- 1Leibniz Institute DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany
- 2Integrated Centre of Systems Biology (BRICS), Braunschweig University of Technology, Braunschweig, Germany
- 3Institute of Microbiology, Braunschweig University of Technology, Braunschweig, Germany
- 4Department of Bioinformatics and Biochemistry, Braunschweig University of Technology, Braunschweig, Germany
Strains of Clostridioides difficile cause detrimental diarrheas with thousands of deaths worldwide. The infection process by the Gram-positive, strictly anaerobic gut bacterium is directly related to its unique metabolism, using multiple Stickland-type amino acid fermentation reactions coupled to Rnf complex-mediated sodium/proton gradient formation for ATP generation. Major pathways utilize phenylalanine, leucine, glycine and proline with the formation of 3-phenylproprionate, isocaproate, butyrate, 5-methylcaproate, valerate and 5-aminovalerate. In parallel a versatile sugar catabolism including pyruvate formate-lyase as a central enzyme and an incomplete tricarboxylic acid cycle to prevent unnecessary NADH formation completes the picture. However, a complex gene regulatory network that carefully mediates the continuous adaptation of this metabolism to changing environmental conditions is only partially elucidated. It involves the pleiotropic regulators CodY and SigH, the known carbon metabolism regulator CcpA, the proline regulator PrdR, the iron regulator Fur, the small regulatory RNA CsrA and potentially the NADH-responsive regulator Rex. Here, we describe the current knowledge of the metabolic principles of energy generation by C. difficile and the underlying gene regulatory scenarios.
Clostridioides (Clostridium) Difficile
Clostridioides (Clostridium) difficile (Lawson et al., 2016) was discovered in 1935 as a commensal of healthy newborns (Hall and O’Toole, 1935). It was only in the late 1970s that C. difficile was recognized as a severe pathogen, responsible for antibiotic-related pseudomembranous colitis (Bartlett et al., 1978). In the last 20 years, an emerging number of nosocomial and community-acquired infections with symptoms ranging from mild diarrhea to pseudomembranous colitis and toxic megacolon was documented (Bartlett, 2006; Rupnik et al., 2009; Knight et al., 2015; Lessa et al., 2015). Major risk factors are antibiotic therapy, age and immunosuppression (Bignardi, 1998). The symptoms including severe intestinal damage are believed to be mainly caused by the two large clostridial toxins A (TcdA) and B (TcdB) and the binary toxin Cdt (Carter et al., 2010; Carman et al., 2011). In vitro, the toxins are predominantly produced in the stationary phase. Toxin production directly depends on the metabolic state of C. difficile (Karlsson et al., 2008; Neumann-Schaal et al., 2015; Hofmann et al., 2018). The number of genome-sequenced C. difficile strains with short-read sequences has increased up to 7000 (https://enterobase.warwick.ac.uk/, Alikhan et al., 2018), while the number of closed genomes remains at about 50 (e.g. Sebaihia et al., 2006; He et al., 2010; Riedel et al., 2017). Currently, several high quality annotations of the C. difficile genome are available, which serve as a solid basis for a systematic investigation of the transcriptome, proteome, metabolome, and for the construction of genome-scale metabolic models (Sebaihia et al., 2006; Monot et al., 2011; Larocque et al., 2014; Pettit et al., 2014; Dannheim et al., 2017a,b; Jenior et al., 2017; Kashaf et al., 2017).
Fermentation Pathways
Stickland Metabolism
C. difficile harbors multiple pathways to utilize amino acids and sugars as energy sources (Mead, 1971; Elsden et al., 1976; Elsden and Hilton, 1979). The fermentation of amino acids via the so-called Stickland pathway occurs in three stages that ultimately couple the oxidation and reduction of amino acids to the formation of ATP (Stickland, 1934; Nisman, 1954). The first step is the transamination of an amino acid to its corresponding 2-oxo-acid (O’Neil and DeMoss, 1968; Barker, 1981) which yields NADH once coupled to the glutamate dehydrogenase reaction (Figure 1). In addition, serine, threonine, methionine, and cysteine are subject to deamination (Hofmeister et al., 1993; Morozova et al., 2013). The second part is either an oxidative or a reductive pathway. In the oxidative pathway, the formed 2-oxo acid gets oxidized by ferredoxin with the formation of a CoA thioester and the release of CO2 (Mai and Adams, 1994; Heider et al., 1996; Lin et al., 2015). The final steps of the pathway (encoded by vorCBA, iorBA) include the cleavage of the CoA thioester with subsequent ATP formation (Nisman, 1954; Valentine and Wolfe, 1960; Cary et al., 1988; Wiesenborn et al., 1989; Musfeldt and Schönheit, 2002). In the reductive pathway, the 2-oxo acid is reduced employing NADH with the formation of a 2-hydroxy acid (Hetzel et al., 2003; Martins et al., 2005; Kim et al., 2006). The dehydratation to enoyl-CoA (Dickert et al., 2002; Kim et al., 2004, 2005, 2008) in a CoA transferase reaction follows this (Kim et al., 2006). The reduction of the enoyl-CoA to acyl-CoA is catalyzed by an electron bifurcating acyl-CoA dehydrogenase (see below). The final step of the pathway is the transfer of the coenzyme A to the 2-hydroxy acid of the reductive path releasing the carboxylic acid (Kim et al., 2006) (Figure 1A). Genes of the reductive path are organized in the hadAIBC-acdB-etfBA operon with the exception of the ldhA gene which is localized upstream of the operon in the opposite direction. Interestingly, C. difficile revealed only a limited spectrum of amino acids utilized in the reductive pathway, while multiple amino acids can be used in the oxidative pathway (Figure 1B) (Elsden et al., 1976; Elsden and Hilton, 1978, 1979; Neumann-Schaal et al., 2015; Rees et al., 2016; Dannheim et al., 2017b; Riedel et al., 2017). Some amino acids are degraded by a modified Stickland pathway like the reduction of proline and glycine or the degradation of arginine via ornithine (Schmidt et al., 1952; Mitruka and Costilow, 1967; Hodgins and Abeles, 1969; Turner and Stadtman, 1973; Cone et al., 1976; Kabisch et al., 1999; Jackson et al., 2006; Fonknechten et al., 2009).
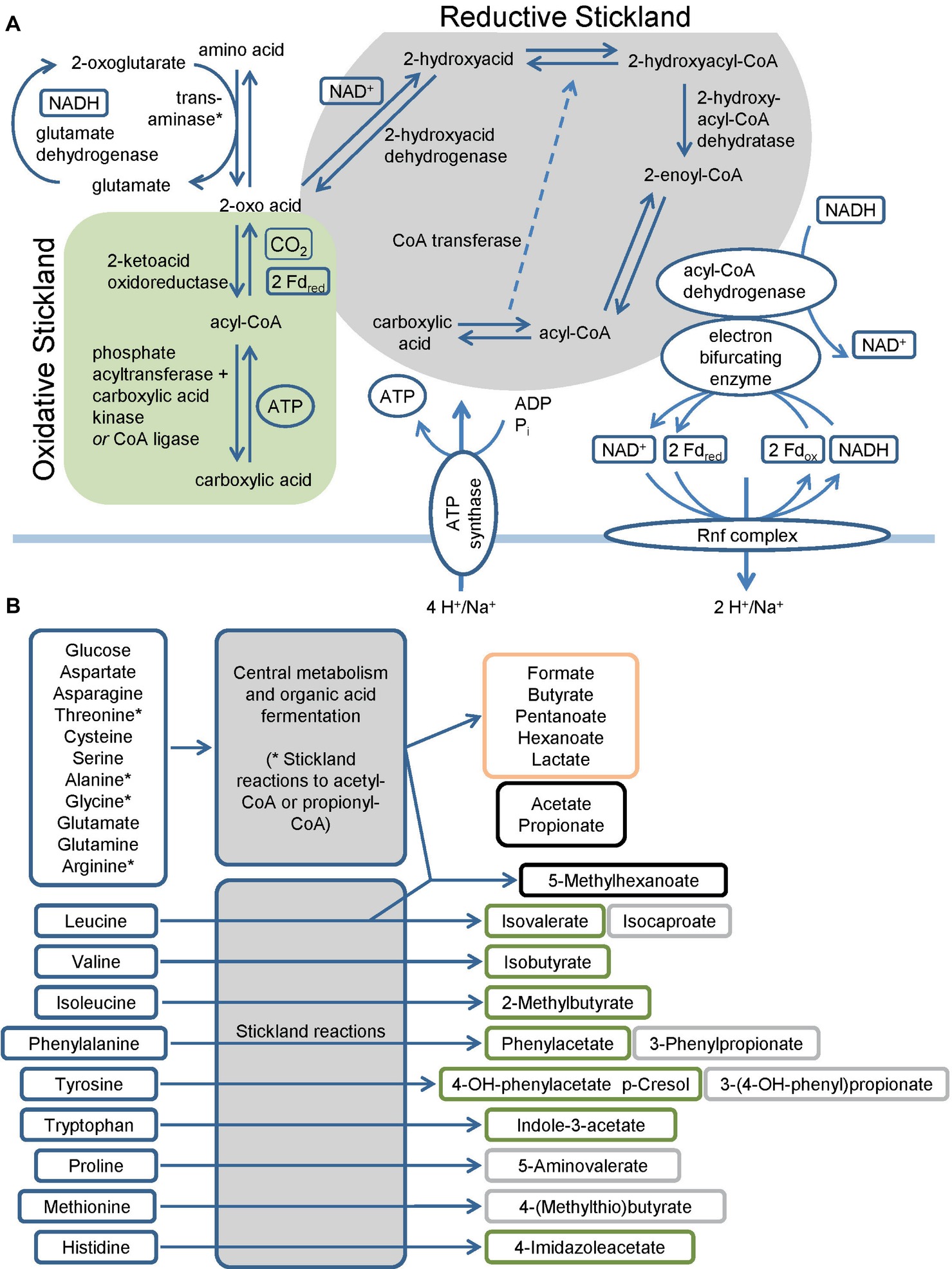
Figure 1. Overview of the fermentation metabolism in Clostridioides difficile. (A) Schematic overview of Stickland reactions showing the reaction steps of classical reductive and oxidative pathways and of the Rnf complex and the connection to the electron bifurcating enzymes. Products are shown at the end of the arrow and in boxes alongside the arrows, * Serine, threonine, methionine and cysteine are also subject to deamination by lyases. (B) Overview of amino acids and glucose as representative sugar and their fermentation products. The figure summarizes published fermentation products and substrates omitting alcohols and intermediates of the pathways for reasons of clarity. Corresponding alcohols are only minor products. (-OH: -hydroxy, Fdox: Ferredoxin oxidized form, Fdred: Ferredoxin reduced form). Green: oxidative Stickland reactions and their products, gray: reductive Stickland reactions and their products, orange: central carbon metabolism-associated fermentation products, black: Stickland products (oxidative and/or reductive) and central carbon metabolism-associated fermentation products.
Central Carbon Metabolism-Associated Fermentation
Besides the branched-chain and aromatic products of the Stickland reactions, C. difficile produces a number of straight-chain organic acids including acetate, lactate, propionate and butyrate (Neumann-Schaal et al., 2015; Rees et al., 2016; Dannheim et al., 2017b). Key metabolites for their formation are pyruvate and acetyl-CoA. With the exception of acetate, reducing equivalents are oxidized during the formation of the organic acids.
Pyruvate, derived from carbohydrates and amino acids, is a key metabolite in both fermentation and the central carbon metabolism. It is fermented in two ways in C. difficile. It can be transformed to propionate via the reductive Stickland pathway as was observed in Clostridium propionicum (Hetzel et al., 2003; Schweiger und Buckel, 1984; Selmer et al., 2002). Or, it can be degraded to acetyl-CoA and produce butyrate via acetoacetyl-CoA and crotonyl-CoA (Figure 2). Clostridia typically use NADH to reduce acetoacetyl-CoA to 3-hydroxybutyryl-CoA (von Hugo et al., 1972; Sliwkowski and Hartmanis, 1984; Aboulnaga et al., 2013). The second reduction step of crotonyl-CoA to butyryl-CoA includes an electron bifurcating step (Aboulnaga et al., 2013) (see below). The enzymes of butyrate fermentation are organized in two operons (bcd-etfBA-crt2-hbd-thlA and ptb1-buk). Other products such as valerate and 5-methylhexanoate can be formed combining acetyl-CoA with propionyl-CoA or isovaleryl-CoA via the identical set of enzymes (Dannheim et al., 2017b). These reactions play a major role as a sink for reducing equivalents when favored substrates such as proline and leucine are not available (Neumann-Schaal et al., 2015).
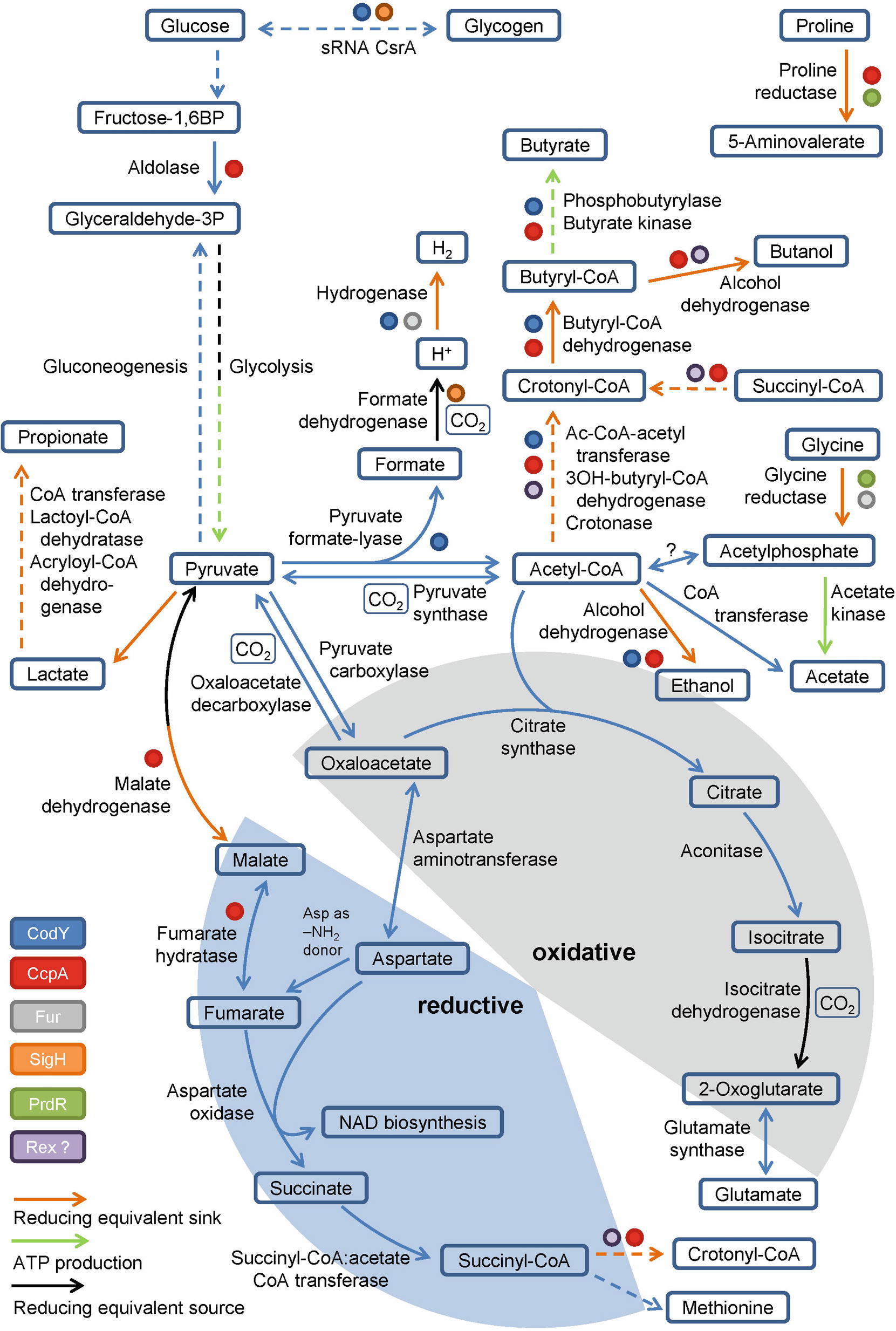
Figure 2. Glycolysis, gluconeogenesis, the fragmented TCA cycle, anaplerotic reactions and global regulators involved in metabolism. Schematic overview of the glycolysis, gluconeogenesis, the fragmented TCA cycle showing the oxidative and the reductive pathway, including anaplerotic reactions as well as global regulators controlling the central metabolism. Global regulators are marked by colored dots, ATP producing and reducing equivalent consuming/producing reactions are marked by colored arrows. (BP: bisphosphate, P: phosphate, Ac: Acetyl, -OH: -hydroxy). Dashed arrows represent multiple reactions.
Electron Bifurcation and the Rnf Complex
Beside substrate-level phosphorylation, C. difficile couples several of the described fermentation pathways to the generation of a sodium/proton gradient using electron bifurcation in combination with the membrane spanning Rnf complex (Figure 1A). The Rnf complex was originally discovered in Rhodobacter capsulatus and catalyzes the reduction of NAD+ by ferredoxin (Schmehl et al., 1993; Biegel and Müller, 2010). Reduced ferredoxins can be produced through several ways. For instance, via ferredoxin-dependent oxidoreductases of the oxidative Stickland pathway or via dehydrogenases coupled to an electron bifurcation complex. Electron bifurcation couples the NADH-dependent reduction of a substrate (often CoA-derivatives) to the reduction of ferredoxin (Bertsch et al., 2013). This unique coupling is possible as the redox potential of enoyl-CoA (around 0 mV) is significantly higher than that of NAD+ (−280 mV) and ferredoxin (−500 mV) (Buckel and Thauer, 2013). Two electrons derived from NADH are distributed to two different electron acceptors, here an enoyl-CoA and ferredoxin. In C. difficile, electron bifurcating enzymes are found in several pathways including the reductive Stickland pathways (Figure 1A) and the butyrate/propionate fermentation pathways (butyryl-CoA und acryloyl-CoA dehydrogenases, Figure 2) (Hetzel et al., 2003; Aboulnaga et al., 2013; Bertsch et al., 2013). Finally, the free energy resulting from redox potential difference between ferredoxin (−500 mV) and NAD+ (−280 mV) is used to transport ions across the membrane (Buckel and Thauer, 2018). The nature of transported ions has not been studied in C. difficile. However, the transport of protons was observed for other clostridia (Biegel and Müller, 2010; Tremblay et al., 2012; Hess et al., 2013; Mock et al., 2015). Already in the 1980s, it was shown that also proline reduction is coupled to proton motive force generation (Lovitt et al., 1986), most likely via a direct interaction of the proline reductase with the Rnf complex. The generated ion gradient is used for either transport processes, motility, or for ATP generation via ATP synthase (Figure 1A).
What is the ATP recovery of the overall process? Usually organic acids are secreted in a protonated state. ATP synthase requires four ions for the generation of one molecule ATP (Dannheim et al., 2017b). The oxidative path reduces two molecules of NAD+ to NADH and phosphorylates 1.5 ADP. The reductive pathway regenerates one molecule NAD+ and produces 0.5 ATP. For leucine as substrate, the redox balance requires the reduction of two molecules leucine per one molecule oxidized leucine (Britz and Wilkinson, 1982; Kim et al., 2005). Overall, this leads to a production of 0.83 molecules ATP per molecule amino acid. Under the same conditions, the formation of acetate from acetyl-CoA yields 1.25 molecules ATP per molecule acetyl-CoA. The fermentation of two molecules acetyl-CoA to butyrate yields 1.75 molecules ATP and regenerates two reducing equivalents (Dannheim et al., 2017b).
Central Carbon Metabolism
Glycolysis and Gluconeogenesis and the Incomplete TCA Cycle
Pyruvate and acetyl-CoA are key metabolites used for a variety of different metabolic reactions in C. difficile. Pyruvate is produced via glycolysis and amino acid degradation (e.g. cysteine or alanine). Interestingly, cysteine and also pyruvate inhibit toxin production in C. difficile (Bouillaut et al., 2015; Dubois et al., 2016) emphasizing the tight connection of metabolism and pathogenicity. While glycolysis and gluconeogenesis follow classical pathways, acetyl-CoA can be produced via the Wood-Ljungdahl-pathway, via pyruvate synthase and via pyruvate formate-lyase.
For Clostridium acetobutylicum an incomplete TCA cycle was described (Amador-Noguez et al., 2011; Crown et al., 2011; Au et al., 2014). Based on genome annotation, C. difficile might also possess a truncated TCA cycle which is still sufficient for the production of biomass precursors and the degradation of nutrients (Dannheim et al., 2017b) (Figure 2). In Clostridium kluyveri, a citrate-(Re)-synthase is catalyzing the acetylation of oxaloacetate to form citrate replacing the common citrate-(Si)-synthase (Li et al., 2007). Citrate is further metabolized to 2-oxoglutarate (α-ketoglutarate), the precursor of the glutamate metabolism. Further oxidation of 2-oxoglutarate to succinyl-CoA is impaired as the 2-oxoglutarate synthase is missing. The reductive path is already interrupted at the level of oxaloacetate since a non-decarboxylating malate dehydrogenase is missing in C. difficile (Dannheim et al., 2017b). However, oxaloacetate is connected to fumarate via pyruvate and malate or aspartate. Aspartate serves as ammonium donor for arginine- and purine biosynthesis with the formation of fumarate. Fumarate can be degraded to pyruvate to refill the pyruvate pool or it can serve as electron acceptor for aspartate oxidase to produce iminosuccinate as a precursor of NAD biosynthesis (Senger and Papoutsakis, 2008; Dannheim et al., 2017b). The resulting succinate is a substrate of succinyl-CoA:acetate CoA transferase. The succinyl-CoA formed is used for methionine biosynthesis or is degraded to butyrate via crotonyl-CoA (Amador-Noguez et al., 2011; Crown et al., 2011; Au et al., 2014). In summary, the only reaction that contributes to the production of NADH in the truncated TCA cycle is the oxidation of isocitrate to 2-oxoglutarate (Figure 2). Other bacteria harboring a complete TCA cycle produce 3 NADH per acetyl-CoA, which is used for proton gradient formation and ATP generation. Since C. difficile is missing the classical electron transport chains, the TCA cycle is mainly used for the production and degradation of various metabolically important intermediates (Sebaihia et al., 2006).
The Wood-Ljungdahl Pathway
Beside the already described reductive Stickland reactions and the butyrate fermentation, the Wood-Ljungdahl pathway also allows re-oxidation of NADH in C. difficile. In this pathway, which is also known as the reductive acetyl-CoA pathway, two molecules CO2 are used as terminal electron acceptors and reduced to acetate (Ragsdale, 1997). First CO2 gets reduced with NADPH to formate or directly into a formyl group by formate dehydrogenase. In a second step catalyzed by the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex the formyl group is reduced to a methyl group and combined with CO and coenzyme A to acetyl-CoA (Ragsdale, 1997). Köpke et al. (2013) showed that the pathway is present in all 28 sequenced C. difficile strains available at that time. Moreover, they showed that the clinical isolate C. difficile 630 and closely related strains are capable of growing autotrophically on CO2 + H2. However, only slight growth was observed probably due to the lack of tryptophan biosynthesis (Sebaihia et al., 2006). Compared to true acetogens like Clostridium ljungdahlii (Köpke et al., 2010), Moorella thermoacetica (Pierce et al., 2008), and Acetobacterium woodii (Poehlein et al., 2012), C. difficile genomes only harbor an orphan acetate kinase gene. No obvious gene for a phosphotransacetylase was detected. However, this reaction might be catalyzed by the phosphotransbutyrylase (Köpke et al., 2013). In summary, fixation of the glycolysis-derived CO2 via the Wood-Ljungdahl pathway might be also a metabolic advantage for C. difficile in the human gut (Köpke et al., 2013).
Pyruvate Utilization via Pyruvate Formate-lyase
C. difficile is utilizing pyruvate via the radical enzyme pyruvate formate-lyase, which forms the products acetyl-CoA and formate in the presence of coenzyme A (Figure 2). Pyruvate formate-lyase (PflD) requires an [4Fe-4S] cluster containing activating enzyme (PflC) for the formation of the catalytic glycyl radical (Crain and Broderick, 2014). The formate generated gets subsequently oxidized to CO2 and an electron by the formate dehydrogenase, a MoCo-containing selenoprotein (Pinske and Sawers, 2016). The electrons formed are transferred to a [NiFe] hydrogenase (Shafaat et al., 2013; Pinske and Sawers, 2016). Overall, the central metabolism in C. difficile mainly serves as an anabolic and catabolic hub and for CO2 fixation, avoiding the generation of NADH due to the lack of classical respiratory chains.
Regulation of C. Difficile Energy Metabolism
Currently, only a partial view of the regulation of the C. difficile energy metabolism at the transcriptional and post-transcriptional level is available (Bouillaut et al., 2015). Clearly, the major regulator is the catabolite control regulator CcpA (Antunes et al., 2012). In Bacillus subtilis the LacI/GalR type regulator forms a complex with phosphorylated form of Hpr or Crh, which in turn is generated in the presence of high cellular glucose or fructose-1,6-bisphosphate concentrations (Fujita, 2009). In C. difficile about 140 genes are directly controlled by CcpA including genes of glycolysis, proline reduction, glycine reduction, butanol and butyrate formation (Figure 2; Antunes et al., 2012). In parallel, CcpA controls the expression of the toxin genes tcdA and tcdB in response to fructose-1,6-bisphosphate without phosphorylated Hpr, providing a strong link between metabolism and toxin production (Antunes et al., 2011). The proline-dependent regulator PrdR activates genes for proline reductase and represses the genes for glycine reductase (Figure 2; Bouillaut et al., 2013). The global regulator CodY provides a link to sporulation and another connection of the metabolism to toxin production (Dineen et al., 2007; Nawrocki et al., 2016; Ransom et al., 2018). In close cooperativity with the SinR and SinR’ proteins and the corresponding genes, CodY controls the toxin off state during the exponential growth phase (Girinathan et al., 2018; Ransom et al., 2018). Interestingly, culture heterogeneity caused by a bistable switch was observed (Ransom et al., 2018). CodY regulates also the energy metabolism via binding to promoters of genes involved in glycogen formation, the pyruvate formate-lyase path to hydrogen and butanol/butyrate generation (Figure 2; Dineen et al., 2010). The sigma factor SigH, controlling the genes of the glycogen metabolism and for formate dehydrogenase, represents another connection of the metabolism with sporulation (Saujet et al., 2011). The function of the NADH/NAD+-responsive regulator Rex in the fermentative metabolism of C. acetobutylicum was described before (Wietzke and Bahl, 2012). Participation in the regulation of butanol and butyrate formation in C. difficile was proposed (Figure 2; Bouillaut et al., 2015). The ferric uptake regulator Fur also directly influences glycine reduction and hydrogen formation from pyruvate in response to low iron condition (Figure 2; Ho and Ellermeier, 2015; Berges et al., 2018). Finally, the small regulatory RNA CsrA serves as carbon storage regulator influencing the genes of glycogen mobilization (Figure 2; Gu et al., 2018). Obviously, a complex regulatory network headed by the pleiotropic regulators CcpA and CodY co-regulates metabolism and toxin production. Similarly, CodY and SigH connect sporulation with the metabolism at the transcriptional level (Martin-Verstraete et al., 2016).
Conclusion and Future Perspectives
In Western countries, hypertoxic C. difficile strains are causing several thousand deaths per year especially after antibiotic treatments. In this context, the mystery of the ecological success of this pathogenic bacterium is closely related to its unique and highly adaptive metabolism. Amino acids as building blocks of proteins are integral parts of our nutrition and thus available in access in our gut. Similarly, sugars from sugar polymers like starch constitute the carbohydrate part of our food. Both are the major energy sources of C. difficile. The versatile organism possesses multiple pathways for amino acid fermentation. However, normal substrate level phosphorylation suffers from very low ATP recoveries and the need to utilize parts of this ATP for ion gradient formation via a reverse ATPase reaction. Thus, smart C. difficile couples amino acid fermentation via electron bifurcation to membrane potential generating processes at the Rnf complex. Similarly, the central metabolism was modified to prevent unnecessary NADH generation, which usually has to be re-oxidized via energetically cost-intensive reactions. The organism uses an incomplete TCA cycle, generating one instead of three NADH. Furthermore, pyruvate formate-lyase instead of pyruvate dehydrogenase produces acetyl-CoA and formate, which gets transformed into protons by formate dehydrogenase and finally to hydrogen by a hydrogenase. Nevertheless, major metabolic fluxes have to be determined. Most likely, additional principles of energy generation will be uncovered. We are at the beginning of an exciting period of systems biology, allowing the integration of the different levels of cellular control represented by transcriptional control, RNA stability, translational control, metabolic control and coordinated degradation.
Currently, we know that this complex metabolism is controlled by a network of regulatory proteins, which directly connects it to toxin formation and sporulation. Major players are the catabolite regulator CcpA, the sporulation sigma factor SigH, the pleiotropic transcription factor CodY, the proline regulator PrdR, the iron responsive Fur and potentially the NADH/NAD+-ratio measuring Rex. Here, SigW and CodY are important players during the onset of sporulation. Similarly, CcpA and CodY regulate toxin gene transcription. A first small regulatory RNA (CsrA) was found involved in flagella formation, toxin production and host cell adherence. Most likely, this is only a small part of the yet unknown regulatory network underlying the efficient adaptation of the metabolism to changing environmental conditions. Novel regulatory principles including new regulators, novel small regulatory RNA and proteins, unknown changes in the protein–protein network with controlled proteolysis, direct metabolic regulation, control of RNA stability to name a few, have still to be elucidated.
Author Contributions
MN-S, DJ, and KS-H wrote the manuscript.
Funding
This work was funded by the Federal State of Lower Saxony, Niedersächsisches Vorab (VWZN2889/3215/3266). We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the Technische Universität Braunschweig.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aboulnaga, E. -H., Pinkenburg, O., Schiffels, J., El-Refai, A., Buckel, W., and Selmer, T. (2013). Effect of an oxygen-tolerant bifurcating butyryl coenzyme A dehydrogenase/electron-transferring flavoprotein complex from Clostridium difficile on butyrate production in Escherichia coli. J. Bacteriol. 195, 3704–3713. doi: 10.1128/JB.00321-13
Alikhan, N. -F., Zhou, Z., Sergeant, M. J., and Achtman, M. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14:e1007261. doi: 10.1371/journal.pgen.1007261
Amador-Noguez, D., Brasg, I. A., Feng, X. -J., Roquet, N., and Rabinowitz, J. D. (2011). Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77, 7984–7997. doi: 10.1128/AEM.05374-11
Antunes, A., Camiade, E., Monot, M., Courtois, E., Barbut, F., Sernova, N. V., et al. (2012). Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 40, 10701–10718. doi: 10.1093/nar/gks864
Antunes, A., Martin-Verstraete, I., and Dupuy, B. (2011). CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79, 882–899. doi: 10.1111/j.1365-2958.2010.07495.x
Au, J., Choi, J., Jones, S. W., Venkataramanan, K. P., and Antoniewicz, M. R. (2014). Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for (13)C metabolic flux analysis. Metab. Eng. 26, 23–33. doi: 10.1016/j.ymben.2014.08.002
Barker, H. A. (1981). Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 50, 23–40. doi: 10.1146/annurev.bi.50.070181.000323
Bartlett, J. G. (2006). Narrative review: the new epidemic of clostridium difficile-associated enteric disease. Ann. Intern. Med. 145, 758–764. doi: 10.7326/0003-4819-145-10-200611210-00008
Bartlett, J. G., Chang, T. W., Gurwith, M., Gorbach, S. L., and Onderdonk, A. B. (1978). Antibiotic-associated pseudomembranous colitis due to toxin-producing Clostridia. N. Engl. J. Med. 298, 531–534. doi: 10.1056/NEJM197803092981003
Berges, M., Michel, A. -M., Lassek, C., Nuss, A. M., Beckstette, M., Dersch, P., et al. (2018). Iron regulation in Clostridioides difficile. Front. Microbiol. 9:3183. doi: 10.3389/fmicb.2018.03183
Bertsch, J., Parthasarathy, A., Buckel, W., and Müller, V. (2013). An electron-bifurcating caffeyl-CoA reductase. J. Biolumin. Chemilumin. 288, 11304–11311. doi: 10.1074/jbc.M112.444919
Biegel, E., and Müller, V. (2010). Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 107, 18138–18142. doi: 10.1073/pnas.1010318107
Bignardi, G. E. (1998). Risk factors for Clostridium difficile infection. J. Hosp. Infect. 40, 1–15. doi: 10.1016/S0195-6701(98)90019-6
Bouillaut, L., Dubois, T., Sonenshein, A. L., and Dupuy, B. (2015). Integration of metabolism and virulence in Clostridium difficile. Res. Microbiol. 166, 375–383. doi: 10.1016/j.resmic.2014.10.002
Bouillaut, L., Self, W. T., and Sonenshein, A. L. (2013). Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854. doi: 10.1128/JB.01492-12
Britz, M. L., and Wilkinson, R. G. (1982). Leucine dissimilation to isovaleric and isocaproic acids by cell suspensions of amino acid fermenting anaerobes: the Stickland reaction revisited. Can. J. Microbiol. 28, 291–300. doi: 10.1139/m82-043
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi: 10.1016/j.bbabio.2012.07.002
Buckel, W., and Thauer, R. K. (2018). Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886. doi: 10.1021/acs.chemrev.7b00707
Carman, R. J., Stevens, A. L., Lyerly, M. W., Hiltonsmith, M. F., Stiles, B. G., and Wilkins, T. D. (2011). Clostridium difficile binary toxin (CDT) and diarrhea. Anaerobe 17, 161–165. doi: 10.1016/j.anaerobe.2011.02.005
Carter, G. P., Rood, J. I., and Lyras, D. (2010). The role of toxin A and toxin B in Clostridium difficile-associated disease: past and present perspectives. Gut Microbes 1, 58–64. doi: 10.4161/gmic.1.1.10768
Cary, J. W., Petersen, D. J., Papoutsakis, E. T., and Bennett, G. N. (1988). Cloning and expression of Clostridium acetobutylicum phosphotransbutyrylase and butyrate kinase genes in Escherichia coli. J. Bacteriol. 170, 4613–4618. doi: 10.1128/jb.170.10.4613-4618.1988
Cone, J. E., Del Río, R. M., Davis, J. N., and Stadtman, T. C. (1976). Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. U.S.A. 73, 2659–2663. doi: 10.1073/pnas.73.8.2659
Crain, A. V., and Broderick, J. B. (2014). Pyruvate formate-lyase and its activation by pyruvate formate-lyase activating enzyme. J. Biolumin. Chemilumin. 289, 5723–5729. doi: 10.1074/jbc.M113.496877
Crown, S. B., Indurthi, D. C., Ahn, W. S., Choi, J., Papoutsakis, E. T., and Antoniewicz, M. R. (2011). Resolving the TCA cycle and pentose-phosphate pathway of Clostridium acetobutylicum ATCC 824: isotopomer analysis, in vitro activities and expression analysis. Biotechnol. J. 6, 300–305. doi: 10.1002/biot.201000282
Dannheim, H., Riedel, T., Neumann-Schaal, M., Bunk, B., Schober, I., Spröer, C., et al. (2017a). Manual curation and reannotation of the genomes of Clostridium difficile 630Deltaerm and Clostridium difficile 630. J. Med. Microbiol. 66, 286–293. doi: 10.1099/jmm.0.000427
Dannheim, H., Will, S. E., Schomburg, D., and Neumann-Schaal, M. (2017b). Clostridioides difficile 630△erm in silico and in vivo: quantitative growth and extensive polysaccharide secretion. FEBS Open Bio 7, 602–615. doi: 10.1002/2211-5463.12208
Dickert, S., Pierik, A. J., and Buckel, W. (2002). Molecular characterization of phenyllactate dehydratase and its initiator from Clostridium sporogenes. Mol. Microbiol. 44, 49–60. doi: 10.1046/j.1365-2958.2002.02867.x
Dineen, S. S., McBride, S. M., and Sonenshein, A. L. (2010). Integration of metabolism and virulence by Clostridium difficile CodY 192, 5350–5362. doi: 10.1128/JB.00341-10
Dineen, S. S., Villapakkam, A. C., Nordman, J. T., and Sonenshein, A. L. (2007). Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66, 206–219. doi: 10.1111/j.1365-2958.2007.05906.x
Dubois, T., Dancer-Thibonnier, M., Monot, M., Hamiot, A., Bouillaut, L., Soutourina, O., et al. (2016). Control of Clostridium difficile physiopathology in response to cysteine availability. Infect. Immun. 84, 2389–2405. doi: 10.1128/IAI.00121-16
Elsden, S. R., and Hilton, M. G. (1978). Volatile acid production from threonine, valine, leucine and isoleucine by Clostridia. Arch. Microbiol. 117, 165–172. doi: 10.1007/BF00402304
Elsden, S. R., and Hilton, M. G. (1979). Amino acid utilization patterns in clostridial taxonomy. Arch. Microbiol. 123, 137–141. doi: 10.1007/BF00446812
Elsden, S. R., Hilton, M. G., and Waller, J. M. (1976). The end products of the metabolism of aromatic amino acids by Clostridia. Arch. Microbiol. 107, 283–288. doi: 10.1007/BF00425340
Fonknechten, N., Perret, A., Perchat, N., Tricot, S., Lechaplais, C., Vallenet, D., et al. (2009). A conserved gene cluster rules anaerobic oxidative degradation of L-ornithine. J. Bacteriol. 191, 3162–3167. doi: 10.1128/JB.01777-08
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Girinathan, B. P., Ou, J., Dupuy, B., and Govind, R. (2018). Pleiotropic roles of Clostridium difficile sin locus. PLoS Pathog. 14:e1006940. doi: 10.1371/journal.ppat.1006940
Gu, H., Qi, H., Chen, S., Shi, K., Wang, H., and Wang, J. (2018). Carbon storage regulator CsrA plays important roles in multiple virulence-associated processes of Clostridium difficile. Microbial. Pathogenesis 121, 303–309. doi: 10.1016/j.micpath.2018.05.052
Hall, I. C., and O’Toole, E. (1935). Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am. J. Dis. Child. 49:390. doi: 10.1001/archpedi.1935.01970020105010
He, M., Sebaihia, M., Lawley, T. D., Stabler, R. A., Dawson, L. F., Martin, M. J., et al. (2010). Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U.S.A. 107, 7527–7532. doi: 10.1073/pnas.0914322107
Heider, J., Mai, X., and Adams, M. W. (1996). Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol. 178, 780–787. doi: 10.1128/jb.178.3.780-787.1996
Hess, V., Schuchmann, K., and Müller, V. (2013). The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biolumin. Chemilumin. 288, 31496–31502. doi: 10.1074/jbc.M113.510255
Hetzel, M., Brock, M., Selmer, T., Pierik, A. J., Golding, B. T., and Buckel, W. (2003). Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270, 902–910. doi: 10.1046/j.1432-1033.2003.03450.x
Ho, T. D., and Ellermeier, C. D. (2015). Ferric uptake regulator fur control of putative iron acquisition systems in Clostridium difficile. J. Bacteriol. 197, 2930–2940. doi: 10.1128/JB.00098-15
Hodgins, D. S., and Abeles, R. H. (1969). Studies of the mechanism of action of
Hofmann, J. D., Otto, A., Berges, M., Biedendieck, R., Michel, A. -M., Becher, D., et al. (2018). Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front. Microbiol. 9:1970. doi: 10.3389/fmicb.2018.01970
Hofmeister, A. E., Grabowski, R., Linder, D., and Buckel, W. (1993). L-serine and L-threonine dehydratase from Clostridium propionicum. Two enzymes with different prosthetic groups. Eur. J. Biochem. 215, 341–349. doi: 10.1111/j.1432-1033.1993.tb18040.x
von Hugo, H., Schoberth, S., Madan, V. K., and Gottschalk, G. (1972). Coenzyme specificity of dehydrogenases and fermentation of pyruvate by Clostridia. Arch. Mikrobiol. 87, 189–202. doi: 10.1007/BF00424880
Jackson, S., Calos, M., Myers, A., and Self, W. T. (2006). Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 188, 8487–8495. doi: 10.1128/JB.01370-06
Jenior, M. L., Leslie, J. L., Young, V. B., and Schloss, P. D. (2017). Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems 2:e00063–17. doi: 10.1128/mSystems.00063-17
Kabisch, U. C., Grantzdorffer, A., Schierhorn, A., Rucknagel, K. P., Andreesen, J. R., and Pich, A. (1999). Identification of
Karlsson, S., Burman, L. G., and Akerlund, T. (2008). Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154, 3430–3436. doi: 10.1099/mic.0.2008/019778-0
Kashaf, S. S., Angione, C., and Lio, P. (2017). Making life difficult for Clostridium difficile: augmenting the pathogen’s metabolic model with transcriptomic and codon usage data for better therapeutic target characterization. BMC Syst. Biol. 11:25. doi: 10.1186/s12918-017-0395-3
Kim, J., Hetzel, M., Boiangiu, C. D., and Buckel, W. (2004). Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 28, 455–468. doi: 10.1016/j.femsre.2004.03.001
Kim, J., Darley, D., and Buckel, W. (2005). 2-Hydroxyisocaproyl-CoA dehydratase and its activator from Clostridium difficile. FEBS J. 272, 550–561. doi: 10.1111/j.1742-4658.2004.04498.x
Kim, J., Darley, D., Selmer, T., and Buckel, W. (2006). Characterization of (R)-2-hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of L-leucine to isocaproate by Clostridium difficile. Appl. Environ. Microbiol. 72, 6062–6069. doi: 10.1128/AEM.00772-06
Kim, J., Darley, D. J., Buckel, W., and Pierik, A. J. (2008). An allylic ketyl radical intermediate in clostridial amino-acid fermentation. Nature 452, 239–242. doi: 10.1038/nature06637
Knight, D. R., Elliott, B., Chang, B. J., Perkins, T. T., and Riley, T. V. (2015). Diversity and evolution in the genome of Clostridium difficile. Clin. Microbiol. Rev. 28, 721–741. doi: 10.1128/CMR.00127-14
Köpke, M., Held, C., Hujer, S., Liesegang, H., Wiezer, A., Wollherr, A., et al. (2010). Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U.S.A. 107, 13087–13092. doi: 10.1073/pnas.1004716107
Köpke, M., Straub, M., and Dürre, P. (2013). Clostridium difficile is an autotrophic bacterial pathogen. PLoS One 8:e62157. doi: 10.1371/journal.pone.0062157
Larocque, M., Chénard, T., and Najmanovich, R. (2014). A curated C. difficile strain 630 metabolic network: prediction of essential targets and inhibitors. BMC Syst. Biol. 8:117. doi: 10.1186/s12918-014-0117-z
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Lessa, F. C., Mu, Y., Bamberg, W. M., Beldavs, Z. G., Dumyati, G. K., Dunn, J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834. doi: 10.1056/NEJMoa1408913
Li, F., Hagemeier, C. H., Seedorf, H., Gottschalk, G., and Thauer, R. K. (2007). Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and isopropylmalate synthase rather than to Si-citrate synthase. J. Bacteriol. 189, 4299–4304. doi: 10.1128/JB.00198-07
Lin, P. P., Mi, L., Morioka, A. H., Yoshino, K. M., Konishi, S., Xu, S. C., et al. (2015). Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 31, 44–52. doi: 10.1016/j.ymben.2015.07.001
Lovitt, R. W., Kell, D. B., and Morris, J. G. (1986). Proline reduction by Clostridium sporogenes is coupled to vectorial proton ejection. FEMS Microbiol. Lett. 36, 269–273. doi: 10.1111/j.1574-6968.1986.tb01708.x
Mai, X., and Adams, M. W. (1994). Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biolumin. Chemilumin. 269, 16726–16732.
Martins, B. M., Macedo-Ribeiro, S., Bresser, J., Buckel, W., and Messerschmidt, A. (2005). Structural basis for stereo-specific catalysis in NAD(+)-dependent (R)-2-hydroxyglutarate dehydrogenase from Acidaminococcus fermentans. FEBS J. 272, 269–281. doi: 10.1111/j.1432-1033.2004.04417.x
Martin-Verstraete, I., Peltier, J., and Dupuy, B. (2016). The regulatory networks that control Clostridium difficile toxin synthesis. Toxins 8:153. doi: 10.3390/toxins8050153
Mead, G. C. (1971). The amino acid-fermenting Clostridia. J. Gen. Microbiol. 67, 47–56. doi: 10.1099/00221287-67-1-47
Mitruka, B. M., and Costilow, R. N. (1967). Arginine and ornithine catabolism by Clostridium botulinum. J. Bacteriol. 93, 295–301.
Mock, J., Zheng, Y., Mueller, A. P., Ly, S., Tran, L., Segovia, S., et al. (2015). Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J. Bacteriol. 197, 2965–2980. doi: 10.1128/JB.00399-15
Morozova, E. A., Kulikova, V. V., Yashin, D. V., Anufrieva, N. V., Anisimova, N. Y., Revtovich, S. V., et al. (2013). Kinetic parameters and cytotoxic activity of recombinant methionine γ-lyase from Clostridium tetani, Clostridium sporogenes, Porphyromonas gingivalis and Citrobacter freundii. Acta Naturae 5, 92–98.
Monot, M., Boursaux-Eude, C., Thibonnier, M., Vallenet, D., Moszer, I., Medigue, C., et al. (2011). Reannotation of the genome sequence of Clostridium difficile strain 630. J. Med. Microbiol. 60, 1193–1199. doi: 10.1099/jmm.0.030452-0
Musfeldt, M., and Schönheit, P. (2002). Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184, 636–644. doi: 10.1128/JB.184.3.636-644.2002
Nawrocki, K. L., Edwards, A. N., Daou, N., Bouillaut, L., and McBride, S. M. (2016). CodY-dependent regulation of sporulation in Clostridium difficile. J. Bacteriol. 198, 2113–2130. doi: 10.1128/JB.00220-16
Neumann-Schaal, M., Hofmann, J. D., Will, S. E., and Schomburg, D. (2015). Time-resolved amino acid uptake of Clostridium difficile 630Δerm and concomitant fermentation product and toxin formation. BMC Microbiol. 15:281. doi: 10.1186/s12866-015-0614-2
O’Neil, S. R., and DeMoss, R. D. (1968). Tryptophan transaminase from Clostridium sporogenes. Arch. Biochem. Biophys. 127, 361–369. doi: 10.1016/0003-9861(68)90237-3
Pettit, L. J., Browne, H. P., Yu, L., Smits, W. K., Fagan, R. P., Barquist, L., et al. (2014). Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15:160. doi: 10.1186/1471-2164-15-160
Pierce, E., Xie, G., Barabote, R. D., Saunders, E., Han, C. S., Detter, J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x
Pinske, C., and Sawers, R. G. (2016). Anaerobic formate and hydrogen metabolism. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0011-2016
Poehlein, A., Schmidt, S., Kaster, A. -K., Goenrich, M., Vollmers, J., Thürmer, A., et al. (2012). An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS One 7:e33439. doi: 10.1371/journal.pone.0033439
Ragsdale, S. W. (1997). The eastern and western branches of the Wood/Ljungdahl pathway: how the east and west were won. BioFactors 6, 3–11. doi: 10.1002/biof.5520060102
Ransom, E. M., Kaus, G. M., Tran, P. M., Ellermeier, C. D., and Weiss, D. S. (2018). Multiple factors contribute to bimodal toxin gene expression in Clostridioides (Clostridium) difficile. Mol. Microbiol. 110, 533–549. doi: 10.1111/mmi.14107
Rees, C. A., Shen, A., and Hill, J. E. (2016). Characterization of the Clostridium difficile volatile metabolome using comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. J. Chromatogr. B 1039, 8–16. doi: 10.1016/j.jchromb.2016.11.009
Riedel, T., Wetzel, D., Hofmann, J. D., Plorin, S. P. E. O., Dannheim, H., Berges, M., et al. (2017). High metabolic versatility of different toxigenic and non-toxigenic Clostridioides difficile isolates. Int. J. Med. Microbiol. 307, 311–320. doi: 10.1016/j.ijmm.2017.05.007
Rupnik, M., Wilcox, M. H., and Gerding, D. N. (2009). Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536. doi: 10.1038/nrmicro2164
Saujet, L., Monot, M., Dupuy, B., Soutourina, O., and Martin-Verstraete, I. (2011). The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 193, 3186–3196. doi: 10.1128/JB.00272-11
Schmehl, M., Jahn, A., Meyer zu Vilsendorf, A., Hennecke, S., Masepohl, B., Schuppler, M., et al. (1993). Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241, 602–615.
Schmidt, G. C., Logan, M. A., and Tytell, A. A. (1952). The degradation of arginine by Clostridium perfringens (BP6K). J. Biolumin. Chemilumin. 198, 771–783.
Schweiger, G., and Buckel, W. (1984). On the dehydration of (R)-lactate in the fermentation of alanine to propionate by Clostridium propionicum. FEBS Lett. 171, 79–84. doi: 10.1016/0014-5793(84)80463-9
Sebaihia, M., Wren, B. W., Mullany, P., Fairweather, N. F., Minton, N., Stabler, R., et al. (2006). The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786. doi: 10.1038/ng1830
Selmer, T., Willanzheimer, A., and Hetzel, M. (2002). Propionate CoA-transferase from Clostridium propionicum. Cloning of gene and identification of glutamate 324 at the active site. Eur. J. Biochem. 269, 372–380. doi: 10.1046/j.0014-2956.2001.02659.x
Senger, R. S., and Papoutsakis, E. T. (2008). Genome-scale model for Clostridium acetobutylicum: Part I. Metabolic network resolution and analysis. Biotechnol. Bioeng. 101, 1036–1052. doi: 10.1002/bit.22010
Shafaat, H. S., Rudiger, O., Ogata, H., and Lubitz, W. (2013). NiFe hydrogenases: a common active site for hydrogen metabolism under diverse conditions. Biochim. Biophys. Acta 1827, 986–1002. doi: 10.1016/j.bbabio.2013.01.015
Sliwkowski, M. X., and Hartmanis, M. G. (1984). Simultaneous single-step purification of thiolase and NADP-dependent 3-hydroxybutyryl-CoA dehydrogenase from Clostridium kluyveri. Anal. Biochem. 141, 344–347. doi: 10.1016/0003-2697(84)90052-6
Stickland, L. H. (1934). Studies in the metabolism of the strict anaerobes (genus Clostridium): the chemical reactions by which Cl. sporogenes obtains its energy. Biochem. J. 28, 1746–1759. doi: 10.1042/bj0281746
Tremblay, P. -L., Zhang, T., Dar, S. A., Leang, C., and Lovley, D. R. (2012). The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. MBio 4, e00406–e00412. doi: 10.1128/mBio.00406-12
Turner, D. C., and Stadtman, T. C. (1973). Purification of protein components of the clostridial glycine reductase system and characterization of protein A as a selenoprotein. Arch. Biochem. Biophys. 154, 366–381. doi: 10.1016/0003-9861(73)90069-6
Valentine, R. C., and Wolfe, R. S. (1960). Purification and role of phosphotransbutyrylase. J. Biolumin. Chemilumin. 235, 1948–1952. Available at: www.jbc.org/content/235/7/1948.full.pdf
Wiesenborn, D. P., Rudolph, F. B., and Papoutsakis, E. T. (1989). Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl. Environ. Microbiol. 55, 317–322.
Keywords: Clostridioides (Clostridium) difficile, metabolism, fermentation, TCA cycle, Wood-Ljungdahl pathway, Stickland reactions
Citation: Neumann-Schaal M, Jahn D and Schmidt-Hohagen K (2019) Metabolism the Difficile Way: The Key to the Success of the Pathogen Clostridioides difficile. Front. Microbiol. 10:219. doi: 10.3389/fmicb.2019.00219
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Rizlan Bernier-Latmani, École Polytechnique Fédérale de Lausanne, SwitzerlandMeera Unnikrishnan, University of Warwick, United Kingdom
Copyright © 2019 Neumann-Schaal, Jahn and Schmidt-Hohagen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kerstin Schmidt-Hohagen, k.schmidt-hohagen@tu-braunschweig.de
 Meina Neumann-Schaal
Meina Neumann-Schaal Dieter Jahn
Dieter Jahn Kerstin Schmidt-Hohagen
Kerstin Schmidt-Hohagen