A Convolutional Neural Network Deep Learning Model Trained on CD Ulcers Images Accurately Identifies NSAID Ulcers
- 1Department of Diagnostic Imaging, Sheba Medical Center, Tel Hashomer, Affiliated to Sackler Medical School, Tel Aviv University, Tel Aviv, Israel
- 2Deep Vision Lab, Sheba Medical Center, Tel Hashomer, Israel
- 3Department of Gastroenterology, Sheba Medical Center, Tel Hashomer, Affiliated to Sackler Medical School, Tel Aviv University, Tel Aviv, Israel
- 4Chr. Hansen A/S, Human Health Innovation, Hoersholm, Denmark
- 5APC Microbiome Ireland, Cork, Ireland
- 6Centre for Gastroenterology, Mercy University Hospital, Cork, Ireland
Background and Study Aims: Deep learning (DL) for video capsule endoscopy (VCE) is an emerging research field. It has shown high accuracy for the detection of Crohn's disease (CD) ulcers. Non-steroidal anti-inflammatory drugs (NSAIDS) are commonly used medications. In the small bowel, NSAIDs may cause a variety of gastrointestinal adverse events including NSAID-induced ulcers. These ulcers are the most important differential diagnosis for small bowel ulcers in patients evaluated for suspected CD. We evaluated a DL network that was trained using CD VCE ulcer images and evaluated its performance for NSAID ulcers.
Patients and Methods: The network was trained using CD ulcers and normal mucosa from a large image bank created from VCE of diagnosed CD patients. NSAIDs-induced enteropathy images were extracted from the prospective Bifidobacterium breve (BIf95) trial dataset. All images were acquired from studies performed using PillCam SBIII. The area under the receiver operating curve (AUC) was used as a metric. We compared the network's AUC for detecting NSAID ulcers to that of detecting CD ulcers.
Results: Overall, the CD training dataset included 17,640 CE images. The NSAIDs testing dataset included 1,605 CE images. The DL network exhibited an AUC of 0.97 (95% CI 0.97–0.98) for identifying images with NSAID mucosal ulcers. The diagnostic accuracy was similar to that obtained for CD related ulcers (AUC 0.94–0.99).
Conclusions: A network trained on VCE CD ulcers similarly identified NSAID findings. As deep learning is transforming gastrointestinal endoscopy, this result should be taken into consideration in the future design and analysis of VCE deep learning applications.
Introduction
Non-steroidal anti-inflammatory drugs (NSAIDS) are among the most commonly used medications in the world, very frequently purchased over the counter. Gastrointestinal adverse events are common with these medications and involve both upper and lower gastrointestinal tract (1). In the small bowel, NSAIDs may cause a variety of mucosal lesions, including mucosal breaks, mucosal edema, diaphragm-like strictures and ulcers that may resemble intestinal inflammatory lesions in patients with Crohn's disease (2). The prevalence of such small bowel abnormalities among NSAIDs users was reported to be as high as 75% in studies that utilized video capsule endoscopy (VCE) for evaluation of the small bowel (3–6).
NSAID-induced enteropathy is the most important differential diagnosis for small bowel ulcers in patients evaluated for suspected Crohn's disease (CD); abstinence from NSAIDs is recommended before performance of VCE, usually for at least 4 weeks (7–9). As NSAID-induced enteropathy is associated with a significant risk of gastrointestinal bleeding (10), several therapeutic agents such as misoprostol and probiotics were evaluated for mucosal protection in NSAIDs users (11, 12).
In recent years, deep learning is becoming frequently used in multiple areas of medicine including radiology, ophthalmology, oncology and gastroenterology (13–16). Convolutional neural network (CNN) techniques demonstrated excellent accuracy for detection of various endoscopic pathologies (16), and we have previously demonstrated their high accuracy for automated identification and classification of VCE-detected small bowel ulcers in CD patients (17, 18). It is expected that DL research for VCE will continue to emerge and will influence clinical practice, ultimately shortening required physicians' time for reading VCE films or possibly abolishing this need altogether. An overview of DL and CNN is presented in the methods section.
NSAIDS-induced enteropathy is commonly encountered in VCE. DL applications for VCE are being rapidly introduced into research and clinics. Most of CNN are trained using images of CD ulcers. However, the ability of DL to identify findings related to NSAID enteropathy have not yet been evaluated.
We evaluated a DL network for the detection of NSAID ulcers on VCE images. Since training a DL network requires a large number of images, and as NSAID enteropathy ulcers are morphologically similar to CD ulcers, we have used a CD ulcers dataset for training the model. Then the machine was solely tested on NSAIDs ulcers dataset that was extracted from a clinical trial in which healthy individuals received NSAID and underwent VCE.
Patients/Material And Methods
Overview of Deep Learning Algorithms for Image Analysis
Deep learning (DL), an emerging technology which has gained much interest in the past decade. It is considered a sub-type of artificial intelligence (AI) (13, 14). AI describes algorithms which perform tasks that usually require “human cognitive”.
DL is almost synonymous with artificial neural networks (ANNs). The structure of ANNs may be considered as a cluster of interconnected linear regression units. This cluster of regression units are joined using non-linear activation functions to create sequential neural layers. Similar to linear regression, each neuron in the network has multiple inputs which are termed weights. These inputs are the outputs of neurons in the previous layer (Figure 1). ANNs may contain many “hidden”/“deep” layers, hence the term “deep learning”.
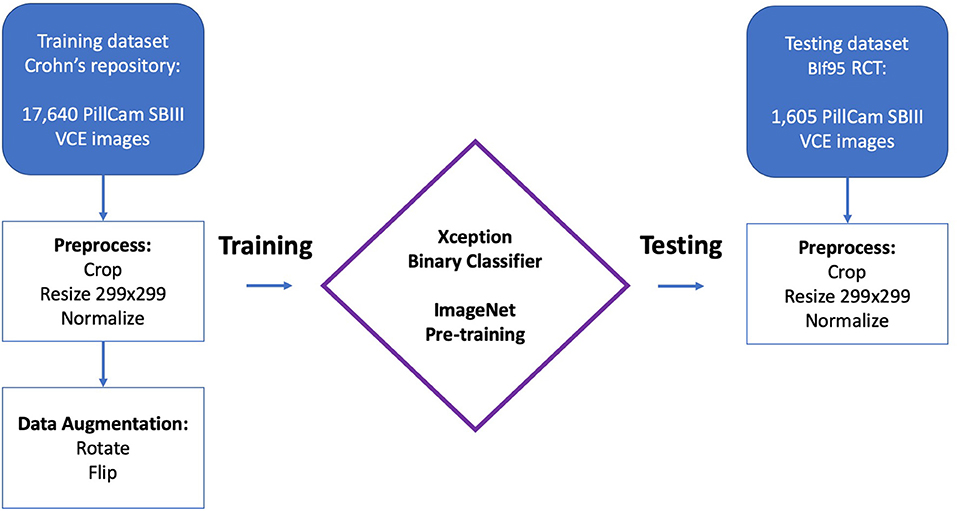
Figure 1. Schematic representation of an artificial neural network (ANN). The network is composed by many linear regression units, termed neurons. These units are structured in sequential layers. Outputs of neurons from the previous layer are the inputs of the neurons in the next layer. The connection between the layers are termed weights, which are similar to the weights of a linear regression unit. Data, such as an image, is propagated through the network, and a final prediction is outputted. An optimized network will produce a desired prediction for each data entry.
The process of training and ANN is essentially tweaking of all the network's weights to achieve an optimized network. Given data, an optimized network will produce a desired prediction. For instance, the network can be given VCE images with either ulcers or normal mucosa. An optimized network could satisfactory differentiate between these two types of images.
Convolutional neural networks (CNN) are a sub-type of ANN designed for image analysis. These algorithms are a very similar to classic ANNs. CNNs were specifically built to recognize repeating patterns in data. Since images contain many repeated patterns, CNNs are optimal for this task.
The CNN is inputted with an image (for example a VCE image), which is an array of pixels. The array is propagated along the deep layers of the CNN until a final neuron outputs a classification prediction (i.e., “ulcer”/“no ulcer”). Each CNN neuron corresponds to a small matrix (i.e., 3X3, 5X5) of weights. The uniqueness of CNN is that these small matrixes are propagated along the image. Thus, each matrix is “shared” across all the regions of the image. This matrix sharing is what allows the pattern recognition of CNNs (Figure 2).
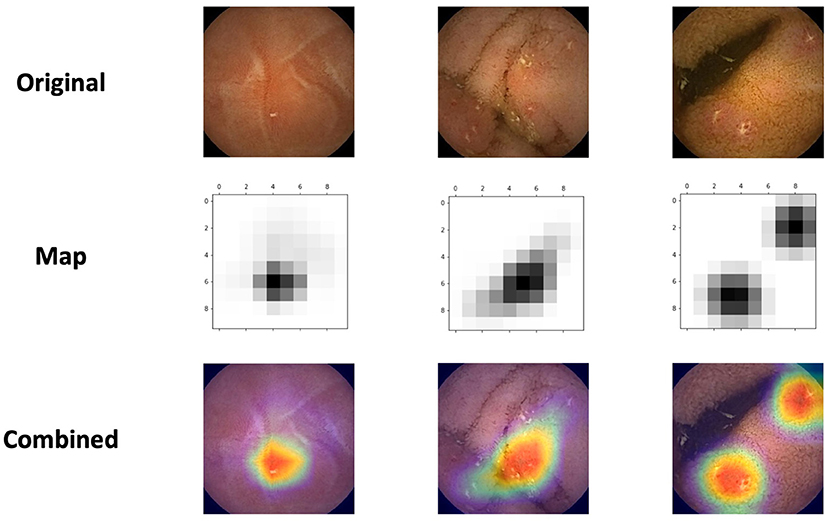
Figure 2. Convolutional neural networks (CNN) are a sub-type of artificial neural networks (ANN). CNNs, are unique in their capacity to identify repeating patterns. Thus, they are ideal for image analysis, as images contain many repeating patterns. CNNs are composed from many small matrixes which are a collection of weights. These small matrices are shared across the array of pixels which constitutes an image.
In the past few years, CNNs are being rapidly introduced into healthcare. Since these algorithms are specifically designed for image analysis, the major involved fields are those dealing with medical images, such as radiology images, endoscopy images, fundoscopy, ECG, etc.
Study Design
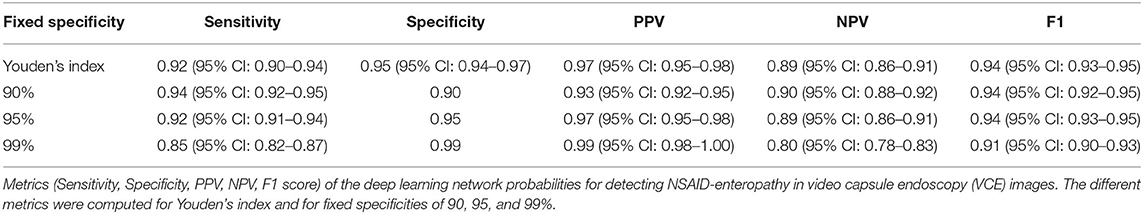
We previously published results of a CNN trained on VCE images of CD patients (17). In the current study, we retrospectively evaluated this CNN for its accuracy in detecting NSAIDs ulcers on VCE images. Thus, the network was solely trained on CD ulcers and solely tested on NSAIDs ulcers.
The network was trained using CD ulcer and normal mucosa images sourced from a repository of VCE diagnosed CD patients as previously described (17). All images were acquired from studies performed using PillCam SBIII (Medtronic Ltd, Dublin, Ireland) and reviewed with Rapid 9 (Medtronic Ltd, Dublin, Ireland) capsule reading software.
NSAIDs induced enteropathy images were extracted from VCEs that were performed during the Bifidobacterium breve (BIf95) randomized controlled trial published by Mortensen at al. (11); the methodology and results of the study are covered in detail elsewhere (11). Briefly, this was a single-site, double-blind, parallel group study that enrolled 75 heathy volunteers given acetylsalicylic acid (ASA) (300 mg) daily for 6 weeks. The participants were randomized to groups given oral capsules of Bif95 or placebo for 6 weeks. Small-intestinal damage was analyzed by serial VCEs for the duration of the study. All VCEs were performed using PillCam SBIII (Medtronic Ltd, Dublin, Ireland) and reviewed with Rapid 9 (Medtronic Ltd, Dublin, Ireland) capsule reading software.
For the purpose of the current study, we only selected the VCEs of patients in the placebo arm, i.e. ASA 300 mg for six weeks with placebo and discarded those who received ASA with Bif95. All studies were reviewed in consensus by VCE experts (UK, AG, RE) and images containing mucosal ulcers were extracted.
Software and Hardware
The models were written in Python (ver. 3.7) utilizing the open-source Keras (ver. 2.1.5) library and the open-source TensorFlow (ver. 1.5.0) library as backend. Models ran on an Intel i7 CPU and two GeForce GTX 1080ti Graphics Cards.
CNN Model
As previously described (17), an Xception CNN (19) was trained to classify capsule images into either images of normal mucosa or images with CD mucosal ulcers. The network's weights were initialized using weights from the 1.2 million everyday color images of ImageNet (20).
Preprocessing of capsule images included cropping of images' borders and legends. Images were then resized into a 299 × 299 matrix and pixels were normalized into the range 0–1 by dividing by 255 (17).
The following hyper-parameters were used for training the network: 2 epochs; batch size 8; Adam optimization with a learning rate of 10−4. Softmax was used as the output activation function.
Class Activation Maps
We used class activation maps (CAM) to analyze which parts of a given image led the network's decisions. This method is helpful for understanding the decision process of the CNN. Of particular interest, we wanted to investigate whether the network identifies NSAIDs ulcers as CD ulcers.
For this purpose, we've applied Gradient-weighted Class Activation Mapping (Grad-CAM). This algorithm uses the gradients of the target label, flowing into the final convolutional layer, to highlight the important regions in the image (21).
Metrics
We assessed the networks performance for detecting images containing NSAIDs mucosal ulcers.
Receiver operating curves (ROC) were plotted for the network's results by varying the operating threshold. The model's metrics included area under the curve (AUC), sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), and F1 score. All metrics were computed for the Youden's index and for fixed specificities of 90, 95 and 99%.
Bootstrapping validations (1,000 bootstrap resamples) were used to calculate 95% confidence intervals (CI) for the metrics.
Results
Study Cohort
Overall, the CD training dataset included 17,640 CE images from 49 patients; 7,391 images with mucosal ulcers and 10,249 images of normal mucosa. Out of 10,249 normal images, 3,577 originated from patients with normal CE and 6,672 from patients with CD (17).
The NSAIDs testing dataset included 1,605 CE images (11). This included 980 images of NSAIDs ulcers and 625 of normal mucosa.
CNN Results
The network's prediction time for the 1,605 images in the testing dataset was 8.9 s. This yields an average of 5.5 ms per single image. For a VCE film with 10,000 images, this amounts to 55 s.
Figure 3 presents the network's activation heatmaps for NSAID ulcers. As can be seen in the heatmaps, a network trained on CD ulcers, clearly detected NSAID ulcers.
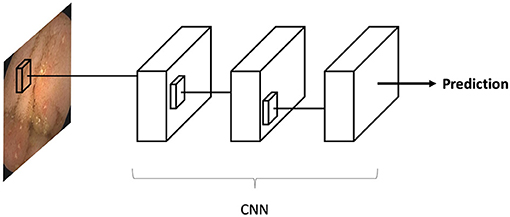
Figure 3. Three examples of the network's activation heatmaps for video capsule endoscopy images of NSAID ulcers.
The receiver operating characteristic curve for predicting NSAIDs ulcers is presented in Figure 4. The network exhibited an AUC of 0.97 (95% CI 0.97–0.98) for identifying images with NSAID mucosal ulcers. This is comparable to the previous results for CD patients, with AUCs of 0.94–0.99.
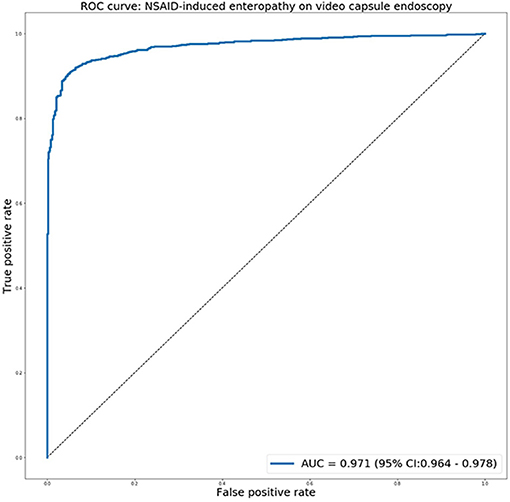
Figure 4. The receiver operating characteristic curve for predicting NSAIDs ulcers using the deep learning network.
A confusion matrix with examples of true and false, negative and positive, images is presented in Figure 5. The false negative example exhibits an ulcer at the edge of the image.
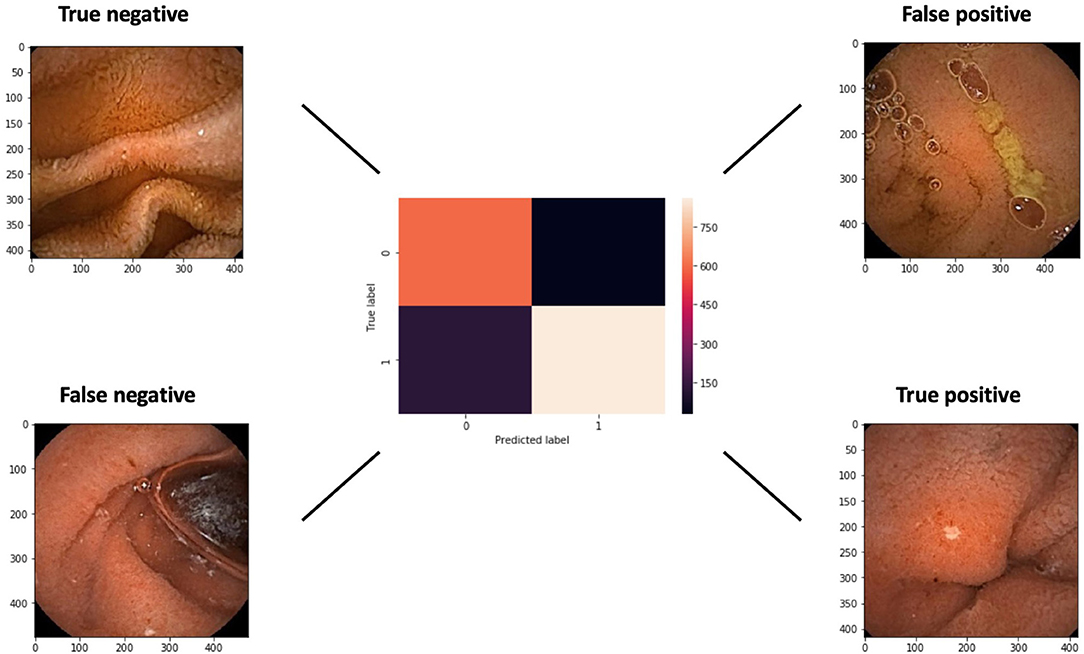
Figure 5. A confusion matrix of the networks results for a cut-off probability of 0.5. Examples of false and true negative and positive images are shown.
For Youden's index, the network showed sensitivity 92% and specificity 95%. For all specificities cut-off values, a high F1 score of >0.90 was shown (Table 1).
Discussion
In the past few years, there has been a surge of publications related to the use of artificial intelligence in gastroenterology and gastrointestinal endoscopy. Several studies have demonstrated the impressive results of detecting ulcers in VCE (16). In recent studies we have found that AI can accurately identify CD ulcers and also rank their severity (17, 18). However, small bowel ulcers are not specific to Crohn's disease, and may be caused by various other etiologies.
NSAID induced enteropathy is a common complication, potentially afflicting >50% of chronic NSAID users and may be detected even in occasional and low-dose NSAID users. NSAID-induced small bowel ulcers are a common cause of occult small bowel bleeding. As NSAID-induced ulcers are morphologically similar to those of CD, this entity should be considered as a potential differential diagnosis for suspected CD.
In order to train a neural network, a large number of images are needed (14). The construction of NSAID ulcers dataset is a complex endeavor. It is also methodologically challenging to generate a ‘clean' dataset of NSAID-induced lesions as other mimicking etiologies are often hard to exclude in real-practice patients. The present study utilized a unique dataset from a clinical trial in which healthy individuals received NSAID and underwent VCE, thereby overcoming the aforementioned methodological constraints. Using healthy individuals, who deliberately ingested aspirin to induce enteric ulcerations as part of a clinical trial is a strong methodological advantage of the present study allowing validated etio-morpholgical comparison with CD ulcers. This stands in contrast with any such future attempt using clinically accrued patients, in whom ascribing ulcers to aspirin or NSAIDs etiology cannot be definitively done in most cases.
The DL network, trained using a large dataset of CD ulcers, achieved an AUC of 0.97 for detecting NSAID-induced ulcers. This prediction capability is similar to the AUC (0.94–0.99) of the network for detecting CD ulcers (17). Our findings corroborate the morphological similarity of CD and NSAID-induced ulcers. Moreover, the network was very fast and efficient in detection of the ulcers, arguably much faster than the expected reading time of a human reader.
Our neural network has proved to be efficient in identifying NSAID induced ulcers; however, it is important to take into account that this system is trained solely on CD ulcers and thereby will identify both types of ulcers as the same entity. The current algorithm will be able to identify ulcers in general and will not be able to specifically determine whether it is a NSAID or a CD induced ulcer. Recently, DL has been implemented in the clinic for GI applications (22). When using this tool, physicians should be aware of the interactions between various pathologies as well as the limitations of AI.
This study has several limitations. This is a retrospective analysis of a prospectively collected data. Prospective interventional studies are needed to evaluate the retrospective results to show real-world usefulness. The study includes a limited number of images, particularly for NSAID induced ulcers. Moreover, DL is a rapidly emerging field, and new algorithms are continuously developed. In this work we've used the Xception model since it was used in a previous related work (17). Yet, this model has several limitations: 1. Xception CNN model network is compatible only with the TensorFlow backend, 2. Xception require 299 × 299 pixel inputs, 3. Xception image weights are between 90 and 100MB. Other models may show better results. Lastly, future projects should prospectively investigate networks in real-life scenarios.
The described method presents a classification model for still VCE images. Future studies should expand this method for entire video analysis to provide a wholistic prediction and to replicate clinical scores such as the Lewis or the CDEIS scores. Moreover, clinical data could be integrated into the final layers of the neural network to augment the visual data.
Conclusion
A network trained on VCE CD ulcers similarly identified NSAID enteropathy findings. As DL is transforming gastrointestinal endoscopy, this result should be taken into consideration in the future design and analysis of VCE deep learning applications.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Cork University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
EK had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. UK, EK, and SB-H: concept and design. EK, UK, BM, AD, SS, YB, EK, AG, RY, MB, FS, RE, and SB-H: acquisition, analysis or interpretation of data, critical revision of the manuscript for important intellectual content, and administrative, technical, or material support. UK, EK, and SS: drafting of the manuscript. EK: algorithm design and statistical analysis. UK, EK, SB-H, and RE: supervision. All authors contributed to the article and approved the submitted version.
Funding
The original study was funded by CHR Hansen.
Conflict of Interest
UK: speaker and advisory fees- ABBVIE, Jannsen, MSD, Takeda, Medtronic. Research support- Janssen Takeda Medtronic. SB-H: speaker and advisory fees for Takeda, Abbvie, Janssen, Celltrion, GSK, Pfizer and research support from Takeda, Celltrion, Janssen, and Abbvie. SB-H: received advisory and/or research support from Abbvie, MSD, Janssen, Celltrion, Takeda, GSK, Pfizer. RE: received advisory and/or research support from Abbvie, Janssen, Takeda and Medtronic. FS: co-founder of Alimentary Health Ltd, 4D pharma Cork and Atlantia Food Clinical Trials and has been scientific adviser to Kaleido Biosciences Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lanas A, Perez-Aisa MA, Feu F, Ponce J, Saperas E, Santolaria S, et al. A nationwide study of mortality associated with hospital admission due to severe gastrointestinal events and those associated with non-steroidal anti-inflammatory drug use. Am J Gastroenterol. (2005) 100:1685–93. doi: 10.1111/j.1572-0241.2005.41833.x
2. Lim YJ, Yang C-H. Non-steroidal anti-inflammatory drug-induced enteropathy. Clin Endosc. (2012) 45:138–44. doi: 10.5946/ce.2012.45.2.138
3. Maiden L, Thjodleifsson B, Seigal A, Bjarnason II, Scott D, Birgisson S, et al. Long-term effects of non-steroidal anti-inflammatory drugs and cyclooxygenase-2 selective agents on the small bowel: a cross-sectional capsule enteroscopy study. Clin Gastroenterol Hepatol. (2007) 5:1040–5. doi: 10.1016/j.cgh.2007.04.031
4. Maiden L. Capsule endoscopic diagnosis of non-steroidal anti-inflammatory drug-induced enteropathy. J Gastroenterol. (2009) 44(Suppl. 19):64–71. doi: 10.1007/s00535-008-2248-8
5. Graham DY, Opekun AR, Willingham FF, Qureshi WA. Visible small-intestinal mucosal injury in chronic NSAID users. Clin Gastroenterol Hepatol. (2005) 3:55–9. doi: 10.1016/s1542-3565(04)00603-2
6. Goldstein JL, Eisen GM, Lewis B, Gralnek IM, Zlotnick S, Fort JG, et al. Video capsule endoscopy to prospectively assess small bowel injury with celecoxib, naproxen plus omeprazole, and placebo. Clin Gastroenterol Hepatol. (2005) 3:133–41. doi: 10.1016/s1542-3565(04)00619-6
7. Sturm A, Maaser C, Calabrese E, Annese V, Fiorino G, Kucharzik T, et al. ECCO-ESGAR guideline for diagnostic assessment in IBD part 2: IBD scores and general principles and technical aspects. J Crohns Colitis. (2019) 13:273–84. doi: 10.1093/ecco-jcc/jjy114
8. Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, et al. ECCO-ESGAR guideline for diagnostic assessment in IBD part 1: initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. (2019) 13:144–64. doi: 10.1093/ecco-jcc/jjy113
9. Pennazio M, Spada C, Eliakim R, Keuchel M, May A, Mulder CJ, et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy. (2015) 47:352–76. doi: 10.1055/s-0034-1391855
10. Lim D-H, Jung K, Lee SB, Park IK, Cha HJ, Park JH, et al. Non-steroidal anti-inflammatory drug-induced enteropathy as a major risk factor for small bowel bleeding: a retrospective study. BMC Gastroenterol. (2020) 20:178. doi: 10.1186/s12876-020-01329-5
11. Mortensen B, Murphy C, O'Grady J, Lucey M, Elsafi G, Barry L, et al. Bifidobacteriumbreve Bif195 protects against small-intestinal damage caused by acetylsalicylic acid in healthy volunteers. Gastroenterology. (2019) 157:637–646.e4. doi: 10.1053/j.gastro.2019.05.008
12. Taha AS, McCloskey C, McSkimming P, McConnachie A. Misoprostol for small bowel ulcers in patients with obscure bleeding taking aspirin and non-steroidal anti-inflammatory drugs (MASTERS): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Gastroenterol Hepatol. (2018) 3:469–76. doi: 10.1016/S2468-1253(18)30119-5
13. Klang E. Deep learning and medical imaging. J Thorac Dis. (2018) 10:1325–8. doi: 10.21037/jtd.2018.02.76
14. Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional neural networks for radiologic images: a radiologist's guide. Radiology. (2019) 290:590–606. doi: 10.1148/radiol.2018180547
15. Barash Y, Klang E. Automated quantitative assessment of oncological disease progression using deep learning. Ann Transl Med. (2019) 7:S379. doi: 10.21037/atm.2019.12.101
16. Soffer S, Klang E, Shimon O, Nachmias N, Eliakim R, Ben-Horin S, et al. Deep learning for wireless capsule endoscopy: a systematic review and meta-analysis. Gastrointest Endosc. (2020) 92:831–839.e8. doi: 10.1016/j.gie.2020.04.039
17. Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, et al. Deep learning algorithms for automated detection of Crohn's disease ulcers by video capsule endoscopy. Gastrointest Endosc. (2020) 91:606–13.e2. doi: 10.1016/j.gie.2019.11.012
18. Yiftach B, Liran A, Soffer S, Reuma YM, Oranit S, Shomron B-H, et al. Ulcer severity grading in video-capsule images of Crohn's disease patients: an ordinal neural network solution. Gastrointest Endosc. (2020) 93:187–92. doi: 10.1016/j.gie.2020.05.066
19. Chollet F. Xception: deep learning with depth wise separable convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI (2017). p. 1800–7.
20. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. (2015) 115:211–52. doi: 10.1007/s11263-015-0816-y
21. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of 2017 IEEE International Conference on Computer Vision (ICCV). Honolulu, HI (2017). p. 618–26.
22. Medtronic. GI GeniusTM Intelligent Endoscopy Module [Internet]. Available online at: https://www.medtronic.com/covidien/en-us/products/gastrointestinal-artificial-intelligence/gi-genius-intelligent-endoscopy.html (accessed Aug 21, 2020).
Keywords: deep learning, capsule endoscopy, Crohn disease, anti-inflammatory agents, non-steroidal, NSAID
Citation: Klang E, Kopylov U, Mortensen B, Damholt A, Soffer S, Barash Y, Konen E, Grinman A, Yehuda RM, Buckley M, Shanahan F, Eliakim R and Ben-Horin S (2021) A Convolutional Neural Network Deep Learning Model Trained on CD Ulcers Images Accurately Identifies NSAID Ulcers. Front. Med. 8:656493. doi: 10.3389/fmed.2021.656493
Received: 20 January 2021; Accepted: 10 August 2021;
Published: 27 August 2021.
Edited by:
Reena Sidhu, Sheffield Teaching Hospital, United KingdomReviewed by:
Nazri Mustaffa, Universiti Sains Malaysia (USM), MalaysiaRadhakrishnan Delhibabu, VIT University, India
Copyright © 2021 Klang, Kopylov, Mortensen, Damholt, Soffer, Barash, Konen, Grinman, Yehuda, Buckley, Shanahan, Eliakim and Ben-Horin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eyal Klang, eyal.klang@mountsinai.org
†These authors have contributed equally to this work and share first authorship
 Eyal Klang
Eyal Klang Uri Kopylov
Uri Kopylov Brynjulf Mortensen4
Brynjulf Mortensen4  Anders Damholt
Anders Damholt Shelly Soffer
Shelly Soffer Martin Buckley
Martin Buckley Fergus Shanahan
Fergus Shanahan