- 1Key Laboratory of Dairy Science, Ministry of Education, Northeast Agricultural University, Harbin, China
- 2Food Science and Nutrition Unit, Department of Animal Science, Faculty of Agriculture, University of Benin, Benin City, Nigeria
- 3Department of Food Science, Food College, Northeast Agricultural University, Harbin, China
Enterococcus durans KLDS6.0930 has previously been shown to have probiotic potential. However, being a potential clinical pathogen, it becomes necessary to evaluate its safety status for novel potential probiotic use. The purpose of this study is to systematically evaluate the safety of E. durans KLDS6.0930 based on its genomics, phenotypic characteristics and oral toxicity. The complete genome of E. durans KLDS6.0930 was sequenced and analyzed for safety-related genes. Antibiotic susceptibility and the production of harmful metabolites were tested. A 28-day repeated oral dose toxicity test was implemented in rats. In vitro, E. durans KLDS6.0930 was resistant to five antibiotics, with intrinsic resistances to four antibiotics and no identified genes for the last. E. durans KLDS6.0930 was not hemolytic and virulence factors were non-functional in its genome. E. durans KLDS6.0930 produced a small amount of tyramine and phenethylamine; genes encoding tyramine decarboxylase were identified. In addition, genotype and phenotype analyses showed that the strain did not have the ability to generate D-lactic acid, indole, or nitroreductase. In vivo, E. durans KLDS6.0930 did not induce adverse effects on the organs, hematological and serum biochemical parameters, or cecal bacterial populations in the oral toxicity test. These results indicate that E. durans KLDS6.0930 can be safely used as a potential probiotic for human consumption and animal feed.
Introduction
Enterococcus, a large genus of lactic acid bacteria (LAB), includes 54 species characterized as gram-positive, facultative anaerobic, and non-spore forming (Foulquié Moreno et al., 2006; Van Tyne and Gilmore, 2014). Enterococci are long-standing and non-pathogenic commensal bacteria in the gastrointestinal tract (GIT) of humans and animals (Byappanahalli et al., 2012; Weng et al., 2013; Jahan et al., 2015). They are among the first LAB to colonize the neonatal GIT (Fanaro et al., 2003) and could be involved in the development of the human microbiome (Dominguez-Bello et al., 2010). Enterococci are also widely distributed in foods and the environment, playing important roles in shelf-life improvement and the development of both flavor and texture when present in fermentation foods such as certain cheeses and fermented sausages (Rea et al., 2004; Gaglio et al., 2016; Santos et al., 2017). Furthermore, some Enterococcus spp. have been shown beneficial properties, such as immune system regulation, normal intestinal microflora maintenance, antitumor activity, antimicrobial activity, antioxidant activity, and lowering cholesterol levels (Pieniz et al., 2014; Molina et al., 2015; Guo et al., 2016; Li P. et al., 2017). They are therefore frequently used as probiotics to promote human and animal health, or treat diseases such as diarrhea, irritable bowel syndrome, or antibiotic-associated diarrhea (Franz et al., 2011).
Despite these benefits, some enterococci are also considered as opportunistic zoonotic pathogens, associated with nosocomial infections like urinary tract infections (the second most common causative agent), bacteraemia (the third most commonly isolated agent), surgical site infections, bloodstream infections, endocarditis and diarrhea (Schaberg et al., 1991; Foulquié Moreno et al., 2006). In addition, production of hazardous compounds by some enterococcal strains can be toxic in food production (Franz et al., 2001; Choi and Woo, 2013; Camargo et al., 2014). Due to properties of enterococci as both beneficial organisms and nosocomial pathogens, verifying the safety of a novel probiotic strain is of utmost importance. However, simple safety evaluations of enterococci are difficult because they usually carry multiple antibiotic resistances and virulence factors (EFSA, 2008). For this reason, comprehensive safety assessment of Enterococcus must be taken into consideration.
E. durans KLDS6.0930 strain evaluated in this study was originally isolated from traditional naturally fermented cream from Inner Mongolia in China. Previous in vitro and in vivo studies indicated that E. durans KLDS6.0930 possesses potential probiotic properties included acid and bile tolerance, adherence to Caco-2 cells and a cholesterol-lowering effect (Guo et al., 2016). The genome of E. durans KLDS6.0930 was the first whole genome sequence of E. durans and has been used as representative genome by GenBank. Genetic elements with potential probiotic properties were exploited in our previous report (Liu et al., 2016). However, to guarantee the safe use of this strain as a potential probiotic or bioprotective adjunct culture, it is necessary to comprehensively evaluate any potential adverse effects. Thus, the aim of this study is to fully assess the safety status of E. durans KLDS6.0930 using genomic data, phenotypic assays and oral toxicity studies.
Materials and Methods
Bacterial Strains and Culture Conditions
E. durans KLDS6.0930 was isolated from traditional naturally fermented cream from Inner Mongolia in China and was made available at the Key Laboratory of Dairy Science (KLDS), Northeast Agricultural University (NEAU), Harbin, China. E. durans KLDS6.0930 was maintained in M17 broth (Oxoid Ltd, Basingstoke, UK) with 10% glycerol and stored at −20°C. Before use, E. durans KLDS6.0930 was activated through three propagation steps in M17 broth at 37°C for 24 h. Bacteria were harvested by centrifugation at 5,000 g for 10 min. Cell pellets were then washed twice with sterile normal saline and resuspended at the desired concentration in sterile normal saline. Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were incubated in Luria-Bertani (LB) broth and Tryptic Soy Broth (TSB) under aerobic conditions at 37°C, respectively.
Genome Sequencing and Taxonomy
E. durans KLDS6.0930 genomic DNA was extracted using the DNeasy Tissue kit (Qiagen, Hilden, Germany) following the manufacturer's instruction. Detailed information on the genome sequencing study can be found in our previous report (Liu et al., 2016). The 16S rRNA gene sequence was compared with closely related sequences from GenBank using BLASTN and aligned using CLUSTALW software (Thompson et al., 1997). The phylogenetic tree was reconstructed using the neighbor-joining method with bootstrap analysis based on 1,000 resamplings using MEGA 7.0 package software (Kumar et al., 2016). The average nucleotide identity (ANI) of the two genomic sequences was evaluated by ANI calculator using the OrthoANIu algorithm (Yoon et al., 2017). The availability of the whole genome of E. durans KLDS6.0930 allowed the development of the ribosomal multilocus sequence typing (rMLST) scheme, which was based on 49 genes encoding the bacterial ribosome protein subunits (rps genes), as performed in rMLST database website (http://pubmlst.org/rmlst/). The ribosomal protein sequences retrieved from the genomes were aligned with CLUSTALW software, while neighbor-joining method was used to infer the phylogenetic tree.
Genomic Analysis of Safety-Related Genes
The predicted genes of E. durans KLDS6.0930 were compared with the comprehensive antibiotic resistance database (CARD, http://arpcard.mcmaster.ca/) (Mcarthur et al., 2013) and virulence factors database (VFDB, http://www.mgc.ac.cn/VFs/main.htm) (Chen et al., 2016) for identifying antibiotic resistance and virulence factor. Furthermore, the ResFinder 3.0 (https://cge.cbs.dtu.dk//services/ResFinder/) (Zankari et al., 2012) and PathogenFinder 1.1 (https://cge.cbs.dtu.dk/services/PathogenFinder/) (Cosentino et al., 2013) were used for identifying the acquired antibiotic resistance genes and pathogenicity factors, respectively. Clustered regularly interspersed short palindromic repeats (CRISPR) and prophage sequences were identified by CRISPR Finder (http://crispr.i2bc.paris-saclay.fr/Server/) (Grissa et al., 2007) and PHASTER (http://phaster.ca/) (Arndt et al., 2016), respectively. The predicted genes were annotated against the restriction enzyme database (REBASE, http://tools.neb.com/genomes/) (Roberts et al., 2015) for searching restriction enzyme. The gene sequences associated to the adverse metabolites such as amino acid decarboxylases, nitroreductase, and D-lactate dehydrogenase, were referenced from Genbank and inquired the genome by BLASTN.
Antibiotic Susceptibility Assay
Minimum inhibitory concentrations (MICs) for antibiotics (ampicillin, penicillin, vancomycin, erythromycin, tetracycline, gentamicin, kanamycin, clindamycin, ciprofloxacin, trimethoprim, and rifampicin) against E. durans KLDS6.0930 were determined using a 96-well plate gradient dilution method (Guo et al., 2016a). Susceptibility was determined on the criteria adopted by European Food Safety Authority (EFSA) for the assessment of bacterial resistance to antibiotics (EFSA, 2012a).
Hemolytic Activity
The hemolytic activity of E. durans KLDS6.0930 was tested by incubating bacteria on blood agar (7% v/v sheep blood) for 48 h at 37°C (Pieniz et al., 2014). α-hemolysis (green-hued halo around the colonies) or β-hemolysis(clear halo around colonies) implied positive hemolytic activity. γ-hemolysis (absence of clearing zone surrounding colonies) was classified as a negative result. S. aureus ATCC 25923 was used as a positive control.
Biofilm Formation
Biofilm formation was evaluated using a previously described method (Kopit et al., 2014) with some modifications. Briefly, bacteria was grown overnight in M17 broth at 37°C. After 24 h, 180 μL fresh M17 broth and 20 μL cell suspension were combined in a sterile 96-well polystyrene microtiter plate. A negative control using 200 μL M17 broth alone was also added. S. aureus ATCC 25923 was used as a positive control. After incubation of the strain at 37°C for 24 h, wells were washed two times with sterile saline, inverted for 15 min to dry and stained with 1% (w/v) crystal violet for 15 min. The wells were then rinsed again with sterile saline and filled with 200 μL of acetone and ethanol solution (20:80, v/v). Absorbance at 595 nm was measured by using a microplate reader (Model 680 BIO-RAD, China). The cut-off OD (ODc) was defined as three standard deviations above the mean OD of the negative control. The ability to form biofilm was classified according to Stepanovic et al. (2000) as follows: OD ≤ ODc as non-biofilm-producer (0), ODc < OD ≤ 2 OD as weak biofilm producer (+), 2 ODc < OD ≤ 4 ODc as moderate biofilm producer (++), and OD ≥4 ODc as strong biofilm producer (+++).
Detection of Harmful Metabolites
Biogenic amines produced by E. durans KLDS6.0930 were quantified by high performance liquid chromatography (HPLC). Pretreatment and dansyl chloride derivatization of samples were performed as previously reported (Dadáková et al., 2009; Lorencová et al., 2012). HPLC measurements were performed on Waters Alliance HPLC system (Waters e2695, USA) equipped with a binary pump and a UV/Vis detector (λ = 245 nm). Samples were separated on a C-18 column (250 mm × 4.6 mm, 5 μm, Japan). Separation and examination of biogenic amines were performed according to Smělá et al. (2004). Chromatograms of biogenic amines (phenylethylamine, tryptamine, cadaverine, putrescine, histamine, tyramine, spermine, and spermidine) in standard solution are presented in Figure S1.
The optical purity of the resulting lactic acid was estimated using an L/D-lactic acid enzymatic test kit following the manufacturer's instructions (Megazyme, Ireland). The ability of E. durans KLDS6.0930 to produce indole was evaluated using Kovacs's indole kit (Hopebio-Technology Co., Ltd, China). A red ring was regarded as positive. E. coli ATCC25922 was used as a positive control. The activity of nitroreductase was determined using the assay kit according to the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute, China). E. coli ATCC25922 was used as a positive control.
Animals
Specific Pathogen Free (SPF) Sprague-Dawley (SD) rats aged 6–7 weeks were supplied by Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All rats were randomly put into plastic cages in an animal chamber under controlled environmental conditions with a 12-h light/dark cycle at a temperature of 23 ± 2°C and a relative humidity of 50 ± 20%. The rats were acclimatized to the laboratory conditions for 1 week before the start of the experiment and permitted with free access to drinking water and standard diet. The experimental protocol was approved by the guidelines of Animal Care and Use Committee of Northeast Agricultural University (SRM-06).
Subacute Oral Toxicity
This study was performed according to Organization for Economic Co-operation and Development (OECD) test guidelines (OECD, 2008). Male and female rats were randomly divided into two groups of six rats for each gender. Rats in the treatment group were orally gavaged with 1 mL E. durans KLDS6.0930 at 1 × 109 CFU/kg body weight (BW) once daily for 28 days. The control group was administered with 1 mL sterile normal saline. During the treatment period, all rats were observed daily for clinical signs of morbidity and mortality. BW and feed intake were monitored weekly. At the end of the dosing period, all rats were deprived of food and water for 16 h before humane sacrifice under diethyl ether anesthesia.
Hematological and Serum Biochemistry Analyses
Hematological variables were analyzed in all rats at the end of the treatment period. Blood samples were collected in heparinized tubes to assess red blood cell count (RBC), white blood cell count (WBC), hemoglobin (HGB), platelet count (PLT), mean corpuscular volume (MCV), mean corpuscular hemoglobin concentration (MCHC), neutrophils, monocytes, lymphocytes, and eosinophils using an automatic hematological analyzer (Nihon Kohden, Japan).
Whole blood was centrifuged to obtain serum for analysis on an automatic biochemistry analyzer (Toshiba, Japan) for the following parameters: aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), total bilirubin (TBIL), total protein (TP), albumin (ALB), glucose (GLU), triglycerides (TG), total cholesterol (TC), high density lipoprotein cholesterol (HDL), low density lipoprotein cholesterol (LDL), urea (UREA), creatinine (CRE), sodium (Na), chloride (Cl), calcium (Ca), and inorganic phosphorus (P).
Gross Necropsy and Histopathological Examination
Organs including heart, spleen, liver, kidney, lung, brain, adrenal thymus, epididymides (male rats), testes (male rats), ovaries (female rats), and uterus (female rats) were collected for gross necropsy and weighed. The organ index was calculated as a percentage of organ weight to terminal BW. Selected organs (heart, spleen, liver, kidney, lung, brain, and jejunum) were excised and fixed in 10% neutral buffered formalin for histopathology. The samples were then embedded in paraffin, cut into 5 μm thickness slices, and stained with hematoxylin-eosin for microscopic examination.
Cecal Microbiota
Total microbiota genomic DNA was extracted from cecal contents using a QIAamp DNA stool mini kit (Qiagen, Germany). Isolated DNA was examined by agarose gel electrophoresis and quantified using an ND-2000C spectrophotometer (NanoDrop, USA). Modified fusion primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), containing a 6-bp error-correcting barcode were used to amplify the V4 hypervariable region of the bacterial 16S rRNA gene. The PCR products were quantified using an Agilent DNA 1000 Kit on an Agilent 2100 Bioanalyser (Agilent Technologies, USA), pooled together and sequenced using the Illumina Miseq (Illumina, USA). In silico analysis of sequenced data was performed using quantitative insights into microbial ecology (QIIME) (Caporaso et al., 2010).
Cecal Colonization
E. durans KLDS6.0930 at 2 × 108 CFU/mL was fluorescently labeled with carboxyfluorescein diacetate succinimidyl ester (cFDA-SE) (Beyotime, China) according to the manufacturer's instructions. Male rats were randomly divided in two groups of six rats each. The control group received 1 mL normal saline. The experimental group was treated with 2 × 108 CFU E. durans KLDS6.0930 labeled with cFDA-SE at by gavage in 1 mL of sterile normal saline. After administration for 1 day, test rats were anesthetized with diethyl ether and sacrificed for cecum tissue collection. Cecal contents were carefully removed, and the cecal wall was rinsed with 1 mL sterile normal saline. Samples were stored in the dark and subjected to flow cytometry (BD LSR Fortessa) using a 488 nm laser.
Statistical Analysis
All experiments were conducted at least in triplicate using independent assays. Values are expressed as means ± the standard deviation (SD). Statistical significance of data comparisons was calculated by an unpaired Student's t-test in the subacute toxicity assays. Values of P < 0.05 were considered to be statistically significant.
Results
Taxonomic Identification and Defense Systems
At the genomic level, the ANI value between the strain KLDS6.0930 and E. durans ATCC6056 was 99.62%. This agrees with the phylogenetic analysis based on 16S rRNA gene sequence (Figure 1A) and the concatenation of 49 ribosomal protein sequences (Figure 1B). These taxonomic results indicated that strain KLDS6.0930 should be assigned to species E. durans.
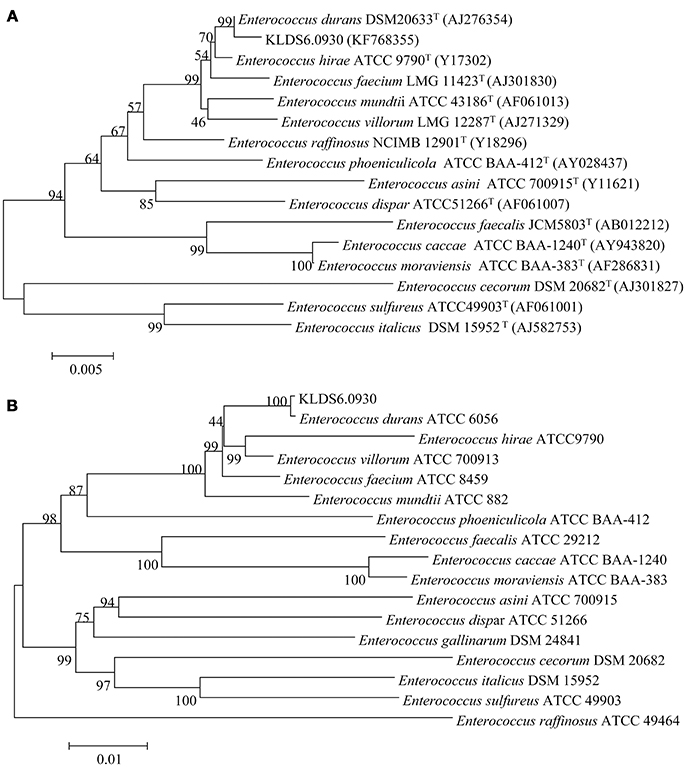
Figure 1. Phylogenetic trees of 16S rRNA gene (A) and concatenated 49 ribosomal protein sequences (B) using MEGA 7.0 software with the neighbor-joining method. Bootstrap values based on 1,000 resampled datasets are shown at branch nodes.
Two circular plasmids that do not encode any genes associated with true behavioral risk factors were detected in E. durans KLDS6.0930 (Figure 2A). Only a prophage region (GC content, 34.84%; length, 34.9 kb) with phage attachment sites (attL and attR) in the E. durans KLDS6.0930 chromosome (GC content, 38.00%; length, 2.87 Mb) was identified. This region contained 36 proteins including integrase, transposase, terminase, phage-like protein, tail protein, fiber protein, lysis protein, and hypothetical protein (Figure 2B).
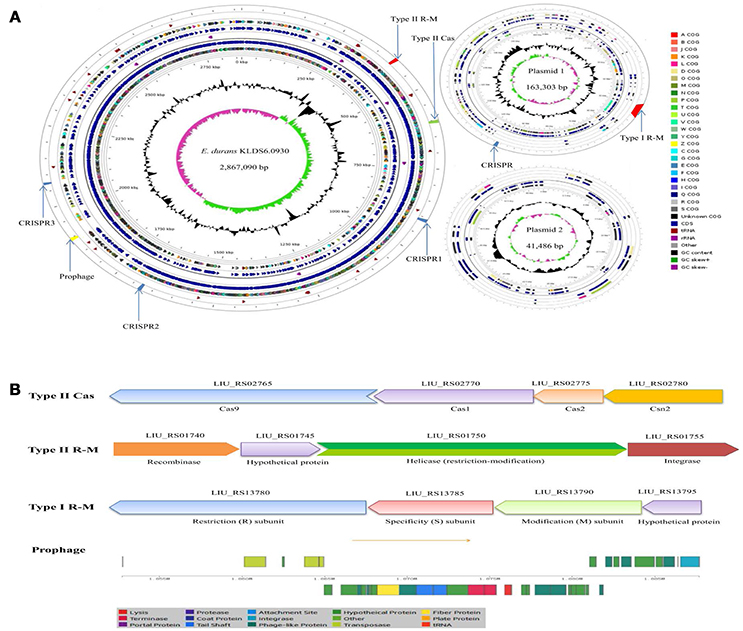
Figure 2. Circular genome map and defense systems of Enterococcus durans KLDS6.0930. (A) Circular genome map. From periphery to center: Defense system and prophage, tRNA, Protein coding genes (CDSs) on forward strand colored according to the assigned COG classes, Genes on forward strand, rRNA, Genes on reverse strand, CDSs on reverse strand colored according to the assigned COG classes, Genome position in kbp, GC content; GC skew (G-C)/(G + C); (B) Organizations of the Type II cas, Type II R-M, Type I R-M, and prophage operons.
In silico analysis showed that the defense system of E. durans KLDS6.0930 confers immunity against mobile elements. Results obtained from the CRISPR finder database displayed that three potential CRISPR arrays are distributed on the chromosome and one confirmed CRISPR array is located on plasmid 1 (Figure 2A). Additionally, a gene cluster (cas9, cas1, cas2, csn2) encoding Type II-A CRISPR associated enzymes (Cas) was found in the genome of E. durans KLDS6.0930 (Figure 2B). Genes encoding restriction enzymes and methyltransferases in E. durans KLDS6.0930 were determined by comparison against the REBASE database. As shown in Figures 2A,B, one Type II restriction-modification (R-M) system and one Type I R-M were observed in the genome and plasmid 1, respectively. These data indicated that the defense systems may endow E. durans KLDS6.0930 with distinct genetic stability.
Antibiotic Resistance and Associated Genes
Antibiotic resistance-related genes in E. durans KLDS6.0930 were identified by searching the CARD using the perfect and strict algorithms (Table 1). The genome of E. durans KLDS6.0930 was found to contain one curated aminoglycoside resistance gene and 12 putative resistance genes associated with resistance to beta-lactams (4), lincosamide (2), tetracycline (2), rifampin (1), aminocoumarin (1), trimethoprim (1), and peptide (1). Regarding the possibility of acquired resistance by horizontal gene transfer (HGT), the ResFinder software did not detect any acquired antibiotic resistance genes.
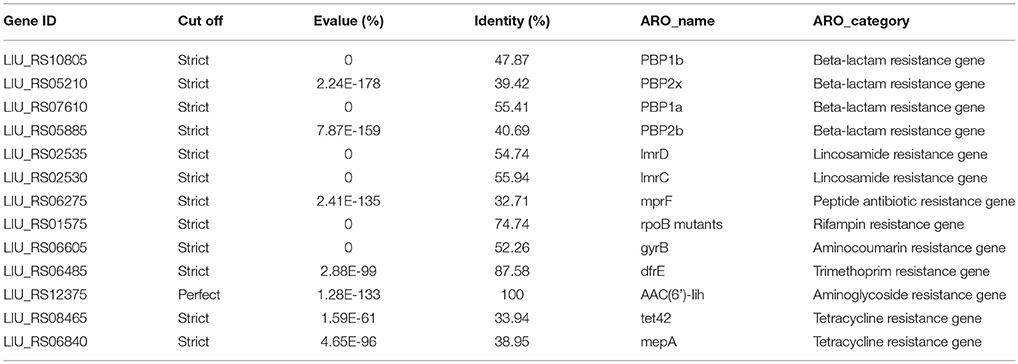
Table 1. Putative antibiotics resistance genes in the Enterococcus durans KLDS6.0930 genome by searching with the CARD.
Susceptibility of E. durans KLDS6.0930 to antibiotics was determined by measuring MICs, and the results were compared to the cut-off values for Enterococcus species as defined by EFSA (2008, 2012a). E. durans KLDS6.0930 was found to be resistant to gentamicin, kanamycin, clindamycin, ciprofloxacin, and rifampicin but was susceptible to ampicillin, penicillin, vancomycin, trimethoprim, and erythromycin (Table 2). The MIC of tetracycline was equal to the cut-off value.
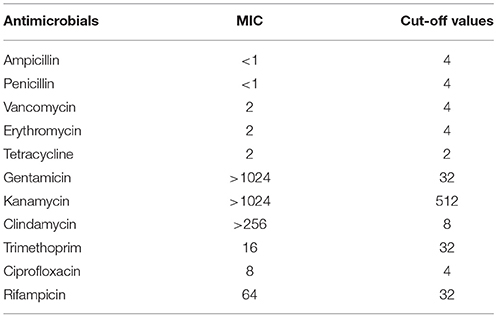
Table 2. Minimum inhibitory concentrations (MICs) of Enterococcus durans KLDS6.0930 toward antimicrobials and the microbiological cut-off values.
Putative Virulence Factors and Assigned Phenotype
Virulence genes contribute the pathogenicity to microorganisms. In order to mine possible virulence genes in the genome of E. durans KLDS6.0930, protein coding sequences (CDSs) were aligned to VFDB using BLASTP. The results showed that E. durans KLDS6.0930 genome and plasmid 1 carry a total of 45 putative virulence factors (Table S1), most of them are cell surface factors associated with host or surface adhesion and promoting biofilm formation.
The genome of E. durans KLDS6.0930 contains two genes encoding efa (LIU_RS03220 and LIU_RS03820), which contributes to adherence to extracellular matrix proteins and may cause endocarditis (Lowe et al., 1995). However, in the absence of other virulence factors, its presence poses no threat, since this gene was also found in starter E. faecium strains, which have a long record of safe use in food (Eaton and Gasson, 2001).
E. durans KLDS6.0930 harbors genes encoding two types of pili, PilA and PilB. A three-gene locus (pilA, pilE, and pilF), encoding PilA-type pilus structures was present in plasmid 1 of E. durans KLDS6.0930. Non-health care-associated E. faecium strains have been reported to express PilA, PilE, and PilF as cell wall-anchored proteins without forming pilus structures (Hendrickx et al., 2010). An operon of three genes, ebpA, ebpB, and ebpC, encoding the pilus subunits of PilB, was found in the genome of E. durans KLDS6.0930. However, the transcriptional regulator ebpR that is the promoter for the co-transcribed ebp operon is a pseudogene in the strain E. durans KLDS6.0930. Thus, pili were not detected on the surface of E. durans KLDS6.0930.
Biofilm formation is thought to be associated with the operon bopABCD, which appears to be regulated by the Fsr system through quorum sensing (Creti et al., 2006; Di Rosa et al., 2006). The operon bopABCD was found in the genome of E. durans KLDS6.0930, but bopD involved in transcriptional regulation of biofilm formation is a pseudogene. Moreover, fsr was absent in E. durans KLDS6.0930 genome. A biofilm formation assay showed that E. durans KLDS6.0930 was a weak biofilm producer, while S. aureus ATCC 25923 was classified as a strong biofilm producer (Table S2).
The term virulence factor refers to elements allowing a microorganism to colonize a host, contributing to the initiation and development of infection processes. Thus, it applies to secreted proteins, cell surface structures, and hydrolytic enzymes that contribute not only to bacterial pathogenicity but also to adhesion and protection (Wassenaar et al., 2015). Remarkably, many probiotic-related traits could be associated to virulence factors; therefore, in silico analyses must be considered in the appropriate context.
The aforementioned cell surface factors are involved in cell adhesion and intestinal colonization, which are considered potentially important for the initiation of infections in pathogens (Lowe et al., 1995; Mohamed and Huang, 2007; Sillanpää et al., 2010). As such, they are not offensive virulence factors, therefore, they could not be considered really harmful in probiotics. On the contrary, the ability to adhere to the intestinal wall is considered a desirable property for probiotics since it can allow the colonization of the GIT. Cell adhesion is a multistep process involving contact between the bacterial cell membrane and interacting surfaces. Despite the aforementioned cell surface factors, the genome of E. durans KLDS6.0930 also carries two fibronectin-binding proteins (LIU_RS07910 and LIU_RS10480) and an S-layer protein (LIU_RS11695), which may contribute to its adhesion. In addition, our previous study (Guo et al., 2016) showed that E. durans KLDS6.0930 could adhere to Caco-2 cells in vitro. As described below, we also demonstrated that E. durans KLDS6.0930 could colonize the cecum in vivo.
Interestingly, genes encoding exoenzymes and toxins such as gelatinase (gelE), serine protease (sprE), hyaluronidase (hyl), and cytolysin (cyl), which are commonly found in hospital-associated pathogenic strains of this species, were devoid of E. durans KLDS6.0930. In silico finding for cytolysin was confirmed by a phenotypic assay. After incubation on blood agar plates for 48 h, E. durans KLDS6.0930 did not exhibit any hemolytic effects (Figure 3). Notably, E. durans KLDS6.0930 also lacks IS16, a marker for hospital-associated strains (Werner et al., 2011) and esp, a marker of pathogenicity island (van Schaik et al., 2010).
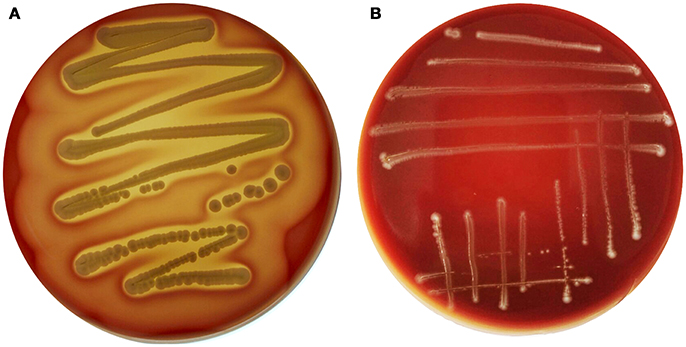
Figure 3. Hemolysin activity of Enterococcus durans KLDS6.0930. (A) The positive control strain, Staphylococcus aureus ATCC 25923, and (B) E. durans KLDS6.0930.
Using Pathogen Finder, a program that performs comparisons between pathogenic and non-pathogenic bacteria using whole genome sequence data, E. durans KLDS6.0930 was predicted to be non-pathogen. Combining this result with the virulence factor analysis indicates that E. durans KLDS6.0930 is a non-toxigenic strain.
Putatively Harmful Metabolites and Related Determinants
The biosynthetic pathway of different biogenic amines is shown in Figure 4. Genes encoding tyramine decarboxylase (TDC) were present in the genome of E. durans KLDS6.0930, including a separate TDC gene (LIU_RS02800) and a TDC gene cluster containing a TDC (LIU_RS00045), Na+/H+ antiporter (LIU_RS00035), tyrosine antiporter (LIU_RS00040), and tyrosine-tRNA ligase (LIU_RS00050).
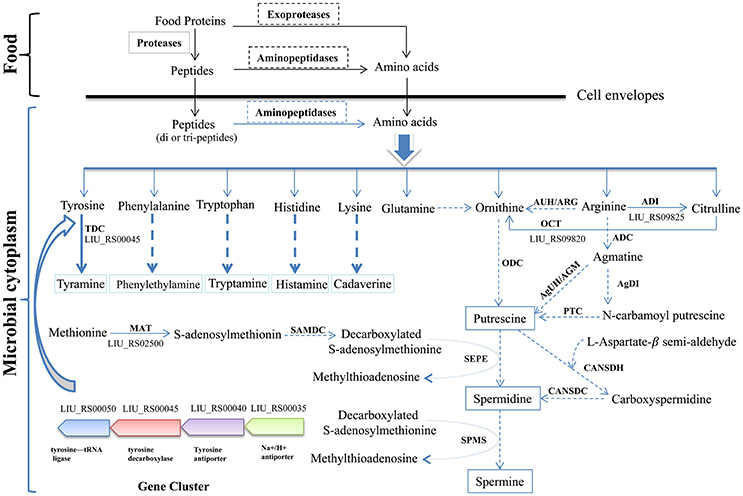
Figure 4. Biosynthesis pathways of different biogenic amines by Enterococcus durans KLDS6.0930. Solid arrows represent that the biosynthesis pathways of biogenic amine is effective, correspondingly, the dashed arrows imply invalid; ODC, ornithine decarboxylase; ARG/AUH, arginase/arginine ureohydrolase; ADI, arginine deiminase; ADC, arginine decarboxylase; AgDI, agmatine deiminase; PTC, putrescine transcarbamoylase; AGM/AgUH, agmatinase/agmatine ureohydrolase; OCT, Ornithine carbamoyl transferase; CANSDH, carboxynorspermidine dehydrogenase; CANSDC, carboxynorspermidine decarboxylase; MAT, methionine adenosyl transferase; SAMDC, S-adenosylmethionine decarboxylase; SEPE, spermidine synthase; SPMS, spermine synthase; TDC, tyramine decarboxylase.
In addition, polyamines such as putrescine, spermidine, spermine and agmatine are formed via different pathways using ornithine and arginine as the starting substrates (Figure 4). The transformation from arginine to ornithine can occur via two pathways. The first pathway is the direct conversion of ornithine from arginine by arginase/arginine ureohydrolase (ARG/AUH); the responsible gene was not found in the genome of E. durans KLDS6.0930. The second pathway is the conversion of arginine into citrulline and followed by catalysis to ornithine via arginine deiminase (ADI) and ornithine carbamoyl transferase (OCT). LIU_RS09825 and LIU_RS09820 encode these two enzymes, respectively, which indirectly catalyze the biosynthesis of ornithine accompanied by the generation of adenosine triphosphate (ATP) (Benkerroum, 2016). However, no genes encoding any of the enzymes for polyamine biosynthesis, except methionine adenosyltransferase (MAT), were found in the genome of E. durans KLDS6.0930. Therefore, the E. durans KLDS6.0930 genome only possesses the potential for tyramine production. HPLC results further confirmed that tyramine and phenylethylamine were produced by E. durans KLDS6.0930, at levels of 51.25 ± 4.38 and 10.48 ± 1.26 mg/L, respectively.
Only two genes (LIU_RS02290 and LIU_RS08060) encoding L-lactate dehydrogenase were identified in the genome of E. durans KLDS6.0930; genes encoding D-lactate dehydrogenase were not characterized. The optical purity of the L/D-lactic acid enzymatic assay showed that lactic acid produced by this strain consisted of 100% L-lactic acid. The E. durans KLDS6.0930 genome does not harbor any genes related to tryptophan metabolism (Figure S2), which can form indigo. Accordingly, this strain did not generate a red ring in the indigo matrix reaction cultures, compared with the positive control strain E. coli ATCC 25922 (Figure 5). LIU_RS10160, LIU_RS07350, and LIU_RS07350, encoding nitroreductase family proteins, were found in the genome of E. durans KLDS6.0930, though they were pseudogenes without any function. Correspondingly, no nitroreductase activity was detected in the fermented supernatant.
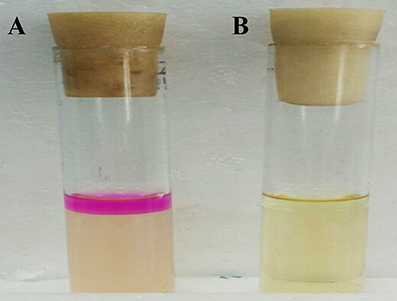
Figure 5. Detection result of the indole produced by Enterococcus durans KLDS6.0930. (A) The positive control strain, Escherichia coli ATCC 25922, and (B) E. durans KLDS6.0930.
Repeated Dose 28-Day Oral Toxicity Study
General Health Status and Body Weight Changes
General behavior and physical activity of rats were not abnormal for both groups during the 28-days observation period. In terms of weight gain and daily food consumption, there was no statistically significant difference in both genders of the treatment group when compared to the control group (Table S3).
Macroscopic Examination and Histopathology
No gross pathological findings were observed in rats of both two groups. Statistical analysis showed that the relative organ weights included heart, liver, spleen, lung, kidney, brain, adrenal, thymus, testis, epididymis, uterus, and ovary were not significantly different between the treatment group and the control group in both sexes (Table 3). Furthermore, light micrograph images of heart, liver, spleen, lung, kidney, brain, and jejunum are presented in Figure 6, there was no obvious histopathological abnormality (inflammation or necrosis) in these examined organs of rats from the treatment group.
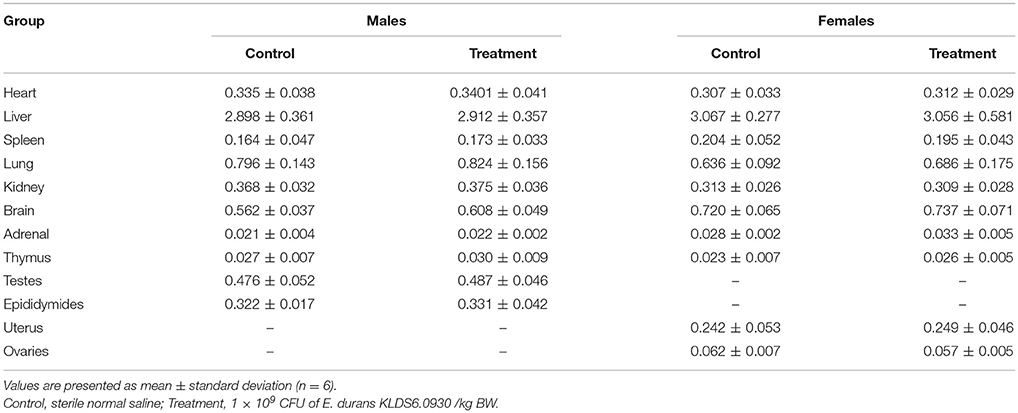
Table 3. Relative organ weights (%) of male and female rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days.
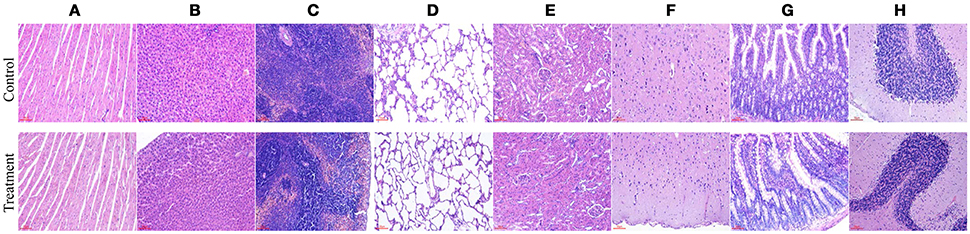
Figure 6. Representative photomicrographs of organs of rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days. Control, sterile normal saline; Treatment, 1 × 109 CFU of E. durans KLDS6.0930 /kg body weight (BW); (A) heart, (B) liver, (C) spleen, (D) lung, (E) kidney, (F) brain, (G) jejunum, (H) cerebellum. Bar = 50 μm.
Hematology Assay and Serum Biochemistry Profile
Hematological and serum biochemistry parameters are reported in Tables 4, 5, respectively. Hematology revealed no significant alterations were found between the treatment and control groups in both sexes. Serum biochemistry parameters showed that administration of E. durans KLDS6.0930 also did not result in any treatment-related effects on all rats of the treatment group compared with the control group. In summary, treatment with E. durans KLDS6.0930 did not cause any biological deviations outside of the normal ranges.
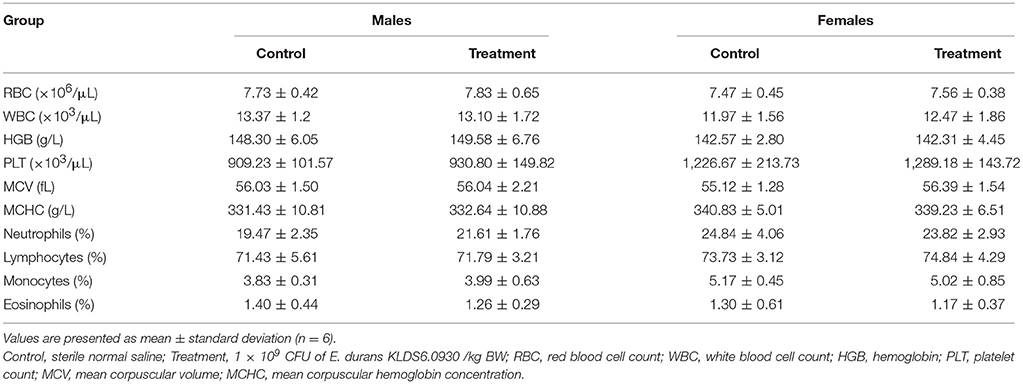
Table 4. Hematological parameters of male and female rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days.
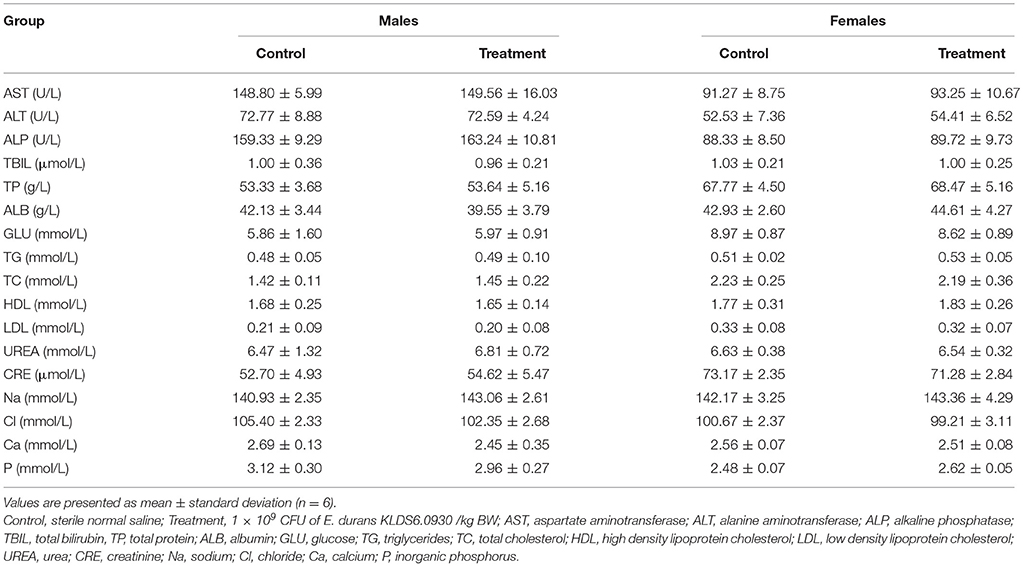
Table 5. Serum biochemical parameters of male and female rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days.
Cecal Microbiota
The effect of E. durans KLDS6.0930 on cecal microbial diversity was determined by amplicon sequencing and analyzed by α-diversity and β-diversity. There were no significant differences in α-diversity, which include Simpson index, Shannon index, observed species and Chao1 index (Figure 7) between the treatment and control groups in male rats. This implied that E. durans KLDS6.0930 treatment did not change the diversity and abundance of cecal microbiota of male rats.
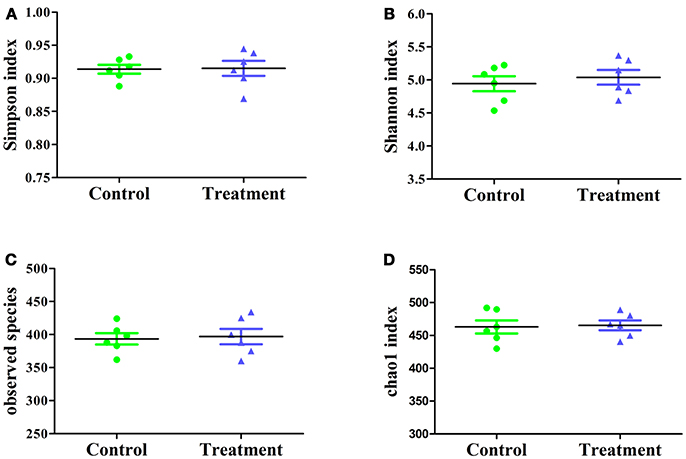
Figure 7. Cecal microbiota α-diversity in male rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days. Control, sterile normal saline; Treatment, 1 × 109 CFU of E. durans KLDS6.0930 /kg BW. (A) Simpson index, (B) Shannon index, (C) observed species, (D) Chao1 index.
Additionally, the sample distance based on weighted UniFrac analysis is shown in Figure 8. The differences in the inter-class distances of the treatment and control groups were not significant. Moreover, the distance between all samples was close, indicating that the differences in cecal microbial β-diversity of the treatment and control groups of male rats was very small. This agrees with the results for α-diversity, suggesting the E. durans KLDS6.0930 treatment did not disrupt the normal cecal microbiota. Similar results from both groups in female rats were obtained as well (data not shown). Therefore, changes in cecal microbiota due to E. durans KLDS6.0930 treatment are not harmful.
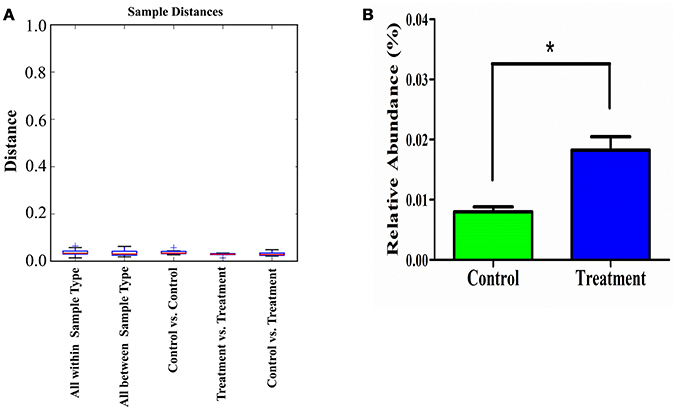
Figure 8. Weighted unifrac distance (β-diversity) of the cecal microbiota (A) and relative abundance of Enterococcus (B) in male rats after oral administration of Enterococcus durans KLDS6.0930 for 28 days. Control, sterile normal saline; Treatment, 1 × 109 CFU of E. durans KLDS6.0930 /kg BW. Statistical analysis was conducted by unpaired two-tailed student's t-test. *P < 0.05.
As shown in Figure 8, E. durans KLDS6.0930 administration specifically enriched the abundance of enterococci in treated rats, indirectly indicating E. durans KLDS6.0930 was present in the cecal content. Furthermore, we used cFDA-SE to trace E. durans KLDS6.0930 in the cecum (Figure S3). One day after gavage, the results from flow cytometry confirmed that E. durans KLDS6.0930 labeled with cFDA-SE was detectable in the cecum of rats, collating our previous results that E. durans KLDS6.0930 is highly tolerant to acid and bile and adherent to Caco-2 cells in vitro via the necessary genetic elements (Guo et al., 2016; Liu et al., 2016). These findings indicate that E. durans KLDS6.0930 can survive and colonize the cecum.
Discussion
Enterococci have gained notoriety over the past few decades as leading nosocomial pathogens, but have recently been investigated as potential probiotic agents (Ogier and Serror, 2008). Concerns about the use of enterococci as a double-edged sword have led to an increasing number of studies intended to evaluate the safety of certain enterococci strains (Kopit et al., 2014; Zhang et al., 2016; Peng et al., 2017).
Our previous studies suggest that E. durans KLDS6.0930 may be a potential probiotic strain. According to the FAO/WHO, probiotic microorganisms must be non-pathogenic and non-toxic (FAO/WHO, 2012). In this study, E. durans KLDS6.0930 was exhaustively analyzed for safety using multiple methods.
E. durans KLDS6.0930 carries potential CRISPR arrays and Type II-A CRISPR associated enzymes. A previous study showed that CRISPR serves as a formidable defense against phage invasion as well as restricting the dissemination of antibiotic resistance genes (Marraffini and Sontheimer, 2008). In addition, E. durans KLDS6.0930 also contains R-M systems, providing another form of genome defense through self-recognition vs. non-self-recognition of methylation signatures (Price et al., 2016). Supporting these, only one prophage was found in the genome of E. durans KLDS6.0930. In addition, no functional genes for the portal protein, one of the essential phage proteins, were found, suggesting that the prophage was defective (Isidro et al., 2004). These defense systems may play key roles in the stability of the E. durans KLDS6.0930 genome.
E. durans KLDS6.0930 lacks the established markers associated with the clinical strains, including esp, hyl, and IS16, which were specified by the EFSA as targets for the safety evaluation of E. faecium strains intended as additives for animal feed (EFSA, 2012b). On the other hand, enterococci that contain a type II-A CRISPR system are reported to be non-pathogenic (Palmer and Gilmore, 2010). As noted above, E. durans KLDS6.0930 carries a type II-A CRISPR, further supporting its non-pathogenicity.
Emerging evidence indicates that human intestinal bacteria can be a reservoir for antibiotic resistance genes that can be transferred to human pathogens. For a bacterial strain to qualify as a probiotic candidate, its susceptibility or resistance to a range of antibiotics must be assessed (EFSA, 2012a). Antibiotic resistance assays showed that E. durans KLDS6.0930 was resistant to aminoglycosides, clindamycin, and rifampicin, and genes associated with resistance to aminoglycosides, clindamycin, and rifampin resistance were detected. The resistance profile of E. durans KLDS6.0930 requires further investigation to determine the nature of the antibiotic resistance (intrinsic resistance or acquired resistance). According to the present EFSA guidelines on the characterization of microbiological feed additives (EFSA, 2018), a non-intrinsic resistance combined with the presence of a resistance-encoding gene disqualifies the strain as a feed additive in the European Union (EU). Initially, it may be necessary to distinguish between acquired and intrinsic resistances when there is limited information on MICs within the relevant taxonomical unit (EFSA, 2012a). In this case, the structural nature and genetic basis of the antibiotic resistance must be demonstrated by analyzing a representative selection of strains belonging to that taxonomical unit. As there are limited data on antibiotic resistances of E. durans, we analyzed the genetic basis of antibiotic resistances in E. durans KLDS6.0930 genome by comparing it with eight other sequenced E. durans genomes. Table S4 shows that genes associated with resistance to aminoglycosides, clindamycin and rifampin were identified in all analyzed E. durans genomes. Furthermore, no mobile elements, such as transposases or insertion sequences, were found in the flanking regions of these antibiotic resistance genes. This implied that these antibiotic resistance genes are ubiquitous in the genome of E. durans, meaning that resistances to aminoglycosides, clindamycin, and rifampin are intrinsic resistances. However, these antibiotic resistances of all other E. durans strains should be further investigated.
Generally, the phenotype does not completely reflect the genotype. Enterococcal isolates are often resistant to β-lactam antibiotics, likely due to the expression of low-affinity penicillin-binding protein 5 (PBP5) (Fontana et al., 1996). Four genes encoding PBPs (LIU_RS05885, LIU_RS07610, LIU_RS08470, LIU_RS10805) were found in E. durans KLDS6.0930 genome. The genes are annotated as PBP2b, PBP1b, PBP1a, and PBP2x, and have a low identity with PBP5 according to BLASTP. Despite the presence of these PBPs, however, E. durans KLDS6.0930 is sensitive to penicillin and ampicillin, a phenomenon also observed in E. hirae R17 (Peng et al., 2017).
E. durans KLDS6.0930 was sensitive to trimethoprim but contained the trimethoprim resistance gene (LIU_RS06485). Interestingly, the MIC value of tetracycline for E. durans KLDS6.0930 was equal to the cut-off value for Enterococcus species. Based on EFSA guidelines (EFSA, 2012a), strains are considered susceptible when the MIC value is equal to or lower than the reference cut-off value. Therefore, E. durans KLDS6.0930 is susceptible to tetracycline, nevertheless, two tetracycline resistance genes were found in the genome. Expression of these genes may have been sufficiently low or induced only under specific conditions, such as environmental stimuli or signals. Additional studies are needed to resolve this discrepancy, as differences between the antibiotic resistance phenotype and genotype may result from modification of genes identified in silico after transcription, pseudogenes, or misidentification due to only partial similarities to known resistance genes. The presence of these resistance genes, therefore, does not represent a dangerous characteristic of the bacterium.
Biogenic amines have a strong impact on maintenance of normal physiological function of the human body. Nonetheless, they may be hazardous to human health if their levels in foods or beverages reach a critical threshold (EFSA, 2011). Therefore, it is essential to evaluate enterococcal strains for their ability to produce biogenic amines. Genes for biogenic amine production were generally absent based on the genomic data of E. durans KLDS6.0930, except those encoding tyramine. Tyramine, phenylethylamine, histamine, tryptamine, and cadaverine are directly catalyzed by amino acid decarboxylases from their respective precursor amino acids (Benkerroum, 2016). Two non-homologous TDC-coding genes were found in the E. durans KLDS6.0930 genome. It is likely that these genes confer the potential of tyramine biosynthesis to the strain. Tyramine, together with tryptamine and phenylethylamine, are classified as vasoactive amines, as high levels can provoke dangerous hypertension (Mohedano et al., 2014). Fortunately, HPLC showed that E. durans KLDS6.0930 produced 51.25 ± 4.38 mg/L tyramine, lower than the toxic level of tyramine (100–800 mg/kg) in foods (ten Brink et al., 1990).
Usually, phenylethylamine production is associated with tyramine production, as demonstrated for some Enterococci (Bover-Cid et al., 2001). E. durans KLDS6.0930 also produced 10.48 ± 1.26 mg/L phenethylamine, under the threshold value of 30 mg/kg (ten Brink et al., 1990). However, E. durans KLDS6.0930 was not found to encode any of the genes related to phenethylamine. A previous study showed that a decarboxylase can decarboxylate different structural homologs (Benkerroum, 2016). Here, the TDC of Enterococcus can accept tyrosine and phenylalanine as substrates (Marcobal et al., 2006, 2012; EFSA, 2011), as they are aromatic amino acids with similar structures.
As a part of extensive safety assessments, in vivo studies can provide valuable information. The physiology of rats behaves similarly in some situations to that of humans, making rats a suitable model for toxicity studies. A 28-day repeated oral dose toxicity study was performed to assess the safety of E. durans KLDS6.0930 in a rat model. No significant strain-related toxigenic symptoms in the physical appearance of male or female rats were observed in any of the groups. Changes in food consumption, BW and organ weight are considered indicators of toxic effects of a test material. Hematological parameters are commonly used to detect inflammation and infections. Serum biochemical parameters are usually used to determine of organ-related problems. There were no significant differences in food consumption, BW, organ weight, serum biochemical parameters, or hematological parameters between the control and treatment groups, implying that E. durans KLDS6.0930 administration for 28 days did not cause any adverse effects. Histopathology showed that there were no anomalies or changes related to repeated oral application of E. durans KLDS6.0930 for 28 days. Notably, our results suggested that a daily dose of 1 × 109 CFU of E. durans KLDS6.0930 /kg BW daily intake was safe and did not give rise to any treatment-related toxicity effects.
To support host health, cecal bacterial populations should be analyzed when evaluating the safety of probiotics. Parisa et al. (2016) showed that Lactobacillus strains decreased pathogenic bacterial populations such as Salmonella and E. coli while increasing beneficial bacterial populations such as lactobacilli and bifidobacteria in the cecal contents of rats through the colony counting method (Parisa et al., 2016). Mukerji et al. (2016) revealed that AB-LIFE® increased L. plantarum counts in a dose-dependent manner by qPCR method. However, these methods focus on individual gut microbes and are not nearly as extensive and comprehensive enough when compared to next-generation sequencing. The effect of E. durans KLDS6.0930 on cecal microbiota diversity was evaluated by sequencing the 16S rRNA V4 region. No significant differences in α-diversity and β-diversity were detected between the control and treatment groups, indicating that E. durans KLDS6.0930 did not impact cecal microbiota diversity and abundance in healthy male and female rats. This result is consistent with our previous study (Li B. et al., 2017), indicating that E. durans KLDS6.0930 administration is safely modulates gut microbiota without causing detrimental effects on diversity.
In summary, we present a safety assessment of E. durans KLDS6.0930 using multiple methods. The results in vitro demonstrated that E. durans KLDS6.0930 does not raise safety concerns regarding acquired antibiotic resistance genes, virulence factors, D-lactic acid, indole production, or nitroreductase activity, as supported by phenotypic and genomic data. The production levels of tyramine and phenethylamine were found to be well below established limits to cause harm to consumers. Furthermore, the in vivo studies showed that E. durans KLDS6.0930 is not toxic to male or female rats. These findings confirm that E. durans KLDS6.0930 is a non-pathogenic strain and can be used as a potential probiotic for human consumption and animal feed.
Author Contributions
GH and BL designed the study. BL, MZ, DJ, SC, and SS performed experiments. BL, FL, MZ, LZ, and SE analyzed data. SE, BL, SC, and SS wrote manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (Grant No. 2017YFD0400303).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank SE and SS for revising the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01943/full#supplementary-material
References
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Benkerroum, N. (2016). Biogenic amines in dairy products: origin, incidence, and control means. Compr. Rev. Food Sci. Food Saf. 15, 801–826. doi: 10.1111/1541-4337.12212
Bover-Cid, S., Hugas, M., Izquierdo-Pulido, M., and Vidal-Carou, M. C. (2001). Amino acid-decarboxylase activity of bacteria isolated from fermented pork sausages. Int. J. Food Microbiol. 66, 185–189. doi: 10.1016/S0168-1605(00)00526-2
Byappanahalli, M. N., Yan, T., Hamilton, M. J., Ishii, S., Fujioka, R. S., Whitman, R. L., et al. (2012). The population structure of Escherichia coli isolated from subtropical and temperate soils. Sci. Total Environ. 417–418, 273–279. doi: 10.1016/j.scitotenv.2011.12.041
Camargo, C. H., Brudernascimento, A., Lee, S. H. I., Júnior, A. F., Kaneno, R., and Rall, V. L. M. (2014). Prevalence and phenotypic characterization of Enterococcus spp. isolated from food in Brazil. Braz. J. Microbiol. 45:111. doi: 10.1590/S1517-83822014000100016
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis-−10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Choi, J. M., and Woo, G. J. (2013). Molecular characterization of high-level gentamicin-resistant Enterococcus faecalis from chicken meat in Korea. Int. J. Food Microbiol. 165, 1–6. doi: 10.1016/j.ijfoodmicro.2013.02.016
Cosentino, S., Voldby Larsen, M., Møller Aarestrup, F., and Lund, O. (2013). PathogenFinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8:e77302. doi: 10.1371/journal.pone.0077302
Creti, R., Koch, S., Fabretti, F., Baldassarri, L., and Huebner, J. (2006). Enterococcal colonization of the gastro-intestinal tract: role of biofilm and environmental oligosaccharides. BMC Microbiol. 6:60. doi: 10.1186/1471-2180-6-60
Dadáková, E., Krížek, M., and Pelikánová, T. (2009). Determination of biogenic amines in foods using ultra-performance liquid chromatography (UPLC). Food Chem. 116, 365–370. doi: 10.1016/j.foodchem.2009.02.018
Di Rosa, R., Creti, R., Venditti, M., D'Amelio, R., Arciola, C. R., Montanaro, L., et al. (2006). Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett. 256, 145–150. doi: 10.1111/j.1574-6968.2006.00112.x
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., and Fierer, N. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Eaton, T. J., and Gasson, M. J. (2001). Molecular screening of enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67, 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001
EFSA (2008). Scientific opinion of the panel on biological hazards on a request from the european food safety authority on foodborne antimicrobial resistance as a biological hazard. EFSAJ. 765, 2–87. doi: 10.2903/j.efsa.2008.765
EFSA (2011). Scientific Opinion on risk based control of biogenic amine formation in fermented foods. EFSAJ. 9:2393. doi: 10.2903/j.efsa.2011.2393
EFSA (2012a). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSAJ. 10:2740. doi: 10.2903/j.efsa.2012.2740
EFSA (2012b). Guidance on the safety assessment of Enterococcus faecium in animal nutrition. EFSAJ. 10:2682 doi: 10.2903/j.efsa.2012.2682
EFSA (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSAJ. 16:5206. doi: 10.2903/j.efsa.2018.5206
Fanaro, S., Chierici, R., Guerrini, P., and Vigi, V. (2003). Intestinal microflora in early infancy: composition and development. Acta Pãdiatr. Suppl. 91, 48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x
FAO/WHO (2012). Guidelines on probiotics: 10 years later. J. Clin. Gastroenterol. 46(Suppl.), S1–S2. doi: 10.1097/MCG.0b013e318269fdd5
Fontana, R., Ligozzi, M., Pittaluga, F., and Satta, G. (1996). Intrinsic penicillin resistance in enterococci. Microb. Drug Resist. 2, 209–213. doi: 10.1089/mdr.1996.2.209
Foulquié Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and Vuyst, L. D. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Franz, C. M., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Franz, C. M., Muscholl-silberhorn, A. B., Yousif, N. M., Vancanneyt, M., Swings, J., and Holzapfel, W. H. (2001). Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67, 4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001
Gaglio, R., Couto, N., Marques, C., de Fatima Silva Lopes, M., Moschetti, G., Pomba, C., et al. (2016). Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses. Int. J. Food Microbiol. 236, 107–114. doi: 10.1016/j.ijfoodmicro.2016.07.020
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, 52–57. doi: 10.1093/nar/gkm360
Guo, L., Li, T., Tang, Y., Yang, L., and Huo, G. (2016). Probiotic properties of Enterococcus strains isolated from traditional naturally fermented cream in China. Microb. Biotechnol. 9, 737–745. doi: 10.1111/1751-7915.12306
Hendrickx, A. P. A., Schapendonk, C. M., and Luit-Asbroek, M. V. (2010). Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiology 156, 2649–2659. doi: 10.1099/mic.0.041392-0
Isidro, A., Henriques, A. O., and Tavares, P. (2004). The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology 322, 253–263. doi: 10.1016/j.virol.2004.02.012
Jahan, M., Zhanel, G. G., Sparling, R., and Holley, R. A. (2015). Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 199, 78–85. doi: 10.1016/j.ijfoodmicro.2015.01.013
Kopit, L. M., Kim, E. B., Siezen, R. J., Harris, L. J., and Marco, M. L. (2014). Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Appl. Environ. Microbiol. 80:1899. doi: 10.1128/AEM.03859-13
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, B., Jin, D., Etareri, E. S., Li, N., Yan, F., Zhao, L., et al. (2017). Safety assessment of Lactobacillus helveticus KLDS1.8701 based on whole genome sequencing and oral toxicity studies. Toxins 9:301. doi: 10.3390/toxins9100301
Li, P., Niu, Q., Wei, Q., Zhang, Y., Ma, X., Kim, S. W., et al. (2017). Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus faecalis as alternatives to antibiotics. Sci. Rep. 7:41395. doi: 10.1038/srep41395
Liu, F., Li, B., Du, J., Yu, S., Li, W., Evivie, S. E., et al. (2016). Complete genome sequence of Enterococcus durans KLDS6.0930, a strain with probiotic properties. J. Biotechnol. 217, 49–50. doi: 10.1016/j.jbiotec.2015.11.012
Lorencová, E., Bunková, L., Matoulková, D., Dráb, V., Pleva, P., Kubán, V., et al. (2012). Production of biogenic amines by lactic acid bacteria and bifidobacteria isolated from dairy products and beer. Int. J. Food Sci. Technol. 47, 2086–2091. doi: 10.1111/j.1365-2621.2012.03074.x
Lowe, A. M., Lambert, P. A., and Smith, A. W. (1995). Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect. Immun. 63, 703–706.
Marcobal, A., Rivas, B. D. L., Landete, J. M., Tabera, L., and Muñoz, R. (2012). Tyramine and Phenylethylamine biosynthesis by food bacteria. Crit. Rev. Food Sci. Nutr. 52:448. doi: 10.1080/10408398.2010.500545
Marcobal, A., Rivas, B. D. L., and Muñoz, R. (2006). First genetic characterization of a bacterial β-phenylethylamine biosynthetic enzyme in Enterococcus faecium RM58. FEMS Microbiol. Lett. 258:144. doi: 10.1111/j.1574-6968.2006.00206.x
Marraffini, L. A., and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
Mcarthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Mohamed, J. A., and Huang, D. B. (2007). Biofilm formation by enterococci. J. Med. Microbiol. 56, 1581–1588. doi: 10.1099/jmm.0.47331-0
Mohedano, M. L., López, P., Spano, G., and Russo, P. (2015). “Controlling the formation of biogenic amines in fermented foods,” in Advances in Fermented Foods and Beverages, ed W. Holzapfel (Cambridgeshire: Woodhead Publishing), 273–310.
Molina, M. A., Díaz, A. M., Hesse, C., Ginter, W., Gentilini, M. V., Nuñez, G. G., et al. (2015). Immunostimulatory effects triggered by Enterococcus faecalis CECT7121 probiotic strain involve activation of dendritic cells and interferon-gamma production. PLoS ONE 10:e0127262. doi: 10.1371/journal.pone.0127262
Mukerji, P., Roper, J. M., Stahl, B., Smith, A. B., Burns, F., Rae, J. C., et al. (2016). Safety evaluation of AB-LIFE(®) (Lactobacillus plantarum CECT 7527, 7528 and 7529): antibiotic resistance and 90-day repeated-dose study in rats. Food Chem. Toxicol. 92:117. doi: 10.1016/j.fct.2016.03.018
OECD (2008). Section 4 (Part 407), Repeated Dose 28-Day Oral Toxicity Study in Rodents. Guidline for the Testing of Chemicals. OECD.
Ogier, J. C., and Serror, P. (2008). Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126, 286–290. doi: 10.1016/j.ijfoodmicro.2007.08.012
Palmer, K. L., and Gilmore, M. S. (2010). Multidrug-resistant enterococci lack CRISPR-cas. MBio 1, 516–524. doi: 10.1128/mBio.00227-10
Parisa, S., Mohammad, F. J., Liang, J. B., Ramasamy, K., Chin, S. C., and Wan, H. Y. (2016). Safety assessment of two new Lactobacillus strains as probiotic for human using a rat model. PLoS ONE 11:e0159851. doi: 10.1371/journal.pone.0159851
Peng, Z., Li, M., Wang, W., Liu, H., Fanning, S., Hu, Y., et al. (2017). Genomic insights into the pathogenicity and environmental adaptability of Enterococcus hirae R17 isolated from pork offered for retail sale. Microbiol. Open 6:e00514. doi: 10.1002/mbo3.514
Pieniz, S., Andreazza, R., Anghinoni, T., Camargo, F., and Brandelli, A. (2014). Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 37, 251–256. doi: 10.1016/j.foodcont.2013.09.055
Price, V. J., Huo, W., Sharifi, A., and Palmer, K. L. (2016). CRISPR-cas and restriction-modification act additively against conjugative Antibiotic resistance plasmid transfer in Enterococcus faecalis. Msphere 1, e00064–e00016. doi: 10.1128/mSphere.00064-16
Rea, M. C., Franz, C. M. A. P., Holzapfel, W. H., and Cogan, T. M. (2004). Development of enterococci and production of tyramine during the manufacture and ripening of cheddar cheese. Irish J. Agric. Food Res. 43, 247–258. Available online at: https://www.jstor.org/stable/25562522
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2015). REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, D234–D236. doi: 10.1093/nar/gku1046
Santos, S. C., Fraqueza, M. J., Elias, M., Barreto, A. S., and Semedo-Lemsaddek, T. (2017). Traditional dry smoked fermented meat sausages: characterization of autochthonous enterococci. LWT Food Sci. Technol. 79, 410–415. doi: 10.1016/j.lwt.2017.01.042
Schaberg, D. R., Culver, D. H., and Gaynes, R. P. (1991). Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91, 72S−75S. doi: 10.1016/0002-9343(91)90346-Y
Sillanpää, J., Nallapareddy, S. R., Singh, K. V., Prakash, V. P., Fothergill, T., Tonthat, H., et al. (2010). Characterization of the pilus-encoding operon of and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1, 236–246. doi: 10.4161/viru.1.4.11966
Smělá, D., Pechová, P., Komprda, T., Klejdus, B., and Kubán, V. (2004). Chromatographic determination of biogenic amines in dry salami during the fermentation and storage. Chem. Pap. 98, 432–437. Available online at: http://www.chemicke-listy.cz/ojs3/index.php/chemicke-listy/article/view/2126
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., and Svabic-Vlahovic, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
ten Brink, B., Damink, C., Joosten, H. M., and Jh, H. I. T. V. (1990). Occurrence and formation of biologically active amines in foods. Int. J. Food Microbiol. 11, 73–84. doi: 10.1016/0168-1605(90)90040-C
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Top, J., Riley, D. R., Boekhorst, J., Vrijenhoek, J. E., Schapendonk, C. M., et al. (2010). Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. doi: 10.1186/1471-2164-11-239
Gilmore, M. S. (2014). Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu. Rev. Microbiol. 68, 337–356. doi: 10.1146/annurev-micro-091213-113003
Wassenaar, T. M., Zschüttig, A., Beimfohr, C., Geske, T., Auerbach, C., Cook, H., et al. (2015). Virulence genes in a probiotic E. coli product witha recorded long history of safe use. Eur. J. Microbiol. Immunol. 5, 81–93. doi: 10.1556/EuJMI-D-14-00039
Weng, P. L., Ramli, R., Shamsudin, M. N., Cheah, Y. K., and Hamat, R. A. (2013). High genetic diversity of Enterococcus faecium and Enterococcus faecalis clinical isolates by pulsed-field gel electrophoresis and multilocus sequence typing from a hospital in Malaysia. BioMed. Res. Int. 2013:938937. doi: 10.1155/2013/938937
Werner, G., Fleige, C., Geringer, U., Schaik, W. V., Klare, I., and Witte, W. (2011). IS element IS 16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 11:80. doi: 10.1186/1471-2334-11-80
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1–6. doi: 10.1007/s10482-017-0844-4
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67:2640–2644. doi: 10.1093/jac/dks261
Keywords: probiotic, Enterococcus durans, safety evaluation, genome, oral toxicity study
Citation: Li B, Zhan M, Evivie SE, Jin D, Zhao L, Chowdhury S, Sarker SK, Huo G and Liu F (2018) Evaluating the Safety of Potential Probiotic Enterococcus durans KLDS6.0930 Using Whole Genome Sequencing and Oral Toxicity Study. Front. Microbiol. 9:1943. doi: 10.3389/fmicb.2018.01943
Received: 23 April 2018; Accepted: 31 July 2018;
Published: 21 August 2018.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Julio Villena, Centro de Referencia para Lactobacilos, ArgentinaAtte Von Wright, University of Eastern Finland, Finland
Copyright © 2018 Li, Zhan, Evivie, Jin, Zhao, Chowdhury, Sarker, Huo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guicheng Huo, guichenghuo@126.com
Fei Liu, 13126870616@163.com
† These authors have contributed equally to this work
 Bailiang Li1†
Bailiang Li1† Meng Zhan
Meng Zhan Smith E. Evivie
Smith E. Evivie Guicheng Huo
Guicheng Huo