- 1Department of Biochemistry, St. Ann’s College for Women, Hyderabad, India
- 2Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 3Department of Agricultural Microbiology, Aligarh Muslim University, Aligarh, India
- 4Department of Microbiology, PSGVP Mandal’s, Arts, Science and Commerce College, Shahada, India
- 5Institute of Bioproduct Development, Universiti Teknologi Malaysia (UTM), Skudai, Malaysia
- 6School of Chemical and Energy Engineering, Faculty of Engineering, Universiti Teknologi Malaysia (UTM), Skudai, Malaysia
- 7City of Scientific Research and Technological Applications, New Borg El-Arab, Egypt
- 8Biology Department, Faculty of Mathematics and Natural Science, Udayana University, Bali, Indonesia
Acidobacteria represents an underrepresented soil bacterial phylum whose members are pervasive and copiously distributed across nearly all ecosystems. Acidobacterial sequences are abundant in soils and represent a significant fraction of soil microbial community. Being recalcitrant and difficult-to-cultivate under laboratory conditions, holistic, polyphasic approaches are required to study these refractive bacteria extensively. Acidobacteria possesses an inventory of genes involved in diverse metabolic pathways, as evidenced by their pan-genomic profiles. Because of their preponderance and ubiquity in the soil, speculations have been made regarding their dynamic roles in vital ecological processes viz., regulation of biogeochemical cycles, decomposition of biopolymers, exopolysaccharide secretion, and plant growth promotion. These bacteria are expected to have genes that might help in survival and competitive colonization in the rhizosphere, leading to the establishment of beneficial relationships with plants. Exploration of these genetic attributes and more in-depth insights into the belowground mechanics and dynamics would lead to a better understanding of the functions and ecological significance of this enigmatic phylum in the soil-plant environment. This review is an effort to provide a recent update into the diversity of genes in Acidobacteria useful for characterization, understanding ecological roles, and future biotechnological perspectives.
Introduction
Prokaryotes, the unseen majority sustaining life on Earth, are involved in a multitude of interactions and biogeochemical processes having global ecological relevance, including decomposition, mineralization, storage, and release of nutrients (Sikorski, 2015). The number of prokaryotes present in a gram of soil can be between 106 and 109 cells (Bulgarelli et al., 2013). It is apparent from the above data that there is an abundant density of bacteria in the soil, however, the discrepancy is that only less than 1% bacteria from natural environments, including soil, could be cultivated by using conventional culturing techniques (Crits-Christoph et al., 2018; Chaudhary et al., 2019). A large segment of the microbial community gets consistently overlooked during routine microscopic analysis, indicating them to be novel and hitherto unrecognized with ambiguous physiology and growth requirements (Youssef et al., 2015). A rational investigation stemming from the recognition of this discrepancy is the identification of these refractive bacteria which escape conventional enrichment and isolation procedures. In this direction, the 16S rRNA gene, the gold standard for phylogenetic analysis, proves to be a useful tool for studying the evolution-based taxonomy of microbial communities from various econiches (Rosselli et al., 2016). The rare biosphere comprises of several identified novel bacterial lineages, which appear to be profoundly branching within the bacterial tree and remain unaffiliated with any known bacterial phyla and are termed as candidate phylum (or candidate division) (Solden et al., 2016).
Molecular studies in the soil revealed the presence of unseen, hidden, recalcitrant bacteria referred to as difficult-to-culture, hitherto-unculturable, or yet-to-be cultivated bacteria. In contrast to standard fast-growing bacteria, these underexplored bacterial groups remain elusive to conventional microbiological cultivation techniques, mostly because their optimized growth conditions are still underexplored. The proportion of such bacteria exceeds far more than that of the culturable ones. Among them, Acidobacteria constitutes the most abundant phylum whose members dominate soil bacterial communities (Vartoukian et al., 2010; Chaudhary et al., 2019). Data from 16S rRNA gene inventories signify the vastness and breadth of the phylum with genetic and metabolic diversity (Janssen, 2006). Study of these hitherto-unculturable constituents of soil microbial population through culture-dependent and -independent approaches revealed the essence of their bioactivity and their ecological functions in plant-soil ecosystems (Huber et al., 2016; Lladó et al., 2018; Elmagzob et al., 2019). The underexplored phylum Acidobacteria thus provides an enthralling and seamless source of biological diversity, offering new avenues for exploitation. Henceforth, the most recent advances mining the difficult-to-culture soil acidobacterial diversity for biotechnological advances and comprehensive understanding of the genetic diversity and ecophysiological profiles of Acidobacteria in the plant-soil econiche has been scrupulously presented in this review.
Underexplored Phylum Acidobacteria
Acidobacteria represents an enigmatic phylum with its members copiously distributed in different ecosystems (Mushinski et al., 2018). Acidobacteria is ubiquitous in diverse terrestrial environments ranging from tundra soils to desert soils, from peatland soils and sediments to grasslands, forests, and agricultural lands (Janssen, 2006; Eichorst et al., 2018) and constitute about 5–70% of the soil microbial populace (Huber et al., 2016). The members are recalcitrant, rendering this phylum to be feebly understood (Hugenholtz et al., 1998; Huber et al., 2016). The recalcitrance of most Acidobacteria members to grow on conventional growth media can be attributed to their oligotrophic nature or ecological K-strategy (Ward et al., 2009; Kielak et al., 2016a). Thus, the use of low nutrient media, modified incubation conditions including elevated CO2, low pH, prolonged incubation periods, and supplementing growth media with amendments like antioxidants, rhizosphere extracts, etc., have led to the isolation of several acidobacterial species (Stevenson et al., 2004; Sait et al., 2006; da Rocha et al., 2009; Tanaka et al., 2017).
Based on the analysis of major 16S rRNA gene sequence clades, the phylum Acidobacteria is phylogenetically classified into 26 subdivisions (Barns et al., 2007). Among these, only seven subdivisions (namely the subdivisions 1, 3, 4, 6, 8, 10, and 23) are represented by taxonomically described members (Dedysh and Yilmaz, 2018; Eichorst et al., 2018). Although more than 12,000 distinct phylotypes and more than 6,500 species-level operational taxonomic units have been reported so far for this predominant soil bacterial group, yet it is described by only 56 cultivable species belonging to 28 genera (Overmann et al., 2017; Vieira et al., 2017; Dedysh and Yilmaz, 2018). All cultured Acidobacteria members are Gram-negative, non-spore formers, and exhibit an oligotrophic mode of nutrition (Fierer et al., 2005; George et al., 2011; Dedysh and Damsté, 2018). Most of the members are acidophilic chemoheterotrophs growing aerobically under mesophilic conditions (Dedysh and Damsté, 2018). To study this phylum, culture-independent approaches have been more successful rather than culture-dependent approaches. Genomic studies provided insights into the genetic make-over of about ten acidobacterial genomes only (Ward et al., 2009; Kielak et al., 2016a). However, owing to the difficulty in cultivating its members, the in-depth ecological purview of this enigmatic phylum has remained evasive. Since information regarding global distribution patterns and apparent ecological roles of Acidobacteria is inadequate, microbial ecologists are deeply engaged to unfurl this obscure phylum.
The Requirement for Holistic Approaches for Studying Phylum Acidobacteria
Acidobacteria phylogenetic diversity, richness, abundance, ubiquity, especially in soil ecosystems, pin down their roles in various biogeochemical cycles and broad metabolic versatility (Naether et al., 2012). Although their presence and abundance are confirmed through culture-independent studies but their ecological functions, interrelations with environmental parameters and interactions with other soil microbial communities remain obscure. Significant variations have been encountered during isolation of Acidobacteria strains belonging to different lineages and getting cultivated under specific sets of physicochemical conditions or nearly a narrow range of conditions. This suggests the use of various strategies for the successful recovery of ecologically different Acidobacteria groups.
Acidobacteria diversity and dominance is quite pronounced along with high overall ecological and phylogenetic diversity in contrast to its low cultivation success due to radically different culture laboratory conditions. Although nine different bacterial phyla viz., Proteobacteria, Acidobacteria, Actinobacteria, Verrucomicrobia, Bacteroidetes, Chloroflexi, Planctomycetes, Gemmatimonadetes, and Firmicutes are known to dominate in soil (Janssen, 2006; da Rocha et al., 2009), the phylum Acidobacteria represents the most predominant not-yet-cultured, or difficult-to-culture group of bacteria (Janssen, 2006; Foesel et al., 2014), occupying a significant fraction of the soil microbial community. Henceforth, the determination of their ecophysiological roles becomes quite imperative to understand their functional status in complex bacterial communities. Thus, the ecological roles of Acidobacteria have been studied by analyzing the genetic components obtained by complete and/or draft whole-genome sequencing for culturable species and metagenome sequencing for unculturable species.
16S rRNA Analyses and Metagenomics in Studying Phylum Acidobacteria
Meta-microbiomic (16S rRNA gene-based) studies analyze only one specific gene and not the entire genomes of the community members. The 16S rRNA gene sequences, obtained from various environments by employing culture-dependent and culture-independent approaches, provide deeper insights into the species structure and taxonomic diversity of the phylum Acidobacteria. Studies focusing on 16S rRNA genes provide taxonomic status in any bacterial community, while metagenomics can provide both taxonomic and functional profiles of the microbiota. Metagenomic DNA extracted from the environment is often amplified using group or species-specific primers targeting 16S rRNA genes (Kalam et al., 2017a). Metagenomics has facilitated in an apt understanding of the diversity, abundance, genomic make-up, and ecological roles of acidobacterial members in various ecosystems (Tyson et al., 2004; Venter et al., 2004; Parsley et al., 2011). To obtain a complete community structure, 16S rRNA analyses and metagenomics are often used in conjunction (Figure 1).
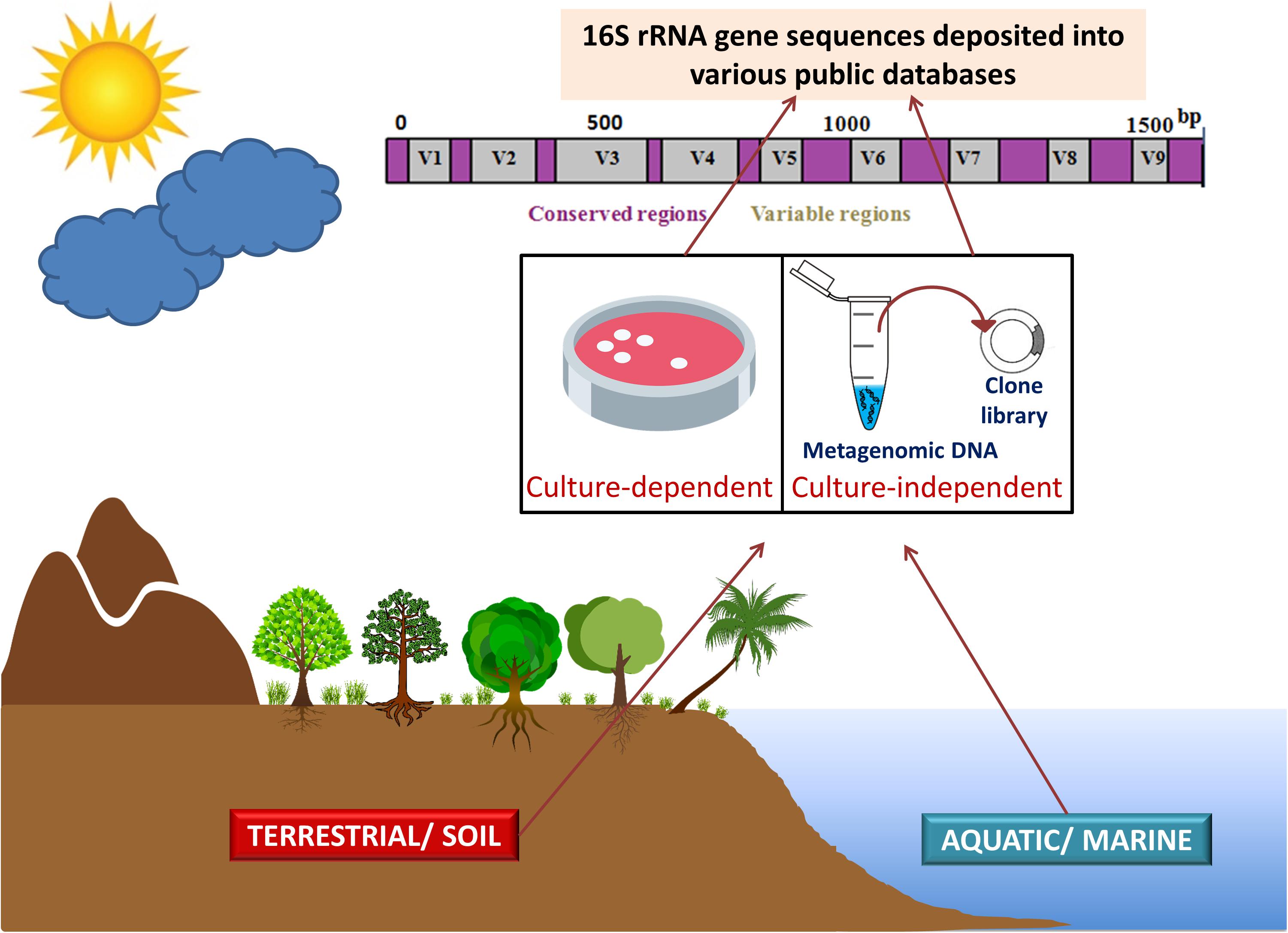
Figure 1. 16S rRNA gene sequencing and metagenomics in studying phylum Acidobacteria. Acidobacterial 16S rRNA gene sequences (from culture-dependent studies) and metagenomic DNA information (from culture-independent studies) obtained from various environments (terrestrial ecosystems including soils and aquatic or marine ecosystems) are deposited in various public databases. The amalgamation of 16S rRNA analyses and metagenomics provides deeper insights into the taxonomic and functional diversity of the phylum Acidobacteria. Figure designed using images from pinclipart.com.
During recent years, the sequencing of collective community genomes employing metagenomics has led to a significant breakthrough in understanding the enigmatic phylum Acidobacteria. Subsequent advent and development of several next-generation sequencing (NGS) platforms further enhanced the metagenomic sequencing efficiencies. Metagenomic approaches, however, at few places fail to retrieve genome scaffolds of sizable length due to soil community complexity, intricacy, and absence of genomes (Tringe et al., 2005; Kowalchuk et al., 2007). Recovering genomic information from the soil environment requires the use of large insert strategies (Rondon et al., 2000). Metagenomic DNA being high in molecular weights requires meticulous isolation strategies along with novel cloning and screening methods to facilitate the recovery of large DNA fragments from difficult-to-culture bacterial genomes. Large genomic fragments might contain intact metabolic pathways. Acidobacterial genome fragments were successfully recovered from the environment by using a large metagenomic insert (Liles et al., 2003). Henceforth, such approaches provide important platforms for exploring the hidden biotechnological potential of hitherto uncultured bacteria leading to the discovery of novel organo-chemical compounds (Daniel, 2004).
Genetic Insights Into the Phylum Acidobacteria
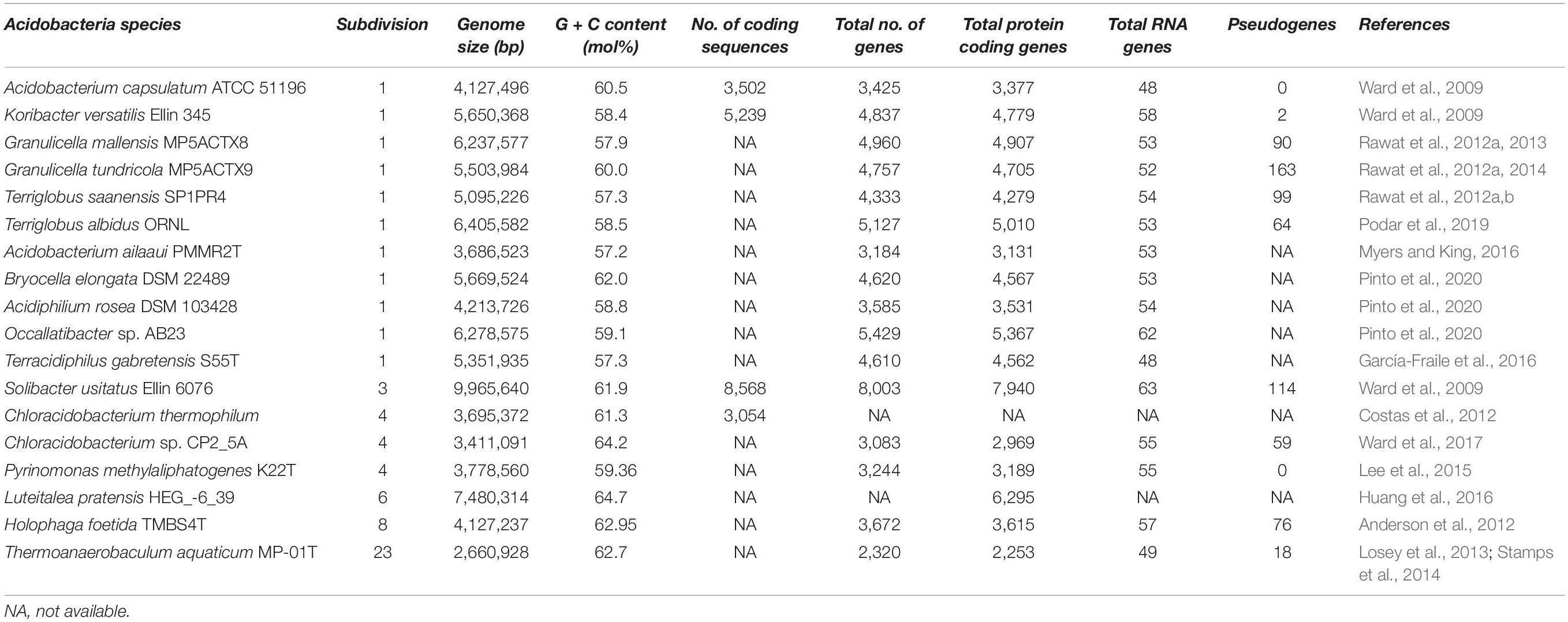
Since the recovery of the first Acidobacteria member and subsequent advances in sequencing technologies has provided a platform to study individual Acidobacteria members at the genomic level. It is quite unfortunate that the number of thoroughly studied genomes of this important phylum is very few, despite their numerical abundance in many environments. Genomic studies have unveiled the hidden physiological and metabolic versatility of Acidobacteria members. Comprehensive studies targeting functional characteristics encoded in acidobacterial genomes may provide new vistas into ecological perspectives of phylum Acidobacteria. Ward et al. (2009) conducted detailed genomic studies for the first time, with three acidobacterial strains [two from subdivision 1 viz., Acidobacterium capsulatum and Candidatus Koribacter versatilis (strain Ellin 345) and one from subdivision 3 viz., Candidatus Solibacter usitatus (Ellin 6076)]. Further genomic profiling and comparative genomic studies (Männistö et al., 2012; Rawat et al., 2012a; Lee et al., 2015; Eichorst et al., 2018, 2020) provided more profound insights into the acidobacterial genome and ecophysiology.
Insights into these genomes reveal large genome size (up to 10 Mbp) and a higher percentage of paralogous genes, which might endow the bacterial strains with potential ecological functions (Challacombe et al., 2011). In the same breath, Acidobacteria genomes also possess a comprehensive physiological set of genes that allows them to adapt to various ecological niches. An overview of the general genomic features of Acidobacteria members and their overall genetic and genomic make-up are respectively represented in Table 1 and Figure 2. Data available from published acidobacterial genome sequences reveal the presence of several genes involved in regulating carbon, nitrogen, and sulfur cycles, and those required for degrading different complex polysaccharides. All cultured Acidobacteria species are reported to produce exopolysaccharide (EPS); the presence of eps gene clusters in the genomes further supports this data. The acidobacterial genomes also contain a substantial proportion of genes encoding for various transporters, to tide over stress and starvation, and for the biosynthesis of cellulose, N-acyl-homoserine lactones, polyketides, siderophores, hopanoids, and mobile genetic elements (Ward et al., 2009; Kielak et al., 2016a; Crits-Christoph et al., 2018).
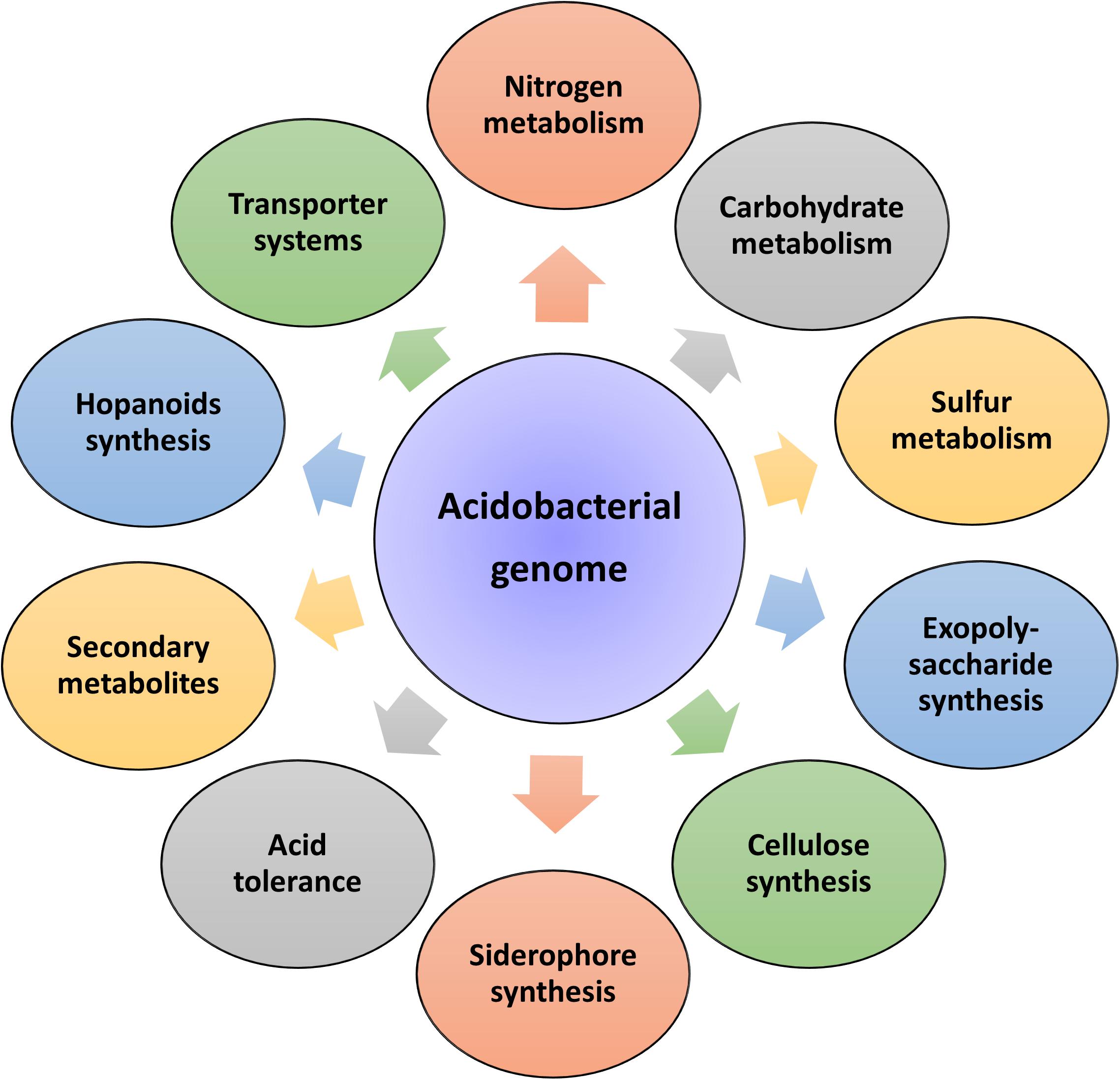
Figure 2. Genetic diversity of the phylum Acidobacteria. Sequencing of acidobacterial genomes and metagenomes revealed a large repertoire of genes responsible for regulating diverse physiological and metabolic functions.
Genes for Carbon Metabolism
Each bacterium carries its enzyme machinery catalyzing the breakdown of diverse carbohydrates and nitrogen-containing compounds, which could be used as an identifying characteristic for differentiating varied bacterial species. Acidobacterial genomes possess genes encoding enzymes for the degradation of complex carbohydrate polymers viz., xylan, cellulose, hemicelluloses, pectin, starch, and chitin; amino acids, alcohols, and metabolic intermediates (Ward et al., 2009; Belova et al., 2018). In addition to these, gene modules for diverse carbohydrate breakdown, utilization, and biosynthesis within carbohydrate-active enzymes (CAZy) family are also present (Männistö et al., 2012; Rawat et al., 2012a), spanning across 131 glycoside hydrolase (GH) families (Gilbert, 2010). Putative chitinases belonging to GH18 and GH19 family were also identified in the genomes of a few select Acidobacteria (Rawat et al., 2012a). Gene calling and annotation studies of Acidobacteria Group 1 Acidipila sp. strain EB88 genome indicated that it was rich in glycolytic enzymes and contained about 85 glycoside hydrolases in 48 families (Domeignoz-Horta et al., 2019).
The acidobacterial genomes are flexible and novel in their carbon metabolizing activity. Few select acidobacterial genomes exhibit anaplerotic CO2 fixation. Strikingly, homologs of phosphoenolpyruvate carboxylase and isocitrate dehydrogenase have also been detected across several Acidobacteria genomes (Lee et al., 2015; Eichorst et al., 2018). Studies suggest a crucial role of these genes for carbon metabolism in various nutritional pathways as well as a significant role in desiccation resistance, as evidenced by the Terriglobus saanensis genome profile (Männistö et al., 2011). A significant contribution is made by acidobacterial enzyme machinery in regulating the carbon biogeochemical cycle (King and Weber, 2007). Since Acidobacteria are endowed with the potential to degrade polymeric carbonaceous complexes, they act as decomposers in soil and actively participate in the cycling of organic matter arising from plants, fungi, and insects (Dedysh and Damsté, 2018).
Genes for Nitrogen Metabolism
Acidobacteria is well equipped with genes catalyzing the metabolism of inorganic and organic sources of nitrogen (Eichorst et al., 2018). They can effectively reduce nitrate, nitrite, and possibly nitric oxide, as could be evidenced by genomic data, supporting their active participation in nitrogen nutrient circuits. Homolog candidate genes for nitrate reductase (nirA) have been identified in Ellin 345, Geobacter fermentans, and Terriglobus aquaticum (Rajeev et al., 2015), while those for nitrate transport (nrtABCD) have also been observed in certain acidobacterial strains. The presence of genes encoding dinitrogenase (nifD and nifK) and dinitrogenase reductase (nifH) have also been reported in one acidobacterial genome (Ward et al., 2009). However, experimental evidence regarding nitrogen fixation by Acidobacteria is missing (Kielak et al., 2016a). Insights into the core genomes provide a wealth of data revealing the presence of putative homologs encoding for extracellular peptidases, which play significant roles in soils by mobilizing ammonium and other intermediates of N-cycle (Bach et al., 2001; Eichorst et al., 2018). Serine endopeptidases are also found to be widely distributed in most Acidobacteria genomes, indicating their proteolytic activity in soil (Brankatschk et al., 2011). Homologs for various extracellular metalloendopeptidases too span across select Acidobacteria genomes, facilitating them for N-uptake during mineral scarcity. Recent studies on the draft genome of Acidobacteria Group 1 Acidipila sp. strain EB88, isolated from forest soil, indicated that the genome lacked the genes required for organic acid uptake but was equipped with the genes essential for amino acid, ammonium, and nitrate uptake (Domeignoz-Horta et al., 2019).
Genes for Sulfur Metabolism
Chloroacidobacterium thermophilium, a photoheterotrophic member of Acidobacteria subdivision 4, requires reduced sulfur for its growth (Tank and Bryant, 2015). Draft metagenome-assembled genomes of Acidobacteria subdivisions 1 and 3 from peat soil revealed the presence of putative genes for dissimilatory sulfur metabolism (dsrAB, dsrC, dsrD, dsrN, dsrT, dsrMKJOP, aprBA, qmoABC, supP, hppA, sat) which function under anoxic conditions (Hausmann et al., 2018). The presence of dsrAB genes encoding dissimilatory (bi)sulfite reductase suggests the capacity of some Acidobacteria members to perform dissimilatory sulfate/sulfite reduction (Huang et al., 2016; Wasmund et al., 2017; Anantharaman et al., 2018). Certain acidobacterial genomes also encode the dsrL gene, which is specific to the sulfur oxidation pathway (Hausmann et al., 2018). The presence of dsrL is unusual as it is generally found in sulfur oxidizers rather than in sulfate-reducing microorganisms. However, acidobacterial genomes lack any other genes involved in oxidative sulfur metabolism (Hausmann et al., 2018).
Genes Encoding Transporters
Different acidobacterial strains harbor an array of genes encoding different ion channels, high-affinity ABC transporters, several other secretory porters, and transport proteins especially required for tiding on oligotrophic conditions (Paulsen et al., 1998; Ward et al., 2009). Genes encoding iron permease FTR1 and FTR2 family proteins and iron transporter genes viz., Mn2+ and Fe2+ transporters are also present in the Acidobacteria genome. The feoAB gene encoding a high-affinity ferrous iron transport protein is invariably present in acidobacterial genomes (Velayudhan et al., 2000). The genome of Terriglobus saanensis (isolated from the Tundra region) contains abundant genes involved in carbohydrate metabolism and transport (Männistö et al., 2011). Few Acidobacteria genomes also possess multiple copies of genes for siderophore transport viz., tonB, exbB, and exbD (Postle and Kadner, 2003; Llamas and Bitter, 2006; Ward et al., 2009). In addition to these, pvuE (candidate gene for vibrioferrin transporter) and feoB (candidate gene for enterobactin transporter) are also found in some Acidobacteria genomes (Ward et al., 2009). Acidobacteria members also have genes for the amino acid-polyamine-organocation (APC) superfamily of transport proteins along with dicarboxylate/amino-acid: cation symporter family of secondary transport proteins (Eichorst et al., 2018).
Genes Regulating Cellulose Synthesis
Acidobacterium capsulatum genome contains an operon with a complete set of genes required for cellulose biosynthesis (Ward et al., 2009). Bacterial cellulose is associated with an array of functions facilitating survival in the soil through biofilm formation, retaining moisture under conditions of stress, thus promoting aeration, contributing to soil aggregate formation (Ude et al., 2006; White et al., 2006). Acidobacteria possesses the remarkable capacity to synthesize cellulose, which would promote biofilms to get adhered easily to ferric iron-containing substrates producing biofilm-ferric iron-reducing “bioshrouds” in acidic environments (Johnson et al., 2008). Cellulose biosynthesis via bcs operon has been reported in Terriglobus saanensis as evidenced through genome sequencing data (Rawat et al., 2012b; de Castro et al., 2013). The genome of Acidipila sp. strain EB88 contained genes for biosynthesis and export of capsule and cellulose (Zhang et al., 2018).
Genes for Oxygen and Hydrogen Utilization
Genomes of particular soil acidobacterial strains possess the potential to respire oxygen at atmospheric and microoxic levels due to the presence of affinity oxidases. Thus, such strains possess an extra selective advantage in the soil microenvironment, where low oxygen concentrations exist (Morris and Schmidt, 2013; Eichorst et al., 2018). The genome of Acidipila sp. strain EB88 contains a low-affinity group A heme-copper oxygen oxidase and five high-affinity cbb3 terminal oxidases that enables it to grow under both hypoxic and hyperoxic conditions (Morris and Schmidt, 2013; Domeignoz-Horta et al., 2019). Few Acidobacteria strains possess hydrogen scavenging property due to the presence of nickel-iron [NiFe] hydrogenases (Greening et al., 2015). The occurrence of multiple structural genes (hhyS, hhyL, hhyE) and maturation genes (hypABCDEF) required for hydrogenase activity further supports acidobacterial hydrogen utilization (Eichorst et al., 2018).
Genes Regulating Secondary Metabolite Biosynthesis
Acidobacteria genomes contain biosynthetic gene clusters that encode a vast repertoire of polyketide and non-ribosomal peptide synthases (Crits-Christoph et al., 2018). Additionally, genes regulating the synthesis of diverse secondary metabolites and other natural compounds like siderophores, antifungals, antibiotics, antivirals, antitumor agents, and antinematodal agents have been reported in Acidobacteria genomes (Challis, 2005; Ward et al., 2009; Parsley et al., 2011; Hadjithomas et al., 2015; Crits-Christoph et al., 2018).
Genes Regulating Stress and Starvation Response
Addiction modules encode various genes, including those required for plasmid maintenance, and has been documented in certain Acidobacteria (Ward et al., 2009). During stress or starvation, the addiction modules operate rapidly and inhibit DNA and protein synthesis (Kroll et al., 2010). This mechanism of stress and starvation tolerance in Acidobacteria enables tiding over environmental oligotrophic nutritional conditions. The prokaryotic transcriptional regulator sigma factor is commonly utilized to control the expression of several gene sets in response to various stresses, including starvation, oxidative stress, heat stress, and exposure to heavy metals that enables the microorganisms to adapt to a stressful environment (Rhodius et al., 2005; Challacombe et al., 2011). The genomes of Candidatus Solibacter usitatus [Ellin 6076] and Candidatus Koribacter versatilis [Ellin 345] contain a vast repertoire of sigma E (σE) homologs (70 and 28 homologs, respectively) that are induced during starvation and other stress conditions (Challacombe et al., 2011).
The presence of hydrogenases enables specific acidobacterial strains to consume atmospheric hydrogen (Greening et al., 2015), which could be a strategy to overcome starvation (Eichorst et al., 2018). Certain Acidobacteria members are equipped with the genes responsible for the dissimilatory reduction of nitrite to ammonia (nrfHA), which not only provides energy supply but also aids in the detoxification of nitrosative stress (Rajeev et al., 2015; Eichorst et al., 2018). A very recent study by Pinto et al. (2020) reported that carotenoid production by Occallatibacter sp. (belonging to Acidobacteria subdivision 1) can confer tolerance to environmental oxidative stress. Thus, the production of carotenoids and related compounds may offer competitive benefits to soil Acidobacteria.
Genes Regulating Acid Tolerance
Most Acidobacteria members prefer acidic conditions (3.0–6.5 pH) for their growth (Sait et al., 2006; Ward et al., 2009). Also, lower pH levels support a higher abundance of Acidobacteria (Männistö et al., 2007; Lladó et al., 2018). Microorganisms equipped with acid resistance (AR) systems are likely to survive in highly acidic conditions (Sun et al., 2012). Certain moderately acidophilic acidobacterial strains viz., A. capsulatum, Ellin 345 (K. versatilis), and Ellin 6076 (S. usitatus) contain candidate genes in the AR3 (arginine-dependent AR) system indicating the presence of an acid tolerance system (Ward et al., 2009; Kielak et al., 2016a). However, the strains lack the genes involved in other inducible AR systems, viz., AR1 (oxidative AR), AR2 (glutamate-dependent AR), and AR4 (lysine-dependent AR).
Genes for Synthesis of Hopanoids
Hopanoids are pentacyclic triterpenoid bacterial membrane lipids facilitating cell membrane permeability, especially during extreme environmental conditions (Damsté et al., 2017). Genes related to hopanoid biosynthesis (shc gene) have been detected in Cadidatus “K. versatilis” belonging to subdivision 1 isolated from soil (Joseph et al., 2003). The presence of C30 hopanoids and bacteriohopane polyols was reported in multiple Acidobacteria subdivisions, evident from the occurrence of respective biosynthesis genes (hpnA, hpnG, hpnH) in acidobacterial genomes (Damsté et al., 2017).
Genes for Exopolysaccharide Biosynthesis
Genomic mining studies suggest that Acidobacteria belonging to subdivision 1 can encode EPS biosynthesis genes (Ward et al., 2009). Additionally, several cultured Acidobacteria species are known to secrete EPS (Eichorst et al., 2007; Pankratov and Dedysh, 2010; Whang et al., 2014; Kielak et al., 2017). Gene prediction, along with functional annotation data, identifies a gene cluster encoding capsular polysaccharide in the genomes of three acidobacterial strains isolated from tundra soil. Granulicella mallensis genome possessed the epsH gene involved in exopolysaccharide synthesis (Rawat et al., 2012a).
Genes for Acyl-Homoserine Lactones (AHL)
Quorum sensing molecules, like acyl-homoserine lactones (AHL), aid in coordinating gene expression in bacterial populations (Parsek et al., 1999). Genomes of Acidobacteria contain a considerable fraction of genes encoding for biosynthesis of N-acyl homoserine lactones (Ward et al., 2009).
Genes for Mobile Genetic Elements
Mobile genetic elements like transposons, bacteriophages, insertion sequence (IS) elements, and integrative and conjugative elements (ICEs) present in several Acidobacteria are known to confer shape and plasticity to the acidobacterial genome. These elements are speculated to mediate horizontal gene transfer, aid in the evolution and ecological success of Acidobacteria (Frost et al., 2005). A recent meta-study by Eichorst et al. (2018) identified 35 putative prophages across 19 acidobacterial genomes. Insertion sequence families encoding their mobility patterns also have been reported to be spanning across Acidobacteria genomes (Challacombe and Kuske, 2012). Multiple genes encoding mobile elements, including transposases and IS elements, have been identified in the genome of Candidatus “Solibacter usitatus” Ellin 6076. Additionally, the genome also harbors genes encoding phage integrase family proteins and several other proteins containing a retroviral integrase catalytic region domain, catalyzing site-specific recombinations (Challacombe and Kuske, 2012).
Excavating the Ecological Roles of Phylum Acidobacteria in Soil
Despite the recent progress in the field of acidobacterial ecology, there is still a paucity of complete information regarding their ecophysiological roles. Significant ecological functions have been reported in forest soil for Acidobacteria members (García-Fraile et al., 2016; Liu et al., 2016). Acidobacteria members in plant-soil ecosystems play pivotal ecological roles, including modulation of biogeochemical cycles and influencing plant growth. The key findings from relevant studies on soil Acidobacteria, highlighting their salient genomic features and ecological roles, are summarized in Table 2. An overview of their ecological roles in the plant-soil ecosystems is diagrammatically represented in Figure 3 and is discussed below.
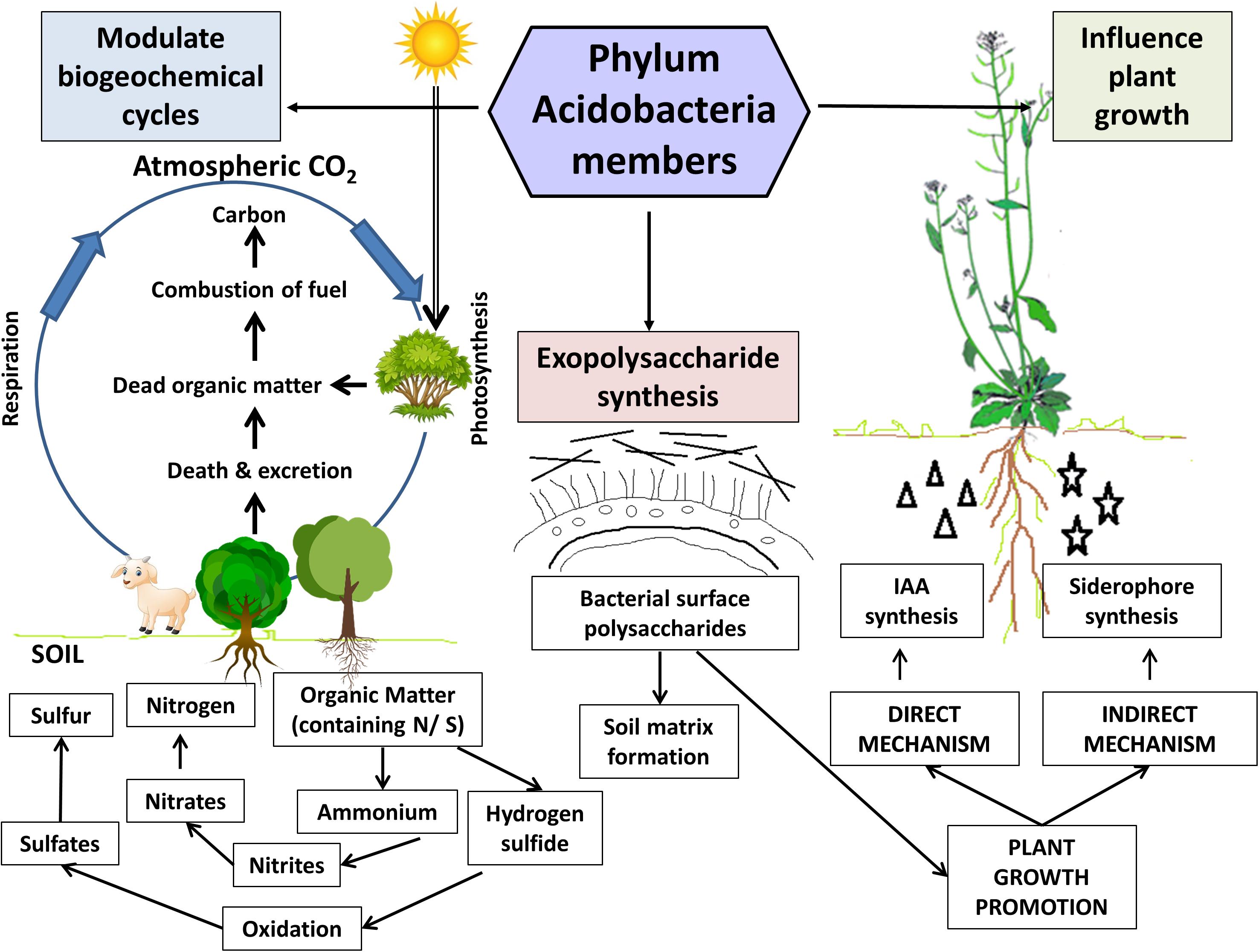
Figure 3. Ecological roles of Acidobacteria members in the soil. Acidobacteria members in the plant-soil ecosystem facilitate modulation of critical biogeochemical cycles viz., carbon, nitrogen, and sulfur cycles. Exopolysaccharides (produced by almost all cultured members) aid in soil matrix formation and contribute to plant growth promotion by facilitating water and nutrient uptake by plants. The representatives belonging to Granulicella and Acidocapsa genera exhibit in vitro plant growth promotion traits in the model plant Arabidopsis thaliana. These acidobacterial strains effectively produced the phytohormone indole-3-acetic acid (IAA) and siderophores, which significantly enhanced the plant growth parameters (shoot and root biomass). Figure designed using images from pinclipart.com.
Acidobacteria as “Keystone Taxa” in Soil Ecosystems
A recent study by Banerjee et al. (2018) has reviewed and re-defined “keystone taxa” in microbial ecology. Microbial keystone taxa have been often ascribed as “ecosystem engineers” as they are the unequivocal drivers of microbial community structure and function in different ecosystems. Acidobacteria was reported to be one of the keystone bacterial taxa in soil associated with the decomposition of soil organic matter (SOM), implying their significance in carbon turnover (Banerjee et al., 2016). Computational inferences obtained from analyses of various terrestrial ecosystems and habitats indicate several subdivisions of phylum Acidobacteria represent the keystone taxa in grasslands (subdivision 4), forest or woodlands (subdivision 4), agricultural soils (subdivision 17), and plant-associated microbiota (subdivisions 1, 3, and 6) (Banerjee et al., 2016, 2018; Jiang et al., 2017; Li et al., 2017). Speculations suggest that these microbial residents exert beneficial effects by selectively modulating the ecological processes of host ecosystems. These bacteria offer two crucial ecological services viz., SOM decomposition, and denitrification, thereby enhancing carbon stability (Banerjee et al., 2018). Successful manipulation of these soil dwellers might augment the key ecological process of soil carbon sequestration. Since Acidobacteria members have been identified as structural and functional keystones in plant-soil microbiomes and agroecosystems, they can be exploited to enhance crop performance and productivity.
Modulation of Biogeochemical Cycles
The gamut of living organisms invariably depends on the supply of essential elements such as oxygen, hydrogen, carbon, nitrogen, phosphorus, and sulfur for growth and survival. In any ecosystem, albeit biogeochemical cycles or nutrient cycles are regulated by the biotic and abiotic components of that system, but microorganisms play a key role in modulating them (Falkowski et al., 2008). The critical metabolic processes viz., nitrogen metabolism, carbon fixation, and methane metabolism, sulfur metabolism operating in microbes effectively control global biogeochemical cycling.
Genomic studies highlight the carbohydrate utilization potential exhibited by Acidobacteria members (Ward et al., 2009; Kielak et al., 2016a). The presence of genes encoding enzymes for the degradation of complex carbohydrate polymers like cellulose, hemicellulose, chitin, xylan, and lignin derivatives signifies their active participation in the carbon circuit as decomposers in soil (Ward et al., 2009; Wegner and Liesack, 2017; Banerjee et al., 2018; Belova et al., 2018) and cycling of organic matter derived from plants, fungi, and insects (King and Weber, 2007; García-Fraile et al., 2016; Dedysh and Damsté, 2018).
Soil bacteria convert the inorganic nitrogen to organic compounds (Canfield et al., 2010), which are, in turn, utilized by the plants and other microbes. In plant-soil ecosystems, Acidobacteria communities represent an important microbial guild, central in nitrogen cycling. Genomic evidence coerces the crucial role of Acidobacteria in N-cycling in soils (Ward et al., 2009). Specialized microorganisms metabolize diverse sulfur compounds via redox reactions and contribute to driving the global sulfur cycle (Wasmund et al., 2017). Acidobacteria genome harbors genes regulating sulfur metabolism that were found to be expressed in native peat soils and upregulated in diverse anoxic conditions, indicating their active role in modulation of the environmental sulfur cycle (Hausmann et al., 2018). Hausmann et al. (2016) reported that consortia of diverse sulfate-reducing microorganisms, including Acidobacteria members, drive the sulfur reduction in peatlands and Acidobacteria members (Holophaga) responded positively to sulfate stimulation in peat soil microcosms.
Exopolysaccharide Production
Exopolysaccharides are bacterial polysaccharides synthesized extracellularly (Nwodo et al., 2012), which play a critical role in the soil matrix and aggregate formation (Bogino et al., 2013). EPS plays a significant role in the development of mature biofilm by acting as bridges between cell surfaces (Bura et al., 1998). In the rhizosphere zone, EPS production by bacterial populations contributes toward nutrient and water uptake by plant roots through modification of the physicochemical properties of rhizosphere soil (Degens et al., 1994; Kalam et al., 2017b) and aids in establishing interactions with the root appendages for successful plant-microbe interactions (Bianciotto et al., 2001, 2009).
Several cultured Acidobacteria species are known to secrete EPS (Eichorst et al., 2007; Kielak et al., 2017). Genomic analyses indicate that Acidobacteria belonging to subdivision 1 can encode EPS biosynthesis genes (Ward et al., 2009). These EPSs might protect Acidobacteria, endowing them the ability to survive for prolonged periods in soil (Kielak et al., 2017). Recently, the characterization of EPSs derived from two subdivisions 1 acidobacterial strain (Granulicella sp.) has indicated them to be possessing potential environmental, bioremediation, and biotechnological applications (Kielak et al., 2017). These EPSs could be used as natural eco-friendly binders and formulants in the biofertilizer industry.
Acidobacterial genome profiling validates the presence of cellulose synthesis genes along with related accessory proteins. Studies postulate several Acidobacteria members to possess the capability to establish biofilms, exhibit resistance toward desiccation, and aid in the formation of soil aggregates (Kielak et al., 2016a). However, physiological data confirming actual EPS production by Acidobacteria and validating their ecophysiological roles remains obscure.
Acidobacteria as Plant Growth Promoting Rhizobacteria (PGPR)
Plant root microbiome harbors beneficial microbes, which offer an eco-friendly alternative to improve plant growth and protect against phytopathogens (Bulgarelli et al., 2013; Philippot et al., 2013). Plant growth promoting rhizobacteria (PGPR) have been defined as free-living soil bacteria dwelling in the rhizosphere and endowed with the potential to stimulate plant growth and crop yield (Dutta and Podile, 2010; Sayyed et al., 2019; Zope et al., 2019). A plethora of reviews exhaustively documents almost all aspects of PGPR (Ahmad et al., 2008; Lugtenberg and Kamilova, 2009; Etesami et al., 2015; Parray et al., 2016; Backer et al., 2018). PGPR, through their direct and indirect effects, can bring about substantial plant growth promotion (PGP). They act directly by facilitating nitrogen absorption and assimilation, mineral solubilization, production of phytohormones (Ahmad et al., 2008; Parray et al., 2016; Shaikh et al., 2018; Kalam et al., 2020). Indirectly, PGPR, through their biocontrol mechanisms, produce siderophores (Wani et al., 2016; Sayyed et al., 2019), lytic enzymes (Jadhav et al., 2020a, b), antibiotics (Vinay et al., 2016; Reshma et al., 2018; Kenawy et al., 2019), 1-aminocyclopropane-1-carboxylic acid (ACC) (Glick, 2014; Goswami et al., 2016; Sagar et al., 2020), guarding host plants against pathogens (Shaikh et al., 2018).
Several studies reported Acidobacteria to be avid rhizosphere colonizers (Lee et al., 2008; da Rocha et al., 2010, 2013). The first evidence for PGP by Acidobacteria subdivision 1 members was provided by Kielak et al. (2016b). Their experiments confirmed interactions between three acidobacterial strains belonging to the genera Granulicella and Acidicapsa and the host plant Arabidopsis thaliana. Determination of in vitro PGP traits provided evidence that all the test strains were active producers of phytohormones IAA and siderophore. There was a significant increase in root growth parameters, although shoot biomass variations were non-significant in comparison to the controls. All three strains were able to adhere to roots, form a biofilm, and grow along the root surface. The pioneering study provided the first time with a direct confirmation regarding the Acidobacteria-plant interactions and PGP by Acidobacteria members. Acidobacteria subdivision 1 member dominates among other subdivisions, followed by subdivisions 4, 3, 8, and 23. Their preponderance in the soil environment in comparison with other subdivisions and their genetic profiles surmise their active roles in plant-soil ecosystems. Also, analysis of biosynthetic gene pathways from few publicly available genomes of soil-inhabiting acidobacterial isolates suggests the presence of genes linked to the production of secondary metabolites involved in plant growth (Parsley et al., 2011; Hadjithomas et al., 2015).
The diversity and composition analysis of broomcorn millet rhizosphere using 16S rDNA sequencing indicated Acidobacteria to be a core bacterial component among rhizobacterial assemblages, comprising 10.7% of the total operational taxonomic units (OTUs) obtained (Na et al., 2019). In another interesting study, Kalam et al. (2017a) explored the effects of three PGPR strains viz., Sphingobacterium sp., Variovorax sp., and Roseomonas sp. on crop rhizospheric population densities of acidobacterial members. They reported that the equivalent cell numbers of Acidobacteria members gradually increased over time with a simultaneous increase in plant growth promotion by PGPR inocula. The study speculated the existence of beneficial interactions between the triad of difficult-to-culture bacteria, PGPR, and host plants. Additional experiments in plant-soil ecosystems are still required for unveiling the exact ecological roles of rhizospheric Acidobacteria members.
Conclusion
Soil represents a luxurious pool of microorganisms, including the shadow biosphere dwellers, the difficult-to-culture bacteria which might be strategic sources for novel and successful biotechnological products. Acidobacteria is an important and among the most abundant difficult-to-culture phylum harboring the soil ecosystem. Acidobacteria plays significant ecological roles, as evidenced through their active participation in key carbon, nitrogen, and sulfur biogeochemical circuits. At the same time, the production of EPS further strengthens their ecophysiological functions in establishing plant-soil beneficial interactions. Since this divergent phylum predominates rhizosphere microbial communities, these bacteria might be a significant component of the crop’s natural environment. Initiatives for manipulating crop rhizosphere with acidobacterial populations to increase plant growth in laboratory and greenhouse studies could be considered to be future challenges and thrust areas for research. The correlation and information presented in this review might prove to be relevant for future modeling of experiments to expose the potential biotechnological roles of Acidobacteria members and exploit them as prospective alternatives to agrochemicals, facilitating sustainable agriculture.
Author Contributions
SK and AB contributed equally to the conception and design of the review and wrote the first draft of the manuscript. IA and RS critically reviewed and edited the manuscript. HE-E, DD, and NS wrote portions of the manuscript. All authors read and approved the final version of the manuscript.
Funding
HE-E and DD would like to thank the MOHE and UTM for HICoE, Grant No. R.J1300.7846.4J262.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SK acknowledges Principal Sister Amrutha, St. Ann’s College for Women, Hyderabad, Telangana, India, for her incessant support and encouragement. AB acknowledges University Grants Commission (UGC), New Delhi, India, for Senior Research Fellowship (SRF).
References
Ahmad, F., Ahmad, I., and Khan, M. S. (2008). Screening of free-living rhizospheric bacteria for their multiple plant growth-promoting activities. Microbiol. Res. 163, 173–181. doi: 10.1016/j.micres.2006.04.001
Anantharaman, K., Hausmann, B., Jungbluth, S. P., Kantor, R. S., Lavy, A., Warren, L. A., et al. (2018). Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 12, 1715–1728. doi: 10.1038/s41396-018-0078-0
Anderson, I., Held, B., Lapidus, A., Nolan, M., Lucas, S., Tice, H., et al. (2012). Genome sequence of the homoacetogenic bacterium Holophaga foetida type strain (TMBS4(T)). Stand. Genomic Sci. 6, 174–184. doi: 10.4056/sigs.2746047
Bach, H. J., Hartmann, A., Schloter, M., and Munch, J. C. (2001). PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J. Microbiol. Methods 44, 173–182. doi: 10.1016/s0167-7012(00)00239-6
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Barns, S. M., Cain, E. C., Sommerville, L., and Kuske, C. R. (2007). Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73, 3113–3116. doi: 10.1128/aem.02012-06
Belova, S. E., Ravin, N. V., Pankratov, T. A., Rakitin, A. L., Ivanova, A. A., Beletsky, A. V., et al. (2018). Hydrolytic capabilities as a key to environmental success: chitinolytic and cellulolytic Acidobacteria from Acidic sub-arctic soils and boreal peatlands. Front. Microbiol. 9:2775. doi: 10.3389/fmicb.2018.02775
Bianciotto, V., Andreotti, S., Balestrini, R., Bonfante, P., and Perotto, S. (2001). Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol. Plant Microbe Interact. 14, 255–260. doi: 10.1094/mpmi.2001.14.2.255
Bianciotto, V., Andreotti, S., Balestrini, R., Bonfante, P., and Perotto, S. (2009). Extracellular polysaccharides are involved in the attachment of Azospirillum brasilense and Rhizobium leguminosarum to arbuscular mycorrhizal structures. Eur. J. Histochem. 45, 39–50. doi: 10.4081/1612
Bogino, P. C., Oliva, M. D. L. M., Sorroche, F. G., and Giordano, W. (2013). The role of bacterial biofilms and surface components in plant-bacteria associations. Int. J. Mol. Sci. 14, 15838–15859. doi: 10.3390/ijms140815838
Brankatschk, R., Töwe, S., Kleineidam, K., Schloter, M., and Zeyer, J. (2011). Abundances and potential activities of nitrogen cycling microbial communities along a chronosequence of a glacier forefield. ISME J. 5, 1025–1037. doi: 10.1038/ismej.2010.184
Bulgarelli, D., Schlaeppi, K., Spaepen, S., van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Bura, R., Cheung, M., Liao, B., Finlayson, J., Lee, B. C., Droppo, I. G., et al. (1998). Composition of extracellular polymeric substances in the activated sludge floc matrix. Water Sci. Technol. 37, 325–333. doi: 10.2166/wst.1998.0657
Canfield, D. E., Glazer, A. N., and Falkowski, P. G. (2010). The evolution and future of Earth’s nitrogen cycle. Science 330, 192–196.
Challacombe, J., and Kuske, C. (2012). Mobile genetic elements in the bacterial phylum Acidobacteria. Mob. Genet. Elements 2, 179–183. doi: 10.4161/mge.21943
Challacombe, J. F., Eichorst, S. A., Hauser, L., Land, M., Xie, G., and Kuske, C. R. (2011). Biological consequences of ancient gene acquisition and duplication in the large genome of Candidatus Solibacter usitatus Ellin6076. PLoS One 6:e24882. doi: 10.1371/journal.pone.0024882
Challis, G. L. (2005). A widely distributed bacterial pathway for siderophore biosynthesis independent of non-ribosomal peptide synthetases. Chembiochem 6, 601–611. doi: 10.1002/cbic.200400283
Chaudhary, D. K., Khulan, A., and Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 9:6666. doi: 10.1038/s41598-019-43182-x
Costas, A. M. G., Liu, Z. F., Tomsho, L. P., Schuster, S. C., Ward, D. M., and Bryant, D. A. (2012). Complete genome of Candidatus Chloracidobacterium thermophilum, a chlorophyll-based photoheterotroph belonging to the phylum Acidobacteria. Environ. Microbiol. 14, 177–190. doi: 10.1111/j.1462-2920.2011.02592.x
Crits-Christoph, A., Diamond, S., Butterfield, C. N., Thomas, B. C., and Banfield, J. F. (2018). Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444. doi: 10.1038/s41586-018-0207-y
da Rocha, U. N., Plugge, C. M., George, I., van Elsas, J. D., and van Overbeek, L. S. (2013). The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS One 8:e82443. doi: 10.1371/journal.pone.0082443
da Rocha, U. N., van Elsas, J. D., and van Overbeek, L. S. (2010). Real-time PCR detection of Holophagae (Acidobacteria) and Verrucomicrobia subdivision 1 groups in bulk and leek (Allium porrum) rhizosphere soils. J. Microbiol. Methods 83, 141–148. doi: 10.1016/j.mimet.2010.08.003
da Rocha, U. N., Van Overbeek, L., and Van Elsas, J. D. (2009). Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 69, 313–328. doi: 10.1111/j.1574-6941.2009.00702.x
Damsté, J. S. S., Rijpstra, I., Dedysh, S. N., Foesel, B. U., and Villanueva, L. (2017). Pheno- and genotyping of hopanoid production in Acidobacteria. Front. Microbiol. 8:968. doi: 10.3389/fmicb.2017.00968
Daniel, R. (2004). The soil metagenome–a rich resource for the discovery of novel natural products. Curr. Opin. Biotechnol. 15, 199–204. doi: 10.1016/j.copbio.2004.04.005
de Castro, V. H. L., Schroeder, L. F., Quirino, B. F., Kruger, R. H., and Barreto, C. C. (2013). Acidobacteria from oligotrophic soil from the Cerrado can grow in a wide range of carbon source concentrations. Can. J. Microbiol. 59, 746–753. doi: 10.1139/cjm-2013-0331
Dedysh, S. N., and Damsté, J. S. S. (2018). Acidobacteria. (Chichester: John Wiley and Sons Ltd), 1–10. doi: 10.1002/9780470015902.a0027685
Dedysh, S. N., and Yilmaz, P. (2018). Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 68, 3796–3806. doi: 10.1099/ijsem.0.003062
Degens, B. P., Sparling, G. P., and Abbott, L. K. (1994). The contribution from hyphae, roots, and organic carbon constituents to the aggregation of a sandy loam under long-term clover-based and grass pastures. Eur. J. Soil Sci. 45, 459–468. doi: 10.1111/j.1365-2389.1994.tb00531.x
Domeignoz-Horta, L. A., DeAngelis, K. M., and Pold, G. (2019). Draft genome sequence of Acidobacteria group 1 Acidipila sp. strain EB88, isolated from forest soil. Microbiol. Resour. Announc. 8:e1464-18. doi: 10.1128/MRA.01464-18
Dutta, S., and Podile, A. R. (2010). Plant growth-promoting rhizobacteria (PGPR): the bugs to debug the root zone. Crit. Rev. Microbiol. 36, 232–244. doi: 10.3109/10408411003766806
Eichorst, S. A., Breznak, J. A., and Schmidt, T. M. (2007). Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73, 2708–2717. doi: 10.1128/aem.02140-06
Eichorst, S. A., Trojan, D., Huntemann, M., Clum, A., Pillay, M., Palaniappan, K., et al. (2020). One complete and seven draft genome sequences of subdivision 1 and 3 Acidobacteria isolated from soil. Microbiol. Resour. Announc. 9:e01087-19. doi: 10.1128/MRA.01087-19
Eichorst, S. A., Trojan, D., Roux, S., Herbold, C., Rattei, T., and Woebken, D. (2018). Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 20, 1041–1063. doi: 10.1111/1462-2920.14043
Elmagzob, A. A. H., Ibrahim, M. M., and Zhang, G. F. (2019). Seasonal diversity of endophytic bacteria associated with Cinnamomum camphora (L.) Presl. Diversity 11:112. doi: 10.3390/d11070112
Etesami, H., Alikhani, H. A., and Hosseini, H. M. (2015). ““Indole-3-acetic acid and 1-aminocyclopropane-1-carboxylate deaminase: Bacterial traits required in rhizosphere, rhizoplane and/or endophytic competence by beneficial bacteria,”,” in Bacterial Metabolites in the Sustainable Agroecosystem, ed. D. K. Maheswari (Cham: Springer), 183–258. doi: 10.1007/978-3-319-24654-3_8
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fierer, N., Jackson, J. A., Vilgalys, R., and Jackson, R. B. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71, 4117–4120. doi: 10.1128/aem.71.7.4117-4120.2005
Foesel, B. U., Nägele, V., Naether, A., Wüst, P. K., Weinert, J., Bonkowski, M., et al. (2014). Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ. Microbiol. 16, 658–675. doi: 10.1111/1462-2920.12162
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
García-Fraile, P., Benada, O., Cajthaml, T., Baldrian, P., and Lladó, S. (2016). Terracidiphilus gabretensis gen. nov., sp. nov., an abundant and active forest soil acidobacterium important in organic matter transformation. Appl. Environ. Microbiol. 82, 560–569. doi: 10.1128/aem.03353-15
George, I. F., Hartmann, M., Liles, M. R., and Agathos, S. N. (2011). Recovery of as-yet-uncultured soil Acidobacteria on dilute solid media. Appl. Environ. Microbiol. 77, 8184–8188. doi: 10.1128/aem.05956-11
Gilbert, H. J. (2010). The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153, 444–455. doi: 10.1104/pp.110.156646
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth-promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2:1127500. doi: 10.1080/23311932.2015.1127500
Greening, C., Carere, C. R., Rushton-Green, R., Harold, L. K., Hards, K., Taylor, M. C., et al. (2015). Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc. Natl. Acad. Sci. U.S.A. 112, 10497–10502. doi: 10.1073/pnas.1508385112
Hadjithomas, M., Chen, I. M. A., Chu, K., Ratner, A., Palaniappan, K., Szeto, E., et al. (2015). IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio 6:e00932-15. doi: 10.1128/mBio.00932-15
Hausmann, B., Knorr, K. H., Schreck, K., Tringe, S. G., del Rio, T. G., Loy, A., et al. (2016). Consortia of low-abundance bacteria drive sulfate reduction-dependent degradation of fermentation products in peat soil microcosms. ISME J. 10, 2365–2375. doi: 10.1038/ismej.2016.42
Hausmann, B., Pelikan, C., Herbold, C. W., Köstlbacher, S., Albertsen, M., Eichorst, S. A., et al. (2018). Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 12, 1729–1742. doi: 10.1038/s41396-018-0077-1
Huang, S., Vieira, S., Bunk, B., Riedel, T., Spröer, C., and Overmann, J. (2016). First complete genome sequence of a subdivision 6 Acidobacterium strain. Genome Announc. 4:e00469-16. doi: 10.1128/genomeA.00469-16
Huber, K. J., Geppert, A. M., Wanner, G., Fösel, B. U., Wüst, P. K., and Overmann, J. (2016). The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil. Int. J. Syst. Evol. Microbiol. 66, 2971–2979. doi: 10.1099/ijsem.0.001131
Hugenholtz, P., Goebel, B. M., and Pace, N. R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998
Jadhav, H. P., Sayyed, R. Z., Shaikh, S. S., Bhamre, H. M., Kumari, S., and El-Enshasy, H. A. (2020a). Statistically designed bioprocess for enhanced production of alkaline protease in Bacillus cereus HP_RZ17. J. Sci. Ind. Res. 79, 491–498.
Jadhav, H. P., Sonawane, M. S., Khairnar, M. H., and Sayyed, R. Z. (2020b). Production of alkaline protease by rhizospheric Bacillus cereus HP_RZ17 & Paenibacillus xylanilyticus HP_RZ19. Environ. Sustain. 3, 5–13. doi: 10.1007/s42398-020-00096-z
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/aem.72.3.1719-1728.2006
Jiang, Y., Li, S., Li, R., Zhang, J., Liu, Y., Lv, L., et al. (2017). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109, 145–155. doi: 10.1016/j.soilbio.2017.02.010
Johnson, D. B., Yajie, L., and Okibe, N. (2008). “Bioshrouding”—a novel approach for securing reactive mineral tailings. Biotechnol. Lett. 30, 445–449. doi: 10.1007/s10529-007-9574-4
Joseph, S. J., Hugenholtz, P., Sangwan, P., Osborne, C. A., and Janssen, P. H. (2003). Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69, 7210–7215. doi: 10.1128/aem.69.12.7210-7215.2003
Kalam, S., Basu, A., and Ankati, S. (2017b). “Plant Root–Associated Biofilms in Bioremediation,” in Biofilms in Plant and Soil Health, eds I. Ahmad and F. M. Husain (Chichester: John Wiley and Sons, Ltd), 337–355. doi: 10.1002/9781119246329.ch18
Kalam, S., Basu, A., and Podile, A. R. (2020). Functional and molecular characterization of plant growth promoting Bacillus isolates from tomato rhizosphere. Heliyon 6:e04734. doi: 10.1016/j.heliyon.2020.e04734
Kalam, S., Das, S. N., Basu, A., and Podile, A. R. (2017a). Population densities of indigenous Acidobacteria change in the presence of plant growth-promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 57, 376–385. doi: 10.1002/jobm.201600588
Kenawy, A., Dailin, D. J., Abo-Zaid, G. A., Abd Malek, R., Ambehabati, K. K., Zakaria, K. H. N., et al. (2019). “Biosynthesis of antibiotics by PGPR and their roles in biocontrol of plant diseases,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, Rhizobacteria in Biotic Stress Management, Vol. II, ed. R. Z. Sayyed (Singapore: Springer), 1–35. doi: 10.1007/978-981-13-6986-5_1
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016a). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kielak, A. M., Castellane, T. C., Campanharo, J. C., Colnago, L. A., Costa, O. Y., Da Silva, M. L. C., et al. (2017). Characterization of novel Acidobacteria exopolysaccharides with potential industrial and ecological applications. Sci. Rep. 7:41193. doi: 10.1038/srep41193
Kielak, A. M., Cipriano, M. A., and Kuramae, E. E. (2016b). Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 198, 987–993. doi: 10.1007/s00203-016-1260-2
King, G. M., and Weber, C. F. (2007). Distribution, diversity, and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118. doi: 10.1038/nrmicro1595
Kowalchuk, G. A., Speksnijder, A. G., Zhang, K., Goodman, R. M., and Van Veen, J. A. (2007). Finding the needles in the metagenome haystack. Microb. Ecol. 53, 475–485. doi: 10.1007/s00248-006-9201-2
Kroll, J., Klinter, S., Schneider, C., Voß, I., and Steinbüchel, A. (2010). Plasmid addiction systems: perspectives and applications in biotechnology. Microb. Biotechnol. 3, 634–657. doi: 10.1111/j.1751-7915.2010.00170.x
Lee, K. C., Morgan, X. C., Power, J. F., Dunfield, P. F., Huttenhower, C., and Stott, M. B. (2015). Complete genome sequence of the thermophilic Acidobacteria, Pyrinomonas methylaliphatogenes type strain K22 T. Stand. Genomic Sci. 10:101. doi: 10.1186/s40793-015-0099-5
Lee, S. H., Ka, J. O., and Cho, J. C. (2008). Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 285, 263–269. doi: 10.1111/j.1574-6968.2008.01232.x
Li, F., Chen, L., Zhang, J., Yin, J., and Huang, S. (2017). Bacterial community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Front. Microbiol. 8:187. doi: 10.3389/fmicb.2017.00187
Liles, M. R., Manske, B. F., Bintrim, S. B., Handelsman, J., and Goodman, R. M. (2003). A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69, 2684–2691. doi: 10.1128/aem.69.5.2684-2691.2003
Liu, J., Sui, Y., Yu, Z., Yao, Q., Shi, Y., Chu, H., et al. (2016). Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 95, 212–222. doi: 10.1016/j.soilbio.2015.12.021
Lladó, S., López-Mondéjar, R., and Baldrian, P. (2018). Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 102, 4331–4338. doi: 10.1007/s00253-018-8950-4
Llamas, M. A., and Bitter, W. (2006). Iron gate: the translocation system. J. Bacteriol. 188, 3172–3174. doi: 10.1128/jb.188.9.3172-3174.2006
Losey, N. A., Stevenson, B. S., Busse, H. J., Damsté, J. S. S., Rijpstra, W. I. C., Rudd, S., et al. (2013). Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 63, 4149–4157. doi: 10.1099/ijs.0.051425-0
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556.
Männistö, M. K., Tiirola, M., and Haggblom, M. M. (2007). Bacterial communities in Arctic fjelds of Finnish Lapland are stable but highly pH-dependent. FEMS Microbiol. Ecol. 59, 452–465. doi: 10.1111/j.1574-6941.2006.00232.x
Männistö, M. K., Rawat, S., Starovoytov, V., and Häggblom, M. M. (2011). Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int. J. Syst. Evol. Microbiol. 61, 1823–1828. doi: 10.1099/ijs.0.026005-0
Männistö, M. K., Rawat, S., Starovoytov, V., and Häggblom, M. M. (2012). Granulicella arctica sp. nov., Granulicella mallensis sp. nov., Granulicella tundricola sp. nov. and Granulicella sapmiensis sp. nov., novel acidobacteria from tundra soil. Int. J. Syst. Evol. Microbiol. 62, 2097–2106. doi: 10.1099/ijs.0.031864-0
Morris, R. L., and Schmidt, T. M. (2013). Shallow breathing: bacterial life at low O2. Nat. Rev. Microbiol. 11, 205–212. doi: 10.1038/nrmicro2970
Mushinski, R. M., Zhou, Y., Gentry, T. J., and Boutton, T. W. (2018). Bacterial metataxonomic profile and putative functional behavior associated with C and N cycle processes remain altered for decades after forest harvest. Soil Biol. Biochem. 119, 184–193. doi: 10.1016/j.soilbio.2018.01.008
Myers, M. R., and King, G. M. (2016). Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat. Int. J. Syst. Evol. Microbiol. 66, 5328–5335. doi: 10.1099/ijsem.0.001516
Na, X., Cao, X., Ma, C., Ma, S., Xu, P., Liu, S., et al. (2019). Plant stage, not drought stress, determines the effect of cultivars on bacterial community diversity in the rhizosphere of broomcorn millet (Panicum miliaceum L.). Front. Microbiol. 10:828. doi: 10.3389/fmicb.2019.00828
Naether, A., Foesel, B. U., Naegele, V., Wüst, P. K., Weinert, J., Bonkowski, M., et al. (2012). Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 78, 7398–7406. doi: 10.1128/aem.01325-12
Nwodo, U. U., Green, E., and Okoh, A. I. (2012). Bacterial exopolysaccharides: functionality and prospects. Int. J. Mol. Sci. 13, 14002–14015. doi: 10.3390/ijms131114002
Overmann, J., Abt, B., and Sikorski, J. (2017). Present and future of culturing bacteria. Annu. Rev. Microbiol. 71, 711–730. doi: 10.1146/annurev-micro-090816-093449
Pankratov, T. A., and Dedysh, S. N. (2010). Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60, 2951–2959. doi: 10.1099/ijs.0.021824-0
Parray, J. A., Jan, S., Kamili, A. N., Qadri, R. A., Egamberdieva, D., and Ahmad, P. (2016). Current perspectives on plant growth-promoting rhizobacteria. J. Plant Growth Regul. 35, 877–902.
Parsek, M. R., Val, D. L., Hanzelka, B. L., Cronan, J. E., and Greenberg, E. P. (1999). Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U.S.A. 96, 4360–4365. doi: 10.1073/pnas.96.8.4360
Parsley, L. C., Linneman, J., Goode, A. M., Becklund, K., George, I., Goodman, R. M., et al. (2011). Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. FEMS Microbiol. Ecol. 78, 176–187. doi: 10.1111/j.1574-6941.2011.01122.x
Paulsen, I. T., Sliwinski, M. K., and Saier, M. H. (1998). Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics, and substrate specificities. J. Mol. Biol. 277, 573–592. doi: 10.1006/jmbi.1998.1609
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pinto, O. H. B., Costa, F. S., Rodrigues, G. R., da Costa, R. A., da Rocha, F. G., Júnior, O. R. P., et al. (2020). Soil Acidobacteria strain AB23 resistance to oxidative stress through production of carotenoids. Microb. Ecol. doi: 10.1007/s00248-020-01548-z [Epub ahead of print].
Podar, M., Turner, J., Burdick, L. H., and Pelletier, D. A. (2019). Complete genome sequence of Terriglobus albidus strain ORNL, an Acidobacterium isolated from the Populus deltoides rhizosphere. Microbiol. Resour. Announc. 8:e01065-19. doi: 10.1128/MRA.01065-19
Postle, K., and Kadner, R. J. (2003). Touch and go: tying TonB to transport. Mol. Microbiol. 49, 869–882. doi: 10.1046/j.1365-2958.2003.03629.x
Rajeev, L., Chen, A., Kazakov, A. E., Luning, E. G., Zane, G. M., Novichkov, P. S., et al. (2015). Regulation of nitrite stress response in Desulfovibrio vulgaris Hildenborough, a model sulfate-reducing bacterium. J. Bacteriol. 197, 3400–3408. doi: 10.1128/jb.00319-15
Rawat, S. R., Männistö, M. K., Bromberg, Y., and Häggblom, M. M. (2012a). Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol. Ecol. 82, 341–355. doi: 10.1111/j.1574-6941.2012.01381.x
Rawat, S. R., Männistö, M. K., Starovoytov, V., Goodwin, L., Nolan, M., Hauser, L., et al. (2012b). Complete genome sequence of Terriglobus saanensis type strain SP1PR4(T), an Acidobacteria from tundra soil. Stand. Genomic Sci. 7, 59–69. doi: 10.4056/sigs.3036810
Rawat, S. R., Männistö, M. K., Starovoytov, V., Goodwin, L., Nolan, M., Hauser, L., et al. (2014). Complete genome sequence of Granulicella tundricola type strain MP5ACTX9(T), an Acidobacteria from tundra soil. Stand. Genomic Sci. 9, 449–461. doi: 10.4056/sigs.4648353
Rawat, S. R., Männistö, M. K., Starovoytov, V., Goodwin, L., Nolan, M., Hauser, L. J., et al. (2013). Complete genome sequence of Granulicella mallensis type strain MP5ACTX8(T), an acidobacterium from tundra soil. Stand. Genomic Sci. 9, 71–82. doi: 10.4056/sigs.4328031
Reshma, P., Naik, M. K., Aiyaz, M., Niranjana, S. R., Chennappa, G., Shaikh, S. S., et al. (2018). Induced systemic resistance by 2,4diacetylphloroglucinol positive fluorescent Pseudomonas strains against rice sheath blight. Indian J. Exp. Biol. 56, 207–212.
Rhodius, V. A., Suh, W. C., Nonaka, G., West, J., and Gross, C. A. (2005). Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. doi: 10.1371/journal.pbio.0040002
Rondon, M. R., August, P. R., Bettermann, A. D., Brady, S. F., Grossman, T. H., Liles, M. R., et al. (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000
Rosselli, R., Romoli, O., Vitulo, N., Vezzi, A., Campanaro, S., De Pascale, F., et al. (2016). Direct 16S rRNA-seq from bacterial communities: a PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Sci. Rep. 6:32165. doi: 10.1038/srep32165
Sagar, A., Sayyed, R. Z., Ramteke, P. W., Sharma, S., Najat, M., Abdallah, M. E., et al. (2020). ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiol. Mol. Biol. Plants 26, 1847–1854. doi: 10.1007/s12298-020-00852-9
Sait, M., Davis, K. E., and Janssen, P. H. (2006). Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72, 1852–1857. doi: 10.1128/aem.72.3.1852-1857.2006
Sayyed, R. Z., Ilyas, N., Tabassum, B., Hashem, A., Abd_Allah, E. F., and Jadhav, H. P. (2019). “Plausible role of plant growth promoting rhizobacteria in future climatic scenario,” in Environmental Biotechnology: For Sustainable Future, eds R. C. Sobti, N. K. Arora, and R. Kothari (Singapore: Springer), 175–197. doi: 10.1007/978-981-10-7284-0_7
Shaikh, S. S., Wani, S. J., Sayyed, R. Z., Thakur, R., and Gulati, A. (2018). Production, purification and kinetics of chitinase of Stenotrophomonas maltophilia isolated from rhizospheric soil. Indian J. Exp. Biol. 56, 274–278.
Solden, L., Lloyd, K., and Wrighton, K. (2016). The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 31, 217–226. doi: 10.1016/j.mib.2016.04.020
Stamps, B. W., Losey, N. A., Lawson, P. A., and Stevenson, B. S. (2014). Genome sequence of Thermoanaerobaculum aquaticum MP-01T, the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Genome Announc. 2:e00570-14. doi: 10.1128/genomeA.00570-14
Stevenson, B. S., Eichorst, S. A., Wertz, J. T., Schmidt, T. M., and Breznak, J. A. (2004). New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70, 4748–4755. doi: 10.1128/aem.70.8.4748-4755.2004
Sun, Y., Fukamachi, T., Saito, H., and Kobayashi, H. (2012). Adenosine deamination increases the survival under acidic conditions in Escherichia coli. J. Appl. Microbiol. 112, 775–781. doi: 10.1111/j.1365-2672.2012.05246.x
Tanaka, Y., Matsuzawa, H., Tamaki, H., Tagawa, M., Toyama, T., Kamagata, Y., et al. (2017). Isolation of novel bacteria including rarely cultivated phyla, Acidobacteria and Verrucomicrobia, from the roots of emergent plants by simple culturing method. Microbes Environ. 32, 288–292. doi: 10.1264/jsme2.me17027
Tank, M., and Bryant, D. A. (2015). Nutrient requirements and growth physiology of the photoheterotrophic Acidobacterium, Chloracidobacterium thermophilum. Front. Microbiol. 6:226. doi: 10.3389/fmicb.2015.00226
Tringe, S. G., Von Mering, C., Kobayashi, A., Salamov, A. A., Chen, K., Chang, H. W., et al. (2005). Comparative metagenomics of microbial communities. Science 308, 554–557. doi: 10.1126/science.1107851
Tyson, G. W., Chapman, J., Hugenholtz, P., Allen, E. E., Ram, R. J., Richardson, P. M., et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43. doi: 10.1038/nature02340
Ude, S., Arnold, D. L., Moon, C. D., Timms-Wilson, T., and Spiers, A. J. (2006). Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 8, 1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Strategies for the culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 309, 1–7.
Velayudhan, J., Hughes, N. J., McColm, A. A., Bagshaw, J., Clayton, C. L., Andrews, S. C., et al. (2000). Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286. doi: 10.1046/j.1365-2958.2000.01987.x
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Vieira, S., Luckner, M., Wanner, G., and Overmann, J. (2017). Luteitalea pratensis gen. nov., sp. nov. a new member of subdivision 6 Acidobacteria isolated from temperate grassland soil. Int. J. Syst. Evol. Microbiol. 67, 1408–1414. doi: 10.1099/ijsem.0.001827
Vinay, J. U., Naik, M. K., Rangeshwaran, R., Chennappa, G., Shaikh, S. S., and Sayyed, R. Z. (2016). Detection of antimicrobial traits in fluorescent pseudomonads and molecular characterization of an antibiotic pyoluteorin. 3 Biotech 6:227. doi: 10.1007/s13205-016-0538-z
Wani, S. J., Shaikh, S. S., and Sayyed, R. Z. (2016). Statistical-based optimization and scale-up of siderophore production process on laboratory bioreactor. 3 Biotech 6:69. doi: 10.1007/s13205-016-0365-2
Ward, L. M., McGlynn, S. E., and Fischer, W. W. (2017). Draft genome sequence of Chloracidobacterium sp. CP2_5A, a phototrophic member of the phylum Acidobacteria recovered from a Japanese hot spring. Genome Announc. 5:e00821-17. doi: 10.1128/genomeA.00821-17
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/aem.02294-08
Wasmund, K., Mußmann, M., and Loy, A. (2017). The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344. doi: 10.1111/1758-2229.12538
Wegner, C. E., and Liesack, W. (2017). Unexpected dominance of elusive Acidobacteria in early industrial soft coal slags. Front. Microbiol. 8:1023. doi: 10.3389/fmicb.2017.01023
Whang, K. S., Lee, J. C., Lee, H. R., Han, S. I., and Chung, S. H. (2014). Terriglobus tenax sp. nov., an exopolysaccharide-producing acidobacterium isolated from rhizosphere soil of a medicinal plant. Int. J. Syst. Evol. Microbiol. 64, 431–437. doi: 10.1099/ijs.0.053769-0
White, A. P., Gibson, D. L., Kim, W., Kay, W. W., and Surette, M. G. (2006). Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188, 3219–3227. doi: 10.1128/jb.188.9.3219-3227.2006
Youssef, N. H., Couger, M. B., McCully, A. L., Criado, A. E. G., and Elshahed, M. S. (2015). Assessing the global phylum level diversity within the bacterial domain: a review. J. Adv. Res. 6, 269–282. doi: 10.1016/j.jare.2014.10.005
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Zope, V. P., El-Enshasy, H. A., and Sayyed, R. Z. (2019). “Plant growth-promoting rhizobacteria: an overview in agricultural perspectives,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, Rhizobacteria in Biotic Stress Management, Vol. II, ed. R. Z. Sayyed (Singapore: Springer), 345–361. doi: 10.1007/978-981-13-6986-5_13
Keywords: acidobacteria, biogeochemical cycles, ecological roles, metagenomics, molecular characterization, plant growth-promoting activities, soil
Citation: Kalam S, Basu A, Ahmad I, Sayyed RZ, El-Enshasy HA, Dailin DJ and Suriani NL (2020) Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Received: 04 July 2020; Accepted: 08 October 2020;
Published: 30 October 2020.
Edited by:
Byung-Kwan Cho, Korea Advanced Institute of Science and Technology, South KoreaReviewed by:
Vasvi Chaudhry, University of Tübingen, GermanySheree J. Finley, Alabama State University, United States
Namil Lee, Korea Advanced Institute of Science and Technology, South Korea
Copyright © 2020 Kalam, Basu, Ahmad, Sayyed, El-Enshasy, Dailin and Suriani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iqbal Ahmad, ahmadiqbal8@yahoo.co.in
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
§ORCID: Anirban Basu, orcid.org/0000-0001-6620-8320; Iqbal Ahmad, orcid.org/0000-0001-8447-4497; R. Z. Sayyed, orcid.org/0000-0002-1553-1213
 Sadaf Kalam
Sadaf Kalam Anirban Basu
Anirban Basu Iqbal Ahmad
Iqbal Ahmad R. Z. Sayyed
R. Z. Sayyed Hesham Ali El-Enshasy
Hesham Ali El-Enshasy Daniel Joe Dailin
Daniel Joe Dailin Ni Luh Suriani
Ni Luh Suriani