- 1Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium
- 2(IB)2 - Interuniversity Institute of Bioinformatics in Brussels (ULB-VUB), Brussels, Belgium
- 3Unité de Chronobiologie Théorique, Service de Chimie Physique, Faculté des Sciences, Université Libre de Bruxelles (ULB), Brussels, Belgium
- 4Systems Bioinformatics, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
Acetobacter pasteurianus 386B is a candidate functional starter culture for the cocoa bean fermentation process. To allow in silico simulations of its related metabolism in response to different environmental conditions, a genome-scale metabolic model for A. pasteurianus 386B was reconstructed. This is the first genome-scale metabolic model reconstruction for a member of the genus Acetobacter. The metabolic network reconstruction process was based on extensive genome re-annotation and comparative genomics analyses. The information content related to the functional annotation of metabolic enzymes and transporters was placed in a metabolic context by exploring and curating a Pathway/Genome Database of A. pasteurianus 386B using the Pathway Tools software. Metabolic reactions and curated gene-protein-reaction associations were bundled into a genome-scale metabolic model of A. pasteurianus 386B, named iAp386B454, containing 454 genes, 322 reactions, and 296 metabolites embedded in two cellular compartments. The reconstructed model was validated by performing growth experiments in a defined medium, which revealed that lactic acid as the sole carbon source could sustain growth of this strain. Further, the reconstruction of the A. pasteurianus 386B genome-scale metabolic model revealed knowledge gaps concerning the metabolism of this strain, especially related to the biosynthesis of its cell envelope and the presence or absence of metabolite transporters.
Introduction
Acetic acid bacteria (AAB) are obligately aerobic bacteria that play an important role in several food fermentation processes, such as vinegar production and cocoa pulp-bean mass fermentation, although they are undesired in wine, cider, and most beer fermentation processes (De Roos and De Vuyst, 2018). AAB can be found in carbohydrate-rich environments, but also in acidic and alcoholic niches (Mamlouk and Gullo, 2013). Their typical metabolic trait is the incomplete oxidation of substrates using a specific respiratory chain. That way, ethanol is converted into acetic acid. Depending on species and strain, also sugar alcohols can be converted, such as glycerol into dihydroxyacetone, D-mannitol into D-fructose, or D-sorbitol into L-sorbose. Even organic acids can be oxidized, such as acetic acid into carbon dioxide and water, which is in fact an overoxidation (Mamlouk and Gullo, 2013).
The cocoa bean fermentation process is fundamental to obtain well-fermented dry cocoa beans, from which cocoa products, such as chocolate, can be made (De Vuyst and Weckx, 2016). Next to two other groups of key microorganisms present during cocoa bean fermentation processes, namely yeasts and lactic acid bacteria, AAB are crucial to produce the necessary acetic acid that contributes to the death of the cocoa bean embryo and subsequent formation of cocoa flavor precursors inside the beans (De Vuyst and Weckx, 2016).
Acetobacter pasteurianus is an AAB species that has been isolated consistently from cocoa bean fermentation processes around the world (Camu et al., 2007; Lefeber et al., 2011; Meersman et al., 2013; Papalexandratou et al., 2013; Miescher Schwenninger et al., 2016; Visintin et al., 2016; Ozturk and Young, 2017). Its apparent adaptation to this fermentation process has been explained by its high ethanol, acid, and heat tolerance (Camu et al., 2007; Illeghems et al., 2013). The strain A. pasteurianus 386B has been isolated from a spontaneous cocoa bean fermentation process performed in Ghana (Camu et al., 2007), and has subsequently been selected as a candidate functional starter culture because of its rapid co-consumption of ethanol and lactate, the production of acetate and acetoin, and the achievement of high cell densities upon fermentation (Lefeber et al., 2010; Moens et al., 2014). To gain more insight into its metabolic potential and niche adaptations, the A. pasteurianus 386B genome has been sequenced and annotated, resulting in a genome encompassing a 2.8-Mb chromosome and seven plasmids with 2,875 protein-encoding genes (Illeghems et al., 2013). Several characteristic metabolic pathways of AAB have been identified in the genome of this strain, for example an incomplete Embden-Meyerhof-Parnas (EMP) pathway, a modified tricarboxylic acid (TCA) cycle, and a truncated respiratory chain. Whereas genome annotation combined with in vitro experiments (Lefeber et al., 2010; Illeghems et al., 2013; Moens et al., 2014), have led to a first general view on the central carbon metabolism of A. pasteurianus 386B, the macromolecule biosynthesis pathways of this strain in particular and of AAB in general are still largely unknown.
Defined media have been used successfully in different microbiological studies to reveal the specific growth requirements of microorganisms and eventual metabolic adaptations to changes in medium composition, unraveling their biosynthesis capacities (Verduyn et al., 1992; van Niel and Hahn-Hägerdal, 1999; Richards et al., 2014). A complementary strategy is to reconstruct the metabolic network of a particular microorganism in silico and assess the accuracy of this reconstruction by comparing the outcome of in silico and in vitro growth experiments in defined media (Teusink et al., 2005). This metabolic network reconstruction process is based on cataloging a set of functionally annotated enzymes and transporters encoded in the genome and coupling them to their respective biochemical reactions via gene-protein-reaction (GPR) associations (Francke et al., 2005; Pitkänen et al., 2010; Thiele and Palsson, 2010). In addition, a (species-specific) biomass reaction has to be added to the model to be able to simulate the biomass production, which is used as a proxy for the specific growth rate of the bacterial cell population (Feist and Palsson, 2010).
So far, genome-scale metabolic models (GEMs) have been reconstructed for many organisms, ranging from bacteria and archaea to fungi, plants, and even human cell lines (Ruppin et al., 2010; Yilmaz and Walhout, 2017). For AAB, the only GEMs that are currently available are those for Gluconobacter oxydans 621H, an industrially important bacterium due to its property of oxidizing a wide range of carbohydrates, and for Komagataeibacter nataicola RZS01, a bacterial cellulose producer (Wu et al., 2014; Zhang et al., 2017). The current study aims to perform a reconstruction of a GEM for A. pasteurianus 386B, which is the first GEM for a species of the genus Acetobacter. Enzymes and transporters related to the consumption of substrates present in the cocoa pulp-bean mass and metabolites produced thereof were specifically targeted. It is expected that this GEM will be useful to perform in silico metabolic flux simulations of A. pasteurianus 386B in response to different environmental conditions to improve the cocoa bean fermentation process.
Materials and Methods
Acetobacter pasteurianus 386B Genome Re-annotation and in silico Genome-Scale Metabolic Reconstruction
The complete genome of A. pasteurianus 386B was sequenced and annotated previously, using a local installation of the bacterial genome annotation system GenDB v2.2 (Meyer, 2003; Illeghems et al., 2013). Since then, different genome annotations of this strain became publicly available in different databases. To perform a thorough re-annotation of the A. pasteurianus 386B genome and to be able to assess differences between the annotation sources, the annotation data were collected in a MySQL database that was built in-house (Figure 1). Publicly available genome annotation sources included the Carbohydrate-Active enZYmes database (CAZy; Lombard et al., 2014), the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2017), the Integrated Microbial Genomes platform of the Joint Genome Institute (JGI IMG; Markowitz et al., 2012), the RefSeq database of the National Center for Biotechnology Information (NCBI; O’Leary et al., 2016), the Pathosystems Resource Integration Center database (PATRIC; Wattam et al., 2017), the proGenomes database (Mende et al., 2017), and TransportDB (Elbourne et al., 2017). The latter database relies on the TC system for annotation, providing a defined ontology to describe transporter functions in analogy to the EC system for enzyme annotation (Saier et al., 2016). Two annotation versions were used in the case of the NCBI RefSeq annotation source, one published in April 2015 (further referred to as NCBI 2015) and one published in April 2017 (further referred as NCBI 2017). Differences in these genome annotations reflect improvements made in the NCBI prokaryotic genome annotation pipeline (Tatusova et al., 2016). Furthermore, the A. pasteurianus 386B genome was re-annotated in-house, using the subcellular localisation predictor CELLO (Yu et al., 2004), eggNOG-mapper (Huerta-Cepas et al., 2017), the enzyme annotation tool PRIAM (Claudel-Renard, 2003), and the tools embedded in InterProScan 5.22-61.0 (Figure 1; Jones et al., 2014).
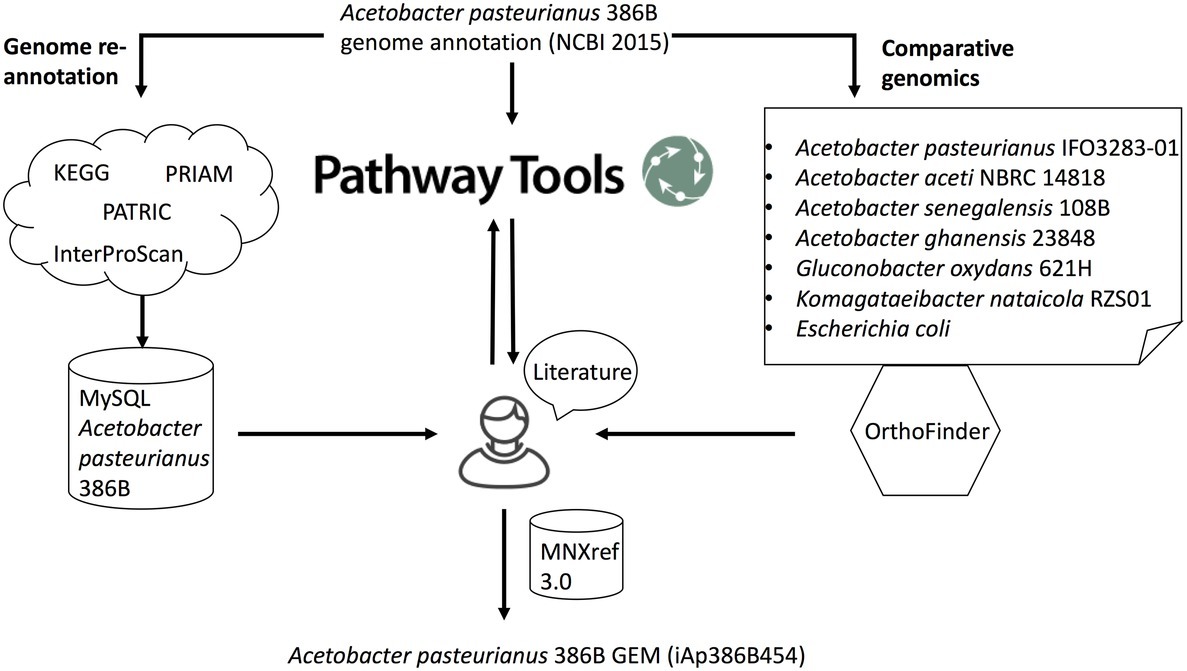
Figure 1. Schematic overview of the metabolic network reconstruction process. Genome re-annotation of A. pasteurianus 386B was performed using a combination of several databases (e.g. KEGG, PATRIC) and tools (e.g., PRIAM, InterProScan). Functional annotations were stored in a MySQL database. Comparative genomics using OrthoFinder allowed to identify orthogroups of protein sequences between A. pasteurianus 386B and other phylogenetically related bacterial species. Pathway Tools was used to predict metabolic pathways as well as the reactions and gene-protein-reaction associations that constitute these pathways. This information was stored in a Pathway/Genome database (PGDB). Edges entering the human curator node represent the information sources that were used to guide manual curation. Using additional information from the literature, the A. pasteurianus 386B PGDB was curated and its information transferred to a new genome-scale metabolic model of A. pasteurianus 386B, iAp386B454, using the MNXref 3.0 namespace of reactions and metabolites.
As a reference set of predicted protein-encoding genes, the NCBI 2015 genome annotation was used, containing 2,854 protein-encoding genes, as it is the same that was used by SRI International to reconstruct the A. pasteurianus 386B Pathway/Genome Database (PGDB) available on BioCyc (Caspi et al., 2016). Subsequently, for each protein-encoding gene, functional annotations from the different annotation sources were added to the MySQL database. In addition, the A. pasteurianus 386B PGDB was manually curated in the Pathway Tools software, assisted by different automatic refinements using the Pathologic tool inside this software (Figure 1; Karp et al., 2016). Features associated to a predicted MetaCyc pathway (Caspi et al., 2016), such as the taxonomic range, pathway score, pathway variants and pathway description, were manually assessed and compared to information available in the literature to decide whether the pathway in question should be omitted.
Also, comparative genomics was performed using OrthoFinder, allowing to predict orthogroups from protein-encoding genes in whole genomes (Figure 1; Emms and Kelly, 2015). Protein sequences of a selection of bacterial species obtained from NCBI RefSeq were compared, namely A. pasteurianus 386B, A. pasteurianus IFO3283-01, A. aceti NBRC 14818, A. senegalensis 108B, A. ghanensis LMG 23848T, Gluconobacter oxydans 621H, Komagataeibacter nataicola RZS01, and Escherichia coli str. K-12 substr. MG1655 (further referred to as E. coli). For the latter three bacterial strains, genome-scale metabolic models have been reconstructed before (Orth et al., 2011; Wu et al., 2014; Zhang et al., 2017).
Finally, based on the information contained in the in-house built MySQL database, the A. pasteurianus 386B PGDB, and the outcome of the comparative genomics analysis, the A. pasteurianus 386B GEM was reconstructed and manually curated using the MNXref 3.0 namespace as a biochemical reaction repository (Figure 1; Bernard et al., 2014; Moretti et al., 2016). Characterized enzymes and associated protein sequences available in the literature were used to perform sequence alignment searches using blastp (Altschul et al., 1990). The best blast hits in the A. pasteurianus 386B genome are indicated by their percentage similarity, percentage identity, and percentage coverage (Supplementary Table S1).
MetaCyc reaction components and GPR associations of biosynthesis pathways in the curated A. pasteurianus 386B PGDB were transferred to the A. pasteurianus 386B GEM by mapping the MetaCyc reaction components to MNXref 3.0 reaction identifiers. Reactions described in the literature but not present in the MNXref 3.0 reaction repository were manually added to the model. Enzyme names and enzyme commission (EC) numbers were assigned to each reaction, based on the ExplorEnz database (McDonald et al., 2009). The mass and charge balances were checked for all reactions in the resulting A. pasteurianus 386B GEM. The co-factor specificity of these reactions was manually curated, based on functional annotations and literature sources. Reaction reversibility constraints were manually curated, taking into account the directionality defined in the MetaCyc database, and to prevent reaction flux cycles from occurring in the GEM. The list of GPR associations was saved in a spreadsheet file (Supplementary File S2). The A. pasteurianus 386B GEM, named iAp386B454, was reconstructed and analyzed using the COBRAPy package version 0.11.3 (Ebrahim et al., 2013). Next, it was exported in the Systems Biology Markup Language (SBML) level 3 format (Supplementary File S3) and validated using the online SBML Validator (Bornstein et al., 2008). Reconstructed metabolic pathways were visualized using Escher (King et al., 2015).
Evaluation of the Manual Curation Process and Comparison to Other Reconstructions
The presumed increase in functional annotation quality of the genes included in iAp386B454 was evaluated based on the EC numbers of enzyme-encoding genes, as the EC system provides a defined enzymatic reaction classification (McDonald et al., 2009). EC numbers of reactions associated to 304 selected genes in the A. pasteurianus 386B GEM were compared to the EC numbers of the genes in the original annotation sources stored in the MySQL database. A number of classes were defined based on the hierarchical EC system that expressed the agreement between the EC number of a gene in the GEM and that in the annotation source. Then, for each annotation source, genes were assigned to the predefined classes. Preference was given to assign a gene to a more precise class if at least one of the EC numbers of the gene in the annotation source did fulfill the requirement set upon that class. Enzyme systems, for which the reaction is catalyzed by a complex containing more than one enzyme (e.g., pyruvate dehydrogenase), were excluded from the analysis, as these reactions inherited different EC numbers from their gene constituents.
The manually reconstructed A. pasteurianus 386B GEM was compared to GEMs obtained from a number of automatic GEM reconstruction tools. These included the RAST and ModelSEED tools in the KBase software (Henry et al., 2010; Overbeek et al., 2014; Arkin et al., 2018), CarveMe (Machado et al., 2018), MetaNetX (Ganter et al., 2013), and Pathway Tools (Karp et al., 2016). The NCBI 2015 annotation version of A. pasteurianus 386B was used for the reconstructions, which were performed without reaction gap-filling. COBRAPy was used to parse the SBML files of the reconstructed GEMs, except for SBML files obtained from Pathway Tools, which were parsed using CBMPy version 0.7.19 (Olivier et al., 2005). Model properties were inferred from the reconstructions and compared to iAp386B454.
Biomass Reaction
A biomass reaction was defined for A. pasteurianus 386B, using a combination of genomic and literature data (Supplementary File S4; Supplementary Figure S1). Protein, DNA, RNA, lipid, fatty acid, peptidoglycan, and carbohydrate mass fractions were taken from a GEM of K. nataicola RZS01 (Zhang et al., 2017). Stoichiometric coefficients of the biomass reaction were obtained by converting the mass fraction of the different macromolecules into a molar fraction, using their estimated molar masses. For each macromolecule, a separate biosynthesis reaction was defined. Stoichiometric coefficients of these reactions resulted from estimations of the molar fractions of the macromolecule building blocks. Molar fractions of amino acids for proteins and of nucleotides for DNA and RNA were estimated using the genome sequence of A. pasteurianus 386B, as proposed before (Thiele and Palsson, 2010). For DNA, molar fractions of building blocks were estimated using the G + C percentage of the genome (in casu 52.86%). For RNA, molar fractions of building blocks were estimated using their respective frequencies in rRNA-, tRNA-, and mRNA-encoding genes. RNA mass fractions were taken from E. coli (Milo and Phillips, 2016). The fatty acid and phospholipid compositions were taken from studies on related Acetobacter species (Yamada et al., 1981; Hanada et al., 2001). The average molar mass of a generic fatty acid was used to estimate the average molar mass of a generic phospholipid. Molar masses of macromolecule building blocks were queried in PubChem and ChEBI (Hastings et al., 2016; Kim et al., 2019). Energy requirements for protein, DNA, and RNA biosyntheses were taken from E. coli (Neidhardt et al., 1990). Genes were manually assigned to the different macromolecule biosynthesis reactions for cellular processes that were not explicitly included in the model, among which tRNA loading, protein elongation, replication, transcription, and translation.
Growth Experiments
Growth experiments were performed with A. pasteurianus 386B in a modified defined medium (Verduyn et al., 1992). This medium contained (per liter): (NH4)2SO4, 5 g; KH2PO4, 1.375 g; MgSO4.7H2O, 0.5 g; EDTA, 15 mg; ZnSO4.7H2O, 4.5 mg; CoSO4.7H2O, 0.35 mg; MnCl2.4H2O, 1.0 mg; CuSO4.5H2O, 0.3 mg; CaCl2.2H2O, 4.5 mg; FeSO4.7H2O, 3.0 mg; MoO3, 0.24 mg; H3BO3, 1.0 mg; KI, 0.1 mg; and 30 mM phosphate buffer (pH 6.0). The pH of the medium was set to 5.0. A filter-sterilized vitamin mixture was added after heat sterilization (121°C, 2.1 bar, 20 min) of the medium. The final vitamin concentrations were (per liter): biotin, 0.0005 mg; calcium pantothenate, 0.01 mg; nicotinic acid, 0.01 mg; myo-inositol, 0.25 mg; thiamine-HCl, 0.01 mg; pyridoxine-HCl, 0.01 mg; and para-aminobenzoic acid, 0.002 mg. Eight different carbon sources (glucose, fructose, mannitol, citric acid, glycerol, lactic acid, ethanol, and sodium acetate) were used at a final concentration of 60 mM to assess if they could sustain growth. The pH of the citric acid, lactic acid (both by using NaOH to increase the pH), and sodium acetate (using HCl to decrease the pH) stock solutions was set to 5.0. To prepare the inocula, A. pasteurianus 386B was grown overnight at 30°C in 10 ml of an undefined medium (pH 5.5), which contained (per liter): lactic acid, 5 g; sodium acetate, 10 g; granulated yeast extract, 5 g; MgSO4.7H2O, 1 g; NH4H2PO4, 20 g; and K2HPO4, 10 g. The stirring rate was set to 160 rpm. The overnight culture was centrifuged (4,000 × g, 20 min, 4°C) and washed with a filter-sterilized saline solution (0.85%, m/v, NaCl). The cells were resuspended in the sterile saline solution and inoculated in 2 ml of the defined medium mentioned above at an optical density at 600 nm (OD600) of 0.01 in triplicate in test tubes with a total volume of 20 ml. A. pasteurianus 386B was allowed to grow at 30°C without shaking for 48 h as the test tubes contained only 2 ml culture medium, resulting in a relatively large surface to medium ratio. A threshold value for the OD600 of 0.1 was used to identify whether or not the strain had grown.
Model Validation
In silico growth experiments were performed using flux balance analysis (FBA). With FBA, an optimization of the flux distribution of a GEM can be performed to maximize an objective function, typically the biomass reaction, thus predicting the specific growth rate of a bacterial cell population (Gottstein et al., 2016). FBA was performed with the A. pasteurianus 386B GEM, setting the biomass reaction as the objective of the simulation. Parameter values of this GEM included the consumption flux values of the different nutrients. Here, the consumption of ammonium as nitrogen source, sulfate as sulfur source, and phosphate as phosphate source were allowed without constraints. For aerobic respiration, an oxygen influx was allowed without constraint. D-glucose, D-mannitol, glycerol, D-lactate, ethanol, and acetate were tested as carbon sources separately to be able to compare the in silico results with the in vitro growth experimental data. The maximum exchange flux of the carbon source was set to 60 C-mmol/gCDW/h for each simulation. This value corresponded with a consumption of 10 mmol/gCDW/h of glucose for E. coli (Varma et al., 1993).
Results and Discussion
Genome Re-annotation
An extensive re-annotation of the genome of A. pasteurianus 386B was performed, based on a combination of several databases and tools, comparative genomics analyses, and manual curations using different software packages (Figure 1). Of a total of 2,875 protein-encoding genes in the original genome annotation of A. pasteurianus 386B (Illeghems et al., 2013), 2,854 genes (NCBI 2015) were re-annotated in the current study, of which 454 genes were included in the A. pasteurianus 386B GEM based on their involvement in the metabolic pathways discussed below. In what follows, metabolic reactions in different parts of the A. pasteurianus 386B metabolism are discussed, based on the evidence that the enzyme-encoding genes were present in the genome.
Central Carbon Metabolism
The incomplete EMP pathway (Supplementary Figure S2) and the pentose phosphate pathway (Supplementary Figure S3) of A. pasteurianus 386B were reconstructed, as described previously (Illeghems et al., 2013). Periplasmic oxidation of D-glucose could be assigned to a pyrroloquinoline quinone (PQQ)-dependent glucose 1-dehydrogenase (EC 1.1.5.2) (Figure 2A). Based on the re-annotation effort of the current study, different gene candidates could be linked to this reaction (e.g., APA386B_2133, Supplementary Table S1), among which genes with a previously unknown function (Illeghems et al., 2013). No transporter could be identified for D-gluconate uptake. However, since the gene encoding cytoplasmic gluconokinase was present in the genome (locus tag APA386B_1158), the D-gluconate transport reaction was nonetheless added to the A. pasteurianus 386B GEM. Cytoplasmic oxidation of D-mannitol could be assigned to a cytoplasmic mannitol 2-dehydrogenase. However, the possibility of periplasmic oxidation of D-mannitol to D-fructose was included in the GEM as well, since extracellular fructose formation from mannitol has been reported previously (Moens et al., 2014).
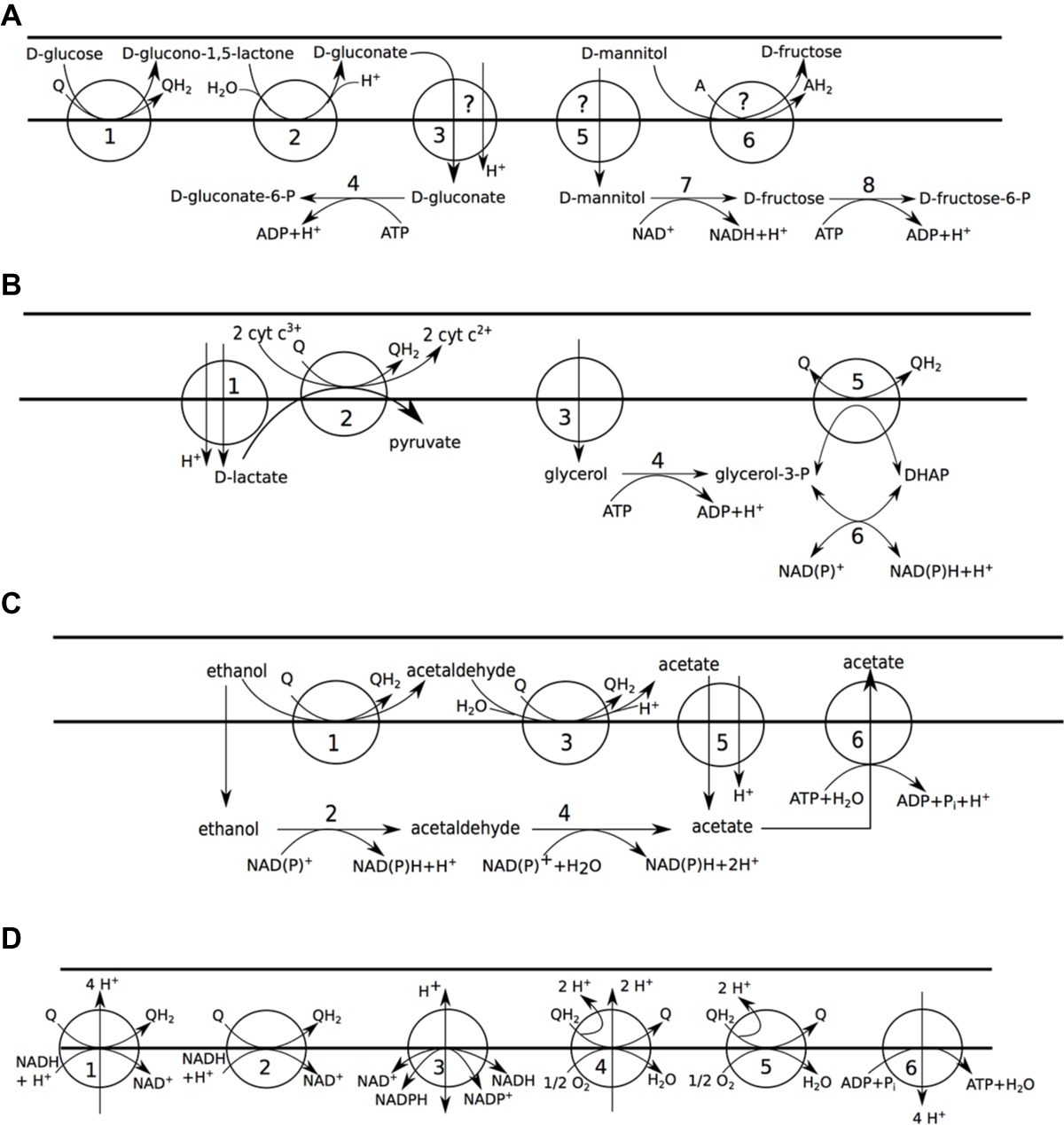
Figure 2. Carbon metabolism of A. pasteurianus 386B. Horizontal bars (from top to bottom) delineate the extracellular environment, the periplasm, and the cytosol, respectively. (A) Carbohydrate oxidation. (1) D-glucose:ubiquinone oxidoreductase (EC 1.1.5.2); (2) gluconolactonase (EC 3.1.1.17); (3) gluconate:H+ symport; (4) gluconokinase (EC 2.7.1.12); (5) mannitol permease; (6) D-sorbitol dehydrogenase (mann_ox_rxn); (7) mannitol 2-dehydrogenase (EC 1.1.1.67); (8) fructokinase (EC 2.7.1.4). (B) Lactate and glycerol oxidation. (1) lactate:H+ symporter; (2) D-lactate dehydrogenase (quinone) (EC 1.1.5.12), D-lactate dehydrogenase (cytochrome) (EC 1.1.2.4); (3) glycerol facilitator; (4) glycerol kinase (EC 2.7.1.30); (5) glycerol-3-phosphate dehydrogenase (EC 1.1.5.3); (6) glycerol-3-phosphate dehydrogenase [NAD(P)+] (EC 1.1.1.94). (C) Ethanol oxidation. (1) alcohol dehydrogenase (quinone) (EC 1.1.5.5); (2) alcohol dehydrogenase [NAD(P)+] (EC 1.1.1.1, 1.1.1.2); (3) aldehyde dehydrogenase (quinone) (EC 1.2.5.2); (4) aldehyde dehydrogenase [NAD(P)+] (EC 1.2.1.3, EC 1.2.1.4); (5) succinate-acetate:H+ symporter; (6) acetate ABC transporter. (D) Aerobic respiration. (1) NADH:ubiquinone reductase (H+-translocating) (EC 7.1.1.2); (2) NADH:ubiquinone reductase (non-electrogenic) (EC 1.6.5.9); (3) proton-translocating NAD(P)+ transhydrogenase (EC 7.1.1.1); (4) ubiquinol oxidase (H+-transporting) (EC 7.1.1.3); (5) ubiquinol oxidase (electrogenic, non-H+-transporting) (EC 7.1.1.7); (6) H+-transporting two-sector ATPase (EC 7.1.2.2).
A gene encoding a lactate permease (TC 2.A.14), more precisely a lactate:H+ symporter, was identified in the genome, which was homologous to the LctP transporter in E. coli (Dong et al., 1993; Núñez et al., 2002). Most probably A. pasteurianus 386B oxidizes D-lactate using a D-lactate dehydrogenase that is dependent on quinone (EC 1.1.5.12) or cytochrome c (EC 1.1.2.4), thus linking this reaction directly to the respiratory chain (Figure 2B). Overflow of lactate to acetoin has been found and metabolic flux analysis has revealed that acetoin is solely produced by decarboxylation of (2S)-2-acetolactate (Adler et al., 2014; Moens et al., 2014). Therefore, this metabolic route, for which the genes were present, and containing the enzymatic reactions of acetolactate synthase (EC 2.2.1.6) and acetolactate decarboxylase (EC 4.1.1.5), was added to the A. pasteurianus 386B GEM (Supplementary Figure S4). Finally, an acetoin dehydrogenase complex (EC 2.3.1.190) was identified that could be involved in the oxidation of acetoin. A glycerol uptake transporter (TC 1.A.8) was identified in the genome that was homologous to the GlpF transporter of E. coli (Fu et al., 2000). Similarly as for lactate, oxidation of glycerol could feed electrons to the respiratory chain (Figure 2B); alternatively, glycerol may be used for glycerolipid biosynthesis.
A. pasteurianus 386B could oxidize ethanol to acetic acid in the periplasm by means of a membrane-bound PQQ-dependent alcohol dehydrogenase or in the cytoplasm by means of NAD(P)+-dependent dehydrogenases (Figure 2C). Periplasmic acetate could be imported by an acetate/succinate:H+ symporter, for which the encoding gene was identified (APA386B_1116) and which was homologous to the E. coli SatP transporter (Sá-Pessoa et al., 2013). Alternatively, excess cytoplasmic acetate may be exported by a primary active ABC transporter (TC 3.A; APA386B_103) (Nakano et al., 2006). A reaction cycle that regulates acetate overflow metabolism, involving acetic acid, acetyl phosphate, and acetyl-CoA, has been described in E. coli (De Mey et al., 2007; Valgepea et al., 2010). Putative orthologs for these enzymes were retrieved in A. pasteurianus 386B, namely genes encoding acetate kinase (EC 2.7.2.1), phosphate acetyltransferase (EC 2.3.1.8), and acetate CoA ligase (EC 6.2.1.1).
A major metabolic adaptation of acetic acid-resistant Acetobacter species is their modified tricarboxylic acid (TCA) cycle (Supplementary Figure S4), containing succinyl-CoA:acetate CoA-transferase (EC 2.8.3.18) and malate:quinone oxidoreductase (EC 1.1.5.4) (Mullins et al., 2008). Two enzymes involved in anaplerotic reactions were encoded in the genome, namely a reversible NAD+-dependent malic enzyme (EC 1.1.1.38), interconverting malate and pyruvate, and phosphoenolpyruvate carboxylase (EC 4.1.1.31) that could carboxylate phosphoenolpyruvate to oxaloacetate. However, in the NCBI 2017 genome annotation version of A. pasteurianus 386B, the latter enzyme-encoding gene was annotated as a pseudogene, containing a frameshift mutation.
Aerobic Respiration
A. pasteurianus is an obligate aerobe that has evolved a truncated aerobic respiratory chain (Figure 2D). In A. pasteurianus 386B, a single gene cluster cyaBACD (APA386B_1578 – APA386B_1581) was found that encodes cytochrome ba3 ubiquinol oxidase (UOX ba3; EC 7.1.1.3). This heme-copper terminal oxidase probably contains a heme A moiety, since the genes encoding heme O (ctaB; APA386B_608) and heme A (ctaA; APA386B_1565) synthase, which are remnants of a cytochrome c oxidase gene cluster, have been shown to be still functional in A. pasteurianus (Matsutani et al., 2014). Additionally, A. pasteurianus 386B contained genes encoding two cytochrome bd-type oxidases, being cytochrome bd oxidase (APA386B_472 – APA386B_473) and its homolog, cyanide insensitive oxidase (CIO; APA386B_1010 – APA386B_1111). Since the reaction catalyzed by both of these terminal oxidases has the same stoichiometry, an identical reaction (EC 7.1.1.7) was added to the GEM. However, it has been shown that these enzymes have different kinetic parameters, indicating a physiological distinct role (Miura et al., 2013). Furthermore, two sets of genes for a cytochrome bc1 complex (E.C. 7.1.1.8) were found in the genome of A. pasteurianus 386B, as is also the case for A. aceti (Sakurai et al., 2011).
Next to ubiquinone, NAD(P)+ is involved in respiratory chain reactions. Two types of NADH:ubiquinone reductases were encoded in the A. pasteurianus 386B genome, namely one that is proton-translocating (EC 7.1.1.2) and one that is not (EC 1.6.5.9). Furthermore, a membrane-bound proton-translocating NAD(P)+ transhydrogenase (EC 7.1.1.1) was present in the genome. Finally, the genes encoding ATP synthase (EC 7.1.2.2) were present in two gene clusters (APA386B_1266 – APA386B_1270 and APA386B_1604 – APA386B_1608), which is also the case for Rhodospirillum rubrum (Falk and Walker, 1988).
Acetobacter pasteurianus 386B Grows in a Defined Medium
From all carbon sources tested during the in vitro growth experiments, A. pasteurianus 386B was only able to grow on a defined medium containing lactic acid as the sole carbon source added. Thus, except for the possible need for some micronutrients, A. pasteurianus 386B had no specific auxotrophies and was able to form all its biomass compounds from ammonium as the sole nitrogen source, sulfate as the sole sulfur source, phosphate as the sole phosphate source, and lactic acid as the sole carbon source.
Biosynthesis Pathways
In what follows, biosynthesis pathways are described that were included in the A. pasteurianus 386B GEM and for which there was genetic evidence based on the genome re-annotation. These pathways allowed to simulate the growth of A. pasteurianus 386B in silico using the defined medium conditions mentioned above. Focus is on those steps of the pathways for which there was evidence that the gene-protein-reaction associations in A. pasteurianus 386B were different from the reference pathways described in the literature.
Fatty Acid and Phospholipid Biosynthesis
The major fatty acids in bacterial cells of the genus Acetobacter are cis-vaccenic acid (C18:1), palmitic acid (C16:0), and stearic acid (C18:0) (Yamada et al., 1981). Further, the presence of a detectable amount of myristic acid (C14:0) distinguishes the genus Acetobacter from the genus Gluconobacter (Yamada et al., 1981). The fatty acid biosynthesis pathway for saturated fatty acids in A. pasteurianus 386B was similar to the one described in E. coli (Janßen and Steinbüchel, 2014). For unsaturated fatty acid biosynthesis, the FabA (EC 4.2.1.59/5.3.3.14) and FabB (EC 2.3.1.41) enzymes are critical for their formation in E. coli (Feng and Cronan, 2009). Although cis-vaccenic acid (C18:1) was the major fatty acid in the fatty acid profile of A. pasteurianus, the genes encoding FabA and FabB could not be found in the A. pasteurianus 386B genome. Even though unsaturated fatty acid biosynthesis in A. pasteurianus 386B seemed to be unclear, reactions of the FabA/FabB pathway were nonetheless added to the model (Supplementary Figure S5). As each fatty acid elongation cycle can be described by a stoichiometric reaction (Janßen and Steinbüchel, 2014), lumped reactions were added to the GEM to describe the formation of myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0) and cis-vaccenic acid (C18:1).
Bacterial cell membranes are composed of amphiphilic lipids, most commonly glycerophospholipids (Sohlenkamp and Geiger, 2016). Here, the phospholipid biosynthesis pathway of Sinorhizobium meliloti was taken as a reference (Geiger et al., 2013). In A. pasteurianus 386B, glycerol 3-phosphate could be produced by a quinone-dependent (EC 1.1.5.3) or NAD(P)+-dependent (EC 1.1.1.94) glycerol-3-phosphate dehydrogenase. The A. pasteurianus 386B genome encoded the PlsX/PlsY/PlsC system for phosphatidic acid biosynthesis, as described before (Parsons and Rock, 2013). Although long-chain acyl-acyl carrier protein (acyl-ACP) are the end-products of fatty acid biosynthesis and are initially transferred to a phosphate moiety by phosphate:acyl-ACP acyltransferase (PlsX), no generic reaction was available in MNXRef 3.0. Therefore, two reactions (EC 3.6.1.7 and EC 3.1.2.14) were added to the GEM to simulate acyl phosphate and acyl-ACP formation to represent the link between fatty acid biosynthesis and glycerophospholipid biosynthesis. Subsequent acylation of glycerol 3-phosphate is performed by the membrane-bound glycerol-3-phosphate acyltransferase (PlsY; EC 2.3.1.n3). Then, 1-acyl-glycerol-3-phosphate acyltransferase (PlsC; EC 2.3.1.51) forms phosphatidic acid (Paoletti et al., 2007; Yoshimura et al., 2007; Hara et al., 2008). Acetobacter aceti contains an unusually high amount of phosphatidylcholine in its membrane, attributed to its acetic acid resistance (Hanada et al., 2001). In A. pasteurianus 386B, the three methylation steps of the S-adenosyl-L-methionine (SAM)-dependent methylation pathway forming phosphatidylcholine from phosphatidylethanolamine were most probably catalyzed by the same enzyme, namely phosphatidylethanolamine N-methyltransferase (Pmt, EC 2.1.1.17), as the enzyme from A. pasteurianus 386B (encoded by the gene with locus tag APA386B_612) belonged to the same family as the one in Rhodobacter sphaeroides and shared high sequence identity to the Pmt enzyme in A. aceti (Hanada et al., 2001; Geiger et al., 2013).
Amino Acid Biosynthesis
The presence of biosynthesis pathways for proteinogenic amino acids in the A. pasteurianus 386B PGDB was confirmed by the growth of A. pasteurianus 386B in a defined medium containing ammonium as the sole nitrogen source (section “Acetobacter pasteurianus 386B Grows in a Defined Medium”). Two genes were retrieved that encoded potential ammonium ion channels (TC 1.A.11; APA386B_239 or APA386B_740). In addition, an ammonium assimilation pathway for de novo biosynthesis of proteinogenic amino acids was assumed to be present. In E. coli, two pathways are known for ammonium assimilation (van Heeswijk et al., 2013), one occurs via a NADP+-dependent glutamate dehydrogenase (EC 1.4.1.4) that allows to form L-glutamate directly from 2-oxoglutarate and ammonium, the other via the formation of L-glutamate and L-glutamine through a cycle of reactions involving glutamine synthetase (EC 6.3.1.2) and glutamate synthase (EC 1.4.1.13), which produces a net amount of L-glutamate. Only the latter pathway was found in the A. pasteurianus 386B genome (Figure 3A; Supplementary Figure S6).
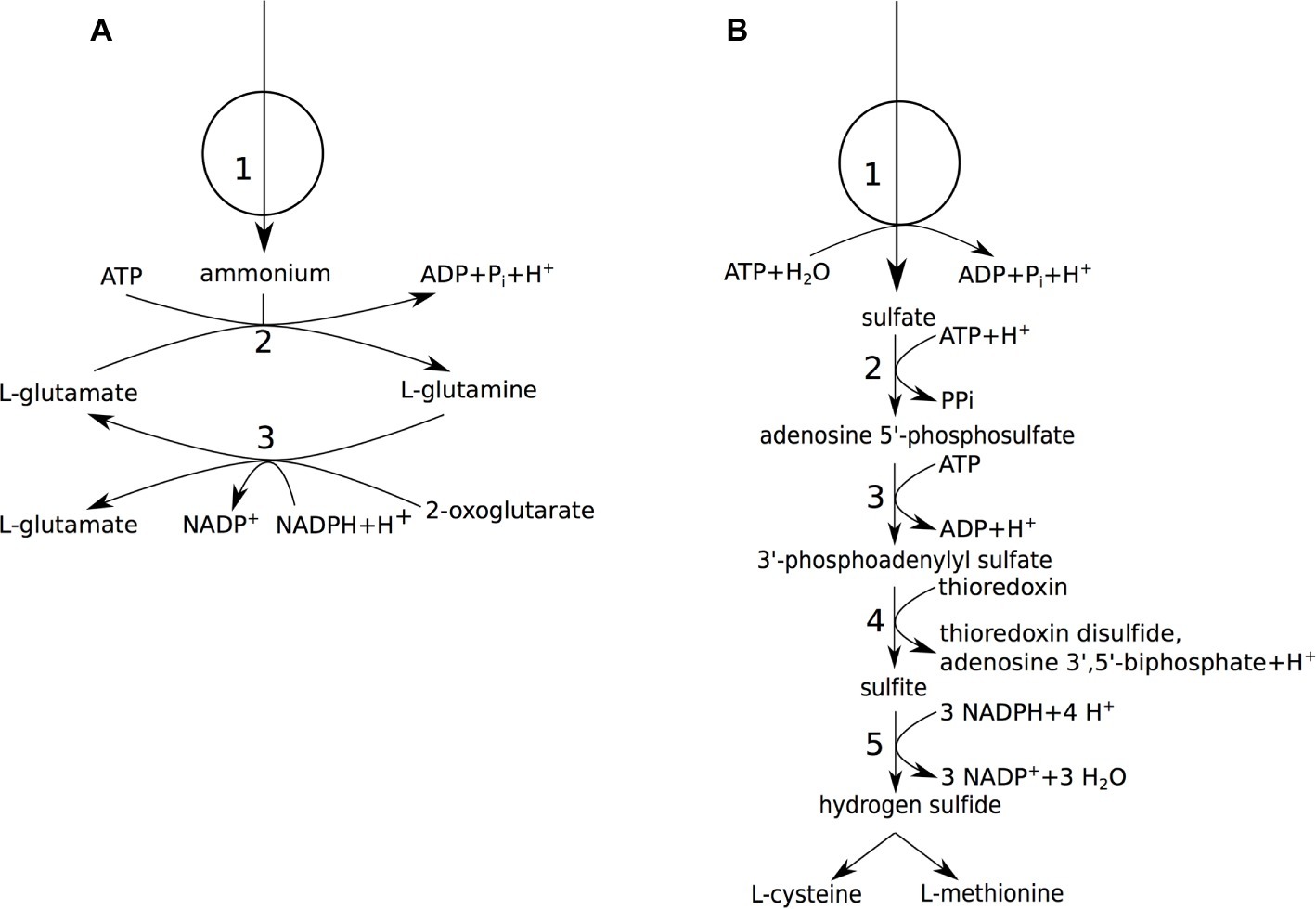
Figure 3. Nitrogen and sulfur metabolism of A. pasteurianus 386B. (A) Ammonium assimilation. (1) ammonium permease; (2) glutamine synthetase (EC 6.3.1.2); (3) glutamate synthase (NADPH-dependent) (EC 1.4.1.13). (B) Sulfate assimilation. (1) sulfate ABC transporter; (2) sulfate adenylyltransferase (EC 2.7.7.4); (3) adenylyl-sulfate kinase (EC 2.7.1.25); (4) phosphoadenylyl-sulfate reductase (thioredoxin-dependent) (EC 1.8.4.8); (5) assimilatory sulfite reductase (NADPH-dependent) (EC 1.8.1.2).
A number of pyridoxal phosphate-dependent aminotransferases were identified based on the genome annotation. These were involved in the formation of L-aspartate and L-alanine and were assigned to specific reactions based on their presence in different orthogroups. Aspartate aminotransferase (EC 2.6.1.1) was linked to three genes, likely encoding this enzyme (APA386B_861, APA386B_862, and APA386B_942). These were not predicted to be homologous to the E. coli aspartate aminotransferase gene (b0928, aspC) but shared considerable sequence identity with curated SwissProt sequences from other bacteria with identical function (>50% identity). The gene encoding glutamate-pyruvate aminotransferase (EC 2.6.1.2; APA386B_991) was a predicted ortholog of the gene in E. coli (b2379, alaC). Finally, the genes encoding aspartate 4-decarboxylase (EC 4.1.1.12; APA386B_482, APA386B_1928) had no E. coli homolog but were related to a bifunctional aspartate aminotransferase and aspartate 4-decarboxylase of Comamonas testosteroni. However, the enzyme characterized had only a minor activity as an aminotransferase (Wang and Lee, 2007). Thus, the A. pasteurianus 386B genes were only associated to the reaction catalyzed by aspartate 4-decarboxylase.
In the biosynthesis pathways of L-valine, L-leucine, and L-isoleucine, a bifunctional enzyme (EC 2.2.1.6) catalyzes the formation of (2S)-2-acetolactate and (S)-2-hydroxy-2-ethyl-3-oxobutanoate, the latter metabolite only involved in L-isoleucine biosynthesis (Barak and Chipman, 2012). The A. pasteurianus 386B genome contained three genes, which likely encode the enzymes for these reactions. The genes with locus tags APA386B_835 and APA386B_836 were most likely involved in amino acid biosynthesis, since these genes each had an E. coli ortholog encoding subunits of a bifunctional acetolactate synthase/acetohydroxybutanoate synthase enzyme complex (ilvI, ilvH). However, the gene with locus tag APA386B_1863 was closely related to two “catabolic” acetolactate synthases, identified in Klebsiella pneumoniae and Lactococcus lactis (Peng et al., 1992; Snoep et al., 1992). This evidence, combined with the fact that the neighboring gene APA386B_1862 was annotated as encoding an acetolactate decarboxylase, forming acetoin from (2S)-2-acetolactate, could be an indication that the physiological role of the APA386B_1863-encoded enzyme would only be related to acetoin formation. Finally, the branched-chain amino acid aminotransferase (EC 2.6.1.42/2.6.1.6; APA386B_1001) could be involved in the last step of the formation of these amino acids.
The aromatic amino acids L-phenylalanine, L-tyrosine, and L-tryptophan share a common initial pathway, which produces chorismate from erythrose 4-phosphate (Yang and Pittard, 2008). Comparative genomics revealed that the gene with locus tag APA386B_1330, encoding a dehydroquinate dehydratase (EC 4.2.1.10), had an ortholog in G. oxydans 621H (locus tag GOX0437) that has been identified as encoding a periplasmic enzyme involved in the oxidation of quinate (Adachi et al., 2008). The cytoplasmic dehydroquinate dehydratase of G. oxydans 621H (locus tag GOX1351) could not be found in the A. pasteurianus 386B genome. In the last reaction step of the biosynthesis of L-tyrosine and L-phenylalanine, an amino group transfer occurs with glutamate as amino group donor. In E. coli, three enzymes have been identified that could catalyze this reaction, namely the aromatic aminotransferase (TyrB), the aspartate aminotransferase (AspC), and the branched-chain amino acid aminotransferase (IlvE), whereby the latter is only involved in the biosynthesis of L-phenylalanine (Yang and Pittard, 2008). In A. pasteurianus 386B, only an ortholog of the gene encoding IlvE was found (APA386B_1001). Thus, the L-tyrosine-forming reaction was tentatively associated to the aspartate aminotransferases encoded in the genome (APA386B_861, APA386B_862, and APA386B_942).
Since A. pasteurianus 386B was able to grow in a defined medium containing sulfate as the sole sulfur source, sulfate assimilation was assumed (Figure 3B). Indeed, import of sulfate ions could occur via a sulfate permease (TC 2.A.53) as well as a probable ortholog of the E. coli cysPUWA ABC sulfate transporter. However, since the stoichiometry of the anion:H+ symporter is not known in Acetobacter, only the ABC transporter was added to the A. pasteurianus 386B GEM. The enzymes of the assimilatory sulfate reduction pathway, which reduce sulfate to hydrogen sulfide, were encoded in the genome. One exception was the flavoprotein subunit of sulfite reductase (EC 1.8.1.2), which was not found in the genome re-annotation. Whereas L-methionine is produced by the transsulfuration pathway in E. coli, only cystathionine 𝛽-lyase (EC 4.4.1.8), but not homoserine O-succinyltransferase (EC 2.3.1.46), could be identified in A. pasteurianus 386B. In Corynebacterium glutamicum, next to the transsulfuration pathway, a direct sulfhydrylation pathway is present that produces L-methionine, of which the key enzyme is O-acetylhomoserine sulfhydrylase (EC 2.5.1.49) (Hwang et al., 2002). The gene with locus tag APA386B_753 was annotated as encoding a putative bifunctional O-acetylhomoserine sulfhydrylase (EC 2.5.1.49)/O-succinylhomoserine sulfhydrylase (EC 2.5.1.48). However, the presence of a homoserine O-acetyltransferase (E.C. 2.3.1.31; APA386B_2138) upstream in the direct sulfhydrylation pathway gave additional evidence to annotate the gene APA386B_753 as encoding an O-acetylhomoserine sulfhydrylase (E.C. 2.5.1.49). Since this pathway was complete, the direct sulfhydrylation pathway was added to the A. pasteurianus 386B GEM instead of the transsulfuration pathway. Finally, L-methionine could be synthesized from L-homocysteine by methionine synthase (EC 2.1.1.13). SAM could be formed from L-methionine by methionine adenosyltransferase (EC 2.5.1.6). This reaction is part of the SAM cycle, which links L-methionine biosynthesis to glycerophospholipid biosynthesis, and involves the key reaction catalyzed by adenosylhomocysteinase (EC 3.3.1.1) (Reddy et al., 2008).
Nucleotide Biosynthesis
Based on the A. pasteurianus 386B PGDB, the nucleotide biosynthesis pathways were curated and added to the A. pasteurianus 386B GEM (Supplementary Figure S7). A broad-substrate-range nucleoside diphosphate kinase (EC 2.7.4.6) was present in the genome of A. pasteurianus 386B that could phosphorylate different nucleoside diphosphate acceptors, using ATP as phosphate donor, to form the respective nucleoside triphosphate products (Armenta-Medina et al., 2014). Similarly, a broad-substrate-range thioredoxin-dependent ribonucleoside-diphosphate reductase (EC 1.17.4.1) could reduce ribonucleoside diphosphate acceptors to their respective deoxyribonucleoside diphosphate forms. The thioredoxin molecule may be recycled by an NADP+-dependent thioredoxin reductase (EC 1.8.1.9). The precursor for purine biosynthesis is 5-phospho-α-D-ribose 1-phosphate (PRPP), which is also the starting point of histidine biosynthesis and involved in tryptophan biosynthesis (Kilstrup et al., 2005). From PRPP, a linear pathway was obtained to produce inosine monophosphate, from which adenosine monophosphate (AMP) or guanosine monophosphate (GMP) could be produced. Two additional adenosine salvage reactions were identified, which were related to side-products formed in other pathways. First, sulfate reduction to hydrogen sulfide could yield adenosine 3′,5′-bisphosphate as a by-product of the initial adenylation of sulfate by ATP. This compound could be recycled to AMP by 3′,5′-bisphosphate nucleotidase (EC 3.1.3.7), which was previously not annotated in the A. pasteurianus 386B genome. Second, the SAM cycle could produce adenosine, which could be recycled to AMP by adenosine kinase (EC 2.7.1.20).
Uridine diphosphate (UDP) production is the result of a linear pathway, in which aspartate is the substrate for pyrimidine biosynthesis and which could be converted into cytidine triphosphate (CTP) via CTP synthase (EC 6.3.4.2) or deoxythymidine triphosphate (dTTP) by a dedicated pathway (Kilstrup et al., 2005). Cytidine monophosphate (CMP) could be salvaged by cytidylate kinase (EC 2.7.4.25), as CMP was a side-product from phospholipid and lipopolysaccharide (LPS) biosynthesis. Consecutive cytidine diphosphate (CDP) reduction by ribonucleoside-diphosphate reductase and phosphorylation by nucleoside diphosphate kinase could produce dCTP.
Peptidoglycan and Lipopolysaccharide Biosynthesis
Biosynthesis pathways of membrane components in E. coli were used as a reference for their reconstruction in A. pasteurianus 386B (Supplementary Figures S8, S9). In general, the biosynthesis of peptidoglycan starts with the formation of glucosamine 6-phosphate by glutamine-fructose-6-phosphate transaminase (EC 2.6.1.16), which is subsequently biochemically activated to form UDP-N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is subsequently converted into UDP-N-acetylmuramic acid (UDP-MurNAc) and used as a basis to attach the first alanine residue by UDP-N-acetylmuramate L-alanine ligase (EC 6.3.2.8), which was previously not annotated in the A. pasteurianus 386B genome. In the next reactions, D-glutamate, meso-diaminopimelate, and D-alanyl-D-alanine are successively attached to UDP-MurNAc (Barreteau et al., 2008). Finally, another molecule of UDP-GlcNAc is linked to UDP-MurNAc to form the peptidoglycan monomer (Vollmer et al., 2008). This last step was included in the macromolecule reaction forming peptidoglycan (Supplementary Figure S1). Whereas it involves in vivo the linkage of the peptidoglycan precursor to undecaprenyl-phosphate (Typas et al., 2012), a growing peptidoglycan chain requires the recycling of undecaprenyl-phosphate, so its biosynthesis was not included in the model.
For lipopolysaccharide biosynthesis, UDP-GlcNAc is an important precursor to form (Kdo)2-lipid A via the Raetz pathway (Whitfield and Trent, 2014). In A. pasteurianus 386B, UDP-2,3-diacylglucosamine pyrophosphatase (EC 3.6.1.54) could form the intermediary lipid X. In contrast to E. coli that has the lpxH gene, in A. pasteurianus 386B and other α-proteobacteria this enzyme is encoded by the lpxI gene, which has a different reaction mechanism (Metzger and Raetz, 2010). Lipid X could be converted into lipid IV and two molecules of CMP-ketodeoxyoctonate (CMP-Kdo) could be transferred to lipid IV by a bifunctional 3-deoxy-D-manno-octulosonic-acid transferase (EC 2.4.99.12 and EC 2.4.99.13). Finally, two fatty acyl chains could be added to form (Kdo)2-lipid A. Whereas E. coli has two distinct enzymes encoded by lpxL and lpxM, one for each fatty acyl chain transfer, of which lpxM is not required for growth (Raetz et al., 2007), A. pasteurianus 386B possessed only one putative ortholog, APA386B_2689, which was most similar to lpxL. Subsequently, a core oligosaccharide unit, of which the composition may differ between microorganisms, is synthesized and attached to lipid A (Raetz and Whitfield, 2002). Here, the pathway present in A. pasteurianus 386B diverged from the one known in E. coli, since only four out of 10 described enzymatic reactions to synthesize the E. coli core oligosaccharide could be linked to putative A. pasteurianus 386B orthologs. Furthermore, two enzymes were missing in the pathway to produce ADP-L-glycero-D-manno-heptose. These reactions were nonetheless added to the model to allow for biomass formation (Supplementary Figure S9). Genes encoding enzymes to form the LPS precursors UDP-glucose and UDP-galactose were present, including the genes encoding phosphoglucomutase (EC 5.4.2.2), UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9), and UDP-glucose 4-epimerase (EC 5.1.3.2). Biosynthesis reactions for the formation of lipoprotein were not added to the A. pasteurianus 386B GEM, since there was no information about its abundance in the biomass composition data used.
Model Validation
In vitro and in silico growth experiments were compared to validate the A. pasteurianus 386B GEM (Table 1). This strain has been routinely cultivated in a cocoa pulp simulation medium containing lactic acid, ethanol, and mannitol as the main carbon sources (Lefeber et al., 2010; Moens et al., 2014). Here, the carbon sources were tested separately to assess their influence on the predicted flux distribution in isolation. For all tested carbon sources, periplasmic proton exchange was necessary to obtain growth in silico.

Table 1. In vitro growth experiments and in silico predicted specific growth rate using different carbon sources.
Growth of A. pasteurianus 386B on lactic acid as the sole carbon source was found in silico as well as in vitro. CO2 and H2O were the sole metabolites secreted by the model, which was in accordance to the experimental results obtained with another A. pasteurianus strain (Adler et al., 2014). A. pasteurianus 386B was not able to grow on ethanol as the sole carbon source, which confirms the growth characteristics of A. pasteurianus and is in contrast to other species of the genus Acetobacter (Cleenwerck et al., 2008). This result was also obtained when acetate was the sole carbon source. These results are probably related to the absence of genes encoding enzymes of the glyoxylate cycle in the A. pasteurianus 386B genome, since this cycle is known to be crucial for growth on C2 sources such as ethanol and acetate (Illeghems et al., 2013).
In contrast to the results of the in vitro growth experiments, in silico growth was possible on D-glucose and D-mannitol as the sole carbon sources. In both cases, FBA predicted their catabolism by the pentose phosphate pathway, leading to the formation of fructose-6-phosphate and glyceraldehyde-3-phosphate. The latter was further catabolized in the lower part of the Embden-Meyerhof-Parnas (EMP) pathway. Due to the absence of a phosphofructokinase enzyme, fructose-6-phosphate was converted into glucose-6-phosphate, leading to a reaction flux cycle involving the pentose phosphate pathway and the upper part of the EMP pathway. A high NADPH+H+ production flux had to be balanced to allow growth, which was mainly performed by a proton-translocating NAD(P)+ transhydrogenase. The contrasting results obtained for D-glucose and D-mannitol might be explained by the fact that no specific transporter could be identified for these metabolites or their oxidation products. In general, functional annotation of transporters is difficult because a limited number of transporters have been functionally characterized (Gelfand and Rodionov, 2008). It is therefore possible that D-glucose and D-mannitol are almost exclusively oxidized in the periplasm, forming D-gluconate and D-fructose, respectively (Moens et al., 2014). In addition, the occurrence of a reaction flux cycle in the FBA prediction could be an indication of the incapability of sugar catabolism to sustain growth of A. pasteurianus 386B.
A similar discrepancy for in vitro and in silico growth was found for glycerol. The predicted specific growth rate on glycerol was the highest of all carbon sources tested, but in silico growth was only possible if the reaction catalyzed by the aerobic glycerol-3-phosphate dehydrogenase (EC 1.1.5.3) was defined as being reversible. Here, a glycerol facilitator could be identified in the genome of A. pasteurianus 386B. No apparent reason for the absence of in vitro growth could be identified in the flux distribution. However, the assumption of the presence of a catabolic glycerol-3-phosphate dehydrogenase may be faulty or glycerol catabolism was not efficient enough for the bacterial cells to grow on glycerol as the sole carbon source.
Evaluation of the Manual Curation Process and Acetobacter pasteurianus 386B GEM Properties
Manual curation of GPR associations in the A. pasteurianus 386B GEM was performed based on different information sources. Hereto, EC numbers were used, as these provide a defined classification of enzymatic reactions. However, GPR associations may be complex and it is thus expected that curation will reveal inconsistencies in the functional annotations provided by the different annotation sources. For genes linked to reactions in the GEM, the differences between the manually curated EC numbers and the EC numbers provided by each annotation source were compared (Figure 4). Some annotation sources, such as eggNOG-mapper and NCBI 2017, did not provide much EC information. EggNOG-mapper annotations are characterized by long annotation notes, mostly descriptive and evading EC classification. For the NCBI annotation, the outcome is related to the RefSeq annotation version used that apparently only contained very few EC numbers. Although overall the EC numbers in the A. pasteurianus 386B GEM corresponded to the ones in the different annotation sources, differences were apparent at the main class or sub-subclass EC classification. The former indicated, for example, the occurrence of multi-functional enzymes, linked to more than one reaction, with EC numbers belonging to different classes. Also, proton-translocating enzymes were recently re-classified in a new EC class of translocases (EC 7). The latter highlighted the subtleties of the manual curation process in defining the substrate and co-factor specificities.
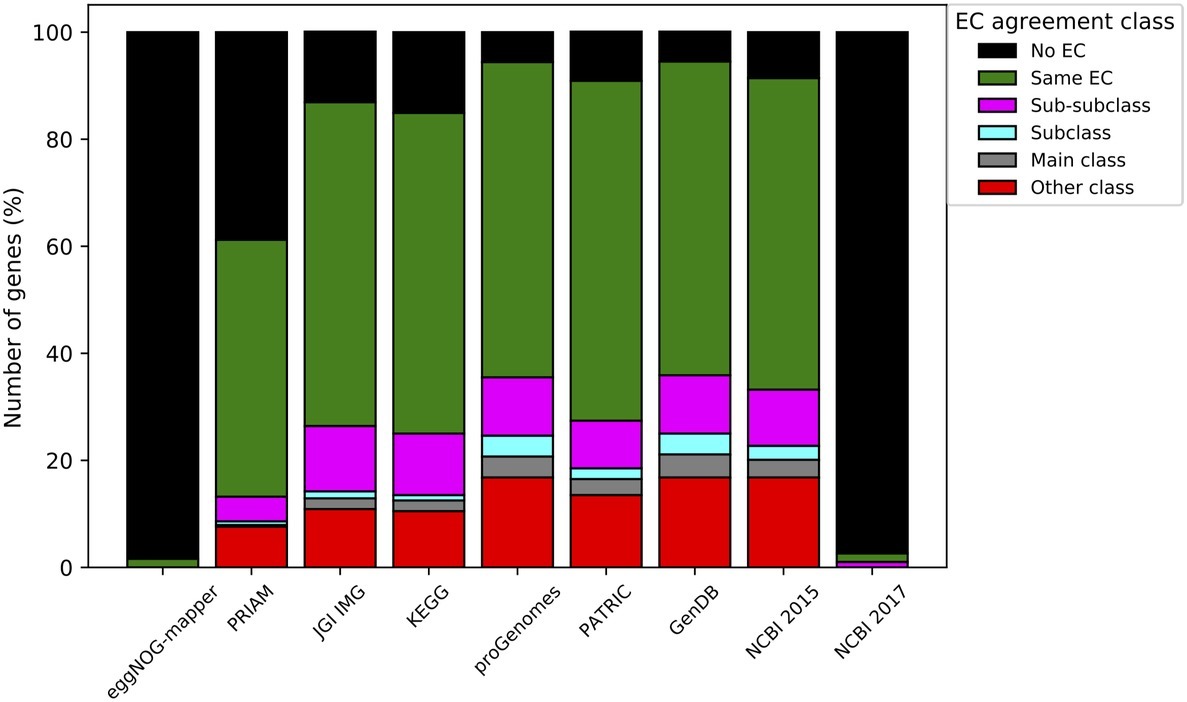
Figure 4. Comparison of EC number annotations of genes in the A. pasteurianus 386B GEM with EC numbers present in other annotation sources. The levels of agreement between annotated EC numbers were classified in a sub-subclass (three first numbers identical), subclass (two first numbers identical), main class (first number identical), and other class (different first number), according to the EC classification system. The number of genes classified in each class is shown for each of the annotation sources considered.
The properties of the resulting A. pasteurianus 386B GEM, the curated A. pasteurianus 386B PGDB, and the models of the automatic reconstruction tools are summarized in Table 2. Compartmentalization differed between the different reconstructions, probably due to a difference in ways to represent a GEM from Gram-negative bacteria. However, since compartmentalization represents important physiological barriers, it most likely influences the GEM simulation results. As could be expected, the number of genes in the automatically reconstructed models was higher than for the A. pasteurianus 386B GEM presented in the current study, as this reconstruction was performed manually and because some pathways were deliberately not included, such as the co-factor biosynthesis pathways. This latter decision was taken based on the assumption that the biosynthesis and degradation fluxes of the reaction co-factors were small compared to their involvement in the metabolic redox reactions. In contrast, the number of orphan reactions was much lower for the A. pasteurianus 386B GEM compared to the other models, providing causal links between reactions and enzyme-encoding genes in the genome. Also, the number of dead-end metabolites was low compared to the automatic reconstructions. Although dead-end metabolites could be an indication of redundancy in the reconstruction, their presence may also reflect the uncertainty in the reactants and products of the different enzymatic reactions in the models, thus providing a compendium of possible reactants and products, as for example in the curated A. pasteurianus 386B PGDB.
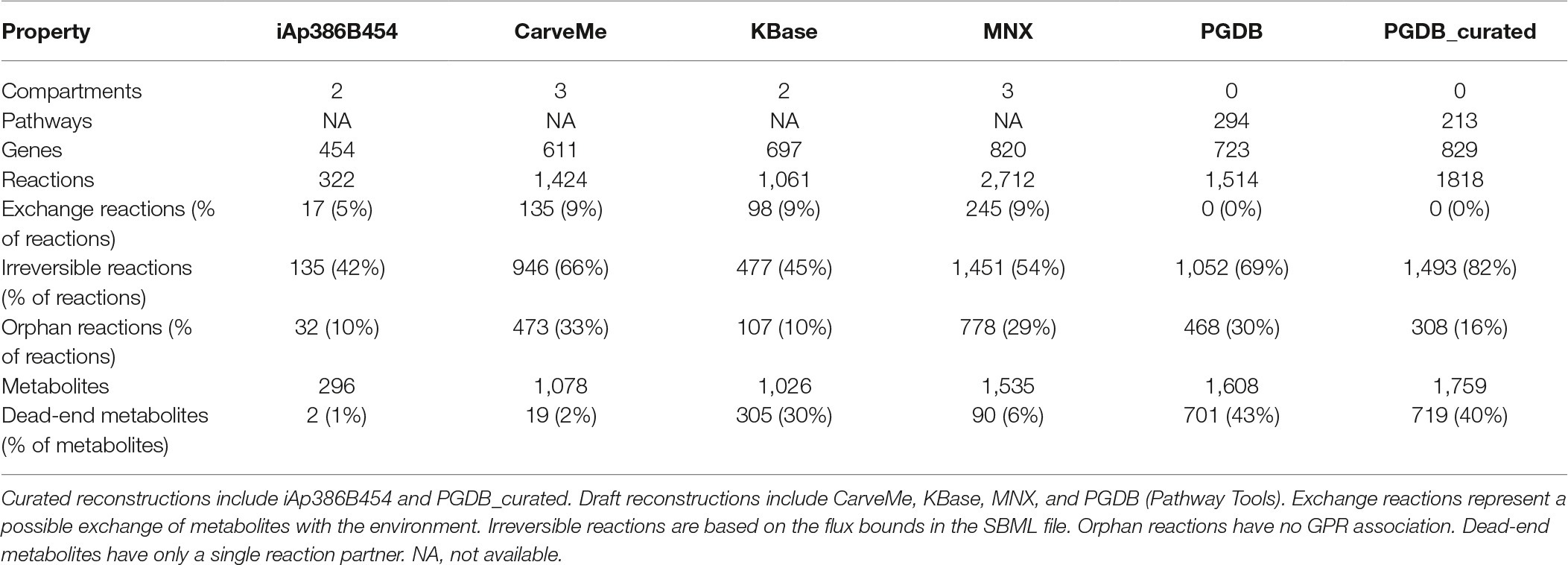
Table 2. Properties of currently available reconstructed genome-scale metabolic models of A. pasteurianus 386B.
Conclusion
During the genome re-annotation of A. pasteurianus 386B, the functional annotation of predicted enzymes and transporters was targeted, as these are critical for an accurate genome-scale metabolic network reconstruction. To improve the quality of the re-annotation, information from multiple, different annotation sources was combined, which proved to be a good strategy to guide manual curation of GPR associations. This methodology was combined with the prediction of orthogroups, using genomes of related species, which was further fine-tuned with information from the literature. Finally, using the Pathway Tools software allowed to set the annotation content in a metabolic pathway context. In this way, possible links between the different biosynthesis pathways, necessary for the biomass formation, were identified.
The re-annotated A. pasteurianus 386B genome was used to compile a curated GEM, named iAp386B454, containing 454 genes, 322 reactions, and 296 metabolites embedded in two cellular compartments. The GEM is available in SBML level 3 format and as a curated Pathway Tools PGDB and represents the first in silico genome-scale metabolic network reconstruction of a species of the genus Acetobacter. The reconstructed model was validated by performing growth experiments in a defined medium, which revealed that lactic acid as the sole carbon source could sustain growth of this strain.
Nevertheless, it became clear that some knowledge gaps remained, for example for the reconstruction of the biosynthesis pathways of cell constituents, especially for the cell envelope. Also, the combination of genome re-annotation and growth experiments could not resolve the presence of all metabolite transporters. The results obtained in this study will help to guide future research to close these knowledge gaps in A. pasteurianus.
Data Availability Statement
The curated A. pasteurianus 386B PGDB of the current study can be found in the BioCyc Tier-2 repository.
Author Contributions
RP and SW designed the study. RP performed the metabolic reconstruction and wrote the manuscript. DG and SW supervised the study, discussed the results, and edited the manuscript. BT contributed to the discussion of the results. LD contributed to the study and edited the manuscript. All authors approved the manuscript.
Funding
The authors acknowledge financial support from the Research Council of the Vrije Universiteit Brussel (OZR234, SRP7, and IOF342 projects). RP is the recipient of a PhD Fellowship strategic basic research of the Research Foundation Flanders (FWO-Vlaanderen; 1S27316N). Part of this work was performed during a research stay of RP at the Systems Bioinformatics Group of the Vrije Universiteit Amsterdam, Amsterdam, the Netherlands, funded by the Research Foundation Flanders (FWO-Vlaanderen; V449017N).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Prof. Dr. Frank Bruggeman and Prof. Dr. Rob Van Spanning of the Vrije Universiteit Amsterdam are acknowledged for their interesting discussions during RP’s research stay.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02801/full#supplementary-material
SUPPLEMENTARY FILE S1, TABLE S1 | Best blast hits of protein sequences of characterized enzymes to the A. pasteurianus 386B genome.
SUPPLEMENTARY FILE S1, FIGUREs S1–S9 | Reactions with EC numbers of the different pathways included in the A. pasteurianus 386B GEM.
SUPPLEMENTARY FILE S2 | Gene-protein-reaction associations of the A. pasteurianus 386B GEM.
SUPPLEMENTARY FILE S3 | SBML file of the A. pasteurianus 386B GEM.
SUPPLEMENTARY FILE S4 | Biomass reaction for A. pasteurianus 386B using a combination of genomic and literature data.
References
Adachi, O., Ano, Y., Toyama, H., and Matsushita, K. (2008). A novel 3-dehydroquinate dehydratase catalyzing extracellular formation of 3-dehydroshikimate by oxidative fermentation of Gluconobacter oxydans IFO 3244. Biosci. Biotechnol. Biochem. 72, 1475–1482. doi: 10.1271/bbb.70778
Adler, P., Frey, L. J., Berger, A., Bolten, C. J., Hansen, C. E., and Wittmann, C. (2014). The key to acetate: metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation-simulating conditions. Appl. Environ. Microbiol. 80, 4702–4716. doi: 10.1128/AEM.01048-14
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569. doi: 10.1038/nbt.4163
Armenta-Medina, D., Segovia, L., and Perez-Rueda, E. (2014). Comparative genomics of nucleotide metabolism: a tour to the past of the three cellular domains of life. BMC Genomics 15:800. doi: 10.1186/1471-2164-15-800
Barak, Z., and Chipman, D. M. (2012). Allosteric regulation in acetohydroxyacid synthases (AHASs) – different structures and kinetic behavior in isozymes in the same organisms. Arch. Biochem. Biophys. 519, 167–174. doi: 10.1016/j.abb.2011.11.025
Barreteau, H., Kovač, A., Boniface, A., Sova, M., Gobec, S., and Blanot, D. (2008). Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 168–207. doi: 10.1111/j.1574-6976.2008.00104.x
Bernard, T., Bridge, A., Morgat, A., Moretti, S., Xenarios, I., and Pagni, M. (2014). Reconciliation of metabolites and biochemical reactions for metabolic networks. Brief. Bioinform. 15, 123–135. doi: 10.1093/bib/bbs058
Bornstein, B. J., Keating, S. M., Jouraku, A., and Hucka, M. (2008). LibSBML: an API library for SBML. Bioinformatics 24, 880–881. doi: 10.1093/bioinformatics/btn051
Camu, N., De Winter, T., Verbrugghe, K., Cleenwerck, I., Vandamme, P., Takrama, J. S., et al. (2007). Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 73, 1809–1824. doi: 10.1128/AEM.02189-06
Caspi, R., Billington, R., Ferrer, L., Foerster, H., Fulcher, C. A., Keseler, I. M., et al. (2016). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480. doi: 10.1093/nar/gkv1164
Claudel-Renard, C. (2003). Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31, 6633–6639. doi: 10.1093/nar/gkg847
Cleenwerck, I., Gonzalez, A., Camu, N., Engelbeen, K., De Vos, P., and De Vuyst, L. (2008). Acetobacter fabarum sp. nov., an acetic acid bacterium from a Ghanaian cocoa bean heap fermentation. Int. J. Syst. Evol. Microbiol. 58, 2180–2185. doi: 10.1099/ijs.0.65778-0
De Mey, M., De Maeseneire, S., Soetaert, W., and Vandamme, E. (2007). Minimizing acetate formation in E. coli fermentations. J. Ind. Microbiol. Biotechnol. 34, 689–700. doi: 10.1007/s10295-007-0244-2
De Roos, J., and De Vuyst, L. (2018). Acetic acid bacteria in fermented foods and beverages. Curr. Opin. Biotechnol. 49, 115–119. doi: 10.1016/j.copbio.2017.08.007
De Vuyst, L., and Weckx, S. (2016). The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J. Appl. Microbiol. 121, 5–17. doi: 10.1111/jam.13045
Dong, J. M., Taylor, J. S., Latour, D. J., Iuchi, S., and Lin, E. C. (1993). Three overlapping lct genes involved in L-lactate utilization by Escherichia coli. J. Bacteriol. 175, 6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993
Ebrahim, A., Lerman, J. A., Palsson, B. O., and Hyduke, D. R. (2013). COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst. Biol. 7:74. doi: 10.1186/1752-0509-7-74
Elbourne, L. D. H., Tetu, S. G., Hassan, K. A., and Paulsen, I. T. (2017). TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324. doi: 10.1093/nar/gkw1068
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Falk, G., and Walker, J. E. (1988). DNA sequence of a gene cluster coding for subunits of the F0 membrane sector of ATP synthase in Rhodospirillum rubrum. Support for modular evolution of the F1 and F0 sectors. Biochem. J. 254, 109–122. doi: 10.1042/bj2540109
Feist, A. M., and Palsson, B. O. (2010). The biomass objective function. Curr. Opin. Microbiol. 13, 344–349. doi: 10.1016/j.mib.2010.03.003
Feng, Y., and Cronan, J. E. (2009). Escherichia coli unsaturated fatty acid synthesis. J. Biol. Chem. 284, 29526–29535. doi: 10.1074/jbc.M109.023440
Francke, C., Siezen, R. J., and Teusink, B. (2005). Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 13, 550–558. doi: 10.1016/j.tim.2005.09.001
Fu, D., Libson, A., Miercke, L. J., Weitzman, C., Nollert, P., Krucinski, J., et al. (2000). Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290, 481–486. doi: 10.1126/science.290.5491.481
Ganter, M., Bernard, T., Moretti, S., Stelling, J., and Pagni, M. (2013). MetaNetX.org: a website and repository for accessing, analysing and manipulating metabolic networks. Bioinformatics 29, 815–816. doi: 10.1093/bioinformatics/btt036
Geiger, O., López-Lara, I. M., and Sohlenkamp, C. (2013). Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1831, 503–513. doi: 10.1016/j.bbalip.2012.08.009
Gelfand, M. S., and Rodionov, D. A. (2008). Comparative genomics and functional annotation of bacterial transporters. Phys. Life Rev. 5, 22–49. doi: 10.1016/j.plrev.2007.10.003
Gottstein, W., Olivier, B. G., Bruggeman, F. J., and Teusink, B. (2016). Constraint-based stoichiometric modelling from single organisms to microbial communities. J. R. Soc. Interface 13:20160627. doi: 10.1098/rsif.2016.0627
Hanada, T., Kashima, Y., Kosugi, A., Koizumi, Y., Yanagida, F., and Udaka, S. (2001). A gene encoding phosphatidylethanolamine N-methyltransferase from Acetobacter aceti and some properties of its disruptant. Biosci. Biotechnol. Biochem. 65, 2741–2748. doi: 10.1271/bbb.65.2741
Hara, Y., Seki, M., Matsuoka, S., Hara, H., Yamashita, A., and Matsumoto, K. (2008). Involvement of PlsX and the acyl-phosphate dependent sn-glycerol-3-phosphate acyltransferase PlsY in the initial stage of glycerolipid synthesis in Bacillus subtilis. Genes Genet. Syst. 83, 433–442. doi: 10.1266/ggs.83.433
Hastings, J., Owen, G., Dekker, A., Ennis, M., Kale, N., Muthukrishnan, V., et al. (2016). ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 44, D1214–D1219. doi: 10.1093/nar/gkv1031
Henry, C. S., DeJongh, M., Best, A. A., Frybarger, P. M., Linsay, B., and Stevens, R. L. (2010). High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982. doi: 10.1038/nbt.1672
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Hwang, B.-J., Yeom, H.-J., Kim, Y., and Lee, H.-S. (2002). Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J. Bacteriol. 184, 1277–1286. doi: 10.1128/JB.184.5.1277-1286.2002
Illeghems, K., De Vuyst, L., and Weckx, S. (2013). Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14:526. doi: 10.1186/1471-2164-14-526
Janßen, H., and Steinbüchel, A. (2014). Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 7:7. doi: 10.1186/1754-6834-7-7
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Karp, P. D., Latendresse, M., Paley, S. M., Krummenacker, M., Ong, Q. D., Billington, R., et al. (2016). Pathway tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. 17, 877–890. doi: 10.1093/bib/bbv079
Kilstrup, M., Hammer, K., Ruhdaljensen, P., and Martinussen, J. (2005). Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29, 555–590. doi: 10.1016/j.fmrre.2005.04.006
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. doi: 10.1093/nar/gky1033
King, Z. A., Dräger, A., Ebrahim, A., Sonnenschein, N., Lewis, N. E., and Palsson, B. O. (2015). Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput. Biol. 11:e1004321. doi: 10.1371/journal.pcbi.1004321
Lefeber, T., Gobert, W., Vrancken, G., Camu, N., and De Vuyst, L. (2011). Dynamics and species diversity of communities of lactic acid bacteria and acetic acid bacteria during spontaneous cocoa bean fermentation in vessels. Food Microbiol. 28, 457–464. doi: 10.1016/j.fm.2010.10.010
Lefeber, T., Janssens, M., Camu, N., and De Vuyst, L. (2010). Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 76, 7708–7716. doi: 10.1128/AEM.01206-10
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Machado, D., Andrejev, S., Tramontano, M., and Patil, K. R. (2018). Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 46, 7542–7553. doi: 10.1093/nar/gky537
Mamlouk, D., and Gullo, M. (2013). Acetic acid bacteria: physiology and carbon sources oxidation. Indian J. Microbiol. 53, 377–384. doi: 10.1007/s12088-013-0414-z
Markowitz, V. M., Chen, I.-M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122. doi: 10.1093/nar/gkr1044
Matsutani, M., Fukushima, K., Kayama, C., Arimitsu, M., Hirakawa, H., Toyama, H., et al. (2014). Replacement of a terminal cytochrome c oxidase by ubiquinol oxidase during the evolution of acetic acid bacteria. Biochim. Biophys. Acta 1837, 1810–1820. doi: 10.1016/j.bbabio.2014.05.355
McDonald, A. G., Boyce, S., and Tipton, K. F. (2009). ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 37, D593–D597. doi: 10.1093/nar/gkn582
Meersman, E., Steensels, J., Mathawan, M., Wittocx, P.-J., Saels, V., Struyf, N., et al. (2013). Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS One 8:e81559. doi: 10.1371/journal.pone.0081559
Mende, D. R., Letunic, I., Huerta-Cepas, J., Li, S. S., Forslund, K., Sunagawa, S., et al. (2017). proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 45, D529–D534. doi: 10.1093/nar/gkw989
Metzger, L. E., and Raetz, C. R. H. (2010). An alternative route for UDP-diacylglucosamine hydrolysis in bacterial lipid A biosynthesis. Biochemistry 49, 6715–6726. doi: 10.1021/bi1008744
Meyer, F. (2003). GenDB – an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31, 2187–2195. doi: 10.1093/nar/gkg312
Miescher Schwenninger, S., Freimüller Leischtfeld, S., and Gantenbein-Demarchi, C. (2016). High-throughput identification of the microbial biodiversity of cocoa bean fermentation by MALDI-TOF MS. Lett. Appl. Microbiol. 63, 347–355. doi: 10.1111/lam.12621
Milo, R., and Phillips, R. (2016). Cell biology by the numbers. New York: Garland Science, Taylor & Francis Group.
Miura, H., Mogi, T., Ano, Y., Migita, C. T., Matsutani, M., Yakushi, T., et al. (2013). Cyanide-insensitive quinol oxidase (CIO) from Gluconobacter oxydans is a unique terminal oxidase subfamily of cytochrome bd. J. Biochem. 153, 535–545. doi: 10.1093/jb/mvt019
Moens, F., Lefeber, T., and De Vuyst, L. (2014). Oxidation of metabolites highlights the microbial interactions and role of Acetobacter pasteurianus during cocoa bean fermentation. Appl. Environ. Microbiol. 80, 1848–1857. doi: 10.1128/AEM.03344-13
Moretti, S., Martin, O., Van Du Tran, T., Bridge, A., Morgat, A., and Pagni, M. (2016). MetaNetX/MNXref – reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 44, D523–D526. doi: 10.1093/nar/gkv1117
Mullins, E. A., Francois, J. A., and Kappock, T. J. (2008). A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J. Bacteriol. 190, 4933–4940. doi: 10.1128/JB.00405-08
Nakano, S., Fukaya, M., and Horinouchi, S. (2006). Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti. Appl. Environ. Microbiol. 72, 497–505. doi: 10.1128/AEM.72.1.497-505.2006
Neidhardt, F. C., Ingraham, J. L., and Schaechter, M. (1990). Physiology of the bacterial cell: A molecular approach. Massachusetts: Sinauer Associates.
Núñez, M. F., Kwon, O., Wilson, T. H., Aguilar, J., Baldoma, L., and Lin, E. C. C. (2002). Transport of L-lactate, D-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem. Biophys. Res. Commun. 290, 824–829. doi: 10.1006/bbrc.2001.6255
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Olivier, B. G., Rohwer, J. M., and Hofmeyr, J.-H. S. (2005). Modelling cellular systems with PySCeS. Bioinformatics 21, 560–561. doi: 10.1093/bioinformatics/bti046
Orth, J. D., Conrad, T. M., Na, J., Lerman, J. A., Nam, H., Feist, A. M., et al. (2011). A comprehensive genome-scale reconstruction of Escherichia coli metabolism–2011. Mol. Syst. Biol. 7:535. doi: 10.1038/msb.2011.65
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucl. Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Ozturk, G., and Young, G. M. (2017). Food evolution: the impact of society and science on the fermentation of cocoa beans. Compr. Rev. Food Sci. F 16, 431–455. doi: 10.1111/1541-4337.12264
Paoletti, L., Lu, Y.-J., and Schujman, G. E., de Mendoza, D., and Rock, C. O. (2007). Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J. Bacteriol. 189, 5816–5824, doi: 10.1128/JB.00602-07
Papalexandratou, Z., Lefeber, T., Bahrim, B., Lee, O. S., Daniel, H.-M., and De Vuyst, L. (2013). Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol. 35, 73–85. doi: 10.1016/j.fm.2013.02.015
Parsons, J. B., and Rock, C. O. (2013). Bacterial lipids: metabolism and membrane homeostasis. Prog. Lipid Res. 52, 249–276. doi: 10.1016/j.plipres.2013.02.002
Peng, H. L., Wang, P. Y., Wu, C. M., Hwang, D. C., and Chang, H. Y. (1992). Cloning, sequencing and heterologous expression of a Klebsiella pneumoniae gene encoding an FAD-independent acetolactate synthase. Gene 117, 125–130. doi: 10.1016/0378-1119(92)90500-O
Pitkänen, E., Rousu, J., and Ukkonen, E. (2010). Computational methods for metabolic reconstruction. Curr. Opin. Biotechnol. 21, 70–77. doi: 10.1016/j.copbio.2010.01.010
Raetz, C. R. H., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Reddy, M. C. M., Kuppan, G., Shetty, N. D., Owen, J. L., Ioerger, T. R., and Sacchettini, J. C. (2008). Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors. Protein Sci. 17, 2134–2144. doi: 10.1110/ps.038125.108
Richards, M. A., Cassen, V., Heavner, B. D., Ajami, N. E., Herrmann, A., Simeonidis, E., et al. (2014). MediaDB: a database of microbial growth conditions in defined media. PLoS One 9:e103548. doi: 10.1371/journal.pone.0103548
Ruppin, E., Papin, J. A., de Figueiredo, L. F., and Schuster, S. (2010). Metabolic reconstruction, constraint-based analysis and game theory to probe genome-scale metabolic networks. Curr. Opin. Biotechnol. 21, 502–510. doi: 10.1016/j.copbio.2010.07.002
Saier, M. H., Reddy, V. S., Tsu, B. V., Ahmed, M. S., Li, C., and Moreno-Hagelsieb, G. (2016). The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379. doi: 10.1093/nar/gkv1103
Sakurai, K., Arai, H., Ishii, M., and Igarashi, Y. (2011). Transcriptome response to different carbon sources in Acetobacter aceti. Microbiology 157, 899–910. doi: 10.1099/mic.0.045906-0
Sá-Pessoa, J., Paiva, S., Ribas, D., Silva, I. J., Viegas, S. C., Arraiano, C. M., et al. (2013). SATP (YaaH), a succinate–acetate transporter protein in Escherichia coli. Biochem. J. 454, 585–595. doi: 10.1042/BJ20130412
Snoep, J. L., Teixeira de Mattos, M. J., Starrenburg, M. J., and Hugenholtz, J. (1992). Isolation, characterization, and physiological role of the pyruvate dehydrogenase complex and α-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 174, 4838–4841. doi: 10.1128/jb.174.14.4838-4841.1992
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Teusink, B., van Enckevort, F. H. J., Francke, C., Wiersma, A., Wegkamp, A., Smid, E. J., et al. (2005). In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 71, 7253–7262. doi: 10.1128/AEM.71.11.7253-7262.2005
Thiele, I., and Palsson, B. Ø. (2010). A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121. doi: 10.1038/nprot.2009.203
Typas, A., Banzhaf, M., Gross, C. A., and Vollmer, W. (2012). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136. doi: 10.1038/nrmicro2677
Valgepea, K., Adamberg, K., Nahku, R., Lahtvee, P.-J., Arike, L., and Vilu, R. (2010). Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst. Biol. 4:166. doi: 10.1186/1752-0509-4-166
van Heeswijk, W. C., Westerhoff, H. V., and Boogerd, F. C. (2013). Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol. Mol. Biol. Rev. 77, 628–695. doi: 10.1128/MMBR.00025-13
van Niel, E. W. J., and Hahn-Hägerdal, B. (1999). Nutrient requirements of lactococci in defined growth media. Appl. Microbiol. Biotechnol. 52, 617–627. doi: 10.1007/s002530051569
Varma, A., Boesch, B. W., and Palsson, B. O. (1993). Biochemical production capabilities of Escherichia coli. Biotechnol. Bioeng. 42, 59–73. doi: 10.1002/bit.260420109
Verduyn, C., Postma, E., Scheffers, W. A., and Van Dijken, J. P. (1992). Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517. doi: 10.1002/yea.320080703
Visintin, S., Alessandria, V., Valente, A., Dolci, P., and Cocolin, L. (2016). Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int. J. Food Microbiol. 216, 69–78. doi: 10.1016/j.ijfoodmicro.2015.09.004
Vollmer, W., Blanot, D., and De Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Wang, N.-C., and Lee, C.-Y. (2007). Enhanced transaminase activity of a bifunctional L-aspartate 4-decarboxylase. Biochem. Biophys. Res. Commun. 356, 368–373. doi: 10.1016/j.bbrc.2007.02.141
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Wu, X., Wang, X., and Lu, W. (2014). Genome-scale reconstruction of a metabolic network for Gluconobacter oxydans 621H. Biosystems 117, 10–14. doi: 10.1016/j.biosystems.2014.01.001
Yamada, Y., Nunoda, M., Ishikawa, T., and Tahara, Y. (1981). The cellular fatty acid composition in acetic acid bacteria. J. Gen. Appl. Microbiol. 27, 405–417. doi: 10.2323/jgam.27.405
Yang, J., and Pittard, J. (2008). “Biosynthesis of the aromatic amino acids” in EcoSal Plus 2013. ed. Stewart, V.
Yilmaz, L. S., and Walhout, A. J. (2017). Metabolic network modeling with model organisms. Curr. Opin. Chem. Biol. 36, 32–39. doi: 10.1016/j.cbpa.2016.12.025
Yoshimura, M., Oshima, T., and Ogasawara, N. (2007). Involvement of the YneS/YgiH and PlsX proteins in phospholipid biosynthesis in both Bacillus subtilis and Escherichia coli. BMC Microbiol. 7:69. doi: 10.1186/1471-2180-7-69
Yu, C.-S., Lin, C.-J., and Hwang, J.-K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Keywords: acetic acid bacteria, Acetobacter pasteurianus, cocoa bean fermentation process, genome-scale metabolic model, genome annotation
Citation: Pelicaen R, Gonze D, Teusink B, De Vuyst L and Weckx S (2019) Genome-Scale Metabolic Reconstruction of Acetobacter pasteurianus 386B, a Candidate Functional Starter Culture for Cocoa Bean Fermentation. Front. Microbiol. 10:2801. doi: 10.3389/fmicb.2019.02801
Edited by:
Lorenzo Morelli, Catholic University of the Sacred Heart, ItalyReviewed by:
Wen-Yong Lou, South China University of Technology, ChinaRasoul Shafiei, University of Isfahan, Iran
Copyright © 2019 Pelicaen, Gonze, Teusink, De Vuyst and Weckx. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Weckx, stefan.weckx@vub.be
 Rudy Pelicaen
Rudy Pelicaen Didier Gonze2,3
Didier Gonze2,3 Bas Teusink
Bas Teusink Luc De Vuyst
Luc De Vuyst Stefan Weckx
Stefan Weckx