolonged inflammatory response (75). Alternatively, Canesso et al. showed that in germ-free Swiss mice, the absence of commensal microbiota enhanced TNF-α and IL-10 production, infiltration of alternatively activated macrophages and mast cells, and impeded neutrophil infiltration (72). These effects likely contributed to high levels of VEGF, type III collagen, and TGF-β, thereby accelerating wound closure and angiogenesis, and reducing in scar tissue formation (72). Further investigations are required to understand the intricate balance between the pro-wound healing commensal microbiota and pro-wound impairing pathogenic polymicrobial biofilms, as well as their role in driving a dysregulated inflammatory response in chronic wounds in humans.
Regulators of Immune Responses in Wounds With Bacterial Infections
Several molecules and pathways have been implicated in driving chronic inflammatory responses in non-healing wounds. Most of these have been identified in the context of overt bacterial infections (e.g., abscess) and do not address issues associated with wound chronicity. It is unclear if similar molecules/pathways contribute to delayed healing in wounds with varying levels of bacterial burden (e.g., contamination, colonization, local infection etc.). Despite these limitations, we have summarized these studies below (Table 3).
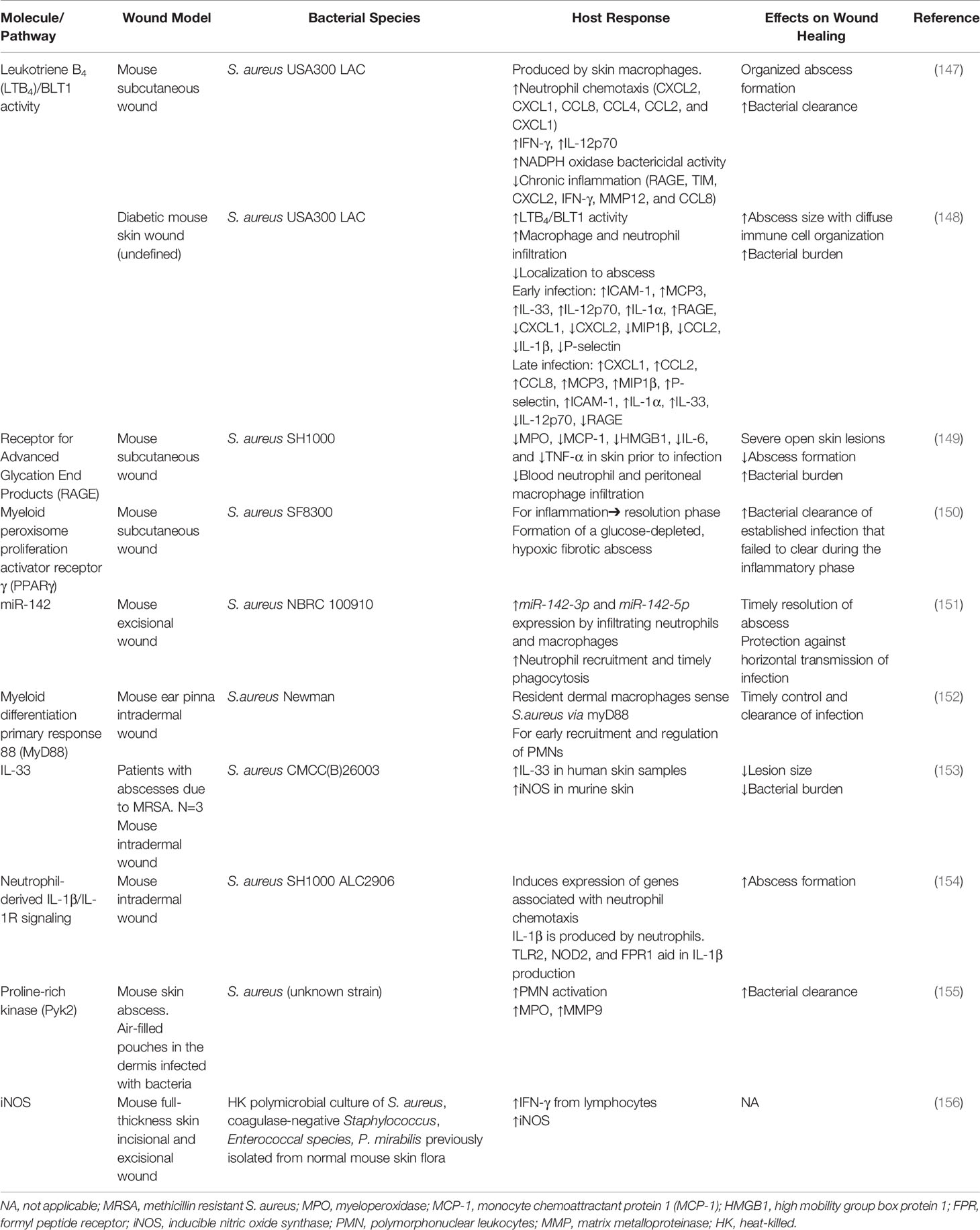
Table 3 Summary of molecules and pathways that regulate the inflammatory responses to bacteria in wounds.
S. aureus is the leading cause of skin and soft tissue infections and is often used in abscess infection models (164, 165). Brandt et al. infected mouse skin with methicillin-resistant S. aureus (MRSA; USA300 LAC) and found high levels of LTB4 was produced by macrophages surrounding the abscess (147). This inflammatory lipid mediator is required to create a chemotactic gradient that directs neutrophil infiltration and helps to form an organized abscess architecture. It also aids in bacterial clearance by upregulation of pro-inflammatory cytokines (IFN-γ and IL-12p70) and increased NADPH oxidase activity (147) and has been shown to modulate keratinocyte activity (166). However, in diabetic mice, very high levels of LTB4 were found to be associated with dysregulated cytokine production and excessive neutrophil recruitment (148). This dysregulation was found to correlate with large nonhealing lesion areas and increased bacterial loads (148). Further, inhibition of LTB4 signaling through its receptor (BLT1) was shown to restore a functional inflammatory response, suggesting this molecule may play an important role in derailing the inflammatory milieu (148).
Advanced glycation end products (AGEs) and its receptor (RAGE) have also been implicated in the regulation of skin inflammation and diabetic pathologies (167, 168). Interestingly, Na et al. found that RAGE knockout mice infected with S. aureus (SH100) experienced less severe skin lesions and increased abscess formation (149). This milder skin damage was associated with increased neutrophil migration and increased bacterial clearance with reduced inflammation (e.g., monocyte chemoattractant protein-1 (MCP-1), high mobility group box protein 1 (HMGB1), IL-6, and TNF-α) (149). Paradoxically, RAGE-/- were also found to have high baseline levels of inflammation prior to infection (149). However, it was speculated that this priming may be protective and help establish rapid innate immune responses in early infection. Collectively, this data suggests that RAGE may be pathogenic in staphylococcal skin infection, particularly in supporting chronic inflammation.
Accumulating evidence suggests appropriate regulation of neutrophil activation is also critical for effective bacterial killing while limiting inflammation. This process is complex and multifactorial. For example, Cho et al. found that neutrophil recruitment and abscess formation is temporally linked to IL-1β/IL-1R activation, which neutrophils produce as part of an autocrine loop. This loop is driven by α-toxin mediated activation of TLR2, NOD2, FPR1 and the ASC/NLRP3 inflammasome (154). Alternatively, Kamen et al. found proline rich kinase 2 (Pyk2) plays an important role in regulating integrin-mediated degranulation responses (155). Further, a number wound healing-related miRNAs have been identified and recent studies suggest that their dysregulation may contribute to wound pathologies (169). Among these, miRNA-142 has been shown to be an inflammation related miRNA that regulates neutrophil recruitment and S. aureus clearance through the inhibition of small GTPase translation (151). Additional studies are required to better elucidate its role in chronic wounds. Interestingly, resident dermal macrophages have been shown to play a central role in regulating both the timely escalation and eventual termination of neutrophil recruitment. Feuerstein et al. showed that this regulation is dependent on MyD88-dependent sensing of staphylococci and the recruitment of Ly6Chigh inflammatory monocytes into the skin (152).
In addition to neutrophils, macrophage responses must be tightly regulated during healing processes. In the inflammatory phase, these cells are proinflammatory (M1) and produce high levels of nitric oxide, reactive oxygen species and other antimicrobial peptides, which can be damaging to the local microenvironment. IL-33 represents a potential target as it plays a central role in activating antibacterial responses by activating the AKT-β-catenin pathway, which induces inducible nitric oxygen synthase (iNOS) and increases NO production (153). Alternatively, Mahoney et al. found lymphocyte derived IFN-γ drives the induction of iNOS in mouse wounds infected with heat-killed polymicrobial culture of S. aureus, coagulase-negative Staphylococcus and Enterococcal species as well as Proteus mirabilis (156). Alternatively, Xu et al. have shown that decreased NADPH oxidase activity and ROS production is associated with decreased infiltration of M2 macrophages and delayed wound healing suggesting a dichotomous role for these bioactive molecules (170). PPAR-γ has been shown to facilitate the M1-M2 transition (150). PPAR-γ has also been shown to play a role in MRSA clearance in chronic wounds by forming a glucose-depleted, hypoxic, fibrotic abscess that hinders bacterial growth (150). Finally, Guo et al. have demonstrated that AGEs contribute to excessive macrophage autophagy, which polarizes macrophages towards an M1 phenotype and supports sustained inflammatory processes (171). Moving forward it will be critical to understand how bacterial bioburden and composition contributes to the activation/inactivation of these pathways and to better understand the downstream consequences of dysregulated M1-M2 phenotypic switching in humans.
Other Modifiers of Host Antibacterial Immune Responses in the Wound Microenvironment
Aging is among one of the most significant predisposing factors to delayed healing and chronic wound formation. Older individuals are more commonly affected by vascular disease, venous insufficiency, unrelieved pressure, and post-surgery wound complications (172). Further, various studies have shown that aging affects all stages of the healing process including delayed re-epithelialization, angiogenesis, and collagen deposition (173–175). Changes skin strength may also result in a more pronounced breakdown of skin epithelial barriers, which may increase the bacterial bioburden in the wound microenvironment (175, 176). At the level of the immune system, advanced age is associated with a hyperinflammatory state (177). Innate cells have delayed infiltration, reduced phagocytic capacity, decreased reactive oxygen and nitrogen species production and impaired intracellular killing (178, 179). Interestingly, a recent study observed no age-related changes in TLR2 and FcγRIII expression, phagocytosis, and bactericidal activity in aged mice with cutaneous S. aureus infection (157). However, they did find that neutrophils had diminished sensitivity to chemokines (e.g., KC, MIP-2, and MCP-1), which reduced their chemotaxis into the wound bed and delayed healing (157). Additional studies are required to evaluate if similar dysfunction is observed in humans.
Diabetes is also associated with delayed healing and chronic wound formation (Table 4). Among the most common manifestations are non-healing foot ulcers (180). In these individuals wound healing is influenced by a predisposition to vascular disease and neuropathy, hypoxia, and hyperglycemia (181). Of particular importance, impaired vascular flow creates a prolonged hypoxic wound microenvironment, which along with hyperglycemia, contributes to oxidative stress (181). Hyperglycemia is also associated with AGE formation, which further delays healing (182). Chronic low-grade inflammatory also defines diabetes pathology and dysregulated healing response in these individuals (183). Emerging evidence suggests altered inflammatory responses to bacteria in the wound microenvironment may contribute to the development of these chronic wounds. In diabetic mice with cutaneous S. aureus infection, there is excessive macrophage and neutrophil infiltration into the wound but poor localization to abscess (148). These mice also have altered inflammatory cytokine and chemokine profiles during early and late stages of infection. Despite forming large abscesses, their structure had diffused immune cell organization and higher bacterial burdens (148). In mice infected with P. aeruginosa, diabetes was associated with prolonged M1 activation, which impaired healing processes by diminishing re-epithelialization, granulation tissue formation and angiogenesis (158). Alternatively, Nguyen et al. found diabetic mice inoculated with S. aureus biofilms had reduced TLR2 and TLR4 mRNA expression and high levels of inflammatory cytokines (IL-1β and TNF-α) (159). They also showed diabetic mice experience poor neutrophil penetration into regions with bacterial aggregates and downregulation of myeloperoxidase activity, a marker of neutrophil oxidative burst (159).
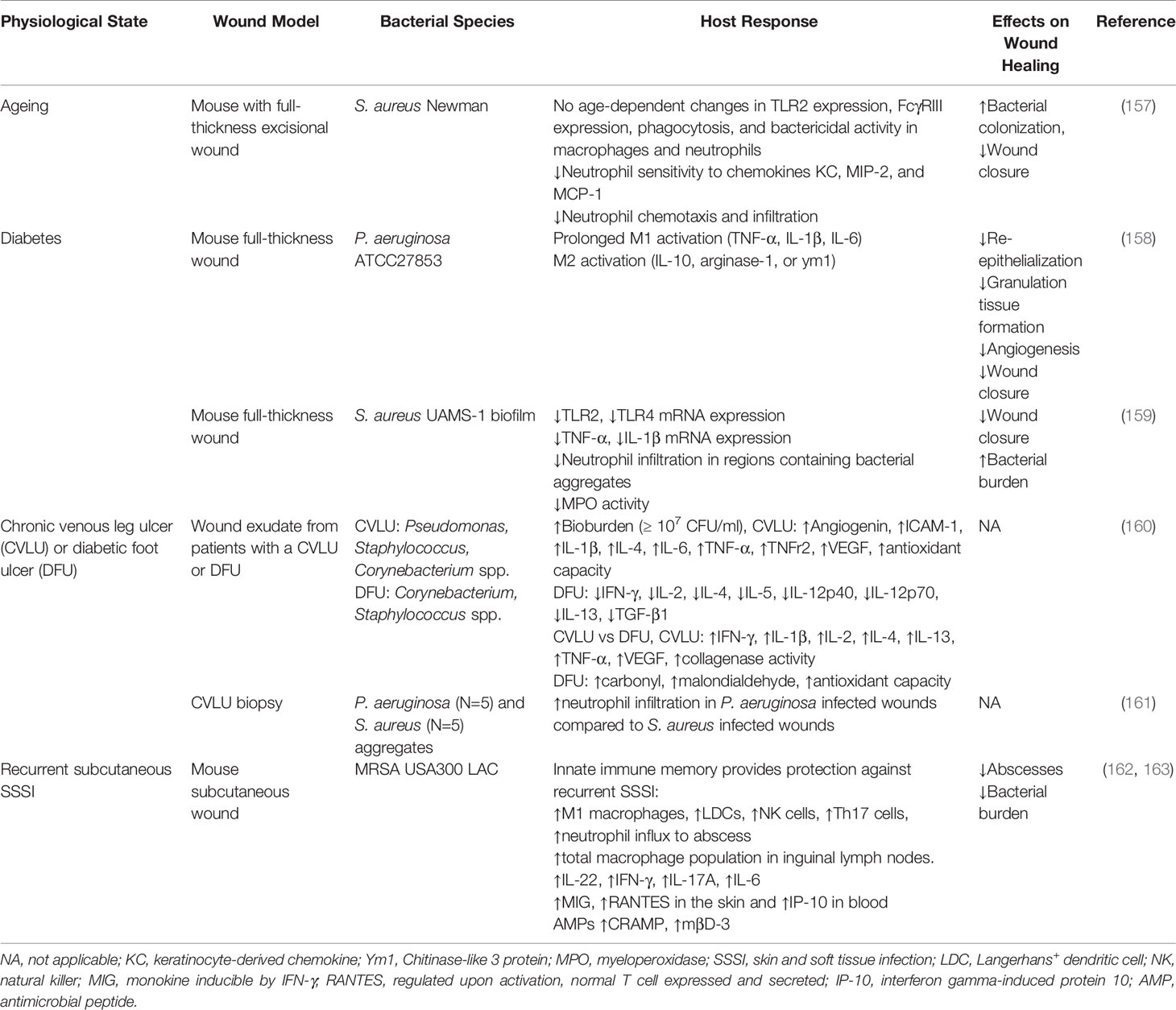
Table 4 Summary of other physiological factors that modify the inflammatory responses to bacteria in wound models.
The type of chronic wound and the diversity/number of bacteria found in the wound may also play an important role in determining the magnitude of the inflammatory responses (Table 4). For example, chronic venous leg ulcer (CVLU) exudate commonly contains Pseudomonas, Staphylococcus, Corynebacterium spp (160). With increasing levels of bacteria (≥107 CFU/ml), angiogenin, ICAM-1, IL-1β, IL-4, IL-6, TNF-α, TNFr2, VEGF, and antioxidant capacity are shown to be elevated (McInnes et al., 2014). Alternatively, diabetic foot ulcers, which lacked P. aeruginosa, have diminished IFN-γ, IL-2, IL-4, IL-5, IL-12p40, IL-12p70, IL-13, and TGF-β1 production with increasing bacterial bioburden (160). In comparison of the inflammatory response between CVLUs and DFUs, McInnes et al. determined that CVLUs have higher levels of IFN-γ, IL-1β, IL-2, IL-4, IL-13, TNF-α, VEGF, and increased collagenase activity compared to DFUs. On the other hand, DFUs showed higher levels of carbonyl, malondialdehyde and antioxidant capacity compared to CVLUs (160). Moreover, Fazli et al. found in chronic venous leg ulcer biopsy samples, P. aeruginosa aggregates displayed amplified neutrophil infiltration compared to S. aureus aggregates in wounds, suggesting that these differences may be due to the intrinsic properties of P. aeruginosa to mount a higher inflammatory response (161). Additional studies are required to further explore these associations and differences.
Past exposures to bacterial infections may also alter local immune responses to wound infections (Table 4). Upon first exposure to a pathogen, innate immune cells can adapt, such that upon re-exposure, they mount a heightened pathogen-specific inflammatory response to boost host defense and provide long-term protection (184). This phenomenon is known as innate immune memory (184). Emerging evidence suggests innate immune memory may provide protection against recurrent staphylococcal skin infection. In mice primed by prior S. aureus infection, lesion severity was reduced by increased M1 macrophage, Langerhans+ DCs (LDC), NK cells, Th17 cells, and neutrophil influx to the abscess (162, 163). Interestingly, cytokines IL-6, IL-17, IL-22, chemokines MIG and RANTES, and antimicrobial peptides CRAMP and mβD-3 contributed to the development of innate immune memory in these mice (162, 163). In the context of chronic wounds, innate immune cells are chronically exposed to bacterial biofilms rather than a first exposure followed by recurrent exposure paradigm. Whether persistent activation of innate immune cells constrains development of this protective innate immune memory or not remains to be elucidated.
Targeting Host-Pathogen Interactions to Restore Healing Processes
Managing and treating chronic wounds can be very challenging. It requires a comprehensive wound assessment and the establishment and implementation of a plan of care. These individualized plans aim to optimize the local wound environment and drive healing using four basic strategies: wound cleansing, debridement, moisture control, and bacterial balance (35). Among these, controlling bacterial bioburden is essential for wound healing and can be done by 1) reducing the levels of bacteria found in the wound and/or by 2) optimizing host immune responses to the infection. For the purposes of this review, we will briefly discuss how standard treatments reduce bacterial bioburden and/or restore immune function. Then, we will discuss emerging therapeutics designed to target interactions between bacteria/biofilms and the host immune response to restore healing processes.
Debridement and Negative Pressure Wound Therapy (NPWT)
Both debridement and NPWT have been shown to affect bacterial bioburden and/or inflammation in the wound bed. Many types of debridement technologies exist including biological (maggot/larval therapy), mechanical, hydrosurgical, chemical, autolytic, enzymatic, surgical, and conservative sharp debridement (35). The purpose of these procedures is to remove necrotic or infected tissue to facilitate healing. In addition to removing infected tissue, debridement has been shown to remove and disrupt mature biofilms. Wolcott et al. found serial debridement to continually remove mature biofilms can be used to increase the efficacy of topical antimicrobials on newly forming/immature biofilms, which are more susceptible to treatment (185). NPWT, also known as vacuum assisted closure (VAC) therapy, improves healing by removing excess exudate, maintaining moisture balance, and increasing blood flow into the wound. It has also been shown to control infection and modulate immune responses. In animal models, NPWT has been shown to have anti-biofilm effects (186–188). It has also been shown to modulate growth factor, cytokine expression, and matrix metalloproteinases to support healing (189, 190). This includes decreasing IL-6, iNOS, TNF-α, IL-1β, MMP-1, and MMP-9 and upregulating VEGF, TGF-β1 and TIMP-1 in patients with diabetic foot ulcers (191, 192).
Antiseptics
Irrigation solutions such as sterile normal saline or sterile water are the simplest wound cleansing methods. Antiseptic agents, such as octenidine dihydrochloride (OCT), polyhexamethylene biguanide (PHMB), povidone-iodine, and super oxidized hypochlorous acid (HOCl) and sodium hypochlorite (NaOCl) are widely used in topical wound therapy in solution form or as functionalized dressings due to their high microbicidal and anti-biofilm properties (193–197). OCT and PHMB have surfactant properties to help break apart biofilms and PHMB is particularly useful due to its low toxicity (198). Silver- and copper- impregnated dressing are also widely used in chronic wound management (199, 200). However, a recent scoping review by Rodriguez-Arguello et al. established mixed results in terms of antimicrobial activity and clinical effectiveness of silver agents (201). Moreover, a recent systematic review that evaluated the efficacy of commercially available topical agents containing silver, iodine, PHMB, or hypochlorous acid concluded that a lack of in vivo evidence makes it difficult to make recommendations for biofilm-infected wounds (202). Little research is available on the effects of these agents on modulation of the immune/host response. In human ex vivo full-thickness skin injury, OCT has been shown to dampen pro-inflammatory and anti-inflammatory cytokines IL-8, IL-33, and IL-10, but not growth factors VEGF and TGF-β1 (203). In an ex vivo porcine skin model, povidone-iodine, silver lactate, and OCT showed antiprotease activity that was dependent on their wound penetration ability (204). These anti-inflammatory properties need to be further investigated in vivo.
Antibiotics
Antibiotics are often also used in the management of chronic wound infections. However, the type of antibiotic prescribed, and the administration route depend on the clinician evaluation and must take into consideration the microbial bioburden, patient clinical condition (e.g., allergies, immunocompetence, comorbidities, and pregnancy), the severity of the infection, and drug toxicity and dosage (35, 205). For instance, contaminated and colonized wounds do not require the use of antibiotics to improve wound outcomes. Alternatively, local infection often involves the use of topical antimicrobials including antibiotics compared to systemic infections that use systemic antibiotics (35, 206).
Topical antibiotics provide a high drug concentration at the infection site and possess low toxicity since the body systemically absorbs a low amount of drug. Moreover, they are easy to apply, and their use can avoid the use of systemic antibiotics. However, topical antibiotics cannot be prescribed to treat deep tissue infections, can affect healing, can cause hypersensitivity, and can select for resistant microorganisms (207, 208). They are often formulated as ointments, gels, creams, and powders, and only a few are available for use (e.g., bacitracin, fusidic acid, gentamicin, mafenide acetate, metronidazole, mupirocin, neomycin, nitrofurazone, polymyxin B, retapamulin, silver sulfadiazine, sulfacetamide Na+). The antibiotic used also depends on the type of wound. Bacitracin, neomycin sulfate, and polymyxin B are frequently used in combination to treat minor skin injuries. Silver sulfadiazine cream is commonly used as a topical antibiotic to treat DFUs and pressure ulcers (209, 210). Gentamicin and sulfacetamide are used to treat secondary infections, colistin (polymyxin E) is used for MDR gram-negative infections and metronidazole is commonly used to treat infections caused by anaerobic microbes and to reduce the odor of wounds (207).
Systemic antibiotics are used in patients with more severe infections. However, in these cases, the resistance profile of the pathogen is closely related to the success of the treatment (206). For instance, vancomycin is the first-line treatment to fight MRSA infections, followed by second-line agents, including linezolid, daptomycin, and quinupristin-dalfopristin. Other examples of systemic antibiotics used to treat chronic wounds include macrolides (e.g., erythromycin, azithromycin, and dirithromycin), β-lactams (e.g., cephalosporin, amoxicillin), penicillinase-resistant penicillins (e.g., cloxacillin, oxacillin), trimethoprim-sulfamethoxazole, fluoroquinolone, tigecycline, and clindamycin (35, 206, 211). The type of antibiotic prescribed is also dependent on the type, location and severity of the wound. For mild and moderate DFUs, narrow-spectrum antibiotics are recommended, especially those active against Gram-positive cocci (211). Alternatively, severe DFUs should be initially treated with broad-spectrum antibiotics, such as carbapenem β-lactams or the combination of β-lactam antibiotics and β-lactamase inhibitors (e.g., piperacillin/tazobactam, ampicillin/sulbactam, ticarcillin/clavulanic acid and amoxicillin/clavulanic acid) (210). The use of intravenous antibiotics is recommended to treat pressure ulcers when there is sign of osteomyelitis (209). In these cases, antibiotics that penetrate the bone are required, such as β-lactams (e.g., penicillin and cephalosporin), fluoroquinolones aminoglycosides, and glycopeptides (e.g., vancomycin), linezolid, and rifampin (212).
Several challenges, such as the formation of multi-species biofilms, are implicated in antibiotic treatment success. Wounds are often infected with polymicrobial biofilms formed by several species of resistant bacteria. These biofilms are commonly resistant to topical and systemic antibiotics, which reduces the effectiveness of the antimicrobial treatment. For instance, Shettigar et al. found the authors showed that 60% of the DFU samples investigated were infected with polymicrobial biofilms, in which the isolated E. faecalis showed higher resistance to antibiotics than non-biofilm grown cells (213). Furthermore, bacteria within biofilms produce several protective components. Among them, the EPS matrix is an important factor that impairs the penetration of antibiotics into the wound bed. For instance, P. aeruginosa EPS contains extracellular DNA and alginate lyase that impairs the diffusion of aminoglycosides (214, 215). Another problem associated with the low permeability of antimicrobial agents through the biofilm structure is the induction of resistance due to the low concentration of antibiotics when they reach the bacterial cells (56).
New Approaches to Treating Chronic Wounds
A variety of new treatments are under development to improve healing and restore tissue homeostasis. Among the most promising are candidates that target or work in conjunction with the innate immune system to improve antibacterial immune responses and/or regulate inflammatory responses. Some of the most promising are highlighted in Figure 3. These pathways and molecules represent viable targets because they can be used to modulate both early and late healing processes. There is also a reduced risk of developing drug resistance. Here, we will discuss the potential use of antimicrobial peptides in targeting bacteria levels/biofilm formation and in modulating immune function. We will also describe other strategies under development that seek to develop smarter and controlled innate immune responses by priming the antibacterial immune responses, restoring inflammatory balance, and selectively inducing an M1-M2 transition.
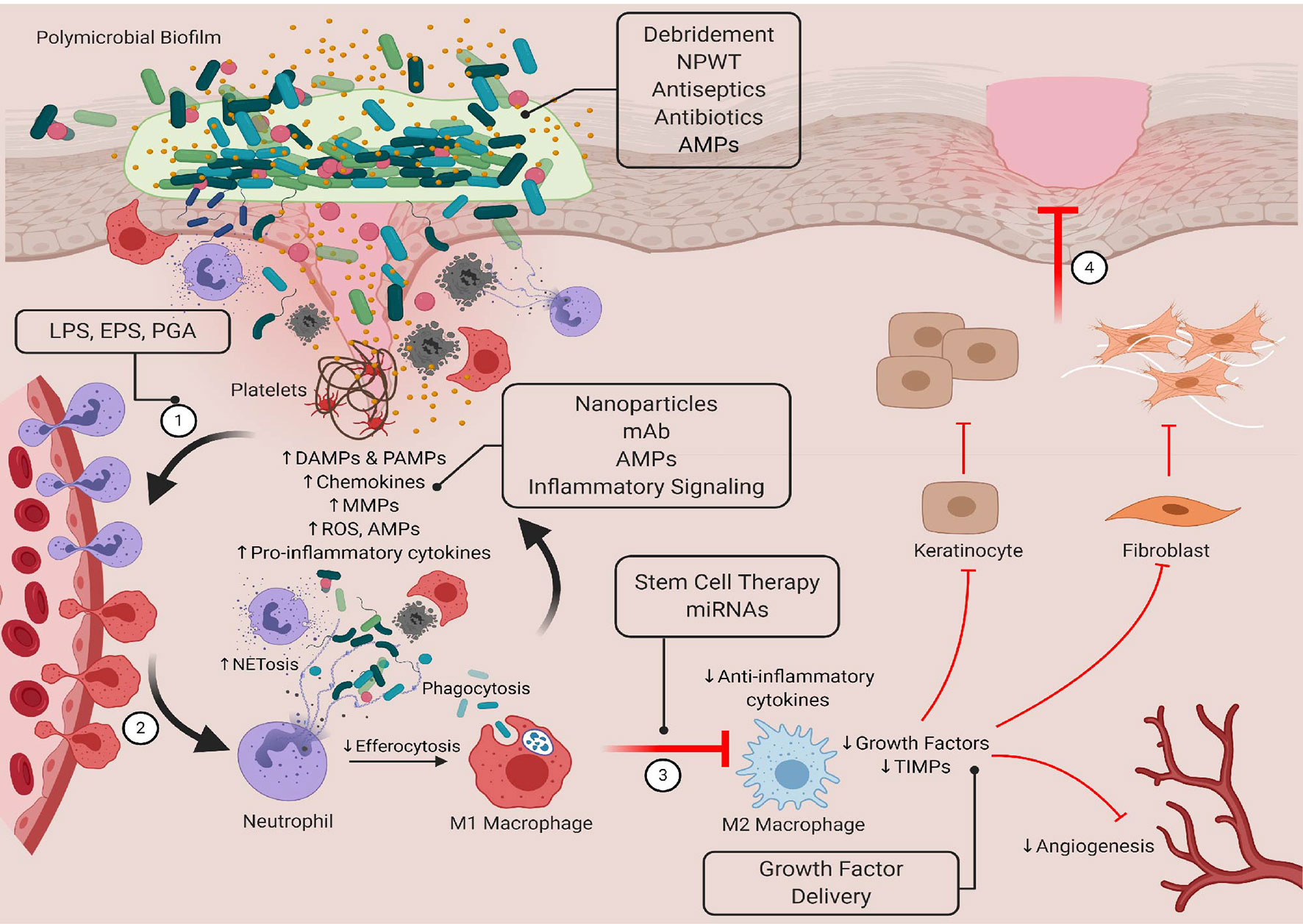
Figure 3 Targeting bacteria-innate immune interactions to restore healing in chronic wounds. Standard therapies such as debridement, NPWT, antiseptics, and antibiotics have been shown to reduce bacterial bioburden in the wound bed, but they do not always restore healing processes. New therapeutics that have both antimicrobial and immunomodulatory properties may be able to overcome the limitations of more traditional treatments. Here, we show novel therapeutics that target these interactions that can be used in early and late stages of healing to restore tissue homeostasis. LPS, lipopolysaccharide; EPS, extracellular polymeric substance; PGA, peptidoglycan; AMP, antimicrobial peptide; mAb, monoclonal antibody; miRNA, microRNA. Created with BioRender.com.
Antimicrobial Peptides
Antimicrobial peptides (AMPs) show tremendous potential for the treatment of severe and chronic infections. These peptides have a broad-spectrum antibacterial activity that usually involves attacking multiple hydrophobic and/or polyanionic targets (216). They have been shown to induce pore formation, disrupt cellular and organelle membrane integrity, inhibit protein and nucleic acid synthesis, block enzymatic activity, inhibit cell wall synthesis, and induce apoptosis through the generation of ROS (217, 218). In addition to their antimicrobial effects, AMPs also modulate immune response to improve bacterial killing by increasing numbers of antigen presenting cells, facilitating the release of NETs, enhancing of phagocytosis, modulating dendritic cell differentiation and T cell activation, suppressing inflammatory signaling and anti-inflammatory cytokines (218). Further, many AMPs have been shown to promote wound healing by modulating of keratinocyte cytokine production and migration, re‐epithelialization and angiogenesis (219). In addition, several natural and synthetic AMPs exhibit strong antibiofilm activities, for example by disrupting bacterial communication networks (quorum sensing), inhibiting bacterial cell adhesion or by stimulating biofilm dispersal (220). Here, we discuss just a few examples of these bioactive molecules.
In humans, AMPs are produced by a variety of cell types including skin epithelia cells. In conditions of health, RNase 5, RNase 7, psoriasin and calprotectin are produced and have antimicrobial activity against both Gram-negative and Gram-positive bacteria (221–223). Alternatively, under conditions of inflammation or infection, β-defensins (h-BD) and LL-37 are selectively induced to mount a wide spectrum of antimicrobial activities including antibiofilm activities (219, 224). In addition to their microbicidal activity, hBD has been shown to regulate inflammatory processes by inhibiting TLR signaling pathways and transcriptionally repressing of pro-inflammatory genes expression (225, 226). Further, hBD-3 has also been shown to act as a ligand for the macrophage receptor CCR2, attracting macrophages to sites of epithelial injury (227). LL37 has also been shown to neutralize the activation of macrophages via LPS and induce proliferation and migration of endothelial cells (228). It also contributes to multiple phases wound repair including the stimulation endothelia cells and fibroblasts (228, 229) stimulation of keratinocytes (230), neovascularization (228) and angiogenesis (231). A number of approaches are currently under development for administering hBD and LL-37 including poly(vinyl alcohol)/cellulose acetate (PVA/CA) films (232), nanoparticles (233, 234), and nanostructured lipid carriers (235). Further, the efficacy of LL-37 cream in treating DFU is currently in clinical trials (https://clinicaltrials.gov).
Interestingly, wound healing is a relatively conserved evolutionarily process and several species including insects such as Drosophila, Caenorhabditis elegans and amphibians have been shown produce their own AMPs. For example, Pseudin-2 isolated from the frog Pseudis paradoxa has been shown to have a broad-spectrum antimicrobial potency and skin biocompatibility against multidrug-resistant (MDR) Pseudomonas aeruginosa (236). It has also been shown to facilitate infected-wound closure by reducing inflammation through suppression of interleukin-1β (IL-1β), IL-6, and tumor necrosis factor alpha (TNF-α) (236). Alternatively, the synthetic peptide A3-APO, derived from natural insect products, has shown promise in in vivo models. It was shown to efficiently ameliorate resistant aerobic and anaerobic intradermal infections, in part by increased recruitment of epithelial macrophages and their immunomodulatory/anti-inflammatory effects (237). Epinecidin-1 (Epi-1), an AMP derived from grouper Epinephelus coioides is also of potential interest. This molecule has been shown to have antibacterial, antifungal, and antiviral activity in vitro and in vivo (238). In mice with MRSA, Epi-1 has been shown to decrease levels of TNF-a, IL-6, and MCP-1, while also regulating monocyte recruitment during wound healing (239). It also enhances wound closure and angiogenesis (239). These molecules and many others are in early stages of development but represent promising antimicrobial and immunomodulatory therapeutics.
Jump-Starting Innate Immune Responses Using Bacteria/Bacterial Components
Topical application of PAMPs isolated from bacteria has been used to stimulate wound healing by initiating inflammatory processes in early stages of healing. In mice, Kostarnoy et al. have shown that application of lipopolysaccharide (LPS), a component of the outer membrane of Gram-negative bacteria and a potent endotoxin, improves healing by accelerating the resolution of inflammation by increasing macrophage infiltration, the expression of proinflammatory cytokines (IL-6, IL-1β, and leukemia inhibitory factor (LIF)), CC-chemokines (CCL2, CCL7, CCL3 and CCL5), growth factors (VEGF, TGF-1β, and FGF-2) in the wound microenvironment and by increasing collagen synthesis in the wound microenvironment (240). Similarly, the exopolysaccharide or extracellular polymeric substance EPS-S3 derived from the marine bacterium Pantoea sp. YU16-S3 has been shown to be a potential biomolecule to promote skin tissue regeneration (241). In vitro, EPS-S3 has been shown to increase dermal fibroblasts and keratinocytes, and macrophage activation (241). In vivo, EPS-S3 increases expression of growth factors and adhesion molecules HB-EGF, FGF, E-cadherin suggesting this exopolysaccharide may modulates wound healing through the Wnt/β-catenin pathway (241). Interestingly, other studies have shown that rats subcutaneously implanted with PVA sponges inoculated with non-viable S. aureus or its peptidoglycan have improved healing responses (242, 243). These responses are associated with increased macrophage, neutrophil and fibroblast infiltration, collagen production, and angiogenesis, which contribute to the formation of reparative tissue.
Improving Antimicrobial Responses
Another area of interest is to develop therapeutics that enhance antimicrobial immune responses. Yu et al. have developed iron oxide nanoparticles (IONPs) that are taken up by macrophages to enhance bactericidal activity against intracellular S. aureus. They do this by increasing the expression of pro-inflammatory M1 markers iNOS, IL-1β, and TNF-α and amplifying ROS production (244). They also found these bactericidal effects could be enhanced by coupling IONPs with vitamin C in a Fenton reaction to augment the formation of ROS in the form of hydroxyl radicals (244). Alternatively, Okumura et al. shown that the pharmacological agent AKB-4924 can promote antibacterial immune responses by stabilizing HIF-1α (245). Intracellular HIF-1α accumulation heightens pro-inflammatory responses by increasing expression of LL-37 and IL-8 in human monocytes and enhancing bactericidal activity in vitro and in vivo (245).
Targeting Inflammatory Balance
As described above, persistent or sustained inflammation is a central drive of dysregulated healing and chronic ulcer formation. Therapeutics that neutralize pro-inflammatory biomolecules such as cytokines, chemokines and bioactive lipids are under investigation to limit tissue damage and restore timely healing processes. Among these, Song et al. have examined the effect of anti-TNF- α monoclonal antibody (MAb) in a primate model of S. aureus-associated skin (246). Systemic administration of anti-TNF-α MAb reduced abscess severity through suppression of circulating proinflammatory IL-8 and IL-12, benefiting host responses to bacterial challenge (246). Alternatively, Brandt et al. have shown that therapeutics that target the LTB4/BLT1 signaling axis can reduce abscess severity and inflammation by limiting neutrophil recruitment, improving chemotaxis, and restoring chemokines and cytokine levels (148). More recently, Vågesjö et al. have developed CXCL12-delivering Lactobacilli to topically administer chemokines with increased bioavailability for wound healing processes (247). In mice models, sustained topical application of CXCL12 increased proliferation of TGF-β expressing macrophages (247) and accelerated wound healing in healthy mice, mice with hyperglycemia, and peripheral ischemia, and in an in vitro human skin disk model (247).
Growth factors also play a critical role in modulating inflammation and inducing cell proliferation, angiogenesis, and granulation tissue formation during wound healing. Diminished levels of growth factors, such as VEGF and FGF-2 have been associated with chronic pressure ulcers (248). To restore this imbalance novel therapeutics have been focused on delivering growth factors to the wound bed to promote healing. In a study on diabetic foot ulcer patients receiving intralesional epidermal growth factor (EGF) therapy, García-Ojalvo et al. demonstrated a reduction in systemic proinflammatory biomolecules C-reactive protein (CRP), IL-6, soluble FAS (sFAS), and CCL3, as well as oxidative capacity and nitrosilative (nitrite/nitrate) stress biomarkers (249). Further, intralesional EGF therapy was shown to increase soluble RAGE (sRAGE), which may have protective effects in diabetic patients (249). Another approach to increase growth factors is to provide bone-marrow mesenchymal stem cells (BM-MSCs). Bai et al. developed an injectable hydrogel made from crosslinking N-chitosan and adipic acid dihydrazide with hyaluronic acid-aldehyde to deliver BM-MSCs into the wound bed (250). In vitro and in vivo investigations in diabetic wound healing showed that the hydrogel was able to stimulate BM-MSC-derived secretion of TGF-β1, VEGF, and FGF-2 as well as inhibiting chronic inflammation through M2 macrophage polarization (250). It also induced granulation tissue formation, collagen deposition, tissue vascularization, and improved wound closure (250). The LeucoPatch uses a similar approach to promote healing. This circular patch is comprised of fibrin, white cells and platelets derived from the patient’s own blood, which concentrates cells and growth factors (e.g., PDGF, TGF-β, EGF and VEGF) to support healing. In an observed-masked randomized controlled trial, it was found to improve healing outcomes and shortened time to healing (251).
Promoting Macrophage Polarization Towards an M2 Phenotype
Given that M1/M2 macrophages play a key role in regulating wound healing, reprogramming macrophage polarization towards an M2 tissue repair phenotype represents an attractive target in later stages of the healing process. Stem cell therapy has emerged as a promising treatment for modulating these processes. In a mouse diabetic wound model, treatment with hyaluronic acid spongy hydrogels impregnated with neurogenically conditioned human adipose stem cells (hASCs) increased neoepidermial thickness and accelerated wound closure (252). Moreover, addition of hASCs in comparison to hydrogel treatment alone increased the M2/M1 macrophage ratio, suggesting that hASCs can promote the transition to the repair phase of healing (252). A recent systematic review by Raghuram et al., identified adipose-derived stem cells, bone marrow-derived stem cells, bone marrow-derived mononuclear cells, epidermally derived mesenchymal stem cells, fibroblast stem cells, keratinocyte stem cells, placental mesenchymal stem cells, and umbilical cord mesenchymal stem cells being used in vitro and in vivo as potential treatments for chronic wounds, however, clinical effectiveness still requires investigation due to heterogeneity of wound etiology (253).
A multitude of micro-RNAs (miRNAs) have been associated with each phase of wound healing, from pro-inflammatory cytokine signaling to proliferation and remodeling, and offer a potential therapeutic strategy for the treatment of chronic wounds (254). Saleh et al. developed adhesive hydrogels containing miR‐223 5p mimic loaded hyaluronic acid-based nanoparticles (255). In vitro, miR-233 5p had the ability to polarize M1 macrophages towards an M2 phenotype, with increased expression of anti-inflammatory gene Arg-1, and suppression of proinflammatory cytokines TNF-α, IL-1β, and IL-6 (255). In vivo experiments on a mouse wound model, miR-233 5p was able to promote tissue vascularization and accelerate wound healing (255).
Discussion
Ageing and immobile individuals as well as those with co-morbid conditions such as diabetes are at high risk for developing non-healing or chronic wounds. These wounds reduce quality of life and increase pain levels, risk for infection and prolong hospital stays. Chronic wounds are also difficult to treat and represent a significant financial burden on all healthcare systems. Globally, as the size of these populations grow, there is an urgent need to understand the pathophysiology of delayed wound healing and to develop effective therapies that repair tissue damage. Critical for the development of these new therapeutics is a comprehensive understanding of the cellular and molecular mechanisms underpinning bacteria-innate immune interactions.
There is still a lot to learn about the microbiological and immunological processes underlying bacteria-innate immune interactions in chronic wounds and their relative contributions to delayed healing. From the microbiological perspective, the use of molecular methods, such as RNA sequencing, has allowed for the identification of a larger diversity bacterial species in the wound. These methodologies have also been used to elucidate microbial activities, behaviors, strategies, and processes during infections (12). However, these approaches are associated with a substantial demand for financial, time and bioinformatic support and cannot be readily transferred into the clinic. Further, they cannot distinguish between living, dead or dormant bacteria and might overlook minority species (52). Moving forward, it will also be critical develop more standardized sampling and analysis to ensure reproducibility across studies (256–258). It is also important to note that, to date, most studies have been performed over an acute timeframe with the longest being over a 28-day period (75). Considering that chronic wounds can take up to 12 months to heal (259), or may not heal at all (55), there is little information about how bacterial populations change over longer time frames. We believe combining single cell analyses (transciptomics, flow, etc.), advanced microscopy and other techniques will provide critical insights into how biofilm structures as well individual cells contribute to chronic wound formation and chronicity across diverse microenvironments and patient populations. Further, we believe more long term longitudinal in vivo studies with larger samples sizes and standardized sample collection/analysis are urgently needed to fully understand the importance of microbial diversity, biofilms and the wound microbiome in chronic wounds infections and to elucidate the impact of aerobic, anaerobic, pathogenic and commensal bacteria in inflammation and wound healing across wound types.
To further our immunological understanding, we require clinically relevant model systems that mimic the complex, dynamic interplay between the wound microbiome, innate immune cells, and the various other factors that contribute to dysregulated healing in chronic wounds (260). To date, many studies have investigated interactions in the context of murine S. aureus abscess models. While S. aureus is a major causative agent of skin and soft-tissue infections, it is a specific type of skin infection. Other studies have evaluated how heat-killed bacteria and planktonic bacteria modulate immune responses but generally only evaluate short term and localized responses. Emerging research has started to evaluate the effects of single-species and polymicrobial biofilms on host immune responses in vivo. However, most of these studies have characterized differential responses to S. aureus and P. aeruginosa planktonic, single-species biofilm, and polymicrobial biofilms (69–71, 73, 74). Given that the wound microbiome is made of a wide diversity of bacterial species in polymicrobial biofilm communities, it is unclear how these findings can be translated into the clinical setting. We also think it is important to note, that much of what we know about the microbial diversity and immune responses in chronic wounds has been derived from models and clinical samples from patients with DFU. In this review, we found a few studies that evaluated interactions in other or non-DFU ulcers such as venous/arterial ulcers and pressure ulcers, but they were somewhat limited in scope (77, 160, 161). Future studies are required to evaluate how host immune responses are modulated by complex polymicrobial biofilms commonly found in wound beds and to better understand if these processes are affected by the wound type/tissue microenvironment.
Author Contributions
All authors contributed to the article and approved the submitted version.
Funding
The preparation of the publication was support by an International Research Seed Grant provided by Carleton University awarded to EC and JO. It was also supported by a Bruyere Research Institute Graduate Studentship Award awarded to ZV.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Blakytny R, Jude E. The molecular biology of chronic wounds and delayed healing in diabetes. Diabetic Med (2006) 23:594–608. doi: 10.1111/j.1464-5491.2006.01773.x
2. Morton LM, Phillips TJ. Wound healing and treating wounds. J Am Acad Dermatol (2016) 74:589–605. doi: 10.1016/j.jaad.2015.08.068
3. Järbrink K, Ni G, Sönnergren H, Schmidtchen A, Pang C, Bajpai R, et al. The humanistic and economic burden of chronic wounds: a protocol for a systematic review. Systematic Rev (2017) 6:15. doi: 10.1186/s13643-016-0400-8
4. Armstrong DG, Wrobel J, Robbins JM. Guest Editorial: are diabetes-related wounds and amputations worse than cancer? Int Wound J (2007) 4:286–7. doi: 10.1111/j.1742-481X.2007.00392.x
5. Tricco AC, Cogo E, Isaranuwatchai W, Khan PA, Sanmugalingham G, Antony J, et al. A systematic review of cost-effectiveness analyses of complex wound interventions reveals optimal treatments for specific wound types. BMC Med (2015) 13:90. doi: 10.1186/s12916-015-0326-3
6. Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis (2004) 17:91–6. doi: 10.1097/00001432-200404000-00004
7. Percival SL, Thomas JG, Williams DW. Biofilms and bacterial imbalances in chronic wounds: anti-Koch. Int Wound J (2010) 7:169–75. doi: 10.1111/j.1742-481X.2010.00668.x
8. Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regeneration (2016) 24:163–74. doi: 10.1111/wrr.12370
9. Metcalf D, Bowler P. Biofilm delays wound healing: A review of the evidence. Burns Trauma (2013) 1:5. doi: 10.4103/2321-3868.113329
10. Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harbor Perspect Biol (2010) 2:a000398–a000398. doi: 10.1101/cshperspect.a000398
11. Malone M, Bjarnsholt T, McBain AJ, James GA, Stoodley P, Leaper D, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care (2017) 26:20–5. doi: 10.12968/jowc.2017.26.1.20
12. Kirketerp-Møller K, Stewart PS, Bjarnsholt T. The zone model: A conceptual model for understanding the microenvironment of chronic wound infection. Wound Repair Regeneration (2020) 28:593–9. doi: 10.1111/wrr.12841
13. Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health (2017) 10:369–78. doi: 10.1016/j.jiph.2016.08.007
14. Bjarnsholt T, Ciofu O, Molin S, Givskov M, Høiby N. Applying insights from biofilm biology to drug development — can a new approach be developed? Nat Rev Drug Discovery (2013) 12:791–808. doi: 10.1038/nrd4000
15. Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID∗ guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect (2015) 21:S1–S25. doi: 10.1016/j.cmi.2014.10.024
16. Hall CW, Mah T-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev (2017) 41:276–301. doi: 10.1093/femsre/fux010
17. Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrobial Agents (2010) 35:322–32. doi: 10.1016/j.ijantimicag.2009.12.011
18. Percival SL, KE H, Williams DW, Hooper SJ, Thomas DW, Costerton JW. A review of the scientific evidence for biofilms in wounds. Wound Repair Regeneration (2012) 20:647–57. doi: 10.1111/j.1524-475X.2012.00836.x
19. Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol (2016) 14:563–75. doi: 10.1038/nrmicro.2016.94
20. Wu Y-K, Cheng N-C, Cheng C-M. Biofilms in Chronic Wounds: Pathogenesis and Diagnosis. Trends Biotechnol (2019) 37:505–17. doi: 10.1016/j.tibtech.2018.10.011
21. Lebeaux D, Chauhan A, Rendueles O, Beloin C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens (2013) 2:288–356. doi: 10.3390/pathogens2020288
22. Martin JM,, Zenilman JM, Lazarus GS. Molecular Microbiology: New Dimensions for Cutaneous Biology and Wound Healing. J Invest Dermatol (2010) 130:38–48. doi: 10.1038/jid.2009.221
23. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev (2012) 70:S38–44. doi: 10.1111/j.1753-4887.2012.00493.x
24. Ursell LK, Haiser HJ, van Treuren W, Garg N, Reddivari L, Vanamala J, et al. The Intestinal Metabolome: An Intersection Between Microbiota and Host. Gastroenterology (2014) 146:1470–6. doi: 10.1053/j.gastro.2014.03.001
25. Wang B, Yao M, Lv L, Ling Z, Li L. The Human Microbiota in Health and Disease. Engineering (2017) 3:71–82. doi: 10.1016/J.ENG.2017.01.008
26. Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell (2016) 164:337–40. doi: 10.1016/j.cell.2016.01.013
27. Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol (2018) 16:143–55. doi: 10.1038/nrmicro.2017.157
28. Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ (2019) 7:e7502. doi: 10.7717/peerj.7502
29. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol (2011) 9:244–53. doi: 10.1038/nrmicro2537
30. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science (2009) 324:1190–2. doi: 10.1126/science.1171700
31. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature (2013) 498:367–70. doi: 10.1038/nature12171
32. Daeschlein G. Antimicrobial and antiseptic strategies in wound management. Int Wound J (2013) 10:9–14. doi: 10.1111/iwj.12175
33. Bowler PG, Duerden BI, Armstrong DG. Wound Microbiology and Associated Approaches to Wound Management. Clin Microbiol Rev (2001) 14:244–69. doi: 10.1128/CMR.14.2.244-269.2001
34. Guo S, DiPietro LA. Factors Affecting Wound Healing. J Dental Res (2010) 89:219–29. doi: 10.1177/0022034509359125
35. Orsted HL, Keast DH, Forest-Laland L, Kuhnke JL, O’Sullivan-Drombolis D, Jin S, et al. Best practice recommendations for the prevention and management of wounds. In: Foundations of Best Practice for Skin and Wound Management. A supplement of Wound Care Canada. (2017) 74 pp. Available at: www.woundscanada.ca/docman/public/health-care-professional/bpr-workshop/165-wc-bpr-prevention-andmanagement-of-wounds/file.
36. Negut I, Grumezescu V, Grumezescu A. Treatment Strategies for Infected Wounds. Molecules (2018) 23:2392. doi: 10.3390/molecules23092392
37. Kingsley A. The wound infection continuum and its application to clinical practice. Ostomy Manage (2003) 49:1–7.
38. Gardner SE, Frantz RA. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs (2008) 10:44–53. doi: 10.1177/1099800408319056
39. Baviera G, Leoni MC, Capra L, Cipriani F, Longo G, Maiello N, et al. Microbiota in healthy skin and in atopic eczema. BioMed Res Int (2014) 2014:436921–1. doi: 10.1155/2014/436921
40. Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Res (2008) 18:1043–50. doi: 10.1101/gr.075549.107
41. Tuttle MS. Association Between Microbial Bioburden and Healing Outcomes in Venous Leg Ulcers: A Review of the Evidence. Adv Wound Care (2015) 4:1–11. doi: 10.1089/wound.2014.0535
42. Smith K, Collier A, Townsend EM, O’Donnell LE, Bal AM, Butcher J, et al. One step closer to understanding the role of bacteria in diabetic foot ulcers: characterising the microbiome of ulcers. BMC Microbiol (2016) 16:54. doi: 10.1186/s12866-016-0665-z
43. Wolcott RDRA, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RDRA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regeneration (2016) 24:163–74. doi: 10.1111/wrr.12370
44. Malone M, Johani K, Jensen SO, Gosbell IB, Dickson HG, Hu H, et al. Next Generation DNA Sequencing of Tissues from Infected Diabetic Foot Ulcers. EBioMedicine (2017) 21:142–9. doi: 10.1016/j.ebiom.2017.06.026
45. Kalan LR, Meisel JS, Loesche MA, Horwinski J, Soaita I, Chen X, et al. Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe (2019) 25:641–55. doi: 10.1016/j.chom.2019.03.006
46. Kpeli GS, Yeboah-Manu D. “Secondary infection of Buruli ulcer lesions,” In: Buruli Ulcer: Mycobacterium Ulcerans Disease. Switzerland: Springer International Publishing. (2019) p. 227–39. doi: 10.1007/978-3-030-11114-4_13
47. Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, et al. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J Invest Dermatol (2016) 136:947–56. doi: 10.1016/j.jid.2016.01.016
48. Kong HH, Andersson B, Clavel T, Common JE, Jackson SA, Olson ND, et al. Performing Skin Microbiome Research: A Method to the Madness. J Invest Dermatol (2017) 137:561–8. doi: 10.1016/j.jid.2016.10.033
49. Nakatsuji T, Chiang H-I, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun (2013) 4:1431. doi: 10.1038/ncomms2441
50. Travis J, Malone M, Hu H, Baten A, Johani K, Huygens F, et al. The microbiome of diabetic foot ulcers: a comparison of swab and tissue biopsy wound sampling techniques using 16S rRNA gene sequencing. BMC Microbiol (2020) 20:163. doi: 10.1186/s12866-020-01843-2
51. Wolcott RD, Dowd SE. A rapid molecular method for characterising bacterial bioburden in chronic wounds. J Wound Care (2008) 17:513–6. doi: 10.12968/jowc.2008.17.12.31769
52. Jneid J, Lavigne JP, la Scola B, Cassir N. The diabetic foot microbiota: A review. Hum Microbiome J (2017) 5–6:1–6. doi: 10.1016/j.humic.2017.09.002
53. Tomic-Canic M, Burgess JL, O’Neill KE, Strbo N, Pastar I. Skin Microbiota and its Interplay with Wound Healing. Am J Clin Dermatol (2020) 21:36–43. doi: 10.1007/s40257-020-00536-w
54. Jahns AC, Lundskog B, Ganceviciene R, Palmer RH, Golovleva I, Zouboulis CC, et al. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: a case-control study. Br J Dermatol (2012) 167:50–8. doi: 10.1111/j.1365-2133.2012.10897.x
55. Lazarus GS. Definitions and Guidelines for Assessment of Wounds and Evaluation of Healing. Arch Dermatol (1994) 130:489. doi: 10.1001/archderm.1994.01690040093015
56. Pereira SG, Moura J, Carvalho E, Empadinhas N. Microbiota of Chronic Diabetic Wounds: Ecology, Impact, and Potential for Innovative Treatment Strategies. Front Microbiol (2017) 8:1791:1791. doi: 10.3389/fmicb.2017.01791
57. Rahim K, Saleha S, Zhu X, Huo L, Basit A, Franco OL. Bacterial Contribution in Chronicity of Wounds. Microbial Ecol (2017) 73:710–21. doi: 10.1007/s00248-016-0867-9
58. Percival SL, Malone M, Mayer D, Salisbury A-M, Schultz G. Role of anaerobes in polymicrobial communities and biofilms complicating diabetic foot ulcers. Int Wound J (2018) 15:776–82. doi: 10.1111/iwj.12926
59. Sadeghpour Heravi F, Zakrzewski M, Vickery K, Armstrong D G, Hu H. Bacterial Diversity of Diabetic Foot Ulcers: Current Status and Future Prospectives. J Clin Med (2019) 8:1935. doi: 10.3390/jcm8111935
60. Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol (2008) 8:43. doi: 10.1186/1471-2180-8-43
61. Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The Neuropathic Diabetic Foot Ulcer Microbiome Is Associated With Clinical Factors. Diabetes (2013) 62:923–30. doi: 10.2337/db12-0771
62. Noor S, Ahmad J, Parwez I, Ozair M. Culture-Based Screening of Aerobic Microbiome in Diabetic Foot Subjects and Developing Non-healing Ulcers. Front Microbiol (2016) 7:1792:1792. doi: 10.3389/fmicb.2016.01792
63. Jneid J, Cassir N, Schuldiner S, Jourdan N, Sotto A, Lavigne J-P, et al. Exploring the Microbiota of Diabetic Foot Infections With Culturomics. Front Cell Infect Microbiol (2018) 8:282:282. doi: 10.3389/fcimb.2018.00282
64. Suryaletha K, John J, Radhakrishnan MP, George S, Thomas S. Metataxonomic approach to decipher the polymicrobial burden in diabetic foot ulcer and its biofilm mode of infection. Int Wound J (2018) 15:473–81. doi: 10.1111/iwj.12888
65. Johani K, BG F, Bjarnsholt T, Lipsky BA, Jensen SO, Yang M, et al. Understanding the microbiome of diabetic foot osteomyelitis: insights from molecular and microscopic approaches. Clin Microbiol Infect (2019) 25:332–9. doi: 10.1016/j.cmi.2018.04.036
66. Jnana A, Muthuraman V, Varghese VK, Chakrabarty S, Murali TS, Ramachandra L, et al. Microbial Community Distribution and Core Microbiome in Successive Wound Grades of Individuals with Diabetic Foot Ulcers. Appl Environ Microbiol (2020) 86(6):e02608–19. doi: 10.1128/AEM.02608-19
67. Min KR, Galvis A, Baquerizo Nole KL, Sinha R, Clarke J, Kirsner RS, et al. Association between baseline abundance of Peptoniphilus, a Gram-positive anaerobic coccus, and wound healing outcomes of DFUs. PloS One (2020) 15:e0227006. doi: 10.1371/journal.pone.0227006
68. Sweere JM, Ishak H, Sunkari V, Bach MS, Manasherob R, Yadava K, et al. The Immune Response to Chronic Pseudomonas aeruginosa Wound Infection in Immunocompetent Mice. Adv Wound Care (2020) 9:35–47. doi: 10.1089/wound.2019.1039
69. Klein P, Sojka M, Kucera J, Matonohova J, Pavlik V, Nemec J, et al. A porcine model of skin wound infected with a polybacterial biofilm. Biofouling (2018) 34:226–36. doi: 10.1080/08927014.2018.1425684
70. Trøstrup H, Lerche CJ, Christophersen LJ, Thomsen K, Jensen PØ, Hougen HP, et al. Chronic Pseudomonas aeruginosa Biofilm Infection Impairs Murine S100A8/A9 and Neutrophil Effector Cytokines – Implications for Delayed Wound Closure? Pathog Dis (2017) 75(7):ftx068. doi: 10.1093/femspd/ftx068
71. Trøstrup H, Lerche CJ, Christophersen LJ, Thomsen K, Jensen PØ, Hougen HP, et al. Pseudomonas aeruginosa biofilm hampers murine central wound healing by suppression of vascular epithelial growth factor. Int Wound J (2018) 15:123–32. doi: 10.1111/iwj.12846
72. Canesso MCC, Vieira AT, Castro TBR, Schirmer BGA, Cisalpino D, Martins FS, et al. Skin Wound Healing Is Accelerated and Scarless in the Absence of Commensal Microbiota. J Immunol (2014) 193:5171–80. doi: 10.4049/jimmunol.1400625
73. Seth AK, Geringer MR, Hong SJ, Leung KP, Galiano RD, Mustoe TA. Comparative Analysis of Single-Species and Polybacterial Wound Biofilms Using a Quantitative, In Vivo, Rabbit Ear Model. PloS One (2012) 7:e42897. doi: 10.1371/journal.pone.0042897
74. Gurjala AN, Geringer MR, Seth AK, Hong SJ, Smeltzer MS, Galiano RD, et al. Development of a novel, highly quantitative in vivo model for the study of biofilm-impaired cutaneous wound healing. Wound Repair Regeneration (2011) 19:400–10. doi: 10.1111/j.1524-475X.2011.00690.x
75. Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Liechty KW, Segre JA, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci (2010) 107:14799–804. doi: 10.1073/pnas.1004204107
76. Kim M-H, Liu W, Borjesson DL, Curry F-RE, Miller LS, Cheung AL, et al. Dynamics of Neutrophil Infiltration during Cutaneous Wound Healing and Infection Using Fluorescence Imaging. J Invest Dermatol (2008) 128:1812–20. doi: 10.1038/sj.jid.5701223
77. van der Laan N, de Leij LFMH, ten Duis HJ. Immunohistopathological appearance of three different types of injury in human skin. Inflammation Res (2001) 50:350–6. doi: 10.1007/PL00000255
78. Thomsen TR, Aasholm MS, Rudkjøbing VB, Saunders AM, Bjarnsholt T, Givskov M, et al. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regeneration (2010) 18:38–49. doi: 10.1111/j.1524-475X.2009.00561.x
79. Horton JM, Gao Z, Sullivan DM, Shopsin B, Perez-Perez GI, Blaser MJ. The Cutaneous Microbiome in Outpatients Presenting With Acute Skin Abscesses. J Infect Dis (2015) 211:1895–904. doi: 10.1093/infdis/jiv003
80. Prieto-Borja L, Pérez-Tanoira R, Levano-Linares D-C, Celdrán A, Mahillo-Fernández I, Esteban J. Sonication of Abdominal Drains: Clinical Implications of Quantitative Cultures for the Diagnosis of Surgical Site Infection. Surg Infect (2016) 17:459–64. doi: 10.1089/sur.2015.268
81. Cuchí E, García LG, Jiménez E, Haro D, Castillón P, Puertas L, et al. Relationship between skin and urine colonization and surgical site infection in the proximal femur fracture: a prospective study. Int Orthopaedics (2020) 44:1031–5. doi: 10.1007/s00264-020-04525-w
82. de Wert LA, Rensen SS, Soons Z, Poeze M, Bouvy ND, Penders J. The cutaneous microbiome in hospitalized patients with pressure ulcers. Sci Rep (2020) 10:5963. doi: 10.1038/s41598-020-62918-8
83. Johnson RC, Ellis MW, Schlett CD, Millar EV, LaBreck PT, Mor D, et al. Bacterial Etiology and Risk Factors Associated with Cellulitis and Purulent Skin Abscesses in Military Trainees. PloS One (2016) 11:e0165491. doi: 10.1371/journal.pone.0165491
84. Vierhout BP, Ott A, Kruithof I, Wisselink G, van Zanten E, Kooistra-Smid AMD, et al. Inguinal microbiome in patients undergoing an endovascular aneurysm repair: Application of next-generation sequencing of the 16S-23S rRNA regions. Med Hypotheses (2019) 132:109358. doi: 10.1016/j.mehy.2019.109358
85. Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci Trans Med (2014) 6:265sr6–6. doi: 10.1126/scitranslmed.3009337
86. Landén NX, Li D, Ståhle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell Mol Life Sci (2016) 73:3861–85. doi: 10.1007/s00018-016-2268-0
87. MacLeod AS, Mansbridge JN. The Innate Immune System in Acute and Chronic Wounds. Adv Wound Care (2016) 5:65–78. doi: 10.1089/wound.2014.0608
88. Ellis S, Lin EJ, Tartar D. Immunology of Wound Healing. Curr Dermatol Rep (2018) 7:350–8. doi: 10.1007/s13671-018-0234-9
89. Larouche J, Sheoran S, Maruyama K, Martino MM. Immune Regulation of Skin Wound Healing: Mechanisms and Novel Therapeutic Targets. Adv Wound Care (2018) 7:209–31. doi: 10.1089/wound.2017.0761
90. Cañedo-Dorantes L, Cañedo-Ayala M. Skin Acute Wound Healing: A Comprehensive Review. Int J Inflammation (2019) 2019:1–15. doi: 10.1155/2019/3706315
91. Nguyen AV, Soulika AM. The Dynamics of the Skin’s Immune System. Int J Mol Sci (2019) 20:1811. doi: 10.3390/ijms20081811
92. Rodrigues M, Kosaric N, Bonham CA, Gurtner GC. Wound Healing: A Cellular Perspective. Physiol Rev (2019) 99:665–706. doi: 10.1152/physrev.00067.2017
93. Nosbaum A, Prevel N, Truong H-A, Mehta P, Ettinger M, Scharschmidt TC, et al. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J Immunol (2016) 196:2010–4. doi: 10.4049/jimmunol.1502139
94. Engelhardt E, Toksoy A, Goebeler M, Debus S, Bröcker E-B, Gillitzer R. Chemokines IL-8, GROα, MCP-1, IP-10, and Mig Are Sequentially and Differentially Expressed During Phase-Specific Infiltration of Leukocyte Subsets in Human Wound Healing. Am J Pathol (1998) 153:1849–60. doi: 10.1016/S0002-9440(10)65699-4
95. Devalaraja RM, Nanney LB, Qian Q, Du J, Yu Y, Devalaraja MN, et al. Delayed wound healing in CXCR2 knockout mice. J Invest Dermatol (2000) 115:234–44. doi: 10.1046/j.1523-1747.2000.00034.x
96. Martins-Green M, Petreaca M, Wang L. Chemokines and Their Receptors Are Key Players in the Orchestra That Regulates Wound Healing. Adv Wound Care (2013) 2:327–47. doi: 10.1089/wound.2012.0380
97. de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol (2016) 16:378–91. doi: 10.1038/nri.2016.49
98. Theilgaard-Mönch K, Knudsen S, Follin P, Borregaard N. The Transcriptional Activation Program of Human Neutrophils in Skin Lesions Supports Their Important Role in Wound Healing. J Immunol (2004) 172:7684–93. doi: 10.4049/jimmunol.172.12.7684
99. Wilgus TA, Roy S, McDaniel JC. Neutrophils and Wound Repair: Positive Actions and Negative Reactions. Adv Wound Care (2013) 2:379–88. doi: 10.1089/wound.2012.0383
100. Kruger P, Saffarzadeh M, Weber ANR, Rieber N, Radsak M, von Bernuth H, et al. Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury. PloS Pathog (2015) 11:e1004651. doi: 10.1371/journal.ppat.1004651
101. Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol (2010) 10:427–39. doi: 10.1038/nri2779
102. Krzyszczyk P, Schloss R, Palmer A, Berthiaume F. The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front Physiol (2018) 9:419. doi: 10.3389/fphys.2018.00419
103. Rodero MP, Licata F, Poupel L, Hamon P, Khosrotehrani K, Combadiere C, et al. In Vivo Imaging Reveals a Pioneer Wave of Monocyte Recruitment into Mouse Skin Wounds. PloS One (2014) 9:e108212. doi: 10.1371/journal.pone.0108212
104. Ishida Y, Gao J-L, Murphy PM. Chemokine receptor CX3CR1 mediates skin wound healing by promoting macrophage and fibroblast accumulation and function. J Immunol (2008) 180:569–79. doi: 10.4049/jimmunol.180.1.569
105. Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood (2012) 120:613–25. doi: 10.1182/blood-2012-01-403386
106. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol (2008) 8:958–69. doi: 10.1038/nri2448
107. Ferrante CJ, Leibovich SJ. Regulation of Macrophage Polarization and Wound Healing. Adv Wound Care (2012) 1:10–6. doi: 10.1089/wound.2011.0307
108. Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol (2010) 10:712–23. doi: 10.1038/nri2852
109. Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE. The phenotype of murine wound macrophages. J Leukocyte Biol (2010) 87:59–67. doi: 10.1189/jlb.0409236
110. Elliott MR, Koster KM, Murphy PS. Efferocytosis Signaling in the Regulation of Macrophage Inflammatory Responses. J Immunol (2017) 198:1387–94. doi: 10.4049/jimmunol.1601520
111. Zhao R, Liang H, Clarke E, Jackson C, Xue M. Inflammation in Chronic Wounds. Int J Mol Sci (2016) 17:2085. doi: 10.3390/ijms17122085
112. Sindrilaru A, Scharffetter-Kochanek K. Disclosure of the Culprits: Macrophages—Versatile Regulators of Wound Healing. Adv Wound Care (2013) 2:357–68. doi: 10.1089/wound.2012.0407
113. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol (2011) 11:723–37. doi: 10.1038/nri3073
114. Wysocki AB, Staiano-Coico L, Grinnell F. Wound Fluid from Chronic Leg Ulcers Contains Elevated Levels of Metalloproteinases MMP-2 and MMP-9. J Invest Dermatol (1993) 101:64–8. doi: 10.1111/1523-1747.ep12359590
115. Bullen EC, Longaker MT, Updike DL, Benton R, Ladin D, Hou Z, et al. Tissue Inhibitor of Metalloproteinases-1 Is Decreased and Activated Gelatinases Are Increased in Chronic Wounds. J Invest Dermatol (1995) 104:236–40. doi: 10.1111/1523-1747.ep12612786
116. Yager DR, Zhang L-Y, Liang H-X, Diegelmann RF, Cohen IK. Wound Fluids from Human Pressure Ulcers Contain Elevated Matrix Metalloproteinase Levels and Activity Compared to Surgical Wound Fluids. J Invest Dermatol (1996) 107:743–8. doi: 10.1111/1523-1747.ep12365637
117. Nwomeh BC, Liang H-XX, Cohen IKK, Yager DR. MMP-8 is the predominant collagenase in healing wounds and nonhealing ulcers. J Surg Res (1999) 81:189–95. doi: 10.1006/jsre.1998.5495
118. Lobmann R, Ambrosch A, Schultz G, Waldmann K, Schiweck S, Lehnert H. Expression of matrix-metalloproteinases and their inhibitors in the wounds of diabetic and non-diabetic patients. Diabetologia (2002) 45:1011–6. doi: 10.1007/s00125-002-0868-8
119. Diegelmann RF. Excessive neutrophils characterize chronic pressure ulcers. Wound Repair Regeneration (2003) 11:490–5. doi: 10.1046/j.1524-475X.2003.11617.x
120. Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci: J Virtual Library (2004) 9:283. doi: 10.2741/1184
121. Mast BA, Schultz GS. Interactions of cytokines, growth factors, and proteases in acute and chronic wounds. Wound Repair Regeneration (1996) 4:411–20. doi: 10.1046/j.1524-475X.1996.40404.x
122. Herrick S, Ashcroft G, Ireland G, Horan M, McCollum C, Ferguson M. Up-regulation of elastase in acute wounds of healthy aged humans and chronic venous leg ulcers are associated with matrix degradation. Lab Invest (1997) 77:281–8.
123. Wlaschek M, Scharffetter-Kochanek K. Oxidative stress in chronic venous leg ulcers. Wound Repair Regeneration (2005) 13:452–61. doi: 10.1111/j.1067-1927.2005.00065.x
124. Schafer M, Werner S. Oxidative stress in normal and impaired wound repair. Pharmacol Res (2008) 58:165–71. doi: 10.1016/j.phrs.2008.06.004
125. Sen CK. Wound healing essentials: Let there be oxygen. Wound Repair Regeneration (2009) 17:1–18. doi: 10.1111/j.1524-475X.2008.00436.x
126. Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, et al. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med (2015) 21:815–9. doi: 10.1038/nm.3887
127. Dunnill C, Patton T, Brennan J, Barrett J, Dryden M, Cooke J, et al. Reactive oxygen species (ROS) and wound healing: the functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int Wound J (2017) 14:89–96. doi: 10.1111/iwj.12557
128. Zamboni P, Izzo M, Tognazzo S, Carandina S, de Palma M, Catozzi L, et al. The overlapping of local iron overload and HFE mutation in venous leg ulcer pathogenesis. Free Radical Biol Med (2006) 40:1869–73. doi: 10.1016/j.freeradbiomed.2006.01.026
129. Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest (2011) 121:985–97. doi: 10.1172/JCI44490
130. Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, et al. Macrophage Dysfunction Impairs Resolution of Inflammation in the Wounds of Diabetic Mice. PloS One (2010) 5:e9539. doi: 10.1371/journal.pone.0009539
131. Klinkert K, Whelan D, Clover AJP, Leblond A-L, Kumar AHS, Caplice NM. Selective M2 Macrophage Depletion Leads to Prolonged Inflammation in Surgical Wounds. Eur Surg Res (2017) 58:109–20. doi: 10.1159/000451078
132. Trøstrup H, Holstein P, Christophersen L, Jørgensen B, Karlsmark T, Høiby N, et al. S100A8/A9 is an important host defence mediator in neuropathic foot ulcers in patients with type 2 diabetes mellitus. Arch Dermatol Res (2016) 308:347–55. doi: 10.1007/s00403-016-1646-7
133. Zhao G, Hochwalt PC, Usui ML, Underwood RA, Singh PK, James GA, et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regeneration (2010) 18:467–77. doi: 10.1111/j.1524-475X.2010.00608.x
134. Zhao G, Usui ML, Underwood RA, Singh PK, James GA, Stewart PS, et al. Time course study of delayed wound healing in a biofilm-challenged diabetic mouse model. Wound Repair Regeneration (2012) 20:342–52. doi: 10.1111/j.1524-475X.2012.00793.x
135. Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, et al. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PloS One (2011) 6:e27317. doi: 10.1371/journal.pone.0027317
136. Watters C, DeLeon K, Trivedi U, Griswold JA, Lyte M, Hampel KJ, et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol (2013) 202:131–41. doi: 10.1007/s00430-012-0277-7
137. Brandenburg KS, Calderon DF, Kierski PR, Czuprynski CJ, McAnulty JF. Novel murine model for delayed wound healing using a biological wound dressing with Pseudomonas aeruginosa biofilms. Microbial Pathogen (2018) 122:30–8. doi: 10.1016/j.micpath.2018.05.043
138. Kim JH, Yang B, Tedesco A, Lebig EGD, Ruegger PM, Xu K, et al. High Levels of Oxidative Stress and Skin Microbiome are Critical for Initiation and Development of Chronic Wounds in Diabetic Mice. Sci Rep (2019) 9:19318. doi: 10.1038/s41598-019-55644-3
139. Bjarnsholt T, Kirketerp-Møller K, Jensen PØ, Madsen KG, Phipps R, Krogfelt K, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regeneration (2008) 16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x
140. Trøstrup H, Laulund ASB, Moser C. Insights into Host–Pathogen Interactions in Biofilm-Infected Wounds Reveal Possibilities for New Treatment Strategies. Antibiotics (2020) 9:396. doi: 10.3390/antibiotics9070396
141. Kirker KR, James GA. In vitro studies evaluating the effects of biofilms on wound-healing cells: a review. APMIS (2017) 125:344–52. doi: 10.1111/apm.12678
142. Ramond E, Jamet A, Coureuil M, Charbit A. Pivotal Role of Mitochondria in Macrophage Response to Bacterial Pathogens. Front Immunol (2019) 10:2461. doi: 10.3389/fimmu.2019.02461
143. Yamada KJ, Kielian T. Biofilm-Leukocyte Cross-Talk: Impact on Immune Polarization and Immunometabolism. J Innate Immun (2019) 11:280–8. doi: 10.1159/000492680
144. Lai Y, di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3–dependent inflammation after skin injury. Nat Med (2009) 15:1377–82. doi: 10.1038/nm.2062
145. Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, et al. Commensal microbiota modulate gene expression in the skin. Microbiome (2018) 6:20. doi: 10.1186/s40168-018-0404-9
146. di Domizio J, Belkhodja C, Chenuet P, Fries A, Murray T, Mondéjar PM, et al. The commensal skin microbiota triggers type I IFN–dependent innate repair responses in injured skin. Nat Immunol (2020) 21:1034–45. doi: 10.1038/s41590-020-0721-6
147. Brandt SL, Klopfenstein N, Wang S, Winfree S, McCarthy BP, Territo PR, et al. Macrophage-derived LTB4 promotes abscess formation and clearance of Staphylococcus aureus skin infection in mice. PloS Pathog (2018) 14:e1007244. doi: 10.1371/journal.ppat.1007244
148. Brandt SL, Wang S, Dejani NN, Klopfenstein N, Winfree S, Filgueiras L, et al. Excessive localized leukotriene B4 levels dictate poor skin host defense in diabetic mice. JCI Insight (2018) 3(17):e120220. doi: 10.1172/jci.insight.120220
149. Na M, Mohammad M, Fei Y, Wang W, Holdfeldt A, Forsman H, et al. Lack of Receptor for Advanced Glycation End Products Leads to Less Severe Staphylococcal Skin Infection but More Skin Abscesses and Prolonged Wound Healing. J Infect Dis (2018) 218:791–800. doi: 10.1093/infdis/jiy007
150. Thurlow LR, Joshi GS, Richardson AR. Peroxisome Proliferator-Activated Receptor γ Is Essential for the Resolution of Staphylococcus aureus Skin Infections. Cell Host Microbe (2018) 24:261–70. doi: 10.1016/j.chom.2018.07.001
151. Tanaka KK, Kim SE, Yano H, Matsumoto G, Ohuchida R, Ishikura Y, et al. MiR-142 Is Required for Staphylococcus aureus Clearance at Skin Wound Sites via Small GTPase-Mediated Regulation of the Neutrophil Actin Cytoskeleton. J Invest Dermatol (2017) 137:931–40. doi: 10.1016/j.jid.2016.11.018
152. Feuerstein R, Seidl M, Prinz M, Henneke P. MyD88 in Macrophages Is Critical for Abscess Resolution in Staphylococcal Skin Infection. J Immunol (2015) 194:2735–45. doi: 10.4049/jimmunol.1402566
153. Li C, Li H, Jiang Z, Zhang T, Wang Y, Li Z, et al. Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin. PloS Pathog (2014) 10:e1003918. doi: 10.1371/journal.ppat.1003918
154. Cho JS, Guo Y, Ramos RI, Hebroni F, Plaisier SB, Xuan C, et al. Neutrophil-derived IL-1β Is Sufficient for Abscess Formation in Immunity against Staphylococcus aureus in Mice. PloS Pathog (2012) 8:e1003047. doi: 10.1371/journal.ppat.1003047
155. Kamen LA, Schlessinger J, Lowell CA. Pyk2 Is Required for Neutrophil Degranulation and Host Defense Responses to Bacterial Infection. J Immunol (2011) 186:1656–65. doi: 10.4049/jimmunol.1002093
156. Mahoney E, Reichner J, Robinson Bostom L, Mastrofrancesco B, Henry W, Albina J. Bacterial Colonization and the Expression of Inducible Nitric Oxide Synthase in Murine Wounds. Am J Pathol (2002) 161:2143–52. doi: 10.1016/S0002-9440(10)64492-6
157. Brubaker AL, Rendon JL, Ramirez L, Choudhry MA, Kovacs EJ. Reduced Neutrophil Chemotaxis and Infiltration Contributes to Delayed Resolution of Cutaneous Wound Infection with Advanced Age. J Immunol (2013) 190:1746–57. doi: 10.4049/jimmunol.1201213
158. Chen S, Li R, Cheng C, Xu J-Y, Jin C, Gao F, et al. Pseudomonas aeruginosa infection alters the macrophage phenotype switching process during wound healing in diabetic mice. Cell Biol Int (2018) 42:877–89. doi: 10.1002/cbin.10955
159. Nguyen KT, Seth AK, Hong SJ, Geringer MR, Xie P, Leung KP, et al. Deficient cytokine expression and neutrophil oxidative burst contribute to impaired cutaneous wound healing in diabetic, biofilm-containing chronic wounds. Wound Repair Regeneration (2013) 21:833–41. doi: 10.1111/wrr.12109
160. McInnes RL, Cullen BM, Hill KE, Price PE, Harding KG, Thomas DW, et al. Contrasting host immuno-inflammatory responses to bacterial challenge within venous and diabetic ulcers. Wound Repair Regeneration (2014) 22:58–69. doi: 10.1111/wrr.12133
161. Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen A, Andersen CB, Givskov M, et al. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Repair Regeneration (2011) 19:387–91. doi: 10.1111/j.1524-475X.2011.00681.x
162. Chan LC, Chaili S, Filler SG, Miller LS, Solis NV, Wang H, et al. Innate Immune Memory Contributes to Host Defense against Recurrent Skin and Skin Structure Infections Caused by Methicillin-Resistant Staphylococcus aureus. Infect Immun (2017) 85(2):e00876–16. doi: 10.1128/IAI.00876-16
163. Chan LC, Rossetti M, Miller LS, Filler SG, Johnson CW, Lee HK, et al. Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc Natl Acad Sci (2018) 115:E11111–9. doi: 10.1073/pnas.1808353115
164. McCaig L, McDonald L, Mandal S, Jernigan D. Staphylococcus aureus –associated Skin and Soft Tissue Infections in Ambulatory Care. Emerg Infect Dis (2006) 12:1715–23. doi: 10.3201/eid1211.060190
165. Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, et al. Methicillin-Resistant S. aureus Infections among Patients in the Emergency Department. New Engl J Med (2006) 355:666–74. doi: 10.1056/NEJMoa055356
166. Sivamani RK. Eicosanoids and Keratinocytes in Wound Healing. Adv Wound Care (2014) 3:476–81. doi: 10.1089/wound.2014.0523
167. Lohwasser C, Neureiter D, Weigle B, Kirchner T, Schuppan D. The Receptor for Advanced Glycation End Products Is Highly Expressed in the Skin and Upregulated by Advanced Glycation End Products and Tumor Necrosis Factor-Alpha. J Invest Dermatol (2006) 126:291–9. doi: 10.1038/sj.jid.5700070
168. Ramasamy R, Yan SF, Schmidt AM. The diverse ligand repertoire of the receptor for advanced glycation endproducts and pathways to the complications of diabetes. Vasc Pharmacol (2012) 57:160–7. doi: 10.1016/j.vph.2012.06.004
169. Mori R, Tanaka K, Shimokawa I. Identification and functional analysis of inflammation-related miRNAs in skin wound repair. Development Growth Differentiation (2018) 60:306–15. doi: 10.1111/dgd.12542
170. Xu Q, Choksi S, Qu J, Jang J, Choe M, Banfi B, et al. NADPH Oxidases Are Essential for Macrophage Differentiation. J Biol Chem (2016) 291:20030–41. doi: 10.1074/jbc.M116.731216
171. Guo Y, Lin C, Xu P, Wu S, Fu X, Xia W, et al. AGEs Induced Autophagy Impairs Cutaneous Wound Healing via Stimulating Macrophage Polarization to M1 in Diabetes. Sci Rep (2016) 6:36416. doi: 10.1038/srep36416
172. Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, et al. Chronic Wound Repair and Healing in Older Adults: Current Status and Future Research. J Am Geriatrics Soc (2015) 63:427–38. doi: 10.1111/jgs.13332
173. Swift ME, Kleinman HK, DiPietro LA. Impaired wound repair and delayed angiogenesis in aged mice. Lab Investigation J Tech Methods Pathol (1999) 79:1479–87.
174. Swift ME, Burns AL, Gray KL, DiPietro LA. Age-related alterations in the inflammatory response to dermal injury. J Invest Dermatol (2001) 117:1027–35. doi: 10.1046/j.0022-202x.2001.01539.x
175. Gosain A, DiPietro LA. Aging and Wound Healing. World J Surg (2004) 28:321–6. doi: 10.1007/s00268-003-7397-6
176. Gomez CR, Boehmer ED, Kovacs EJ. The aging innate immune system. Curr Opin Immunol (2005) 17:457–62. doi: 10.1016/j.coi.2005.07.013
177. Franceschi C, Bonafè M, Valensin S, Olivieri F, de luca M, Ottaviani E, et al. Inflamm-aging: An Evolutionary Perspective on Immunosenescence. Ann New Y Acad Sci (2006) 908:244–54. doi: 10.1111/j.1749-6632.2000.tb06651.x
178. Lu Q, Ceddia MA, Price EA, Ye S-M, Woods JA. Chronic exercise increases macrophage-mediated tumor cytolysis in young and old mice. Am J Physiol-Regulatory Integr Comp Physiol (1999) 276:R482–9. doi: 10.1152/ajpregu.1999.276.2.R482
179. Fulop T, Larbi A, Douziech N, Fortin C, Guérard K-P, Lesur O, et al. Signal transduction and functional changes in neutrophils with aging. Aging Cell (2004) 3:217–26. doi: 10.1111/j.1474-9728.2004.00110.x
180. Armstrong DG, Boulton AJM, Bus SA. Diabetic Foot Ulcers and Their Recurrence. New Engl J Med (2017) 376:2367–75. doi: 10.1056/nejmra1615439
181. Baltzis D, Eleftheriadou I, Veves A. Pathogenesis and treatment of impaired wound healing in diabetes mellitus: new insights. Adv Ther (2014) 31:817–36. doi: 10.1007/s12325-014-0140-x
182. Peppa M, Brem H, Ehrlich P, Zhang J-G, Cai W, Li Z, et al. Adverse Effects of Dietary Glycotoxins on Wound Healing in Genetically Diabetic Mice. Diabetes (2003) 52:2805–13. doi: 10.2337/diabetes.52.11.2805
183. Pradhan L, Nabzdyk C, Andersen ND, LoGerfo FW, Veves A. Inflammation and neuropeptides: the connection in diabetic wound healing. Expert Rev Mol Med (2009) 11:e2. doi: 10.1017/S1462399409000945
184. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and Adaptive Immune Memory: an Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe (2019) 25:13–26. doi: 10.1016/j.chom.2018.12.006
185. Wolcott RD, Rumbaugh KP, James G, Schultz G, Phillips P, Yang Q, et al. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care (2010) 19:320–8. doi: 10.12968/jowc.2010.19.8.77709
186. Yang C, Goss SG, Alcantara S, Schultz G, Lantis Ii JC. Effect of Negative Pressure Wound Therapy With Instillation on Bioburden in Chronically Infected Wounds. Wounds: Compendium Clin Res Pract (2017) 29:240–6.
187. Guoqi W, Zhirui L, Song W, Tongtong L, Lihai Z, Licheng Z, et al. Negative pressure wound therapy reduces the motility of Pseudomonas aeruginosa and enhances wound healing in a rabbit ear biofilm infection model. Antonie Van Leeuwenhoek (2018) 111:1557–70. doi: 10.1007/s10482-018-1045-5
188. Li Z, Yu Q, Wang S, Wang G, Li T, Tang P, et al. Impact of negative-pressure wound therapy on bacterial behaviour and bioburden in a contaminated full-thickness wound. Int Wound J (2019) 16:1214–21. doi: 10.1111/iwj.13197
189. Glass GE, Murphy GF, Esmaeili A, Lai L-MM, Nanchahal J. Systematic review of molecular mechanism of action of negative-pressure wound therapy. Br J Surg (2014) 101:1627–36. doi: 10.1002/bjs.9636
190. Borys S, Hohendorff J, Frankfurter C, Kiec-Wilk B, Malecki MT. Negative pressure wound therapy use in diabetic foot syndrome—from mechanisms of action to clinical practice. Eur J Clin Invest (2019) 49:e13067. doi: 10.1111/eci.13067
191. Karam RA, Rezk NA, Abdel Rahman TM. al Saeed M. Effect of negative pressure wound therapy on molecular markers in diabetic foot ulcers. Gene (2018) 667:56–61. doi: 10.1016/j.gene.2018.05.032
192. Wang T, Li X, Fan L, Chen B, Liu J, Tao Y, et al. Negative pressure wound therapy promoted wound healing by suppressing inflammation via down-regulating MAPK-JNK signaling pathway in diabetic foot patients. Diabetes Res Clin Pract (2019) 150:81–9. doi: 10.1016/j.diabres.2019.02.024
193. Hübner N-O, Kramer A. Review on the Efficacy, Safety and Clinical Applications of Polihexanide, a Modern Wound Antiseptic. Skin Pharmacol Physiol (2010) 23:17–27. doi: 10.1159/000318264
194. Koburger T, Hubner N-O, Braun M, Siebert J, Kramer A. Standardized comparison of antiseptic efficacy of triclosan, PVP-iodine, octenidine dihydrochloride, polyhexanide and chlorhexidine digluconate. J Antimicrobial Chemother (2010) 65:1712–9. doi: 10.1093/jac/dkq212
195. Sopata M, Kucharzewski M, Tomaszewska E. Antiseptic with modern wound dressings in the treatment of venous leg ulcers: Clinical and microbiological aspects. J Wound Care (2016) 25:419–26. doi: 10.12968/jowc.2016.25.8.419
196. Bigliardi PL, Alsagoff SAL, El-Kafrawi HY, Pyon J-K, Wa CTC, Villa MA. Povidone iodine in wound healing: A review of current concepts and practices. Int J Surg (2017) 44:260–8. doi: 10.1016/j.ijsu.2017.06.073
197. Gold MH, Andriessen A, Bhatia AC, Bitter P, Chilukuri S, Cohen JL, et al. Topical stabilized hypochlorous acid: The future gold standard for wound care and scar management in dermatologic and plastic surgery procedures. J Cosmetic Dermatol (2020) 19:270–7. doi: 10.1111/jocd.13280
198. Leaper D, Assadian O, Edmiston CE. Approach to chronic wound infections. Br J Dermatol (2015) 173:351–8. doi: 10.1111/bjd.13677
199. Borkow G, Gabbay J, Dardik R, Eidelman AI, Lavie Y, Grunfeld Y, et al. Molecular mechanisms of enhanced wound healing by copper oxide-impregnated dressings. Wound Repair Regeneration (2010) 18:266–75. doi: 10.1111/j.1524-475X.2010.00573.x
200. Wilkinson LJ, White RJ, Chipman JK. Silver and nanoparticles of silver in wound dressings: A review of efficacy and safety. J Wound Care (2011) 20:543–9. doi: 10.12968/jowc.2011.20.11.543
201. Rodriguez-Arguello J, Lienhard K, Patel P, Geransar R, Somayaji R, Parsons L, et al. A Scoping Review of the Use of Silver-impregnated Dressings for the Treatment of Chronic Wounds. Ostomy Wound Manage (2018) 64:14–31. doi: 10.25270/owm.2018.3.1431
202. Schwarzer S, James GA, Goeres D, Bjarnsholt T, Vickery K, Percival SL, et al. The efficacy of topical agents used in wounds for managing chronic biofilm infections: A systematic review. J Infect (2020) 80:261–70. doi: 10.1016/j.jinf.2019.12.017
203. Nikolić N, Kienzl P, Tajpara P, Vierhapper M, Matiasek J, Elbe-Bürger A. The Antiseptic Octenidine Inhibits Langerhans Cell Activation and Modulates Cytokine Expression upon Superficial Wounding with Tape Stripping. J Immunol Res (2019) 2019:1–11. doi: 10.1155/2019/5143635
204. Pavlík V, Sojka M, Mazúrová M, Velebný V. Dual role of iodine, silver, chlorhexidine and octenidine as antimicrobial and antiprotease agents. PloS One (2019) 14:e0211055. doi: 10.1371/journal.pone.0211055
205. Tzaneva V, Mladenova I, Todorova G, Petkov D. Antibiotic treatment and resistance in chronic wounds of vascular origin. Med Pharm Rep (2016) 89:365–70. doi: 10.15386/cjmed-647
206. Hernandez R. The use of systemic antibiotics in the treatment of chronic wounds. Dermatol Ther (2006) 19:326–37. doi: 10.1111/j.1529-8019.2006.00091.x
207. Lipsky BA, Hoey C. Topical Antimicrobial Therapy for Treating Chronic Wounds. Clin Infect Dis (2009) 49:1541–9. doi: 10.1086/644732
208. Cowling T, Jones S. Topical Antibiotics for Infected Wounds: A Review of the Clinical Effectiveness and Guidelines (2017). Available at: https://www.ncbi.nlm.nih.gov/books/NBK487400/ [Accessed December 29, 2020])
209. Boyko TV, Longaker MT, Yang GP. Review of the Current Management of Pressure Ulcers. Adv Wound Care (2018) 7:57–67. doi: 10.1089/wound.2016.0697
210. Ramirez-Acuña JM, Cardenas-Cadena SA, Marquez-Salas PA, Garza-Veloz I, Perez-Favila A, Cid-Baez MA, et al. Diabetic Foot Ulcers: Current Advances in Antimicrobial Therapies and Emerging Treatments. Antibiotics (2019) 8:193. doi: 10.3390/antibiotics8040193
211. Lipsky BA, Berendt AR, Cornia PB, Pile JC, Peters EJG, Armstrong DG, et al. Infectious Diseases Society of America Clinical Practice Guideline for the Diagnosis and Treatment of Diabetic Foot Infectionsa. Clin Infect Dis (2012) 2012) 54:e132–73. doi: 10.1093/cid/cis346
212. Rao N, Ziran BH, Lipsky BA. Treating Osteomyelitis: Antibiotics and Surgery. Plast Reconstruct Surg (2011) 127:177S–87S. doi: 10.1097/PRS.0b013e3182001f0f
213. Shettigar K, Bhat DV, Satyamoorthy K, Murali TS. Severity of drug resistance and co-existence of Enterococcus faecalis in diabetic foot ulcer infections. Folia Microbiol (2018) 63:115–22. doi: 10.1007/s12223-017-0547-2
214. Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, et al. Extracellular DNA Shields against Aminoglycosides in Pseudomonas aeruginosa Biofilms. Antimicrobial Agents Chemother (2013) 57:2352–61. doi: 10.1128/AAC.00001-13
215. Germoni LAP, Bremer PJ, Lamont IL. The effect of alginate lyase on the gentamicin resistance of Pseudomonas aeruginosa in mucoid biofilms. J Appl Microbiol (2016) 121:126–35. doi: 10.1111/jam.13153
216. Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discovery (2012) 11:37–51. doi: 10.1038/nrd3591
217. Luong HX, Thanh TT, Tran TH. Antimicrobial peptides – Advances in development of therapeutic applications. Life Sci (2020) 260:118407. doi: 10.1016/j.lfs.2020.118407
218. Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ. Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discovery (2020) 19:311–32. doi: 10.1038/s41573-019-0058-8
219. Mangoni ML, McDermott AM, Zasloff M. Antimicrobial peptides and wound healing: biological and therapeutic considerations. Exp Dermatol (2016) 25:167–73. doi: 10.1111/exd.12929
220. Strempel N, Strehmel J, Overhage J. Potential Application of Antimicrobial Peptides in the Treatment of Bacterial Biofilm Infections. Curr Pharm Design (2014) 21:67–84. doi: 10.2174/1381612820666140905124312
221. Eckert RL, Broome A-M, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol (2004) 123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x
222. Nakashige TG, Zhang B, Krebs C, Nolan EM. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol (2015) 11:765–71. doi: 10.1038/nchembio.1891
223. Schröder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci (2006) 63:469–86. doi: 10.1007/s00018-005-5364-0
224. Overhage J, Campisano A, Bains M, Torfs ECW, Rehm BHA, Hancock REW. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun (2008) 76:4176–82. doi: 10.1128/IAI.00318-08
225. Semple F, MacPherson H, Webb S, Cox SL, Mallin LJ, Tyrrell C, et al. Human β-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol (2011) 41:3291–300. doi: 10.1002/eji.201141648
226. Semple F, Webb S, Li H-N, Patel HB, Perretti M, Jackson IJ, et al. Human β-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol (2010) 40:1073–8. doi: 10.1002/eji.200940041
227. Jin G, Kawsar HI, Hirsch SA, Zeng C, Jia X, Feng Z, et al. An Antimicrobial Peptide Regulates Tumor-Associated Macrophage Trafficking via the Chemokine Receptor CCR2, a Model for Tumorigenesis. PloS One (2010) 5:e10993. doi: 10.1371/journal.pone.0010993
228. Ramos R, Silva JP, Rodrigues AC, Costa R, Guardão L, Schmitt F, et al. Wound healing activity of the human antimicrobial peptide LL37. Peptides (2011) 32:1469–76. doi: 10.1016/j.peptides.2011.06.005
229. Duplantier AJ, van Hoek ML. The Human Cathelicidin Antimicrobial Peptide LL-37 as a Potential Treatment for Polymicrobial Infected Wounds. Front Immunol (2013) 4:143. doi: 10.3389/fimmu.2013.00143
230. Tokumaru S, Sayama K, Shirakata Y, Komatsuzawa H, Ouhara K, Hanakawa Y, et al. Induction of Keratinocyte Migration via Transactivation of the Epidermal Growth Factor Receptor by the Antimicrobial Peptide LL-37. J Immunol (2005) 175:4662–8. doi: 10.4049/jimmunol.175.7.4662
231. Koczulla R, von Degenfeld G, Kupatt C, Krötz F, Zahler S, Gloe T, et al. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J Clin Invest (2003) 111:1665–72. doi: 10.1172/JCI17545
232. Felgueiras HP, Teixeira MA, Tavares TD, Homem NC, Zille A, Amorim MTP. Antimicrobial action and clotting time of thin, hydrated poly(vinyl alcohol)/cellulose acetate films functionalized with LL37 for prospective wound-healing applications. J Appl Polymer Sci (2020) 137:48626. doi: 10.1002/app.48626
233. Fumakia M, Ho EA. Nanoparticles Encapsulated with LL37 and Serpin A1 Promotes Wound Healing and Synergistically Enhances Antibacterial Activity. Mol Pharmaceut (2016) 13:2318–31. doi: 10.1021/acs.molpharmaceut.6b00099
234. Bolatchiev A, Baturin V, Bazikov I, Maltsev A, Kunitsina E. Effect of antimicrobial peptides HNP-1 and hBD-1 on Staphylococcus aureus strains in vitro and in vivo. Fundam Clin Pharmacol (2020) 34:102–8. doi: 10.1111/fcp.12499
235. Garcia-Orue I, Gainza G, Girbau C, Alonso R, Aguirre JJ, Pedraz JL, et al. LL37 loaded nanostructured lipid carriers (NLC): A new strategy for the topical treatment of chronic wounds. Eur J Pharmaceut Biopharmaceut (2016) 108:310–6. doi: 10.1016/j.ejpb.2016.04.006
236. Kang HK, Seo CH, Luchian T, Park Y. Pse-T2, an Antimicrobial Peptide with High-Level, Broad-Spectrum Antimicrobial Potency and Skin Biocompatibility against Multidrug-Resistant Pseudomonas aeruginosa Infection. Antimicrobial Agents Chemother (2018) 62(12):e01493–18. doi: 10.1128/AAC.01493-18
237. Ostorhazi E, Voros E, Nemes-Nikodem E, Pinter D, Sillo P, Mayer B, et al. Rapid systemic and local treatments with the antibacterial peptide dimer A3-APO and its monomeric metabolite eliminate bacteria and reduce inflammation in intradermal lesions infected with Propionibacterium acnes and meticillin-resistant Staphylococcus au. Int J Antimicrobial Agents (2013) 42:537–43. doi: 10.1016/j.ijantimicag.2013.08.001
238. Chee PY, Mang M, Lau ES, Tan LT-H, He Y-W, Lee W-L, et al. Epinecidin-1, an Antimicrobial Peptide Derived From Grouper (Epinephelus coioides): Pharmacological Activities and Applications. Front Microbiol (2019) 10:2631:2631. doi: 10.3389/fmicb.2019.02631
239. Huang HN, Rajanbabu V, Pan CY, Chan YL, Wu CJ, Chen JY. Use of the antimicrobial peptide Epinecidin-1 to protect against MRSA infection in mice with skin injuries. Biomaterials (2013) 34:10319–27. doi: 10.1016/j.biomaterials.2013.09.037
240. Kostarnoy AV, Gancheva PG, Logunov DY, Verkhovskaya LV, Bobrov MA, Scheblyakov DV, et al. Topical Bacterial Lipopolysaccharide Application Affects Inflammatory Response and Promotes Wound Healing. J Interferon Cytokine Res (2013) 33:514–22. doi: 10.1089/jir.2012.0108
241. Sahana TG, Rekha PD. A novel exopolysaccharide from marine bacterium Pantoea sp. YU16-S3 accelerates cutaneous wound healing through Wnt/β-catenin pathway. Carbohydr Polymers (2020) 238:116191. doi: 10.1016/j.carbpol.2020.116191
242. Imegwu O, Chang TH, Steinberg JJ, Levenson SM. Staphylococcus aureus peptidoglycan ameliorates cyclophosphamide-induced impairment of wound healing. Wound Repair Regeneration (1997) 5:364–72. doi: 10.1046/j.1460-9568.1997.50411.x
243. Kilcullen JK, Ly QP, Chang TH, Levenson SM, Steinberg JJ. Nonviable Staphylococcus aureus and its peptidoglycan stimulate macrophage recruitment, angiogenesis, fibroplasia, and collagen accumulation in wounded rats. Wound Repair Regeneration (1998) 6:149–56. doi: 10.1046/j.1524-475X.1998.60209.x
244. Yu B, Wang Z, Almutairi L, Huang S, Kim M-H. Harnessing iron-oxide nanoparticles towards the improved bactericidal activity of macrophage against Staphylococcus aureus. Nanomed: Nanotechnol Biol Med (2020) 24:102158. doi: 10.1016/j.nano.2020.102158
245. Okumura CYM, Hollands A, Tran DN, Olson J, Dahesh S, von Köckritz-Blickwede M, et al. A new pharmacological agent (AKB-4924) stabilizes hypoxia inducible factor-1 (HIF-1) and increases skin innate defenses against bacterial infection. J Mol Med (2012) 90:1079–89. doi: 10.1007/s00109-012-0882-3
246. Song XR, Fox F, Gallo MA, Rosenberg A, Jordan R, Shealy D, et al. Effects of 2 Different Anti–Tumor Necrosis Factor–α Agents in a Primate Model of Subcutaneous Abscess Formation. J Infect Dis (2002) 185:204–13. doi: 10.1086/338451
247. Vågesjö E, Öhnstedt E, Mortier A, Lofton H, Huss F, Proost P, et al. Accelerated wound healing in mice by on-site production and delivery of CXCL12 by transformed lactic acid bacteria. Proc Natl Acad Sci (2018) 115:1895–900. doi: 10.1073/pnas.1716580115
248. Jiang L, Dai Y, Cui F, Pan Y, Zhang H, Xiao J, et al. Expression of cytokines, growth factors and apoptosis-related signal molecules in chronic pressure ulcer wounds healing. Spinal Cord (2014) 52:145–51. doi: 10.1038/sc.2013.132
249. García-Ojalvo A, Berlanga Acosta J, Figueroa-Martínez A, Béquet-Romero M, Mendoza-Marí Y, Fernández-Mayola M, et al. Systemic translation of locally infiltrated epidermal growth factor in diabetic lower extremity wounds. Int Wound J (2019) 16:1294–303. doi: 10.1111/iwj.13189
250. Bai H, Kyu-Cheol N, Wang Z, Cui Y, Liu H, Liu H, et al. Regulation of inflammatory microenvironment using a self-healing hydrogel loaded with BM-MSCs for advanced wound healing in rat diabetic foot ulcers. J Tissue Eng (2020) 11:204173142094724. doi: 10.1177/2041731420947242
251. Game F, Jeffcoate W, Tarnow L, Jacobsen JL, Whitham DJ, Harrison EF, et al. LeucoPatch system for the management of hard-to-heal diabetic foot ulcers in the UK, Denmark, and Sweden: an observer-masked, randomised controlled trial. Lancet Diabetes Endocrinol (2018) 6:870–8. doi: 10.1016/S2213-8587(18)30240-7
252. da Silva LP, Santos TC, Rodrigues DB, Pirraco RP, Cerqueira MT, Reis RL, et al. Stem Cell-Containing Hyaluronic Acid-Based Spongy Hydrogels for Integrated Diabetic Wound Healing. J Invest Dermatol (2017) 137:1541–51. doi: 10.1016/j.jid.2017.02.976
253. Raghuram AC, Yu RP, Lo AY, Sung CJ, Bircan M, Thompson HJ, et al. Role of stem cell therapies in treating chronic wounds: A systematic review. World J Stem Cells (2020) 12:659–75. doi: 10.4252/wjsc.v12.i7.659
254. Mulholland EJ, Dunne N, McCarthy HO. MicroRNA as Therapeutic Targets for Chronic Wound Healing. Mol Ther Nucleic Acids (2017) 8:46–55. doi: 10.1016/j.omtn.2017.06.003
255. Saleh B, Dhaliwal HK, Portillo-Lara R, Shirzaei Sani E, Abdi R, Amiji MM, et al. Local Immunomodulation Using an Adhesive Hydrogel Loaded with miRNA-Laden Nanoparticles Promotes Wound Healing. Small (2019) 15:1902232. doi: 10.1002/smll.201902232
256. Yager DR, Kulina RA, Gilman LA. Wound Fluids: A Window Into the Wound Environment? Int J Lower Extremity Wounds (2007) 6:262–72. doi: 10.1177/1534734607307035
257. Voegeli D, Lwaleed B. Back to basics: Histological, microbiological and biochemical sampling in wound care. J Wound Care (2013) 22:650–ssss654. doi: 10.12968/jowc.2013.22.11.650
258. Ramsay S, Cowan L, Davidson JM, Nanney L, Schultz G. Wound samples: moving towards a standardised method of collection and analysis. Int Wound J (2016) 13:880–91. doi: 10.1111/iwj.12399
259. Frykberg RG, Banks J. Challenges in the Treatment of Chronic Wounds. Adv Wound Care (2015) 4:560–82. doi: 10.1089/wound.2015.0635
Keywords: chronic wound, delayed healing, innate immune responses, inflammation, biofilm, host-pathogen interaction, skin microbiome
Citation: Versey Z, da Cruz Nizer WS, Russell E, Zigic S, DeZeeuw KG, Marek JE, Overhage J and Cassol E (2021) Biofilm-Innate Immune Interface: Contribution to Chronic Wound Formation. Front. Immunol. 12:648554. doi: 10.3389/fimmu.2021.648554
Received: 31 December 2020; Accepted: 17 March 2021;
Published: 09 April 2021.
Edited by:
Semih Esin, University of Pisa, ItalyReviewed by:
Giuseppantonio Maisetta, University of Pisa, ItalyGuy Cohen, Dead Sea and Arava Science Center, Israel
Claus Moser, Rigshospitalet, Denmark
Copyright © 2021 Versey, da Cruz Nizer, Russell, Zigic, DeZeeuw, Marek, Overhage and Cassol. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edana Cassol, edana.cassol@carleton.ca
 Zoya Versey
Zoya Versey Waleska Stephanie da Cruz Nizer
Waleska Stephanie da Cruz Nizer Emily Russell1
Emily Russell1 Sandra Zigic
Sandra Zigic Jonah E. Marek
Jonah E. Marek Joerg Overhage
Joerg Overhage Edana Cassol
Edana Cassol