Avicennia Genetic Diversity and Fine-Scaled Structure Influenced by Coastal Proximity of Mangrove Fragments
- 1Ecology and Biodiversity Research Group, Department of Biology, Vrije Universiteit Brussel, Brussels, Belgium
- 2Marine Biology Unit, College of Aquatic and Applied Sciences, Southern Leyte State University, Southern Leyte, Philippines
- 3Romblon State University, Romblon, Philippines
- 4Department of Biological Sciences, Visayas State University, Baybay, Philippines
Avicennia dominated mangrove forests occur from seaward to landward sites and hence are subject to different dynamics within estuarine ecosystems. Regeneration of mangrove forests primarily depends on the extent of propagule spread and subsequent establishment in suitable habitats. The complex nature of estuarine systems induces a wide variety of local conditions for within-site propagule retention and settlement thereby allowing spontaneous regeneration of mangroves. In this study, we estimated the fine-scale spatial genetic structure (FSGS) of Avicennia populations and examined whether their position relative to the seaside or the size of mangrove patches could have influenced the extant local population genetic structure. A kinship-based FSGS was performed using microsatellite markers in 523 A. marina, 189 A. rumphiana and 60 A. alba adult trees of 24 sites in The Philippines. Transects within each estuary were taken both parallel and perpendicular to the coastline or tidal river edge. The extent of local mangrove areas and various human-induced encroachments as such did not show any trend in allele diversity, heterozygosity values or inbreeding levels. However, farther inland situated mangrove patches showed a larger FSGS extent across the neighborhood (up to 75 m) though less diversity along with inbreeding, most likely due to retention of related propagules and lowered chance of external propagule input. Estimation of connectivity along a same coastline stretch supported a unidirectional steppingstone or adjacent migration model for populations of either A. marina, A. alba or A. rumphiana. These were congruent with ocean currents across mangrove estuaries of the Tablas Strait and along Western Leyte, thereby emphasizing the relevance of coastal connectivity for long term persistence. From this study, we conclude that both proximity to open water and narrowness of mangrove patches may affect their captured diversity, inbreeding and fine-scale structure caused by propagule movement within or beyond a local mangrove fragment during recent generations. Higher levels of allele diversity for seaward sites and highest likelihood of migration for adjacent mangroves both add to the importance of coastal connectivity that is the only natural cohesive force on longer term and necessary to counteract short term effects of increasingly encroached mangrove environments.
Introduction
Mangrove ecosystems support a wide variety of aquatic species (Gillanders, 2002; Lefcheck et al., 2019) and provide habitat connectivity to nearby coastal ecosystems of seagrass beds and coral reefs (Nagelkerken et al., 2008). Aside from habitat provisions, mangrove ecosystems also provide abundance of food sources including mangrove litters to aquatic organisms (Lee et al., 2014). Mangrove areas play an important role in the protection of shoreline communities against strong impact of waves, wind, storm and other calamities (Giri et al., 2011; Gedan et al., 2011). A protective capacity of mangrove forests appears strongly correlated with an overall quality of the mangrove vegetation such as forest density, vegetation size and type, diameter of stem and roots and soil elevation (Dahdouh-Guebas et al., 2005; Quartel et al., 2007; Alongi, 2008) to allow vertical accretion by accelerating sedimentation rates, trapping of sediments and direct organic input (Lee et al., 2014).
Despite these benefits, the mangrove ecosystem is encroached by human activities (Richards and Friess, 2016; Bryan-Brown et al., 2020). It was estimated that more than 50% of the original mangrove forests has been destroyed (Feller et al., 2010). Loss was primarily caused by urbanization, conversion of mangrove habitats into commercial and residential areas, considered as an economical form of coastal development. In addition, aquaculture activities have led to mangrove forest loss and fragmentation with shrimp farms alone accounting for a largest mangrove habitat loss globally (Ellison, 2008). Mangrove forest cover worldwide became fragmented or degraded due to over-exploitation for fuelwood and timber production (Valiela et al., 2001), pollution events such as oil spills (Burns et al., 1993), agricultural catchment runoff (Duke et al., 2005), extreme sedimentation (Ellison et al., 1999), and alteration of hydrological regimes (Gordon, 1988). Habitat fragmentation intrinsically encompasses population fragmentation, and potential loss of genetic diversity of the mangrove tree species (Giri et al., 2011; Wee et al., 2015; Hasan et al., 2018). Though continuous conservation efforts have been done, the recovery success of replanted mangrove trees appeared inadequate because factors such as changes in hydrodynamics, salt content, acidity and levels of nutrients may have prevented successful regeneration of mangroves (Sandilyan and Kathiresan, 2012). Therefore, understanding the extant natural processes of spontaneous dispersal, settlement and persistence of mangrove areas in fragmented patches can be helpful for management and conservation priority settings (Ngeve et al., 2017).
Mangrove propagules have no dormant stage and dispersal of propagules are influenced by factors including buoyancy, propagule viability and timely establishment (Rabinowitz, 1978). Population persistence thus exclusively depends on propagule formation, release, distribution and establishment, as clonal growth or vegetative dispersal is absent in mangroves. Tidal influence, ocean currents and wind action predict potential dispersal patterns and colonization of mangrove species over long oceanic distances (Clarke, 1993; Van der Stocken et al., 2015, 2019b). Many studies illustrated a long-distance connectivity of mangrove trees in relation to oceanic currents and directionality especially along coastlines (Mori et al., 2015; De Ryck et al., 2016; Ngeve et al., 2017; Hodel et al., 2018; Van der Stocken et al., 2019a). A steppingstone model of migration between estuaries was often obtained for Avicennia L. species (Do et al., 2019; Wee et al., 2020; Triest et al., 2021b). This adds to the importance of coastal connectivity through gene flow (i.e., propagule flow) that is the only natural cohesive force between estuaries on longer term for a species to maintain its evolutionary units. Barriers to genetic connectivity however may come from land masses and different migration histories (Triest, 2008; Hodel et al., 2018; Wee et al., 2020; Triest et al., 2021a), opposite ocean currents (Mori et al., 2015; Ngeve et al., 2017) or very large rivers (Triest et al., 2018). Overall, the expectation that migration routes followed major oceanic and coastal currents is largely confirmed from genetic diversity and genetic structure approaches at population level.
However, connectivity patterns of spontaneous mangrove establishment at local and fine scale level within an estuary and different habitats thereof, is more complicated. Estuarine landscapes are very diverse and unique in their complexity such that establishment of mangrove propagules depend on suitable habitats resulting from sedimentation patterns of coastal and major river systems, channels or creeks and sandbar dunes. Tidal currents and wind action allow mangrove propagules to disperse and settle in various habitats that range from most seaward protruding open systems, over mudflat areas, along dynamic riverbank systems and up to farthest inland sheltered systems that seldom experience high tide or spring tide. Spontaneous processes of propagule dispersal and formation of a mangrove vegetation are expected to result in a different neighborhood size of individual trees in e.g., exposed seaward versus sheltered landward positioned populations (Triest and Van der Stocken, 2021), depending on river flow (Ngeve et al., 2017; Chablé Iuit et al., 2020), channel structures (Triest et al., 2020), or degree of fragmentation (Hasan et al., 2018). For example, Rhizophora mangle L. along a river showed a fine-scaled genetic structure that did became extended (Chablé Iuit et al., 2020) most likely from onward carried-away propagules alongside the bank.
Fine-scaled genetic structure of populations thus can be indicative of establishment events that happened during most recent generations and therefore, a comparison of mangrove patches in different positions may help in understanding whether local sheltered conditions effectively did promote internal dispersal and allowed cohorts of related propagules to settle within a neighborhood, still detectable from their genetic relatedness (Triest and Van der Stocken, 2021). Alternatively, absence of any fine-scale structure points at a more dynamic system within which mangroves of mixed kinship or unrelatedness became settled. The complexity of hydrodynamics in an estuarine system induces a wide variety of conditions for propagule retention and establishment that allow a mangrove zonation. Avicennia species are among the first to colonize new areas and are fast-growing. Avicennia marina (Forsk.) Vierh. dominated mangrove forests may occur from seaward to landward sites and hence their populations are subject to very different dynamics typical for estuarine ecosystems (Dahdouh-Guebas et al., 2004). Their zonal regeneration primarily depends on the extent of propagule spread and on subsequent establishment in suitable habitats either within close vicinity or beyond. Suffice prior knowledge of polymorphic genetic markers is available of the geographically widespread A. marina and related species (Triest, 2008) to allow resolution within an estuary and at local fine-scale level (Hasan et al.,2018; Do et al., 2019; Triest et al., 2020; Triest and Van der Stocken, 2021).
In this study, at local scale, we test the hypothesis that persistence of Avicennia trees is influenced by their positioning in coastal mangroves. These may show gradients of oceanic influences, from strongly exposed coastal-protruding into open sea-toward far pushed inland estuaries, barely subject to tidal flood. Dispersal and establishment that happened in past few-and overlapping-generations along this gradient of different tidal and sea currents, could have left traces in the locally captured amount of genetic diversity and in the fine-scale spatial genetic structure. We specifically aim to (1) analyze the genetic diversity of Avicennia populations located at different distances from the open sea or tidal influence; (2) estimate and compare the extent of a fine-scale spatial genetic structure of these differently positioned population fragments. Additionally (3), at a regional scale (up to 115 km coastline) we estimate the likelihood of different migration models between populations along a same coastline for each Avicennia species separately and test for influence of coastal currents.
We considered the fine-scale spatial genetic structure of pioneer mangrove species A. marina, A. rumphiana Hallier f. and A. alba Blume, using a linear transect approach in 24 sites of The Philippines along coastlines of Tablas Strait and Western Leyte (Eastern Visayas), representing a gradient of strongly exposed protruding mangroves up to the sheltered landward edge of the estuary. Through this study, short term processes of these populations can be assessed which may then be used in protection priorities and rehabilitation of fragmented mangrove areas.
Materials and Methods
Study Area
For the purpose of this study, data were collected from 24 samples of mangrove populations located in The Philippines (Figure 1) with different characteristics of landward-seaward position, tidal influence, parallel or perpendicular to the coastal or estuarine river edge (Table 1), fragment sizes and of rural encroachment from roads, houses, deforestation and aquaculture (Supplementary Table 1). More specifically, eight populations were alongside Tablas Strait on Oriental Mindoro (Verde Passage, over 8 km) and Tablas Island (Romblon, over 7 km) where ocean currents are directed from East to West although circulation patterns very near to the coastline can be weaker. Another 16 populations were from Western Leyte Island (ca. 115 km) where both ocean and near coast currents are albeit weak and mostly from South to North (May et al., 2011). The ocean current that gets through the Western Leyte study areas partly comes from the North of Leyte through the San Bernardino Strait (influenced by Kuroshio current), whereas ocean currents coming along the Southern part of Leyte through the Surigao Strait (influenced by Northern Equatorial Current) may also affect coastal sites of Western Leyte.
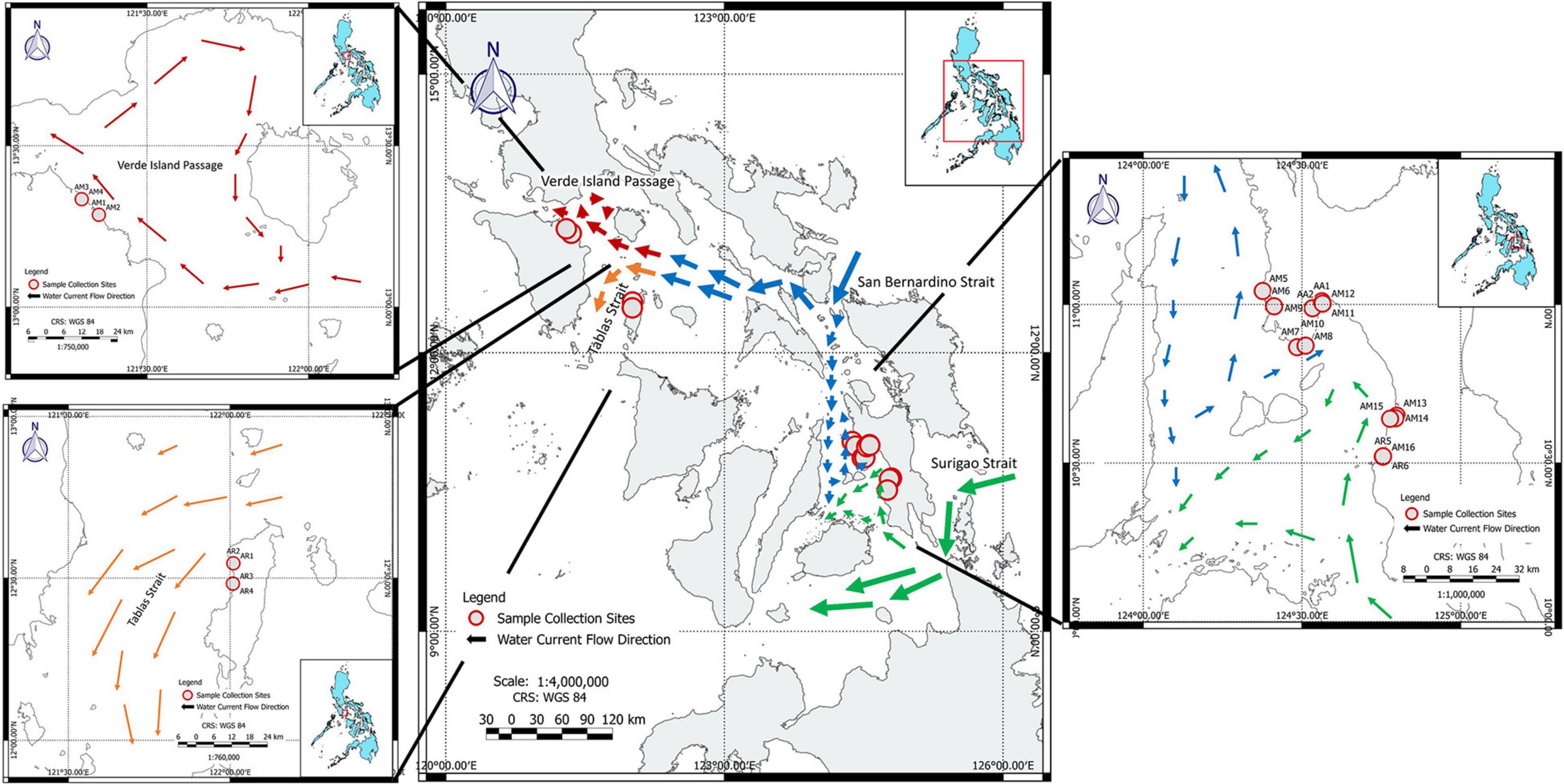
Figure 1. Overview of the studied areas in The Philippines with detailed maps of sampled Avicennia marina (AM), A. rumphiana (AR), and A. alba (AA) collection sites. Population codes are denoted as in Table 1 and in Supplementary Figure 1 with images of all estuaries. Principal ocean currents of the study area are indicated by different arrow lengths, conceptually indicating the strength of currents. Note the weak ocean currents of Western Leyte (Modified from May et al., 2011).
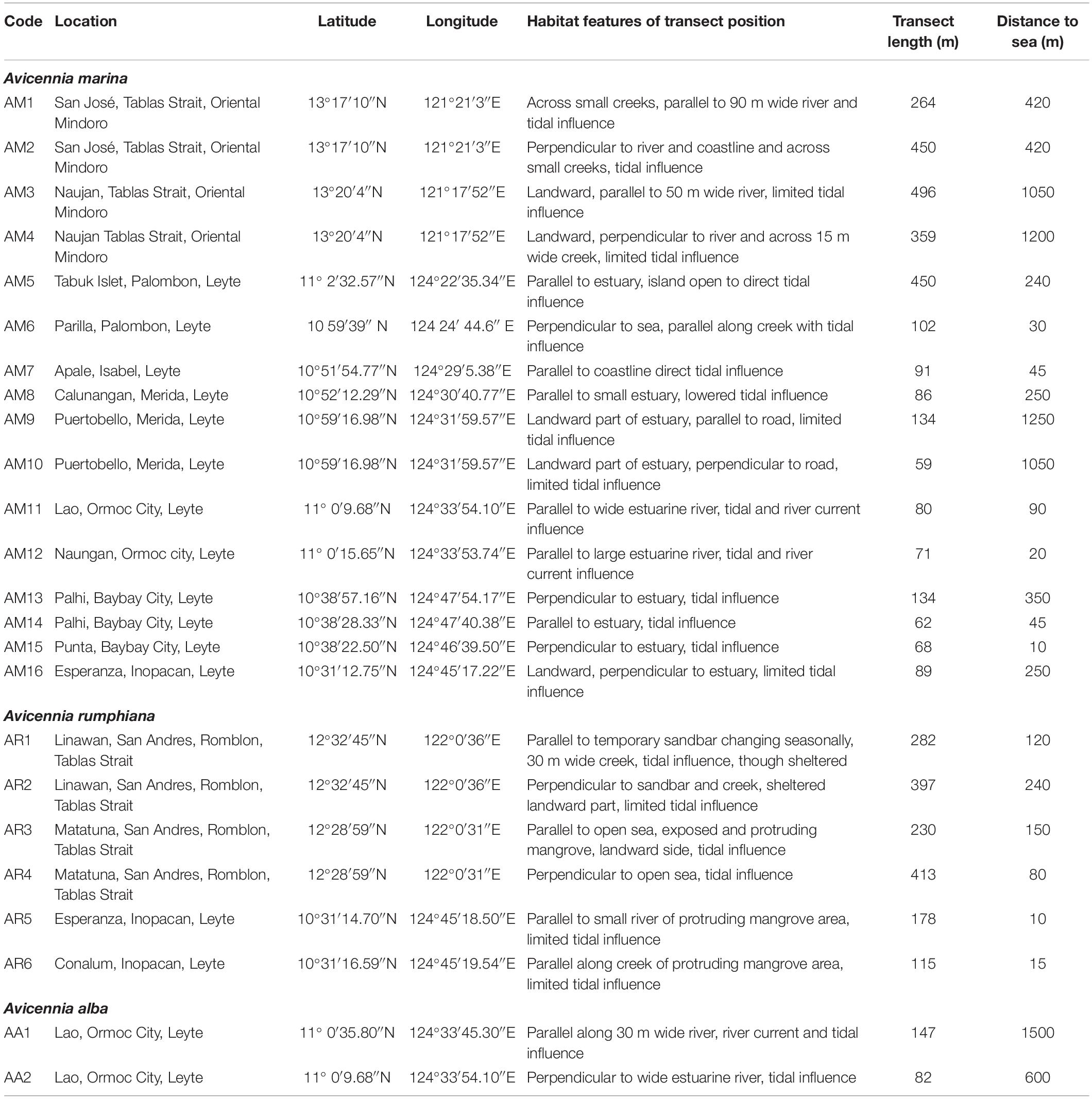
Table 1. Location details of twenty-four Avicennia sites in estuaries along Tablas Strait and Western Leyte (Philippines).
Study Species and Sample Collection
Avicennia marina is the most widely distributed of all mangrove species, common throughout its range and found across the Indo-Pacific (Tomlinson, 2016). It grows both in the lowest and highest portions in the intertidal zone but is generally rare in the mid-intertidal areas. A. marina is shade tolerant and its saplings are often associated with gaps in the forest canopy. A. marina is a pioneer species on newly formed mudflats with a high proportion of sand and has a high tolerance to hypersaline conditions. It is a hardy species in natural conditions and regenerates quickly from coppices. It can grow at a salinity of range of 0-30 ppt (Robertson and Alongi, 1992). A. marina has aerial roots-pneumatophores-that grow up from lateral roots and can form a dense mat that extends many meters from the main stem of the plant (Kathiresan and Bingham, 2001). A. marina has crypto-viviparous propagules that do not enlarge sufficiently to rupture the pericarp while attached to the parent. The propagule is buoyant and upon release it is dispersed in the seawater. In the studied area, this mangrove species is very common. A total of 523 A. marina individual trees were sampled in sixteen locations (Figure 1, Table 1, and Supplementary Table 1). Avicennia rumphiana is endemic to south east Asia (Tomlinson, 2016). Avicennia rumphiana is one of the first to colonize new areas and fast-growing. The IUCN Red List of Threatened Species considers A. rumphiana with a vulnerable status because it has a patchy distribution, is uncommon in some areas and is in general decline. It grows in the upper part of the intertidal zone where it is most vulnerable to human activities and habitat destruction. A total of 189 A. rumphiana individual trees were sampled in six locations (Figure 1, Table 1, and Supplementary Table 1). Avicennia alba is very common and widespread in Southeast Asia, the islands of the South Pacific Ocean, and Australia. It is a pioneering species (Tomlinson, 2016). A total of 60 A. alba trees were sampled in two locations of a same large estuary solely for comparison with A. marina in that same estuary (Figure 1, Table 1, and Supplementary Table 1).
Transects were taken both parallel and perpendicular to the coastline or estuarine river as replicates for capturing variability of the local environmental setting. The 24 transect locations were in a variety of estuarine environments though the majority was encroached by human activities. Transect lengths ranged from 59 to 496 m depending on the narrowness of the mangrove plot (Table 1). Distances were taken between each consecutive individual in each transect. The number of sampled trees was 30 on average per transect though ranged exceptionally from 15 to 52 for few A. rumphiana sites (Table 2). Shortest distance intervals between neighboring trees were considered and mostly were within less than 10 m. Eventual gaps between mangrove patches were included in the total distance because a fine-scaled analysis focuses on pairs of individual trees within distance classes below 100 m, regardless the patch where they occur. At regional level, this sampling resulted in 16 A. marina, 6 A. rumphiana and 2 A. alba populations that allowed for an intra-species analysis along each specific stretch of island coastline.
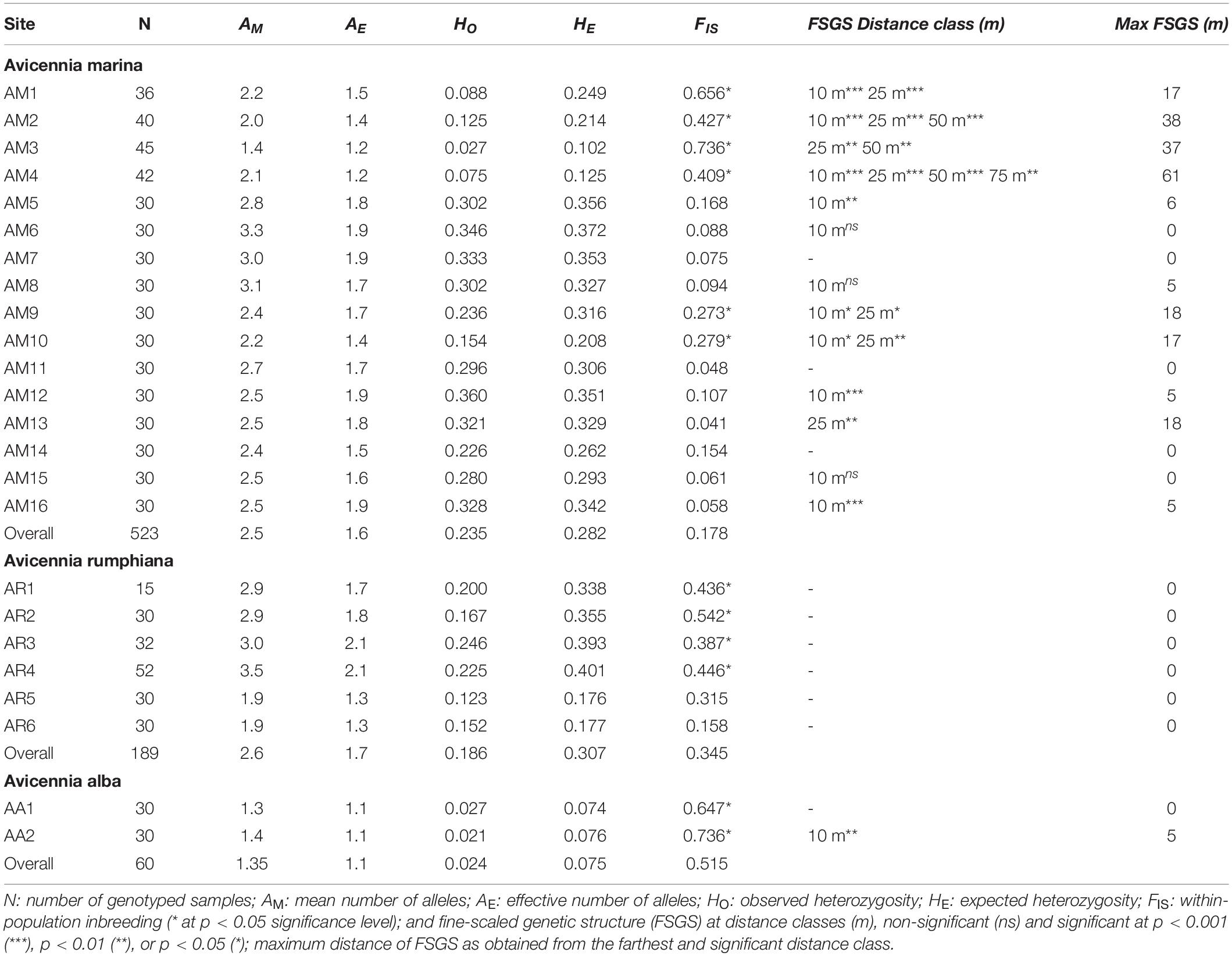
Table 2. Population genetic variables of Avicennia transects in mangrove areas along Tablas Strait and Western Leyte (Philippines).
DNA Extraction and Microsatellite Primers
Genomic DNA was extracted from approximately 20 mg of dried leaf tissue using the E.Z.N.A. SP plant DNA Mini kit (Omega bio-tek, Norcross, GA, United States). The multiplexed polymerase chain reactions (PCR) consisted of thirteen microsatellite markers of which for A. marina twelve were chosen for their allele polymorphism within a population (Avma1, Avma02, Avma03, Avma05, Avma6, Avma8, Avma9, Avma10, Avma14, Avma17, Am81, Am40), hence suitable for fine-scaled analysis. For A. rumphiana, seven microsatellite markers (Avma1, Avma05, Avma8, Avma10, Avma14, Am64, Am81) and for A. alba, ten microsatellite markers cross-amplified (Avma1, Avma02, Avma05, Avma6, Avma8, Avma10, Avma14, Avma17, Am3, Am81). Avma markers were previously developed by Geng et al. (2007) and Am markers by Maguire et al. (2000).
Primers were fluorescence-labeled with 4 different dye-labels (6FAM/VIC/NED/PET) and a primer mix was made by mixing 0.2 μM of each primer together. Multiplex PCR reactions consisted of 6.25 μl master mix (Qiagen Multiplex PCR kit), 1.25 μl primer mix, 2.5μl H2O and 2.5μl of genomic DNA. PCR was performed in a thermal cycler (Bio-Rad MyCycler) with the following conditions: an initial denaturation of 95°C for 15 min followed by 35 cycles of: 30 s denaturation at 95°C, 90 s annealing at 57°C and 80 s elongation at 72°C followed by a final extension of 30 min at 60°C. PCR products were separated on an ABI3730XL sequencer (Macrogen, Seoul, South Korea) and allele sizes were determined with GeneMarker V2.60 (SoftGenetics LLC, State College, United States).
Genetic Analyses
Prior to population and individual-based data analysis we tested for genotypic disequilibrium, presence of potential null alleles and overall resolution of the selected microsatellite markers for either A. marina. A. rumphiana or A. alba. A linkage test between all pairs of loci (1000 permutations) gave no genotypic disequilibrium at the 0.05 level using FSTAT (v.2.9.3) (Goudet, 2001) for any of the three species. No scoring errors, large allele dropouts or null alleles were indicated using MICRO-CHECKER (Van Oosterhout et al., 2004). The probability of identity (PI), namely whether two individuals could share an identical multilocus genotype by chance using GenAlEx v.6.5 (Peakall and Smouse, 2012), gave a cumulative probability of identity for all polymorphic loci in each site of 6.6 10–3 on average in A. marina and 2.4 10–2 in A. rumphiana thereby providing good resolution. This was only 0.2 for the limited samples of A. alba and therefore were excluded from comparative analyses except for estimate of fine-scale genetic structure.
Basic population genetic variables were measured separately for each species and site: total number of alleles (A), mean number of alleles (AM), effective number of alleles (AE), observed heterozygosity (HO), expected heterozygosity (HE), population inbreeding coefficient (FIS) – with 1000 permutations test – using FSTAT and GenAlEx. The genetic structure among sites (FST), inbreeding within sites (FIS), overall inbreeding (FIT) and a pairwise genotypic differentiation matrix (FST) of each species was calculated via AMOVA-FST at 999 random permutations using GenAlEx, thereby allowing to estimate overall connectivity levels as Nm = FST/(1-4FST) under the assumption of an island migration model, most likely to be violated. Therefore, specific hypotheses to estimate gene flow were tested with Migrate-n (Beerli, 2006; Beerli and Palczewski, 2010) from the mutation-scaled population sizes (Theta) and immigration rates (M). The Brownian model was tested locus by locus along with the product of all distributions of all loci. Uni- and bidirectional historical migration/expansion models were tested. Uniform prior distribution settings (min, max, delta) were as follows for Theta = 0.0, 10.0, 0.1 and for M = 0.0, 100, 10.0. The number of recorded steps was 106 at a sampling frequency of 103 after an initial burn-in. The effective number of immigrants per generation (Nem) was calculated as [Theta × M]/4. Specific hypotheses testing on directionality were considered in panmixia, source-sink, adjacent or steppingstone models for the migration between the most seaward located plots of mangrove estuaries situated along a same coastline. The most seaward transect of the replicates from each estuary was used. More precisely, we considered six A. marina populations along Western Leyte, two A. marina populations near the Verde Passage on Oriental Mindoro and two A. rumphiana populations in Tablas Strait on Romblon. The Brownian motion mutation model within each case was adopted for randomly generated subsamples of 20 individuals in a transect, following the above mentioned settings, computing two replicate chains (with different seed), and using the Bezier thermodynamic integration (Beerli and Palczewski, 2010) for calculation of the Bayes factors from marginal likelihoods giving model probabilities.
A Bayesian clustering analysis at individual level for 16 A. marina populations of Western Leyte was carried out in STRUCTURE version 2.3.4 (Pritchard et al., 2000) using an admixture model with correlated allele frequencies. The model ran 10 iterations for each K-value from 1 to 13; the burn-in period was 50,000 with 500,000 Markov chain Monte Carlo (MCMC) repeats. The optimal K was inferred with the ΔK statistic (Evanno et al., 2005) and LnPK using Structure Harvester (Earl and von Holdt, 2012) calculated with StructureSelector (Li and Liu, 2018). We tested for recent bottlenecks in each site under the two-phase model (TPM) with 95% single-step mutations and 5% multiple-step mutations (Wilcoxon’s test 1-tailed) using bottleneck 1.2.02 (Piry et al., 1999).
The overall FIJ kinship coefficient (Loiselle et al., 1995) for all pairs of individuals of within-site comparisons was tested as a first exploratory approach using the longest transect of 450 m (AM5), obtained for an equal number of pairwise comparisons within five classes as well as obtained for three distance class scenarios (5-10-25-50-100 m;10-20-30-40-50 m; and 10-25-50-75-100 m) by SPAGeDi 1.5a (Hardy and Vekemans, 2002) and using the whole sample as a reference. On basis of that comparison of classes and the obtained significant kinship values, we then finally tested the FIJ kinship coefficient again for all within-site comparisons using distance classes up to 10 m, 25 m, 50 m, 75 m, and 100 m and beyond for A. marina and A. alba populations and up to 50 m, 100 m, and 200 m and beyond for A. rumphiana populations. The latter were considered throughout this study to estimate the fine-scale genetic structure (FSGS), i.e., the spatial autocorrelation of individuals of populations, which were all tested for significance with 1000 permutations using the total sample within each species as a reference. We computed the log-slope (-b) of linear regressions between pairwise genetic coefficients and geographical distance over restricted distance with 1000 permutations.
Environmental Data
From google earth images we calculated the proximity of each transect to the open water edge (coastal or tidal riverine), the approximate size of the estuary as could be delineated from the landward contours of high intertidal mangrove fragments (surface area of polygons). We identified the extent (using polygons) of main forms of human-mediated encroachment (roads and houses, cleared and deforestation, aquaculture ponds), the percentage of human altered area, the remaining mangrove forest size (ha) and narrowness at position of Avicennia transects taken (Supplementary Figure 1 and Supplementary Table 1). Spearman rank correlations were estimated between the genetic structure variable (FSGS extent, i.e., the farthest distance of detectable kinship within a transect) and the eight abovementioned estuary features (across species, N = 24). Spearman rank correlations of A. marina populations (N = 16) were additionally estimated between four genetic variables (AE, HE, FIS, FSGS) and eight estuary features using SPSS statistics 27.0.1. Thus, the FSGS correlations were done for all 24 sites of the three species and the genetic diversity variables only for the 16 A. marina mangroves. This choice is justified because FSGS can be used among species, regardless of the amount of diversity, whereas diversity is not comparable between species due to their different microsatellite loci and number of alleles.
To test for an effect on allelic diversity, inbreeding or detectable FSGS distances over which propagules once became established in either a hydrodynamic open habitat or sheltered inland sites, a generalized linear model (GLZ) was performed on 24 sites (including three Avicennia species). This GLZ considered FSGS as a response variable and eight habitat features as predictor variables (Distance to sea, Estuary size, houses/roads surface, deforestation area, aquaculture area, mangrove area, percentage anthropogenic modification in land use, and the narrowness at position of transect). An additional GLZ was performed on 16 sites of A. marina with AE, HE, FIS and FSGS as response variables and the same abovementioned eight habitat features as predictor variables. GLZ models were performed with SPSS statistics 27.0.1.
The genetic variables (AE, HE, FIS) and the extent (mean distance within significant and farthest class) of detectable FSGS were linearly regressed to predictor variables, namely to the distance of sea or tidal river edge (m), to the mangrove surface area (ha) and to the narrowest strip (m) of the mangrove patch due to any encroachment. Both Spearman rank (RS) and Pearson’s r were calculated and tested for significance at p < 0.05. The interaction between two habitat variables (Distance to sea and narrowness) was tested for significance with the genetic variables AE, HE, FIS and with the extent of FSGS.
Results
Basic Allele and Gene Diversity
For A. marina populations, the mean number of alleles (AM) was 2.5 and ranged between 1.4-3.1; an effective number of alleles (AE) reached an overall 1.6 and ranged between 1.2-1.9 (Table 2). A. rumphiana populations showed AM = 2.6 (from 1.9 to 3.5) and AE = 1.7 (from 1.3 to 2.1) and A. alba populations showed AM = 1.4 and AE = 1.1 (Table 2). The overall observed heterozygosity (HO = 0.235) was lower than the expected heterozygosity (HE = 0.282) for A. marina, A. rumphiana (HO = 0.186, HE = 0.307) and A. alba populations (HO = 0.024, HE = 0.075) The within-population inbreeding of A. marina (mean FIS = 0.178; p < 0.001) ranged from 0.041 to 0.736 and was significant for six transects (Table 2). The within-population inbreeding of A. rumphiana (mean FIS = 0.345; p < 0.001) and A. alba (mean FIS = 0.515; p < 0.001) were mostly significant (Table 2).
AMOVA results of A. marina revealed that 45% of the genetic variation was explained among populations, 14% among individuals and 42% within individuals, giving an estimate of FST = 0.445, FIS = 0.248 and FIT = 0.583 (Table 3). AMOVA of A. rumphiana and A. alba gave an estimate of FST = 0.287 and FST = 0.189, respectively (Table 3). Pairwise genetic differentiation FST was significantly different for nearly all population pairs except those within close vicinity in a same seaward protruding mangrove, namely AM7-AM8 and AR3-AR4 (Supplementary Table 2). For both A. marina and A. rumphiana, estimates of genetic divergence were largest between Tablas Strait and Western Leyte populations (Supplementary Table 2).
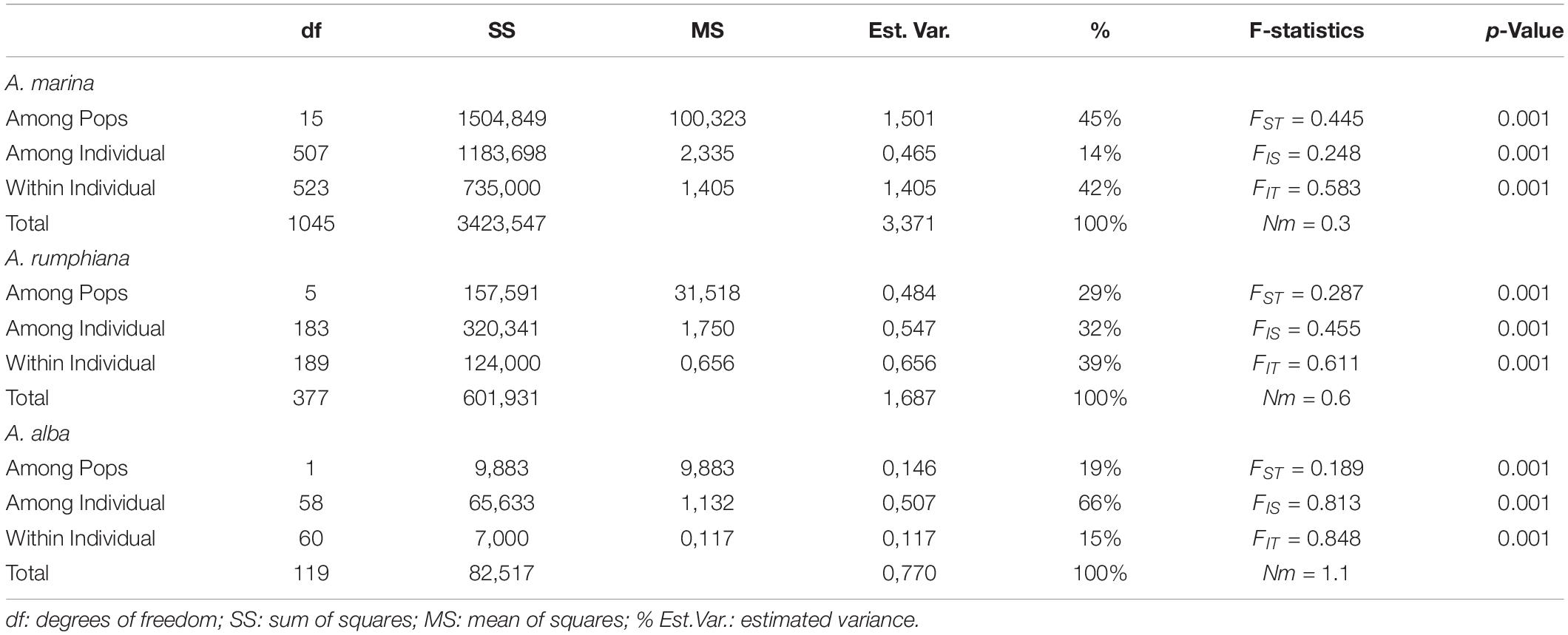
Table 3. Summary of AMOVA and F-statistics of Avicennia marina, Avicennia rumphiana and Avicennia alba mangrove fragments along coasts of Oriental Mindoro, Romblon, and Western Leyte (Philippines).
Coastal Connectivity
The specific testing with migrate-n on directionality for A. marina across mangrove estuaries located along a 115 km stretch of Western Leyte, indicated that panmixia or bidirectional stepping-stone models (from South to North as well as from North to South), appeared less likely than a customized stepping-stone model that considered a South to North migration as well as a local bidirectionality between AM10 and AM11 located in the Ormoc Bay at about 4 km distance within a historically single large mangrove area (Table 4 and Supplementary Figure 1). Highest estimated gene flow values were from AM10 toward AM7 (Nem = 1.44) and from AM16 toward AM14 (Nem = 1.17), at a distance of ca. 18 km and 13 km, respectively. Lowest gene flow estimate was toward a fragmented mangrove area AM10 (Nem = 0.45). Migration of A. marina across mangrove estuaries located along Verde Passage in Oriental Mindoro and of A. rumphiana on Tablas Island were both supported by a unidirectional adjacent model (and both gene flow estimates of Nem = 0.84), respectively from South to North over 8 km and North to South over 7 km, following the main ocean current directionality going through Tablas Strait (Figure 1 and Table 4).
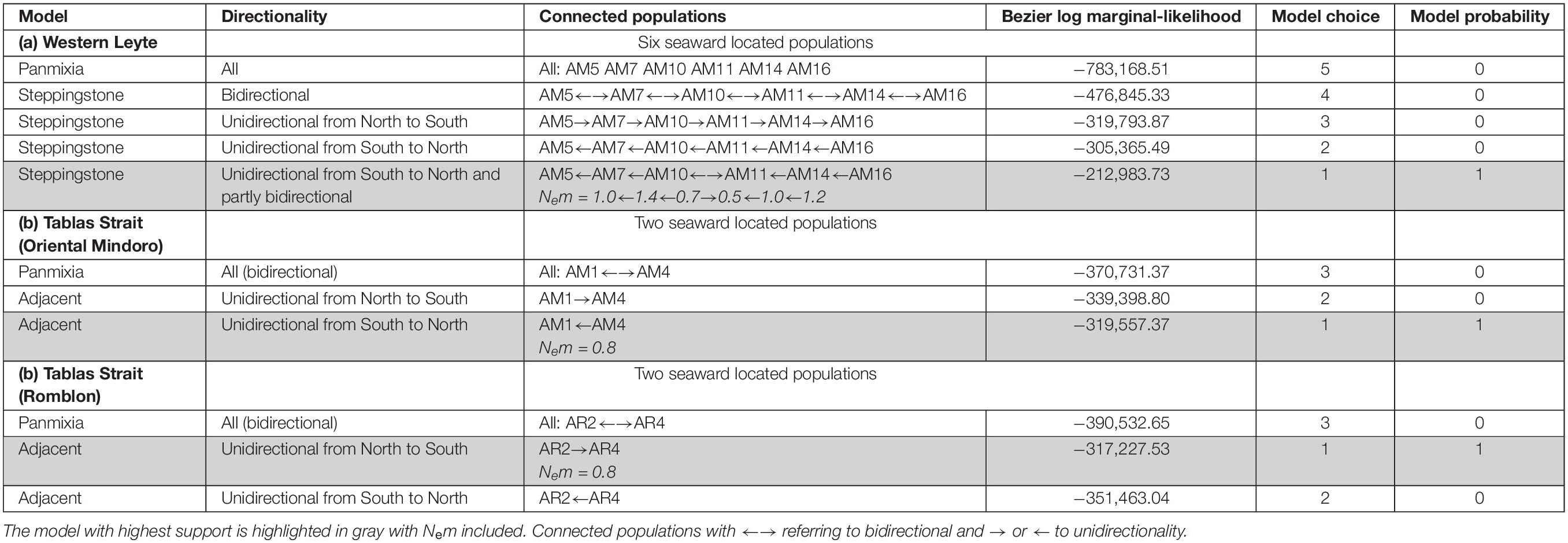
Table 4. Comparison of migration models on the directionality for Avicennia populations across mangrove estuaries of the Tablas Strait (Verde Passage and Romblon) and of Western Leyte (Philippines).
A Bayesian clustering analysis of individual A. marina trees of Western Leyte performed in STRUCTURE indicated a gradient of admixed clusters (Figure 2). Delta K was high for K = 2 (usually this is the case because of large difference with K = 1), very low for all other inferred clusters except for K = 4 (Supplementary Figure 2), referring to a regional substructure of most northern populations (AM5, AM6) and of southern populations (AM12, AM14, AM15, AM16). However, this K = 4 value must be regarded as an estimation relevant for a coastal stretch of Western Leyte with limited cases of assignment of an individual to but a single gene pool. The STRUCTURE outcome at K = 4 was supported from each iteration (low standard deviation), is much closer to the LnP(K) plateau and corresponds to the abovementioned Migrate-n supported models. A significant bottleneck could be detected in few sites of A. marina (AM12, AM13) and A. rumphiana (AR3, AR4).

Figure 2. STRUCTURE results for K = 4 with pie charts for Avicennia marina populations AM5 to AM16 of Western Leyte showing a South-North gradient.
Fine-Scale Genetic Structure
We tested at first an equal number of pairwise comparisons within five classes as well as obtained for three distance class scenarios (5-10-25-50-100 m;10-20-30-40-50 m; and 10-25-50-75-100 m) by SPAGeDi 1.5a (Hardy and Vekemans, 2002) and using the whole sample as a reference. The kinship within each transect reached FIJ = 0.48 in A. marina, FIJ = 0.25 in A. rumphiana and FIJ = 0.16 in A. alba. We then subsequently tested FIJ kinship coefficients for all within-site comparisons using distance classes up to 10 m, 25 m, 50 m, 75 m, 100 m and beyond for A. marina and A. alba populations and up to 50 m, 100 m, 200 m and beyond for A. rumphiana populations. These were considered throughout this study to estimate the fine-scale spatial autocorrelation of individuals of populations, which were all tested for significance with 1000 permutations using the total sample within each species as a reference.
The overall FIJ kinship coefficient for all within-site comparisons of A. marina revealed positive kinship values within distance classes of 10 m (FIJ = 0.072; p < 0.001), 25 m (FIJ = 0.075; p < 0.001), 50 m (FIJ = 0.065; p < 0.001), 75 m (FIJ = 0.055; p < 0.001), and 100 m (FIJ = 0.017; p < 0.001) (Figure 3A). The kinship value (FIJ) decreased significantly over the full distance (b-log slope of regression = −0.051 at p < 0.001). The overall FIJ kinship coefficient for all within-site comparisons of A. rumphiana revealed positive kinship values within shortest distance classes of 50 m (FIJ = 0.033; p < 0.001) and 100 m (FIJ = 0.037; p < 0.001) though not beyond (Figure 3B). The kinship value (FIJ) decreased significantly over the full distance (b-log slope of regression = −0.027 at p < 0.001). The two A. alba populations of a same large estuary showed positive kinship values within distance classes of 25 m (FIJ = 0.034; p < 0.05) though not beyond (Figure 3C). The kinship value (FIJ) decreased significantly over the full distance (b-slope of regression = −0.0007 at p < 0.05).
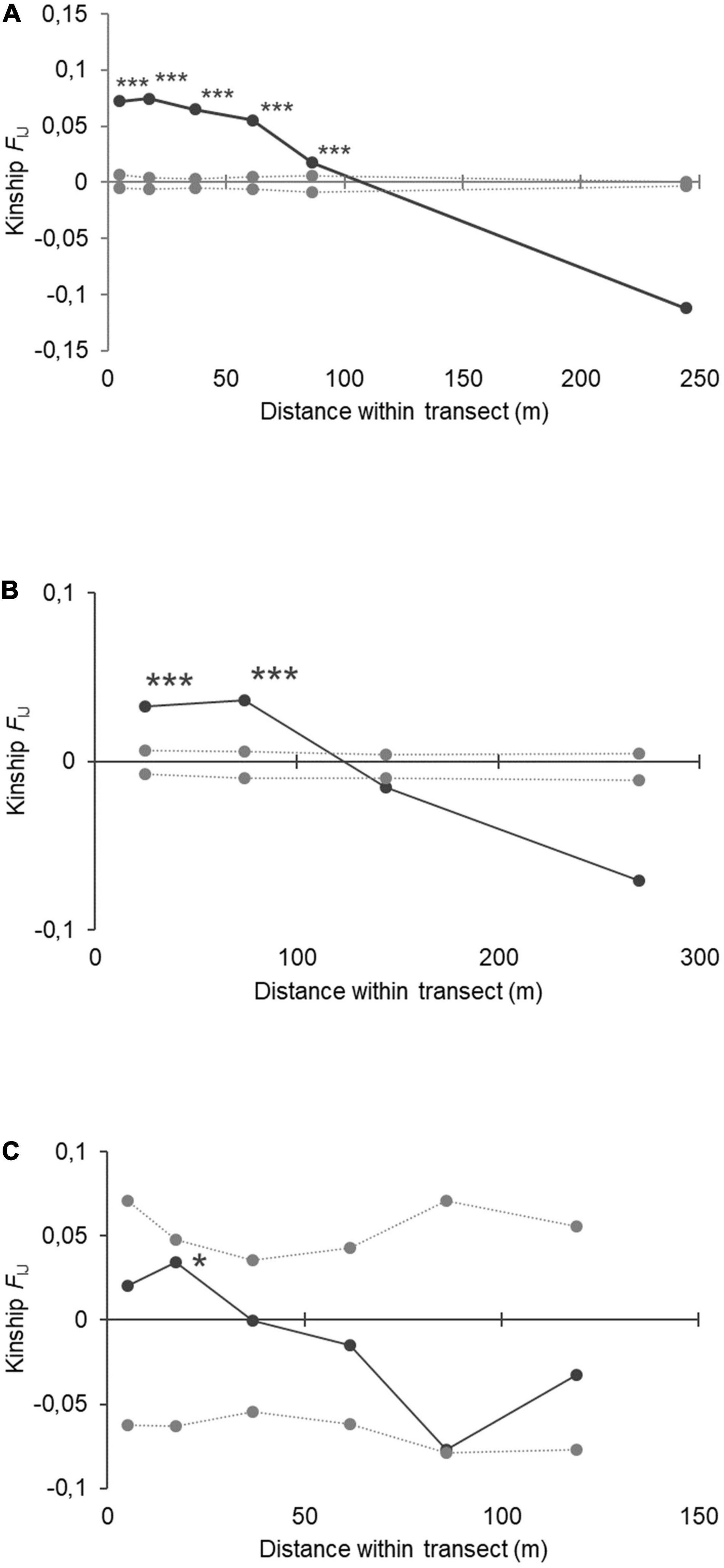
Figure 3. Fine-scale genetic structure within transects for panel (A) Avicennia marina populations (N = 16); (B) Avicennia rumphiana populations (N = 6) and (C) Avicennia alba populations (N = 2) from mangroves in The Philippines. Distance classes showed significant kinship (FIJ) values to 100 m in A. marina and 60 m in A. rumphiana (*** for p < 0.001; * for p < 0.05). The log-slope of the regression over full distance was b = −0.050 (p < 0.001), -0,028(p < 0.001), and -0,019 (p < 0.05) respectively for the three species.
A detailed analysis of the fine-scale genetic structure of each Avicennia transect separately, revealed a range of outcomes with significant positive kinship values over the considered distance classes (Table 2; FSGS Distance class in m). The maximum distance of detectable FSGS (Table 2), i.e., corresponding to the extent of the FSGS (ranging from 0 m to 61 m), was obtained for each transect from the mean distance between individual pairs from that largest distance class as could be generated from SPAGeDi.
Association With Habitat Features
Among the genetic diversity variables, AE and HE were strongly correlated, whereas among the environmental variables, the estuary size was strongly correlated to the various types of disturbances (Supplementary Table 3). From the genetic variables, the FSGS extent (namely the farthest distance of detectable kinship within a transect) was strongest and positively correlated to the ‘distance to sea’ either when considering all 24 populations of the three species or when restricted to 16 A. marina populations only (Supplementary Table 3). FSGS and FIS were positively related to the ‘distance to sea,’ whereas AE and HE were negatively related to the ‘distance to sea.’ AE and HE were negatively related to the ‘narrowness’ of the transect (Supplementary Table 3). The generalized linear model (GLZ) resulted in a similar outcome and supported the ‘distance to sea’ as a significant predictor for the FSGS of both cases (N = 24 and N = 16) and as a significant predictor for all other tested genetic variables AE and HE and FIS of A. marina (N = 16) (Supplementary Table 4). Additionally, the GLZ indicated ‘Aquaculture’ and ‘Percentage anthropogenic disturbance’ as a marginally significant predictor of FSGS across the three species (N = 24). For the case of A. marina (N = 16) also the ‘narrowness’ appeared as a significant predictor for the dependent genetic variables AE and HE (Supplementary Table 4). Linear regression of the relevant genetic variables to the ‘distance from coast’ and ‘narrowness of mangrove fragment’ illustrated that the FSGS appeared more elevated at inland sites than at seaward sites (rs = 0.81, p < 0.001 and r(22) = 0.79, p < 0.001; y = 0.031x + 0.008) across the populations of three Avicennia species (N = 24). For A. marina populations (N = 16), the distance to sea was positively correlated to the within-population inbreeding FIS (rs = 0.60, p = 0.013 and r(14) = 0.62, p = 0.011; y = 0.0003x + 0.1) though not with any other basic genetic variable (Figure 4). However, the narrowness, i.e., width of mangrove patch at the smallest position of the sampled transect (Figure 4), was negatively correlated to the diversity variables namely the number of effective alleles AE (rs = −0.56, p = 0.024 and r(14) = 0.58, p = 0.018; y = −0.002x + 1.9), and heterozygosity HE (rs = −0.49, p = 0.05 and r(14) = 0.55, p = 0.028; y = −0.0007x + 0.37). The test of interaction between two relevant habitat variables (Distance to sea and narrowness) gave non-significance for the genetic variables AE, HE, FIS though a positive interaction for the extent of FSGS (p = 0.01).
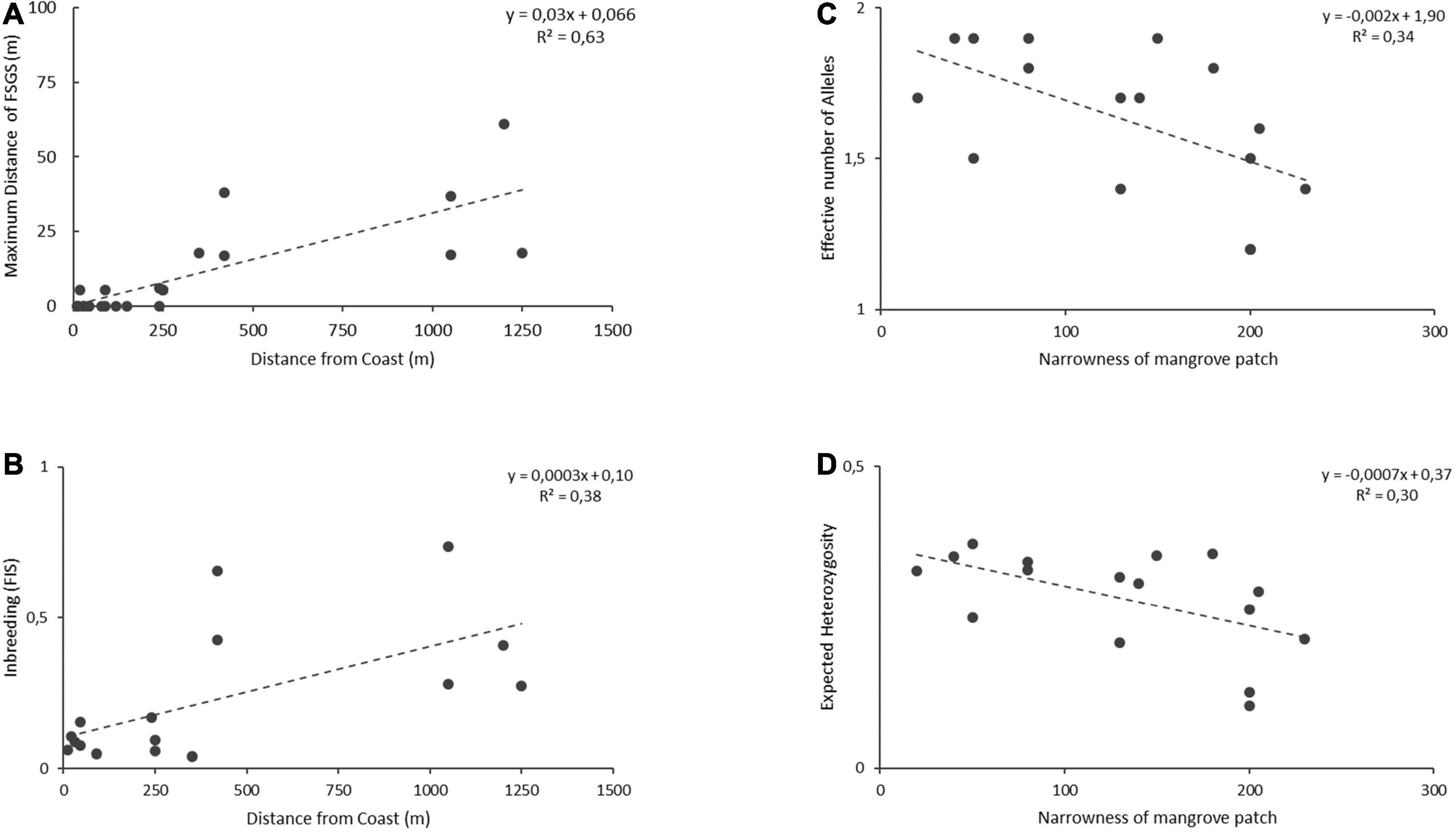
Figure 4. Relationship of the position (distance to coastline) of Avicennia populations (N = 24) and (A) the extent of their fine-scale genetic structure, (B) the within-population inbreeding (FIS). Association between the narrowness of a mangrove patch, and (C) effective number of alleles and (D) expected heterozygosity.
Discussion
Overall Consideration of Estuaries and Hydrodynamics
Avicennia dominated mangrove forests occur within a gradient ranging from strongly exposed protruding vegetations up to the most sheltered landward edge and hence are subject to different dynamics in estuarine ecosystems. Encroachment and fragmentation usually start from landward sides. Knowing that the regeneration of mangrove forests primarily depends on the available propagule pool and their opportunity to establish in suitable habitats, it is expected that this complex nature of estuarine tides and currents will create a variety of conditions for their spread, conservation and establishment of such propagules (Triest et al., 2020). Spontaneous regeneration of Avicennia mangroves may occur either from a propagule pool of related genotypes within close vicinity (Ngeve et al., 2017) or might result from a mixed migrant pool of unrelated origin (Hasan et al., 2018; Do et al., 2019). We considered the fine-scale spatial genetic structure (FSGS) of pioneer mangrove species A. marina, A. rumphiana and A. alba, using a transect approach in 24 sites distributed in The Philippines along coastline stretches of Tablas Strait and Western Leyte. These are representing a series of both open coastal mangroves and sheltered landward edges of the estuaries, though mostly encroached by roads, houses, deforestation and aquaculture ponds. However, neither human-induced changes such as the proportion of aquaculture ponds, deforested areas or hardened structures of roads and houses showed a link to genetic diversity variables of Avicennia populations, nor did the sizes of mangrove fragments. Only the FSGS appeared marginally affected by aquaculture or overall anthropogenic modifications within the estuary. Therefore, perpetual natural forces such as the tidal sea or river influence, of which ‘distance to sea/tidal river’ was used as a proxy in this study, have clearly offset the potential side-effects that could result from proper habitat fragmentation. Dispersal and establishment, which took place over the past few, though overlapping, generations under different degree of tidal and sea currents, could have left detectable traces in the current amount of trapped genetic diversity but especially in the fine-scale spatial genetic structure (Hasan et al., 2018). Overall, levels of allele and gene diversities within populations of A. marina were higher along Western Leyte than along Tablas Strait and were more elevated than in A. rumphiana or A. alba. Therefore, evidence could be obtained from sixteen A. marina populations as much as from the twenty-four populations of three species. We considered the full data of 24 sites of three species mainly to interpret the distance over which an FSGS was apparent, whereas other genetic variables were tested separately for each species, even so for testing migration models.
Connectivity Along Same Coastline
Long-distance dispersal characteristics of A. marina can be attributed to buoyant characteristic of the propagules and the action of hydrodynamic forces, i.e., tidal inundation regimes (Breitfuss et al., 2003) and ocean currents (Steinke and Ward, 2003). Wind may play a lesser part in the dispersal of Avicennia propagules, as their propagules show low surface water contact which reduces the drag force needed to move the propagule (Van der Stocken et al., 2015). Propagules mainly strand and establish close to their parent, although also dispersed over longer distances (Clarke, 1993) and are in fact the only responsible source of historically accumulated gene flow between populations (Duke et al., 1998). Factors such as propagule buoyancy, propagule viability and other environmental influences like tidal influence, ocean currents and wind action determine the success of dispersal and establishment of mangrove species (Rabinowitz, 1978; Clarke, 1993; Van der Stocken et al., 2015). In A. marina, within days upon the release of crypto-viviparous propagule in the seawater, the buoyant pericarp is shed and the seedling sinks. The propagule has an obligate dispersal phase of several weeks before the radicle extends sufficiently for roots to develop. If seedlings do not touch a sediment these may remain viable in seawater for several months (Clarke, 1993) thereby guaranteeing a long-distance dispersal between geographically disjunct estuaries. A. marina propagules may float and remain viable for several days to weeks (Steinke, 1986; Clarke and Myerscough, 1991; Clarke et al., 2001), but floating periods of several months have been reported in other Avicennia species (Rabinowitz, 1978; Alleman and Hester, 2011) though vary among estuaries (Steinke, 1986).
Our estimation of connectivity along a same coastline through comparison of migration models for seaward located A. marina and A. rumphiana populations supported a putative unidirectional dispersal route that was congruent with prevailing ocean currents (May et al., 2011) across mangrove estuaries of the Tablas Strait (northward in Verde Passage and southward along Tablas) and of Western Leyte (northward except for bidirectionality in Ormoc Bay), thereby emphasizing the relevance of coastal connectivity for persistence of mangroves (Van der Stocken et al., 2019b) but also highlighting the importance of proximity of apparently discrete estuaries, even when only over few km distance, e.g., about 7 -18 km as encountered for estuaries in our study. Although an overall outcome pointed at restricted connectivity along a 115 km stretch of Western Leyte, namely the significant pairwise differentiation FST-values and a STRUCTURE analysis giving at least a few gene pools, the connectivity between adjacent mangroves was supported by the steppingstone migration model.
Several studies suggest that dispersal in Avicennia species is likely restricted to a few tens of kilometers (e.g., Clarke, 1993; Duke et al., 1998; Melville and Burchett, 2002). Binks et al. (2018) found evidence for occasional long-distance dispersal up to 100 km, which is comparable to the distance (ca. 115 km when following coastal distance) between the southernmost and northernmost population of Western Leyte considered in this study. Our data clearly demonstrate connectivity within less than 10 km for population pairs of Tablas Strait and indicate the connectivity between populations along Western Leyte, most evidently in a stepping-stone manner though not exclusively, e.g., bidirectional in a historically large mangrove area of the sheltered Ormoc Bay. Our results on A. marina and A. rumphiana add to the emerging evidence that Avicennia species in general tend to follow an adjacent migration, such as the unidirectional way obtained for A. alba in the western part of the Malaysian Peninsula (Wee et al., 2020) and bidirectional ways of A. germinans along each of the Caribbean and Pacific coasts of central America (Ochoa-Zavala et al., 2019). A continuous distribution model instead of steppingstone might even be applicable for individuals of populations that still are physically connected (e.g., genuinely coastal mangroves of AM7-AM8 and AM14-AM15) or that once were distributed over a large extended estuarine landscape without discrete subpopulations, despite the nowadays very visible encroachment and fragmentation (e.g., AM9-AM10).
Repeated bottlenecks or founder effects of the pioneering Avicennia species may have caused the differentiation of populations, similar as noted for A. marina (Maguire et al., 2000; Arnaud-Haond et al., 2006) although we noticed only limited evidence of recent bottlenecks in the study region. In addition, reduced variance between A. rumphiana populations (FST = 0.287) could be the result of both restricted gene flow between remote islands and especially of inbreeding events (Islam et al., 2015). The latter is supported by the within-population inbreeding for A. rumphiana populations (FIS = 0.455). Giang et al., 2003 suggested that low variation of populations could be affected by topography and hydrology, but for our A. rumphiana populations the effect of topography was unlikely since the sites allow favorable dispersal of mangrove propagules within the estuary.
Distance to Sea vs. Human Induced Ecosystem Changes
The extent of mangrove areas (8 – 99 ha) within each estuary as such showed no relationship to allele diversity, heterozygosity values or inbreeding levels. However, the narrowness (20 – 310 m) of the mangrove area where transects were taken, showed a trend of increasing diversity and decreased inbreeding. This seems counter intuitive as one would expect more diversity and less inbreeding in large-sized wide patches. The latter can be explained from the proximity of a given transect to the open sea or river edge where unrelated mixed-origin propagules (i.e., zero FSGS) may establish as well as outbred cohorts within shortest distance near to mother trees (FSGS < 10 m). Farther inland or wider mangrove patches showed a larger FSGS extent across the neighborhood (mostly within 10-25 m and occasionally up to 75 m) though less diversity along with inbreeding, most likely due to a lowered chance of external propagule input when remote from tidal influence. More precisely, when considering FSGS as a proxy for past events of dispersal and propagule establishment, those mangrove patches close to the open water showed higher diversity levels. These coastal sites also showed no or a very local kinship structure that was restricted to the immediate neighborhood (0-10 m). In the contrary, more distant landward sites (>500 m) showed lower allele diversity, more inbreeding though with a kinship structure spreading out over a wider neighborhood (up to 75 m).
Even when an estuary is open for exchanges between the sea and a river or creek, then weak currents might slow down movement of propagules thereby increasing the risk of entrapment to mangrove roots. The restriction on successful dispersal of Avicennia propagules may be attributed to retention of propagules in the pneumatophores of the trees themselves, however low or absent tidal currents of fragmented mangrove patches are primary causes of leaving a trace of elevated kinship values. Such FSGS traces in other studies also estimated within spatial stretches of a few meters up to several hundreds of meters (Mori et al., 2015; Do et al., 2019; Chablé Iuit et al., 2020; Triest et al., 2020) and when populations are sheltered (Triest and Van der Stocken, 2021) or severely fragmented and confined within artificial dikes (Hasan et al., 2018), these kinship values may become enhanced. Rhizophora mangle showed a fine-scale spatial genetic structure up to 90 m in different hydrological estuarine conditions of Caribbean mangroves (Yucatan, Mexico), although up to 240 m along a river (Chablé Iuit et al., 2020). In a high rainfall area of the Cameroon Estuary Complex, Rhizophora racemosa showed no or only limited autocorrelation within 25 m due to strong hydrodynamic situations (Ngeve et al., 2017). Nonetheless, one may not forget that the fine-scale genetic structure and diversity of mangrove populations is also determined by the cumulative effect of insect, wind, and bird pollination (Hermansen et al., 2014; Wee et al., 2015) and not solely by propagule dispersal patterns. The flowers of Avicennia are visited by many species which comprise largely by insects (i.e., honeybees – one of the frequent visitors), bats and birds (Clarke and Myerscough, 1991) showing possibility for cross-pollination and mating of siblings. The elevated levels of inbreeding in many sites should be explained also from a lack of pollen flow and from non-random mating, hence this requires a different design to study.
Fragmentation Context of the Philippines
While gene flow is driven by pollen and propagule dispersal, habitat fragmentation on the other hand limits the process of connectivity. In local isolated patches, void of suffice tidal currents, species dispersal and migration can be hindered, thereby disrupting the gene flow across a landscape. Therefore, it must be suggested to remove the hindrances, e.g., abandoned aquaculture ponds and dikes, that limit the successful dispersal of mangrove propagules to maintain connectivity in the area. Moreover, programs such as establishment of nursery sites for mangrove reforestation and annual planting activities may help to sustain gene flow of the mangrove species in the area. Emerging initiatives on reverting abandoned aquaculture ponds into reforestation sites could also be explored to potentially address increasing habitat fragmentation as the country is among the top ten nations globally with higher mangrove forest fragmentation rates (Bryan-Brown et al., 2020). In these programs, sources of propagules from mangrove stands with high genetic diversity should be considered. In our study, the narrowness of mangrove fragment patches was associated to higher allele and gene diversity caused by several small protruding coastal sites (AM7 and AM8) that apparently harbored more diversity in such open system. AM7 and AM8 were also well-connected and such situations could be suggested as source areas of planting materials. Narrowness or small patches did not cause inbreeding and as such indicated that the fragmentated areas considered were either too recent (a very likely hypothesis that merits testing) or simply not really small enough to have an effect.
In the Philippines, mangrove populations decreased primarily because of pond and shrimp pond conversions (Walters et al., 2008). Because of this, mangrove replanting programs have been initiated through community initiatives, government-sponsored projects and large-scale international development assistance programs. Rehabilitation programs have been especially intensive in the Visayas region where importance of mangrove forests for coastal protection to typhoons are most realized. However, despite the massive rehabilitation programs, the long-term survival rates of mangroves are generally low at 10–20% (Primavera and Esteban, 2008). Site selection and planting materials has been reported to be the crucial component in the success or failures of the rehabilitation activities. The continued disturbance by typhoons affects and degrades the mangrove forests and may represent an overlooked stochastic factor in a search of relationships between population genetic and estuarine habitat features. Recent mangrove reforestation programs working on reversion of disused fishpond lease agreement (FLA) are met with challenges as many disused FLA areas were in the lower intertidal zone or foreshore area with sub-optimal hydrological conditions and frequent occurrence of typhoons disrupting the spontaneous establishment or growth of replanted mangroves (Buitre et al., 2019).
From this study, we conclude that the proximity to the open sea or tidal water rather than the size and type of mangrove fragmentation may affect the captured diversity, inbreeding and fine-scale structure caused by propagule movement within or beyond the mangrove fragment. Higher levels of allele diversity for seaward sites and a highest likelihood of migration for adjacent mangroves both add to the importance of coastal connectivity that is the only natural cohesive force on longer term and necessary to counteract short term effects of increasingly encroached mangrove environments. Therefore, it must be a priority to preserve the exposed coastal mangroves as a diverse source of propagules for connecting estuaries along a coastline. The increasing activities of human-induced encroachment most likely were too recent to show clear effects on genetic variables, but it can be expected that more inbreeding and a hampered propagule flow will lead to depauperated landward mangrove areas for coming generations.
Author’s Note
Permission from the City Environment & Natural Resources Office (for Baybay and Ormoc sampling) and Municipal Environment & Natural Resources Office (for the rest of the sampling areas) were secured prior to the collection of mangrove samples. Letter requests stating the purpose of collection were submitted and signed by the abovementioned agencies. The following offices signed the transport permit for samples of the Romblon: Village Captain (Barangay Captain) through the Committee Chair for Agriculture and Environment; the Municipal Mayor through the Municipal Agriculture Office; and for samples of Mindoro: Regional Office of the Bureau of Fisheries and Aquatic Resources.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
All authors did the conceptualization, carried out the data curation, formal analysis, funding acquisition, investigated and visualized the data, performed the methodology and resources, wrote the original draft of the manuscript, and wrote, reviewed, and edited the manuscript.
Funding
This work was supported by the Vrije Universiteit Brussel (VUB; Grant Number BAS42), and by VLIR (IUC scholarships for VG and AD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely thank Prof. Iris Stiers and Dr. Dennis De Ryck for facilitating and guiding IUC scholarship holders.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.643982/full#supplementary-material
References
Alleman, L. K., and Hester, M. W. (2011). Reproductive ecology of black mangrove (Avicennia germinans) along the Louisiana coast: propagule production cycles, dispersal limitations, and establishment elevations. Estuar. Coast. 34, 1068–1077. doi: 10.1007/S12237-011-9404-8
Alongi, D. M. (2008). Mangrove forests: Resilience, protection from tsunamis, and responses to global climate change. Estuar. Coast. Shelf Sci. 76, 1–13. doi: 10.1016/j.ecss.2007.08.024
Arnaud-Haond, S., Teixeira, S., Massa, S. L., Billot, C., Saenger, P., Coupland, G., et al. (2006). Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 15, 3515–3525. doi: 10.1111/j.1365-294X.2006.02997.x
Beerli, P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345. doi: 10.1093/bioinformatics/bti803
Beerli, P., and Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326. doi: 10.1534/genetics.109.112532
Binks, R. M., Byrne, M., McMahon, K., Pitt, G., Murray, K., and Evans, R. D. (2018). Habitat discontinuities from strong barriers to gene flow among mangrove populations, despite the capacity for long-distance dispersal. Divers. Distrib. 25, 298–309. doi: 10.1111/ddi.12851
Breitfuss, M. J., Connolly, R. M., and Dale, P. E. R. (2003). Mangrove distribution and mosquito control: transport of Avicennia marina propagules by mosquito-control runnels in southeast Queensland saltmarshes. Estuar. Coast. Shelf Sci. 56, 573–579. doi: 10.1016/S0272-7714(02)00207-X
Bryan-Brown, D. N., Connolly, R. M., Richards, D. R., Adame, F., Friess, D. A., and Brown, C. J. (2020). Global trends in mangrove forest fragmentation. Sci. Rep. 10:7117. doi: 10.1038/s41598-020-63880-1
Burns, K. A., Garrity, S. D., and Levings, S. C. (1993). How many years until mangrove ecosystems recover from catastrophic oil spills? Mar. Pollut. Bull. 26, 239–248. doi: 10.1016/0025-326X(93)90062-O
Buitre, M. J. C., Zhang, H., and Lin, H. (2019). The mangrove forests change and impacts from tropical cyclones in the Philippines using time series satellite imagery. Remote Sens. 11:688. doi: 10.3390/rs11060688
Chablé Iuit, L. R., Machkour-M’Rabet, S., Espinoza-Ávalos, J., Hernández-Arana, H. A., López-Adame, H., and Hénaut, Y. (2020). Genetic structure and connectivity of the Red mangrove at different geographic scales through a complex transverse hydrological system from freshwater to marine ecosystems. Diversity 12:48. doi: 10.3390/d12020048
Clarke, P. J., Kerrigan, R. A., and Westphal, C. J. (2001). Dispersal patterns and early growth in 14 tropical mangroves: do early life history traits correlate with patterns of adult distribution. J. Ecol. 89, 648–659. doi: 10.1046/j.0022-0477.2001.00584.x
Clarke, P. J. (1993). Dispersal of Gray Mangrove (Avicennia marina) propagules in Southeastern Australia. Aquat. Bot. 43, 195–204. doi: 10.1016/0304-3770(93)90021-N
Clarke, P. J., and Myerscough, P. J. (1991). Buoyancy of Avicennia marina propagules in South-Eastern Australia. Aust. J. Bot. 39, 77–83. doi: 10.1071/BT9910077
Dahdouh-Guebas, F., De Bondt, R., Abeysinghe, P., Kairo, J., Cannicci, S., Triest, L., et al. (2004). Comparative study of the disjunct zonation pattern of the gray mangrove Avicennia marina (Forsk.) Vierh. in Gazi Bay (Kenya). Bull. Mar. Sci. 74, 237–252.
Dahdouh-Guebas, F., Jayatissa, L. P., Di Nitto, D., Bosire, J. O., Lo Seen, D., and Koedam, N. (2005). How effective were mangroves as a defence against the recent tsunami? Curr. Biol. 15, 443–447. doi: 10.1016/j.cub.2005.06.008
De Ryck, D. J. R., Koedam, N., Van der Stocken, T., van der Ven, R. M., Adams, J., and Triest, L. (2016). Dispersal limitation of the mangrove Avicennia marina at its South African range limit in strong contrast to connectivity in its core East African region. Mar. Ecol. Prog. Ser. 545, 123–134. doi: 10.3354/MEPS11581
Do, B. T. N., Koedam, N., and Triest, L. (2019). Avicennia marina maintains genetic structure whereas Rhizophora stylosa connects mangroves in a flooded, former inner sea (Vietnam). Estuar. Coast. Shelf Sci. 222, 195–204. doi: 10.1016/j.ecss.2019.04.005
Duke, N. C., Benzie, J. A. H., Goodall, J. A., and Ballment, E. R. (1998). Genetic structure and evolution of species in the mangrove genus Avicennia (Avicenniaceae) in the Indo-West Pacific. Evolution 52, 1612–1626. doi: 10.1111/j.1558-5646.1998.tb02242.x
Duke, N. C., Bell, A. M., Pederson, D. K., Roelfsema, C. M., and Nash, S. B. (2005). Herbicides implicated as the cause of severe mangrove dieback in the Mackay region, NE Australia: consequences for marine plant habitats of the GBR World Heritage Area. Mar. Pollut. Bull. 51, 308–324. doi: 10.1016/j.marpolbul.2004.10.040
Earl, D. M., and von Holdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Ellison, A. M., Farnsworth, E. J., and Merkt, R. E. (1999). Origins of mangrove ecosystems and the mangrove biodiversity anomaly. Glob. Ecol. Biogeogr. 8, 95–115. doi: 10.1046/J.1466-822X.1999.00126.X
Ellison, A. M. (2008). Managing mangroves with benthic biodiversity in mind: moving beyond roving banditry. J. Sea. Res. 59, 2–15. doi: 10.1016/j.seares.2007.05.003
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Feller, I. C., Lovelock, C. E., Berger, U., McKee, K. L., Joye, S. B., and Ball, M. C. (2010). Biocomplexity in Mangrove Ecosystems. Annu. Rev. Mar. Sci. 2, 395–417. doi: 10.1146/annurev.marine.010908.163809
Gedan, K. B., Kirwan, M. L., Wolanski, E., Barbier, E. B., and Silliman, B. R. (2011). The present and future role of coastal wetland vegetation in protecting shorelines: answering recent challenges to the paradigm. Clim. Change 106, 7–29. doi: 10.1007/s10584-010-0003-7
Geng, Q. F., Lian, C. L., Tao, M., Li, Q., and Hogetsus, T. (2007). Isolation and characterization of 10 new compound microsatellite markers for a mangrove tree species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Mol. Ecol. Notes 7, 1208–1210. doi: 10.1111/j.1471-8286.2007.01834.x
Giang, L. H., Hong, P. H., Tuan, M. S., and Harada, K. (2003). Genetic variation of Avicennia marina (Forsk.) Vierh. (Avicenniaceae) in Vietnam revealed by microsatellite and AFLP markers. Genes Genet. Syst. 78, 399–407. doi: 10.1266/ggs.78.399
Gillanders, B. M. (2002). Connectivity between juvenile and adult fish populations: do adults remain near their recruitment estuaries? Mar. Ecol. Prog. Ser. 240, 215–223. doi: 10.3354/meps240215
Giri, C., Ochieng, E., Tieszen, L. L., Zhu, Z., Singh, A., Loveland, T., et al. (2011). Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 20, 154–159. doi: 10.1111/j.1466-8238.2010.00584.x
Gordon, D. (1988). Disturbance to Mangroves in Tropical-Arid Western Australia: Hypersalinity and Restricted Tidal Exchange as Factors Leading to Mortality. J. Arid Environ. 15, 117–145. doi: 10.1016/S0140-1963(18)30986-8
Goudet, J. (2001). FSTAT version 2.9.3: a program to estimate and test gene diversities and fixation indices (update from version 1.2, Goudet 1995): a computer program to calculate F-statistic. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Hardy, O. J., and Vekemans, X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. doi: 10.1046/j.1471-8286.2002.00305.x
Hasan, S., Triest, L., Afrose, S., and De Ryck, D. (2018). Migrant pool model of dispersal explains strong connectivity of Avicennia officinalis within Sundarban mangrove areas: Effect of fragmentation and replantation. Estuar. Coast. Shelf Sci. 214, 38–47. doi: 10.1016/j.ecss.2018.09.007
Hermansen, T. D., Britton, D. R., Ayre, D. J., and Minchonton, T. E. (2014). Identifying the real pollinators? Exotic honeybees are the dominant flower visitors and only effective pollinators of Avicennia marina in Australian temperate mangroves. Estuar. Coast. 37, 621–635. doi: 10.1007/s12237-013-9711-3
Hodel, R. G. J., Knowles, L. L., McDaniel, S. F., Payton, A. C., Dunaway, J. F., Soltis, P. S., et al. (2018). Terrestrial species adapted to sea dispersal: differences in propagule dispersal of two Caribbean mangroves. Mol. Ecol. 27, 4612–4626. doi: 10.1111/mec.14894
Islam, M. S., Lian, C. L., Kameyama, N., and Hogetsu, T. (2015). Analysis of the mating system, reproductive characteristics, and spatial genetic structure in a natural mangrove tree (Bruguiera gymnorrhiza) population at its northern biogeographic limit in the southern Japanese archipelago. J. For. Res. 20, 293–300. doi: 10.1007/s10310-014-0473-y
Kathiresan, K., and Bingham, B. L. (2001). Biology of mangroves and mangrove Ecosystems. Adv. Mar. Biol. 40, 81–251. doi: 10.1016/S0065-2881(01)40003-4
Lee, S. Y., Primavera, J. H., Dahdouh-Guebas, F., McKee, K., Bosire, J. O., Cannicci, S., et al. (2014). Ecological role and services of tropical mangrove ecosystems: a reassessment. Glob. Ecol. Biogeogr. 23, 726–743. doi: 10.1111/geb.12155
Lefcheck, J. S., Hughes, B. B., Johnson, A. J., Pfirrmann, B. W., Rasher, D. B., Smyth, A. R., et al. (2019). Are coastal habitats important nurseries? A meta-analysis. Conserv. Lett. 12:e12645. doi: 10.1111/conl.12645
Li, Y. L., and Liu, J. X. (2018). StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 18, 176–177. doi: 10.1111/1755-0998.12719
Loiselle, B., Sork, V. L., Nason, J., and Graham, C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82, 1420–1425. doi: 10.1002/j.1537-2197.1995.tb12679.x
Maguire, T. L., Edwards, K. J., Saenger, P., and Henry, R. (2000). Characterisation and analysis of microsatellite loci in a mangrove species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Theor. Appl. Genet. 101, 279–285. doi: 10.1007/s001220051480
May, P. W., Doyle, J. D., Pullen, J. D., and David, L. T. (2011). Two-way coupled atmosphere-ocean modeling of the PhilEx Intensive Observational Periods. Oceanography 24, 48–57. doi: 10.5670/oceanog.2011.03
Melville, F., and Burchett, M. (2002). Genetic variation in Avicennia marina in three estuaries of Sydney (Australia) and implications for rehabilitation and management. Mar. Pollut. Bull. 44, 469–479. doi: 10.1016/S0025-326X(01)00259-4
Mori, G. M., Zucchi, M. I., and Souza, A. P. (2015). Multiple-geographic-scale genetic structure of two mangrove tree species: the roles of mating system, hybridization, limited dispersal and extrinsic factors. PLoS One 10:e0118710. doi: 10.1371/journal.pone.0118710
Nagelkerken, I., Blaber, S. J. M., Bouillon, S., Green, P., Haywood, M., Kirton, L. G., et al. (2008). The habitat function of mangroves for terrestrial and marine fauna: A review. Aquat. Bot. 89, 155–185. doi: 10.1016/j.aquabot.2007.12.007
Ngeve, M., Van der Stocken, T., Menemenlis, D., Koedam, N., and Triest, L. (2017). Hidden founders? Strong bottlenecks and fine-scale genetic structure in mangrove populations of the Cameroon Estuary complex. Hydrobiologia 803, 189–207. doi: 10.1007/s10750-017-3369-y
Ochoa-Zavala, M., Jaramillo-Correa, J. P., Piñero, D., Nettel-Hernanz, A., and Núñez-Farfán, J. (2019). Contrasting colonization patterns of black mangrove (Avicennia germinans (L.) L.) gene pools along the Mexican coasts. J. Biogeogr. 46, 884–898. doi: 10.1111/jbi.13536
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539. doi: 10.1111/j.1471-8286.2005.01155.x
Piry, S., Luikart, G., and Cornuet, J. M. (1999). BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Heredity 90, 502–503. doi: 10.1093/jhered/90.4.502
Primavera, J. H., and Esteban, J. M. A. (2008). A review of mangrove rehabilitation in the Philippines: successes, failures and future prospects. Wetl. Ecol. Manag. 16, 345–358. doi: 10.1007/s11273-008-9101-y
Pritchard, J. K., Stephens, M., and Donelly, P. S. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Quartel, S., Kroon, A., Augustinus, P. G. E. F., Van Santen, P., and Tri, N. H. (2007). Wave attenuation in coastal mangroves in the Red River Delta, Vietnam. J. Asian Earth Sci. 29, 576–584. doi: 10.1016/j.jseaes.2006.05.008
Rabinowitz, D. (1978). Dispersal properties of mangrove propagules. Biotropica 10, 47–57. doi: 10.2307/2388105
Richards, D. R., and Friess, D. A. (2016). Rates and drivers of mangrove deforestation on Southeast Asia, 2000–2012. Proc. Natl. Acad. Sci. U S A. 113, 344–349. doi: 10.1073/pnas.1510272113
Robertson, A. I., and Alongi, D. M. (1992). Tropical Mangrove Ecosystems. Washington, DC: American Geophysical Union. doi: 10.1029/CE041
Sandilyan, S., and Kathiresan, K. (2012). Mangroves conservation: a global perspective. Biodivers. Conserv. 21, 3523–3542. doi: 10.1007/s10531-012-0388-x
Steinke, T. D. (1986). A preliminary study of buoyancy behavior in Avicennia marina propagules. S. Afr. J. Bot. 52, 559–565. doi: 10.1016/S0254-6299(16)31492-2
Steinke, T. D., and Ward, C. J. (2003). Use of Plastic Drift Cards As as Indicators of Possible Dispersal of Propagules of the Mangrove Avicennia Marina by Ocean Currents. Afr. J. Mar. Sci. 25, 169–176. doi: 10.2989/18142320309504007
Tomlinson, P. B. (2016). The Botany of Mangroves. United Kingdom: Cambridge University Press. doi: 10.1017/CBO9781139946575
Triest, L. (2008). Molecular ecology and biogeography of mangrove trees towards conceptual insights on gene flow and barriers: A review. Aquat. Bot. 89, 138–154. doi: 10.1016/j.aquabot.2007.12.013
Triest, L., and Van der Stocken, T. (2021). Coastal landform constrains dispersal in mangroves. Front. Mar. Sci. [preprint]. doi: 10.3389/fmars.2021.617855
Triest, L., Hasan, S., Mitro, P., De Ryck, D., and Van der Stocken, T. (2018). Geographical Distance and Large Rivers Shape Genetic Structure of Avicennia officinalis in the Highly Dynamic Sundarbans Mangrove Forest and Ganges Delta Region. Estuar. Coast. 41, 908–920. doi: 10.1007/s12237-017-0309-z
Triest, L., Van der Stocken, T., Akinyi, A. A., Sierens, T., Kairo, J., and Koedam, N. (2020). Channel network structure determines genetic connectivity of landward–seaward Avicennia marina populations in a tropical bay. Ecol. Evol. 10, 12059–12075. doi: 10.1002/ece3.6829
Triest, L., Van der Stocken, T., De Ryck, D., Kochzius, M., Lorent, S., Ngeve, M., et al. (2021a). Expansion of the mangrove species Rhizophora mucronata in the Western Indian Ocean launched contrasting genetic patterns. Sci. Rep. 11:4987. doi: 10.1038/s41598-021-84304-8
Triest, L., Van der Stocken, T., Sierens, T., Deus, E. K., Mangora, M. M., and Koedam, N. (2021b). Connectivity of Avicennia marina populations within a proposed marine transboundary conservation area between Kenya and Tanzania. Biol. Cons. 256:109040. doi: 10.1016/j.biocon.2021.109040
Valiela, I., Bowen, J. L., and York, J. K. (2001). Mangrove forests: one of the world’s threatened major tropical environments. BioScience 51, 807–815. doi: 10.1641/0006-3568(2001)051[0807:mfootw]2.0.co;2
Van der Stocken, T., Vanschoenwinkel, B., De Ryck, D. J. R., Bouma, T. J., Dahdouh-Guebas, F., and Koedam, N. (2015). Interaction between water and wind as a driver of passive dispersal in mangroves. PLoS One 10:e0121593. doi: 10.1371/journal.pone.0121593
Van der Stocken, T., Wee, A. K. S., De Ryck, D. J. R., Vanschoenwinkel, B., Friess, D. A., Dahdouh-Guebas, F., et al. (2019b). A general framework for propagule dispersal in mangroves. Biol. Rev. 94, 1547–1575. doi: 10.1111/brv.12514
Van der Stocken, T., Carroll, D., Menemenlis, D., Simard, M., and Koedam, N. (2019a). Global-scale dispersal and connectivity in mangroves. Proc. Natl. Acad. Sci. U S A. 116, 915–922. doi: 10.1073/pnas.1812470116
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Walters, B. B., Rönnbäck, P., Kovacs, J. M., Crona, B., Hussain, S. A., Badola, R., et al. (2008). Ethnobiology, socio-economics and management of mangrove forests: A review. Aquat. Bot. 89, 220–236. doi: 10.1016/j.aquabot.2008.02.009
Wee, A. K. S., Low, S. Y., and Webb, E. L. (2015). Pollen limitation affects reproductive outcome in the bird-pollinated mangrove Bruguiera gymnorrhiza (Lam.) in a highly urbanized environment. Aquat. Bot. 120, 240–243. doi: 10.1016/j.aquabot.2014.09.001
Keywords: Avicennia, fragmentation, genetic structure, connectivity, microsatellites
Citation: Triest L, Del Socorro A, Gado VJ, Mazo AM and Sierens T (2021) Avicennia Genetic Diversity and Fine-Scaled Structure Influenced by Coastal Proximity of Mangrove Fragments. Front. Mar. Sci. 8:643982. doi: 10.3389/fmars.2021.643982
Received: 19 December 2020; Accepted: 06 May 2021;
Published: 08 July 2021.
Edited by:
Benjamin Lee Branoff, United States Environmental Protection Agency, United StatesReviewed by:
Richard Hodel, Smithsonian National Museum of Natural History, United StatesGustavo Maruyama Mori, São Paulo State University, Brazil
Copyright © 2021 Triest, Del Socorro, Gado, Mazo and Sierens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludwig Triest, ltriest@vub.be; ltriest@vub.ac.be
 Ludwig Triest
Ludwig Triest Alieza Del Socorro
Alieza Del Socorro Vincent Jay Gado
Vincent Jay Gado Analyn M. Mazo
Analyn M. Mazo Tim Sierens
Tim Sierens