- 1Department of Microbiology and Immunology, Drexel University College of Medicine, Philadelphia, PA, United States
- 2Center for Molecular Virology and Translational Neuroscience, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, Philadelphia, PA, United States
- 3School of Biomedical Engineering, Science and Health Systems, Drexel University, Philadelphia, PA, United States
- 4Sidney Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA, United States
Discussion
Human immunodeficiency virus type 1 (HIV-1) has been shown to encode a small basic protein known as the transactivator of transcription (Tat) (Rice and Mathews, 1988) and as recently reviewed (Spector et al., 2019). Tat is a multifunctional protein, though it is primarily responsible for recruitment of the host positive transcription elongation factor b (P-TEFb) by interaction with an RNA stem-loop designated the transactivation response (TAR) element, which is encoded by the viral long terminal repeat (LTR) (Dingwall et al., 1989; Li et al., 2011; Khoury et al., 2018). This interaction leads to efficient transactivation of HIV-1 and this function has been shown to be contained within the first exon of Tat (residues 1–58) (Kuppuswamy et al., 1989; Link et al., 2019). A number of other functional properties have been associated with Tat and many of these activities have been noted once the protein resides in extracellular space (Khan et al., 2019). In this regard, Tat has been shown to be secreted through a number of different mechanisms, as previously reviewed (Mele et al., 2018) which are dependent on residues at positions 11 and 49–51 and their interactions with phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2), a lipid in the plasma membrane (Rayne et al., 2010b). Tat secretion is a highly active process, and concentrations of extracellular Tat have been found (200 pg/ml to 6.5 ng/ml) in the cerebral spinal fluid (CSF) of HIV-1-infected patients who are well-suppressed on antiretroviral therapy (Johnson et al., 2013; Henderson et al., 2019). Within the central nervous system (CNS), extracellular Tat can recruit peripheral immune cells that has been shown to lead to low levels of chronic inflammation through activation of bystander cells and release of pro-inflammatory cytokines, such as interleukin (IL) 1 beta (IL-1β), IL-6, and monocyte chemoattractant protein 1 (MCP-1) from monocytes and macrophages (Hofman et al., 1994; Albini et al., 1998; Hudson et al., 2000; Pulliam et al., 2007; Rayne et al., 2010a; Bachani et al., 2013). Additionally, Tat has been shown to be directly neurotoxic through hyper-activation of neurons (Fields et al., 2015), which contributes to the development of HIV-1-associated neurocognitive disorders (HAND) (Gaskill et al., 2017). These functional properties, however, are dependent on the amino acid mutations present at a number of different Tat residues.
HIV-1 is predisposed to genetic variation, which is caused by factors such as the error-prone viral reverse transcriptase in conjunction with an array of selective pressures including the host immune response (Li et al., 2012). Tat, which is subject to genetic variation (Dampier et al., 2016) during the course of HIV disease, may also result in a number of alterations in amino acid residues that have been associated with altered function. One such example of a functional alteration would be a reduction in LTR transactivation (Boven et al., 2007; Ronsard et al., 2017), which can occur with only a single residue mutation, such as position 11 substitutions to either alanine, phenylalanine, or leucine (Yezid et al., 2009), a glutamine substitution at position 50 (Brès et al., 2002), or an alanine substitution at position 51 (Van Duyne et al., 2008). As discussed, HIV-1 Tat has been shown to be encoded by two exons, which after being alternatively spliced and translated, results in a 101 amino acid protein. We (Link et al., 2019), and others (Jeang et al., 1999; Marcello et al., 2001; López-Huertas et al., 2010; van der Kuyl et al., 2018), have demonstrated that the most prevalent Tat length within subtype B HIV-1-infected patient samples has been shown to be Tat 101 (>85%), while other widely used forms of Tat, including Tat 86 and Tat 72, occur at much lower frequencies. It is important to note that Tat 86 is still found in patient sequences albeit at a low occurrence as we have previously shown (Link et al., 2019) (CARES Cohort 15.38/207.01; 7.42% and LANL 58.91/1483; 3.97%) and as reported with the BEEHIVE Cohort (7/291; 2.4%) (van der Kuyl et al., 2018). They have also shown that in an analysis of Tat sequences in LANL from other subtypes that Tat 86 might be more dominant in subtypes D; 43/56 (76%) of subtype D viruses, 3/4 (75%) of subtype H viruses, 22/812 (2.7%) of subtype B viruses, and 3/382 (0.8%) of subtype C viruses; but not in subtypes A (0/130), F (0/32), G (0/6) or J/K (0/15) (van der Kuyl et al., 2018). However, this may be due to the small number of sequences available for each of these subtypes. We observed a similar trend when examining the world-wide distribution of subtype B sequences and found certain countries, such as France, had a reduced number of sequences submitted to LANL, resulting in their most frequent length being Tat 86 (Link et al., 2019). Given this observation, the following discussion has examined functional alterations that have been observed between Tat 86 and Tat 101, primarily from studies using subtype B or C. In order to accurately understand the biological properties that can be influenced by Tat, it would seem important to utilize the most prevalent and biologically relevant protein.
It appears that most HIV-1 Tat investigations have utilized Tat 86, however, the accuracy of this statement has not been examined. In order to quantify the frequency of particular Tat truncations in the literature, a meta-analysis of publicly available publications was performed for the last 10 years. A PubMed search was performed using the following parameters: (HIV-1 Tat) NOT “review”[Publication Type], 2009/01/01 to 2018/12/31, and free full text. At the time this manuscript was drafted, there were 973 publications, which were then read and assessed for their utilization of different forms of Tat (Figure 1). There were an additional 407 publications that were not publicly available and thus were not assessed, due to access limitations. The results included expression of Tat within animal models, plasmids, or purified recombinant proteins, as well as molecular simulations and sequencing studies. One of the more prominent uses of Tat was as a fusion peptide for intracellular trafficking of another protein. This was listed as Tat peptides (PEP), however, this also included a number of additional minor truncations, such as Tat 82 (Dutta and Roy, 2015). Publications that did not explicitly state the length of Tat or the molecular clone it was derived from were listed as Tat length not stated in publication (LNS) [18.29% (178 of 973)] (Supplemental Table 1). There were a few results that examined SIV rather than HIV, and those were not included in Figure 1. As shown, 40.18% (391 of 973) of the publications used Tat 86, while only 15.51% (151 of 973) used Tat 101. Over each of the 10 years there was an average of 24 more Tat 86 papers relative to Tat 101. A paired Student's t-test of the number of publications for Tat 101 compared to Tat 86 each year over this 10-year period was statistically significant (p < 0.0001). The results of this meta-analysis supports the statement that Tat 86 is used more frequently than Tat 101 even though it has been encountered much less frequently in HIV-1-infected patient samples. Furthermore, a surprising number of publications did not clearly describe the length of Tat in the study, which may affect our understanding of Tat functionality.
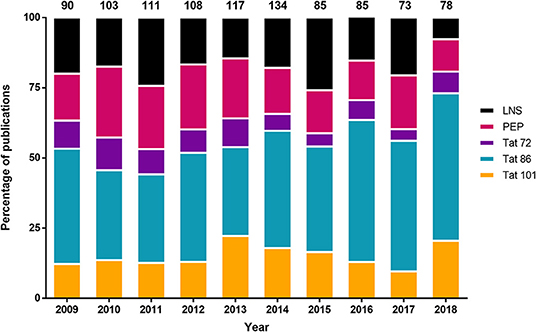
Figure 1. Distribution of the different Tat lengths used in the literature from 2009 to 2018. The 101 residue Tat was not the most frequently utilized Tat length variant in the literature from publicly available publications on PubMed. The parameters used on the PubMed search were: (HIV-1 Tat) NOT “review”[Publication Type], 2009/01/01 to 2018/12/31, and free full text. The number of publications per year were converted into a percentage (y-axis), and the total number of publications was listed above the figure. Refer to Supplemental Table 1 for more information regarding the categories of Tat. Tat length not stated in publication (LNS); Tat peptides (PEP).
Tat is a multifunctional protein and premature truncations such as Tat 72 or 86 have been demonstrated to alter its functional properties. Studies observed that expression of Tat 101 significantly altered mitochondrial DNA transcription, mitochondrial content, and their distribution within peripheral blood lymphocytes in comparison to Tat 72 (Rodríguez-Mora et al., 2015). Additional studies arrived at a similar conclusion where they observed that Jurkat cells expressing Tat 101 had altered cell morphology, proliferation, chemotaxis, polarization, and actin polymerization, but these effects were not present in cells expressing Tat 72 (López-Huertas et al., 2010). Furthermore, the second exon of Tat has also been demonstrated to be critical in reducing host innate responses, such as downregulating interferon-stimulating genes (Kukkonen et al., 2014). When examining the cellular gene modulation of different Tat lengths in THP-1 cells, a monocytic cell line, and in human primary monocyte-derived macrophages, it was found that an HIV-1SF2 molecular clone expressing only the first exon of Tat (Tat 72) significantly increased the RNA and protein of innate response genes, such as Stat-1, MX2, and IRF-7, when compared to Tat 101, which resulted in a downregulation of these genes (Kukkonen et al., 2014). Tat 101 has also been demonstrated to induce ten-fold more IL-2 secretion vs. the amount that Tat 72 was capable of in Jurkat cells (Johnson et al., 2013).
Another example related to the functional properties of the second exon of Tat has been the dependence on the NF-κB-associated motif, ESKKKVE, which localize to residues 86–92 in HIV-1 subtype B Tat and would not be contained in the Tat 72 or Tat 86 versions of the protein. Expression of Tat 101 increased anti-apoptotic proteins, such as BCL2, and was demonstrated to cause a delay in FasL-mediated apoptosis of human peripheral blood lymphocytes as well as Jurkat cells (López-Huertas et al., 2013). This effect was not seen when examining Tat 72, to which the authors concluded was dependent on the NF-κB-associated motif, Tat 86 was not tested. Further characterization of this motif has also demonstrated that the molecular clone HIV-189.6 expressing Tat 72 replicated at significantly lower levels than Tat 101-expressing HIV-189.6 in human primary blood lymphocytes (Mahlknecht et al., 2008). Additional mutations were made within and downstream of the ESKKKVE motif in the Tat 101-expressing virus and also demonstrated the necessity for the glutamic acids (positions 92, 94, and 96) and lysines (positions 88, 89, and 90) in this particular activity. Tat function is commonly assessed via an LTR transactivation assay, but as shown in this publication, LTR transactivation may not be indicative of viral replication if the Tat 101 protein is not used.
These results demonstrate that within the past decade, investigations have favored the use of Tat 86. While this may not alter transactivation function of Tat, particularly within a plasmid expression system (Link et al., 2019), there may be alterations in other mechanisms, as described above. As we have discussed, the biological relevance of Tat length within animal models may be another key distinguishing assay to assess differences in function. Animal models are crucial for understanding Tat, specifically due to Tat being measurably expressed in patients with low-to-undetectable viremia or without detecting other HIV-1 proteins (Johnson et al., 2013; Henderson et al., 2019). Tat animal models often employ direct injection of Tat 72 or 86 into the brain (Aksenov et al., 2001, 2003; Cass et al., 2003; Fitting et al., 2008; Agrawal et al., 2012), however, this is not the only method of Tat exposure. Mice have also been exposed to repeated intranasal administration of Tat 86 and were able to demonstrate trafficking of Tat into the CNS, via the olfactory bulb (Pulliam et al., 2007). However, this model was not examined with Tat 101, and it is unknown if the trafficking patterns would be similar. In order to more closely model neurocognitive impairment within patients, use of a chronic exposure animal model that continuously expresses Tat may be of particular importance in determining the in vivo functional properties of Tat 101 vs. other length Tat proteins.
To our knowledge, there are no transgenic animal models that express Tat 101 (Soontornniyomkij et al., 2016; Langford et al., 2018; Green et al., 2019). The iTat model, which expresses Tat in a doxycycline-dependent manner that models chronic exposure, is a widely utilized model of HAND, however, it also does not express the Tat 101 protein (Fan et al., 2016; Langford et al., 2018). Similarly, the rtTA-Tat mouse model also expresses Tat 86 under the control of a glial fibrillary acidic protein (GFAP) promoter, which has resulted in low levels of chronic inflammation (Bruce-Keller et al., 2008; Dickens et al., 2017). These mice were observed for an entire year, leading to noticeable reductions in brain volume and alterations in synaptic and axonal damage. Another model that mimics chronic HIV-1 exposure was assessed by infecting human cell reconstituted SCID mice and monocyte-derived macrophages. This model utilized a macrophage-tropic HIV-1 molecular clone expressing either Tat 72 or Tat 101, and it was observed that the second exon of Tat was required for efficient replication within macrophages (Neuveut et al., 2003). While molecular alterations caused by genetic variation of Tat, specifically Tat length, have been characterized in in vitro cell culture systems, behavioral alterations have not been characterized. Various behavioral tests are available, such as: open field, elevated plus mazes, marble burying tests, acoustic startle response, and pre-pulse inhibition, which are used for determining anxiety-like responses and correlating body flinching to neuronal damage, respectively (Fitting et al., 2008; Paris et al., 2014, 2015). While behavior alterations have been examined in Tat 72 and 86 in mice and rats, to our knowledge, there have not been investigations to determine if different Tat lengths, specifically Tat 101, will cause detectable alterations in animal behavior.
As previously mentioned, Tat is synthesized by two exons, primarily resulting in a protein of 101 amino acids, however, early clinical isolates of subtype B HIV-1 Tat encoded for a 86 amino acid variant, which led to the prominent use of Tat 72 and 86 in a multitude of studies. Based on this literature meta-analysis, we were able to conclusively demonstrate that although the most biologically relevant, Tat 101 is not the most frequently used variant. There does not appear to be an increase in the number of publications that utilize the Tat 101 (Figure 1), however, as stated above, functional differences of Tat have been observed when using different length variants. Therefore, due to recent publications focused on subtype B Tat length (van der Kuyl et al., 2018; Link et al., 2019), and the functions that are associated with the second exon of Tat, it is the opinion of the authors that future studies utilize both Tat 86 and Tat 101 whenever possible, but if only one can be used to use Tat 101 to ensure biological relevance to the studies performed (Jeang et al., 1999; Marcello et al., 2001; López-Huertas et al., 2010; van der Kuyl et al., 2018; Link et al., 2019).
Author Contributions
AM conceived the study. AM and MN designed the study. AM, WD, JM, BW, and MN prepared and designed the figures and drafted the manuscript. All authors have read and approved the final manuscript.
Funding
The authors were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke (NINDS) R01 NS089435 (PI, MN), the NIMH Comprehensive NeuroAIDS Center (CNAC) P30 MH092177 (Kamel Khalili, PI; BW, PI of the Drexel subcontract involving the Clinical and Translational Research Support Core) and under the Ruth L. Kirschstein National Research Service Award T32 MH079785 (PI, Kamel Khalili and Tricia Burdo; with BW serving as the PI of the Drexel University College of Medicine component and Olimpia Meucci as Co-Director). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00444/full#supplementary-material
Supplemental Table 1. Publication results from a PubMed search for HIV-1 Tat from 2009 to 2018. Reported here are the publications used for the analysis in Figure 1. The parameters used on the PubMed search were: (HIV-1 Tat) NOT “review”[Publication Type], 2009/01/01 to 2018/12/31, and free full text. 973 publications were examined for their utilization of Tat, whether they were: a recombinant protein, expressed within an animal model, derived from a plasmid, or used in a molecular simulation. Publications that did not explicitly state the length of Tat utilized or the molecular clone that it was derived from were listed as Tat length not stated in publication (LNS). The majority of HIV-1 transgenic animal models are Tat 86, however, if it was not explicitly stated it was reported as LNS. There were publications that were not accessible and those were listed as No access to publication (NAP). Additionally, publications that are listed as Tat peptides (PEP) refers to any truncations of Tat that are not 72 or 86 residues in length. Publications that solely examined the sequences of patients (i.e., Did not utilize a molecular clone that would express Tat, a recombinant protein, a transgenic animal model, or an expression vector) were listed as No Tat used in this publication (NTU) and not included in Figure 1 (n = 124). Publications that examined SIV rather than HIV were listed as SIV and were also not included in Figure 1 (n = 3). Abbreviation used in this document are: PEP, Tat peptide; 72, Tat 72; 86, Tat 86; 101, Tat 101; NTU, No Tat used in this publication; LNS, Tat length not stated in publication, and NAP, no access to publication.
References
Agrawal, L., Louboutin, J. P., Reyes, B. A., Van Bockstaele, E. J., and Strayer, D. S. (2012). HIV-1 Tat neurotoxicity: a model of acute and chronic exposure, and neuroprotection by gene delivery of antioxidant enzymes. Neurobiol. Dis. 45, 657–670. doi: 10.1016/j.nbd.2011.10.005
Aksenov, M. Y., Hasselrot, U., Bansal, A. K., Wu, G., Nath, A., Anderson, C., et al. (2001). Oxidative damage induced by the injection of HIV-1 Tat protein in the rat striatum. Neurosci. Lett. 305, 5–8. doi: 10.1016/S0304-3940(01)01786-4
Aksenov, M. Y., Hasselrot, U., Wu, G., Nath, A., Anderson, C., Mactutus, C. F., et al. (2003). Temporal relationships between HIV-1 Tat-induced neuronal degeneration, OX-42 immunoreactivity, reactive astrocytosis, and protein oxidation in the rat striatum. Brain Res. 987, 1–9. doi: 10.1016/S0006-8993(03)03194-9
Albini, A., Ferrini, S., Benelli, R., Sforzini, S., Giunciuglio, D., Aluigi, M. G., et al. (1998). HIV-1 Tat protein mimicry of chemokines. Proc. Natl. Acad. Sci. U. S. A. 95, 13153–13158. doi: 10.1073/pnas.95.22.13153
Bachani, M., Sacktor, N., McArthur, J. C., Nath, A., and Rumbaugh, J. (2013). Detection of anti-tat antibodies in CSF of individuals with HIV-associated neurocognitive disorders. J. Neurovirol. 19, 82–88. doi: 10.1007/s13365-012-0144-8
Boven, L. A., Noorbakhsh, F., Bouma, G., van der Zee, R., Vargas, D. L., Pardo, C., et al. (2007). Brain-derived human immunodeficiency virus-1 Tat exerts differential effects on LTR transactivation and neuroimmune activation. J. Neurovirol. 13, 173–184. doi: 10.1080/13550280701258399
Brès, V., Kiernan, R., Emiliani, S., and Benkirane, M. (2002). Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J. Biol. Chem. 277, 22215–22221. doi: 10.1074/jbc.M201895200
Bruce-Keller, A. J., Turchan-Cholewo, J., Smart, E. J., Geurin, T., Chauhan, A., Reid, R., et al. (2008). Morphine causes rapid increases in glial activation and neuronal injury in the striatum of inducible HIV-1 Tat transgenic mice. Glia 56, 1414–1427. doi: 10.1002/glia.20708
Cass, W. A., Harned, M. E., Peters, L. E., Nath, A., and Maragos, W. F. (2003). HIV-1 protein Tat potentiation of methamphetamine-induced decreases in evoked overflow of dopamine in the striatum of the rat. Brain Res. 984, 133–142. doi: 10.1016/S0006-8993(03)03122-6
Dampier, W., Nonnemacher, M. R., Mell, J., Earl, J., Ehrlich, G. D., Pirrone, V., et al. (2016). HIV-1 genetic variation resulting in the development of new quasispecies continues to be encountered in the peripheral blood of well-suppressed patients. PLoS ONE 11:e0155382. doi: 10.1371/journal.pone.0155382
Dickens, A. M., Yoo, S. W., Chin, A. C., Xu, J., Johnson, T. P., Trout, A. L., et al. (2017). Chronic low-level expression of HIV-1 Tat promotes a neurodegenerative phenotype with aging. Sci. Rep. 7:7748. doi: 10.1038/s41598-017-07570-5
Dingwall, C., Ernberg, I., Gait, M. J., Green, S. M., Heaphy, S., Karn, J., et al. (1989). Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 86, 6925–6929. doi: 10.1073/pnas.86.18.6925
Dutta, R., and Roy, S. (2015). Chronic morphine and HIV-1 Tat promote differential central nervous system trafficking of CD3+ and Ly6C+ immune cells in a murine Streptococcus pneumoniae infection model. J. Neuroinflammation 12:120. doi: 10.1186/s12974-015-0341-5
Fan, Y., Gao, X., Chen, J., Liu, Y., and He, J. J. (2016). HIV tat impairs neurogenesis through functioning as a notch ligand and activation of notch signaling pathway. J. Neurosci. 36, 11362–11373. doi: 10.1523/JNEUROSCI.1208-16.2016
Fields, J. A., Dumaop, W., Crews, L., Adame, A., Spencer, B., Metcalf, J., et al. (2015). Mechanisms of HIV-1 Tat neurotoxicity via CDK5 translocation and hyper-activation: role in HIV-associated neurocognitive disorders. Curr. HIV Res. 13, 43–54. doi: 10.2174/1570162X13666150311164201
Fitting, S., Booze, R. M., and Mactutus, C. F. (2008). Neonatal intrahippocampal injection of the HIV-1 proteins gp120 and Tat: differential effects on behavior and the relationship to stereological hippocampal measures. Brain Res. 1232, 139–154. doi: 10.1016/j.brainres.2008.07.032
Gaskill, P. J., Miller, D. R., Gamble-George, J., Yano, H., and Khoshbouei, H. (2017). HIV, Tat and dopamine transmission. Neurobiol. Dis. 105, 51–73. doi: 10.1016/j.nbd.2017.04.015
Green, M. V., Raybuck, J. D., Zhang, X., Wu, M. M., and Thayer, S. A. (2019). Scaling synapses in the presence of HIV. Neurochem. Res. 44, 234–246. doi: 10.1007/s11064-018-2502-2
Henderson, L. J., Johnson, T. P., Smith, B. R., Reoma, L. B., Santamaria, U. A., Bachani, M., et al. (2019). Presence of Tat and transactivation response element in spinal fluid despite antiretroviral therapy. AIDS 33(Suppl.2), S145–S157. doi: 10.1097/QAD.0000000000002268
Hofman, F. M., Dohadwala, M. M., Wright, A. D., Hinton, D. R., and Walker, S. M. (1994). Exogenous tat protein activates central nervous system-derived endothelial cells. J. Neuroimmunol. 54, 19–28. doi: 10.1016/0165-5728(94)90226-7
Hudson, L., Liu, J., Nath, A., Jones, M., Raghavan, R., Narayan, O., et al. (2000). Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J. Neurovirol. 6, 145–155. doi: 10.3109/13550280009013158
Jeang, K. T., Xiao, H., and Rich, E. A. (1999). Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274, 28837–28840. doi: 10.1074/jbc.274.41.28837
Johnson, T. P., Patel, K., Johnson, K. R., Maric, D., Calabresi, P. A., Hasbun, R., et al. (2013). Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc. Natl. Acad. Sci. U. S. A. 110, 13588–13593. doi: 10.1073/pnas.1308673110
Khan, N., Lakpa, K. L., Halcrow, P. W., Afghah, Z., Miller, N. M., Geiger, J. D., et al. (2019). BK channels regulate extracellular Tat-mediated HIV-1 LTR transactivation. Sci. Rep. 9:12285. doi: 10.1038/s41598-019-48777-y
Khoury, G., Darcis, G., Lee, M. Y., Bouchat, S., van Driessche, B., Purcell, D. F. J., et al. (2018). The molecular biology of HIV latency. Adv. Exp. Med. Biol. 1075, 187–212. doi: 10.1007/978-981-13-0484-2_8
Kukkonen, S., Martinez-Viedma Mdel, P., Kim, N., Manrique, M., and Aldovini, A. (2014). HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology 11:30. doi: 10.1186/1742-4690-11-30
Kuppuswamy, M., Subramanian, T., Srinivasan, A., and Chinnadurai, G. (1989). Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 17, 3551–3561. doi: 10.1093/nar/17.9.3551
Langford, D., Oh Kim, B., Zou, W., Fan, Y., Rahimain, P., Liu, Y., et al. (2018). Doxycycline-inducible and astrocyte-specific HIV-1 Tat transgenic mice (iTat) as an HIV/neuroAIDS model. J. Neurovirol. 24, 168–179. doi: 10.1007/s13365-017-0598-9
Li, L., Aiamkitsumrit, B., Pirrone, V., Nonnemacher, M. R., Wojno, A., Passic, S., et al. (2011). Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. J. Neurovirol. 17, 92–109. doi: 10.1007/s13365-010-0014-1
Li, L., Dahiya, S., Kortagere, S., Aiamkitsumrit, B., Cunningham, D., Pirrone, V., et al. (2012). Impact of Tat genetic variation on HIV-1 disease. Adv. Virol. 2012:123605. doi: 10.1155/2012/123605
Link, R. W., Mele, A. R., Antell, G. C., Pirrone, V., Zhong, W., Kercher, K., et al. (2019). Investigating the distribution of HIV-1 Tat lengths present in the drexel medicine CARES cohort. Virus Res. 272:197727. doi: 10.1016/j.virusres.2019.197727
López-Huertas, M. R., Callejas, S., Abia, D., Mateos, E., Dopazo, A., and Alcamí, J. (2010). Modifications in host cell cytoskeleton structure and function mediated by intracellular HIV-1 Tat protein are greatly dependent on the second coding exon. Nucleic Acids Res. 38, 3287–3307. doi: 10.1093/nar/gkq037
López-Huertas, M. R., Mateos, E., Sánchez Del Cojo, M., Gómez-Esquer, F., Díaz-Gil, G., and Rodríguez-Mora, S. (2013). The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: a potential mechanism for persistent viral production. J. Biol. Chem. 288, 7626–7644. doi: 10.1074/jbc.M112.408294
Mahlknecht, U., Dichamp, I., Varin, A., Van Lint, C., and Herbein, G. (2008). NF-kappaB-dependent control of HIV-1 transcription by the second coding exon of Tat in T cells. J. Leukoc. Biol. 83, 718–727. doi: 10.1189/jlb.0607405
Marcello, A., Zoppé, M., and Giacca, M. (2001). Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 51, 175–181. doi: 10.1080/152165401753544241
Mele, A. R., Marino, J., Chen, K., Pirrone, V., Janetopoulos, C., Wigdahl, B., et al. (2018). Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter. Traffic 19, 655–665. doi: 10.1111/tra.12578
Neuveut, C., Scoggins, R. M., Camerini, D., Markham, R. B., and Jeang, K. T. (2003). Requirement for the second coding exon of Tat in the optimal replication of macrophage-tropic HIV-1. J. Biomed. Sci. 10, 651–660. doi: 10.1007/BF02256316
Paris, J. J., Fenwick, J., and McLaughlin, J. P. (2014). Progesterone protects normative anxiety-like responding among ovariectomized female mice that conditionally express the HIV-1 regulatory protein, Tat, in the CNS. Horm. Behav. 65, 445–453. doi: 10.1016/j.yhbeh.2014.04.001
Paris, J. J., Singh, H. D., Carey, A. N., and McLaughlin, J. P. (2015). Exposure to HIV-1 Tat in brain impairs sensorimotor gating and activates microglia in limbic and extralimbic brain regions of male mice. Behav. Brain Res. 291, 209–218. doi: 10.1016/j.bbr.2015.05.021
Pulliam, L., Sun, B., Rempel, H., Martinez, P. M., Hoekman, J. D., Rao, R. J., et al. (2007). Intranasal tat alters gene expression in the mouse brain. J. Neuroimmune Pharmacol. 2, 87–92. doi: 10.1007/s11481-006-9053-z
Rayne, F., Debaisieux, S., Bonhoure, A., and Beaumelle, B. (2010a). HIV-1 Tat is unconventionally secreted through the plasma membrane. Cell Biol. Int. 34, 409–413. doi: 10.1042/CBI20090376
Rayne, F., Debaisieux, S., Yezid, H., Lin, Y. L., Mettling, C., Konate, K., et al. (2010b). Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J. 29, 1348–1362. doi: 10.1038/emboj.2010.32
Rice, A. P., and Mathews, M. B. (1988). Transcriptional but not translational regulation of HIV-1 by the tat gene product. Nature 332, 551–553. doi: 10.1038/332551a0
Rodríguez-Mora, S., Mateos, E., Moran, M., Martín, M., Á López, J. A., and Calvo, E. (2015). Intracellular expression of Tat alters mitochondrial functions in T cells: a potential mechanism to understand mitochondrial damage during HIV-1 replication. Retrovirology 12:78. doi: 10.1186/s12977-015-0203-3
Ronsard, L., Ganguli, N., Singh, V. K., Mohankumar, K., Rai, T., Sridharan, S., et al. (2017). Impact of genetic variations in HIV-1 Tat on LTR-mediated transcription via TAR RNA interaction. Front. Microbiol. 8:706. doi: 10.3389/fmicb.2017.00706
Soontornniyomkij, V., Kesby, J. P., Morgan, E. E., Bischoff-Grethe, A., Minassian, A., Brown, G. G., et al. (2016). Effects of HIV and methamphetamine on brain and behavior: evidence from human studies and animal models. J. Neuroimmune Pharmacol. 11, 495–510. doi: 10.1007/s11481-016-9699-0
Spector, C., Mele, A. R., Wigdahl, B., and Nonnemacher, M. R. (2019). Genetic variation and function of the HIV-1 Tat protein. Med. Microbiol. Immunol. 208, 131–169. doi: 10.1007/s00430-019-00583-z
van der Kuyl, A. C., Vink, M., Zorgdrager, F., Bakker, M., Wymant, C., Hall, M., et al. (2018). The evolution of subtype B HIV-1 tat in the Netherlands during 1985-2012. Virus Res. 250, 51–64. doi: 10.1016/j.virusres.2018.04.008
Van Duyne, R., Easley, R., Wu, W., Berro, R., Pedati, C., Klase, Z., et al. (2008). Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5:40. doi: 10.1186/1742-4690-5-40
Keywords: HIV-1, Tat, genetic variation, transcription, truncation
Citation: Mele AR, Marino J, Dampier W, Wigdahl B and Nonnemacher MR (2020) HIV-1 Tat Length: Comparative and Functional Considerations. Front. Microbiol. 11:444. doi: 10.3389/fmicb.2020.00444
Received: 16 January 2020; Accepted: 02 March 2020;
Published: 24 March 2020.
Edited by:
Nejat Duzgunes, University of the Pacific, United StatesReviewed by:
Antoinette Van Der Kuyl, University of Amsterdam, NetherlandsBruno Beaumelle, UMR9004 Institut de Recherche en Infectiologie de Montpellier (IRIM), France
Copyright © 2020 Mele, Marino, Dampier, Wigdahl and Nonnemacher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael R. Nonnemacher, mrn25@drexel.edu
 Anthony R. Mele
Anthony R. Mele Jamie Marino
Jamie Marino Will Dampier
Will Dampier Brian Wigdahl
Brian Wigdahl Michael R. Nonnemacher
Michael R. Nonnemacher