- 1Department of Skin & VD, Institute of Medical Sciences, SUM Hospital, Siksha O Anusandhan University, Bhubaneswar, India
- 2Division of Microbiology & NCDs, ICMR-Regional Medical Research Centre, Bhubaneswar, India
- 3Department of Zoology, Maharaja Sriram Chandra Bhanja Deo University, Baripada, India
- 4Department of Zoology, Utkal University, Vani Vihar, Bhubaneswar, India
- 5Centre of Environment, Climate Change and Public Health, RUSA 2.0, Utkal University, Vani Vihar, Bhubaneswar, India
Biofilms form a complex layer with defined structures, that attach on biotic or abiotic surfaces, are tough to eradicate and tend to cause some resistance against most antibiotics. Several studies confirmed that biofilm-producing bacteria exhibit higher resistance compared to the planktonic form of the same species. Antibiotic resistance factors are well understood in planktonic bacteria which is not so in case of biofilm producing forms. This may be due to the lack of available drugs with known resistance mechanisms for biofilms. Existing antibiotics cannot eradicate most biofilms, especially of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species). Insects produce complex and diverse set of chemicals for survival and defense. Antimicrobial peptides (AMPs), produced by most insects, generally have a broad spectrum of activity and the potential to bypass the resistance mechanisms of classical antibiotics. Besides, AMPs may well act synergistically with classical antibiotics for a double-pronged attack on infections. Thus, AMPs could be promising alternatives to overcome medically important biofilms, decrease the possibility of acquired resistance and treatment of multidrug-resistant pathogens including ESKAPE. The present review focuses on insect-derived AMPs with special reference to anti-biofilm-based strategies. It covers the AMP composition, pathways and mechanisms of action, the formation of biofilms, impact of biofilms on human diseases, current strategies as well as therapeutic options to combat biofilm with antimicrobial peptides from insects. In addition, the review also illustrates the importance of bioinformatics tools and molecular docking studies to boost the importance of select bioactive peptides those can be developed as drugs, as well as suggestions for further basic and clinical research.
Introduction
A biofilm is a layer of polymeric organic matter to which microorganisms like fungi and/or bacteria, are attached in a sessile form. Biofilms are characterized by the presence of extracellular polymers, that create a visible slimy layer on a solid surface. The extracellular matrix: glycocalyx, can be made up of polysaccharides, proteins, glycoproteins, lipids, deoxyribonucleic acid, etc., which are collectively called extracellular polymeric substances (Costerton et al., 1987; Anwar et al., 1990; Matz et al., 2004). This glycocalyx provides a matrix for attachment of microbial cells and forms the internal architecture of the biofilm community. Biofilms have been defined as “a structured community of bacterial cells surrounded in a self-produced polymeric matrix and adherent to an inert or living surface” by Costerton et al. (1999) and “surface-associated microbial communities, surrounded by an extracellular polymeric substance matrix” by Hall-Stoodley and Stoodley (2009). A biofilm can also be defined as “a collective of microbial cells attached to a living or non-living surface, fixed within by a medium of extracellular polymeric substance matrix” (Hall-Stoodley et al., 2012). The most recent definitions treat them as “complex, sessile communities of microbes found attached to a surface in an extracellular matrix as aggregates” (Roy et al., 2018); and as a “well−organized structure formed by a bacterial community assemblage that is enclosed in a self−produced matrix in which bacterial cells communicate” (Armbruster and Parsek, 2018). We propose microbial biofilms as “communities of microbial cells with defined structures that attach on biotic or abiotic surfaces and are embedded in self-produced matrices consisting of extracellular polysaccharides, DNA and protein, with interspersed water channels.”
Peptides are biomolecules consisting of linear chains of amino acids that are found in almost every organism. Peptides can be generated as endogenous molecules for endocrine or neuronal signaling, or by degradation of proteins. The latter may have a positive impact on body functions as well as on health. These peptides perform major roles in the metabolic functions of living organisms, and (depending on their sequence) can have antimicrobial, anticancer, antidiabetic, anti-oxidative, immunomodulatory, etc., effects. Certain isolated bioactive peptides are used in the formulation or production of health-promoting food and food supplements, pharmaceuticals, cosmetics, or nutraceuticals. Most commonly, peptides can be generated from proteins by two methods, viz. chemical degradation (acid and alkali) (Andreu and Rivas, 1998), or enzymatic cleavage (Bongers and Heimer, 1994). With those methods, the original complex proteins from a plant or animal are broken down to yield peptides of 2-20 amino acid long residues. Nevertheless, the structure of peptides generated depends upon the native protein folding, degree of hydrolysis, enzyme specificity, and additional conditions of hydrolysis like temperature and time (Nehete et al., 2013). The amino acid sequence and composition are the primary characteristics of these peptides as these determine the specific bioactivity. Single peptides with a specific sequence can also be synthesized or produced by rDNA technology.
Under the above backdrop, the current review is planned to offer three aspects on the subject: (1) bacterial biofilm formation, (2) current therapeutic opportunities against biofilm producing bacteria, and (3) the probable role of insect-derived antimicrobial peptides in combating/reducing the biofilm formation. The first aspect includes bacterial biofilm composition, involvement of genetic virulence factors with reference to ESKAPE pathogens and its impact on human health and diseases. The second aspect discusses biofilm inhibition, dispersal, and eradication strategies along with the pathways and molecular mechanisms involved in biofilm formation. The third part of the review deals with AMPs from insect sources with some exclusive examples, the synergistic role of AMPs and antibiotics, and related in vitro studies and clinical trials. Besides, a section is added to emphasize the role of various bioinformatics tools and molecular modeling and docking analyses to accelerate the peptide-based drug discovery opportunities.
Bacterial Biofilms and Their Formation
A biofilm, as a self-organized extra surface within the bacterial community, changes significantly the bacterial physiology in favor of exogenous stress tolerance and resistance to applied antibiotics or other biocides. Bacterial biofilms appear in both mono- and multilayers, depending on the attachment of the exopolysaccharide (EPS) matrix and the involvement of neighboring bacteria. The process of biofilm formation is complex and usually initiated by attachment of bacteria to a solid surface. Due to their hydrophobic nature, certain dissolved organic molecules accumulate on the solid: water interface, resulting in a film, and then they form small groups of bacteria, known as micro-colonies. EPS such as proteins, glycopeptides, glycolipids, lipopolysaccharides, and extracellular DNA accumulate in the attachment (Shirtliff et al., 2009). After completion of phase II irreversible microbial attachment, a mature biofilm forms, and the micro-colonies assume a distinct phenotype with a different gene expression than their planktonic counterpart (Stoodley et al., 2002). The process of differentiation can be activated by deposition of N-acyl homoserine lactones as a sensing molecule for cell to cell communication (Costerton et al., 1999). Biofilm formation is a good strategy for survival in a nutrient-poor environment suggesting starvation to favor biofilm formation. Besides, higher antibiotic resistance is observed when bacteria are grown or living under starvation. Biofilm development can be an adaptation of microorganisms to aggressive environments (De la Fuente-Núñez et al., 2013). Biofilm formation can happen on an assortment of surfaces, including living tissues and prosthetic implants (Donlan, 2002). About 99% of the microbial world exists in the form of biofilms containing a wide range of microorganisms (Garrett et al., 2008). For instance, over 500 types of microorganisms are found in biofilms in the oral cavity (Whittaker et al., 1996).
Composition of Bacterial Biofilm
Microorganisms constitute 5-35% of the biofilm volume, the remaining part being extracellular matrix. The cellular matrix comprises proteins (e.g., fibrin), essential nutrients, and minerals from the surrounding environment. The extracellular matrix contains 1-2% polysaccharides (e.g., alginate), <1% DNA, <1% RNA, ions and 97% water. The flow of essential nutrients inside a biofilm is maintained through the water compartment. The EPS matrix (0.2-1.0 μm thick) strengthens the interaction of the microorganism’s and protects them from external factors like mechanical stress or antibiotics.
Bacterial Genetic-Virulence Factors in Biofilm Formation, With Special Reference to ESKAPE
Biofilm-producing bacteria show higher (often 10-1000-fold) antibiotic tolerance/resistance to administered antibiotics in the biofilm compared to their planktonic state. Generally, antibiotic resistance factors such as mutations and efflux pumps are well understood in planktonic forms than in biofilm form (Munita and Arias, 2016). Thus, biofilm-associated antibiotic tolerance is assumed to involve alternative mechanisms. The ESKAPE (E. faecalis, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species) pathogens are the leading biofilm-forming microorganisms causing nosocomial infections. Thus, understanding the unknown biofilm drug-resistance mechanisms is crucial for developing effective antimicrobial agents.
Enterococcus faecalis is the Gram-positive (Gm +ve) anaerobic bacterium among all Enterococcus species with resistance to antibiotics like ampicillin and vancomycin. The adhesion of bacterial cell to host tissues is the crucial step in biofilm production, which is mediated by enterococcal surface protein (Esp) involved in cell adherence, colonization, and persistence in the urinary tract, evasion of the immune system by aggregation protein (agg or asa1) and collagen-binding protein (aec) (Mohamed and Huang, 2007). The endocarditis antigens (efaAfs and efaAfm), endocarditis biofilm-associated pili (ebpABC), surface anchor protein, sortage (srt) and secretory antigens (salAB) like virulence factors are also involved in facilitating cell adhesion and biofilm formation (Chauang et al., 2000; Hashem et al., 2017). Enterococcus cells communicate through peptide pheromones cpd, cob, and ccf linking the receptor-donor pathways to transfer the biofilm regulator and promotor virulence genes. Mainly, the hemolytic exotoxins such as cytolysin A (cylA), autolysis (ata), hyaluronidase (hyl), gelatinase (GelE), and serine protease (sprE) are the most important virulence genes affecting host cells through regulating cell lysis and autolysis process (Seno et al., 2005; Thomas et al., 2008; Paganelli et al., 2013; Mottola et al., 2016; Naorem et al., 2020).
Staphylococcus aureus is another ubiquitous opportunistic Gm +ve pathogen, and the human nasal passage being the common route of infection in humans with a high risk of bloodstream infection and bacteremia in a later stage. Based on the flow-cell and microscopic studies, it is evident that initial S. aureus biofilm matrix formation occurs through polysaccharide intercellular adhesin (PIA) or polymeric N-acetyl-glucosamine (PNAG) which are responsible for synthesis, export, and modification of PIA as well as maintaining the structural integrity of the biofilm (Lauderdale et al., 2009; Brooks and Jefferson, 2014). Moreover, extracellular DNA or proteins can support biofilm formation in the absence of PIA (Rohde et al., 2007; Boles et al., 2010). Additionally, studies have also proved that a variety of proteins are available in S. aureus, viz. fibronectin−binding proteins A and B or fnbpAB (Cortés et al., 2011), fibrinogen−binding protein (fib) (Shannon and Flock, 2004), fibrinogen−binding protein clumping factors A and B or clfAB (O’Brien et al., 2002), biofilm−associated protein (bap) (Cucarella et al., 2001), collagen−binding protein (cna), serine−aspartate repeat proteins (Sdr) (Barbu et al., 2014), elastin−binding protein (Ebp) (Campoccia et al., 2009), and laminin−binding protein (eno) (Azara et al., 2017) that may regulate formation of biofilms in a strain- and environment-specific manner (Atshan et al., 2012; Tang et al., 2013; Serray et al., 2016).
Klebsiella pneumoniae is a Gm -ve ubiquitous bacterium, especially found in the intestinal tract of humans, but less common in the nasopharynx (Piperaki et al., 2017). In the pre-antibiotic period, K. pneumoniae was an important cause of pneumonia in the community, especially in alcoholics and diabetics while in past few decades it is established as the leading cause of diseases associated with health care in hospitals (Piperaki et al., 2017). Worldwide, K. pneumoniae infections rise in hospitals with severe antibiotic resistance making it difficult to treat patients, particularly immunocompromised individuals, and considered as the second most common cause of hospital-acquired Gm –ve infection (Candan and Aksöz, 2015; Paczosa and Mecsas, 2016). Schroll et al. (2010) found that type-3 fimbriae promote development of biofilm in catheter-associated infections caused by K. pneumoniae. Recently, Zheng et al. (2018) observed the biofilm formation to be more pronounced among magA (K1), aero+, rmpA+, rmpA2+, allS+, wcaG+, and iutA+ isolates than in isolates that were negative for these virulence factors; and concluded that presence of the wcaG virulence factor gene to be responsible for biofilm formation in K. pneumoniae.
Acinetobacter baumannii, a Gm -ve human pathogen (Family: Moraxellaceae, Class: Proteobacteria of Eubacteria; Evans et al., 2012) causes pneumonia, meningitis, bacteremia, wounds and soft-tissue infection, peritonitis and urinary tract infections (UTIs) (Dahdouh et al., 2017; Shirmohammadlou et al., 2018). Chronic infection, antimicrobial resistance and formation of biofilm in both biotic and abiotic surfaces are its important characteristics (Kongthai and Sitthisak, 2016). The rate of formation of biofilms is about 80-91% in case of A. baumannii (Sung, 2018). Different virulence factor proteins such as the outer membrane protein A (OmpA) with 38 kDa which play a crucial role in the attachment and attack to epithelial cells via contact with fibronectin; biofilm associated protein (Bap) (854 kDa), a cell surface protein responsible for cell to cell interactions and biofilm maturation; chaperon-usher pilus (Csu), are responsible for initiation of biofilm formation on abiotic surfaces (Dahdouh et al., 2017; Chapartegui-González et al., 2018; Ghasemi et al., 2018). In addition, EPS, two-component system (BfmS/BfmR), poly-β-(1,6)-N-acetyl glucosamine (PNAG) also participate in the formation of biofilm, quorum sensing system (Kongthai and Sitthisak, 2016; Ghasemi et al., 2018), virulence and antibiotic resistance (Amala Reena et al., 2017).
Pseudomonas aeruginosa, a rod-shaped Gm -ve bacteria, is also responsible to form biofilms (Tavares, 2000). Several virulence factors such as lipopolysaccharide (LPS), flagella, pili (type IV), exotoxin A, enzyme proteases, alginate, and QS, etc., are responsible for its pathogenicity. LPS perform a key role in the activation of the host’s innate (TLR4, NLRP1, NLRP2, and NLRP3) as well as adaptive immune responses. Eventually it aims to dysregulate the inflammation responses that compensates to high morbidity and mortality (Raetz and Whitfield, 2002). Type IV pili play a critical role in cell adhesion (Raetz and Whitfield, 2002). The T3SS (Type III secretion system) is another virulence factor which transports proteins from the cytoplasm of P. aeruginosa into the cytosol of host cells resulting phagocytosis by bacteria and damage to host tissues (Punsalang and Sawyer, 1973; Frank, 1997). The exotoxic A virulence factor is secreted by type II secretion mechanism, it secretes proteins in to the extracellular matrix conforming enzymes protease, lipase, phospholipase, and alkaline phosphatase which ultimately help the pilus-like apparatus (Passador and Iglewski, 1994).
The family Enterobacteriaceae is a complex group of Gm -ve bacteria basically found in the intestinal tract and urinary tract and most common cause of UTI and lower respiratory tract infections (Peleg and Hooper, 2010; Magiorakos et al., 2017). The virulence factors include different adhesins, hemolysin production and serum resistance, etc. aids to form biofilms in the human intestine that can affect the colon, and have a significant effect on the functioning of the intestinal microbiome and its interaction with the gut (Rossi et al., 2018).
Impact of Biofilm on Human Health
The concept of biofilm is not new in medical microbiology. Antoni van Leeuwenhoek, as early as 1683, observed and described biofilms through his primitive microscope using matter from his own teeth. However, the concept of biofilm was not an essential concept in medical microbiology until early 1970s. Recognition of persistent infection and aggregates of bacteria in cystic fibrosis patients by Høiby (2017), and later evidence of biofilm involvement in pathogenesis in chronic infection and antibiotic treatment failure familiarized the concept in medical microbiology (Bjarnsholt, 2013). Today, it is an emerging field of research in mechanism of antibiotic resistance and antibacterial drug development.
Biofilm production is a part of bacterial survival mechanisms and is associated with several health complications (Figure 1). Currently, both communicable and non-communicable diseases are associated with infection, where biofilm enhances complicacy of disorder in most of the cases. The genetic disorder cystic fibrosis (CF) was the first recognized biofilm infection model and so far, the most thoroughly studied one. CF generally affects the respiratory and digestive systems and are characterized through the production of viscid mucus and chronic infection. Lungs infecting and biofilm predicting strains like S. aureus, H. influenzae, and P. aeruginosa lead to mortality of CF patients as age advances (Lyczak et al., 2002). Biofilm like-structure have primarily been observed in lung lavage samples and apical surface of respiratory epithelia in CF patients which stimulate epithelia to increase the secretion of inflammation medicating factors. Biofilm producing pathogens were continuously isolated from other respiratory disorders such as chronic rhinosinusitis, pharyngitis or sore throat, and pertussis or whooping cough (Vestby et al., 2020).
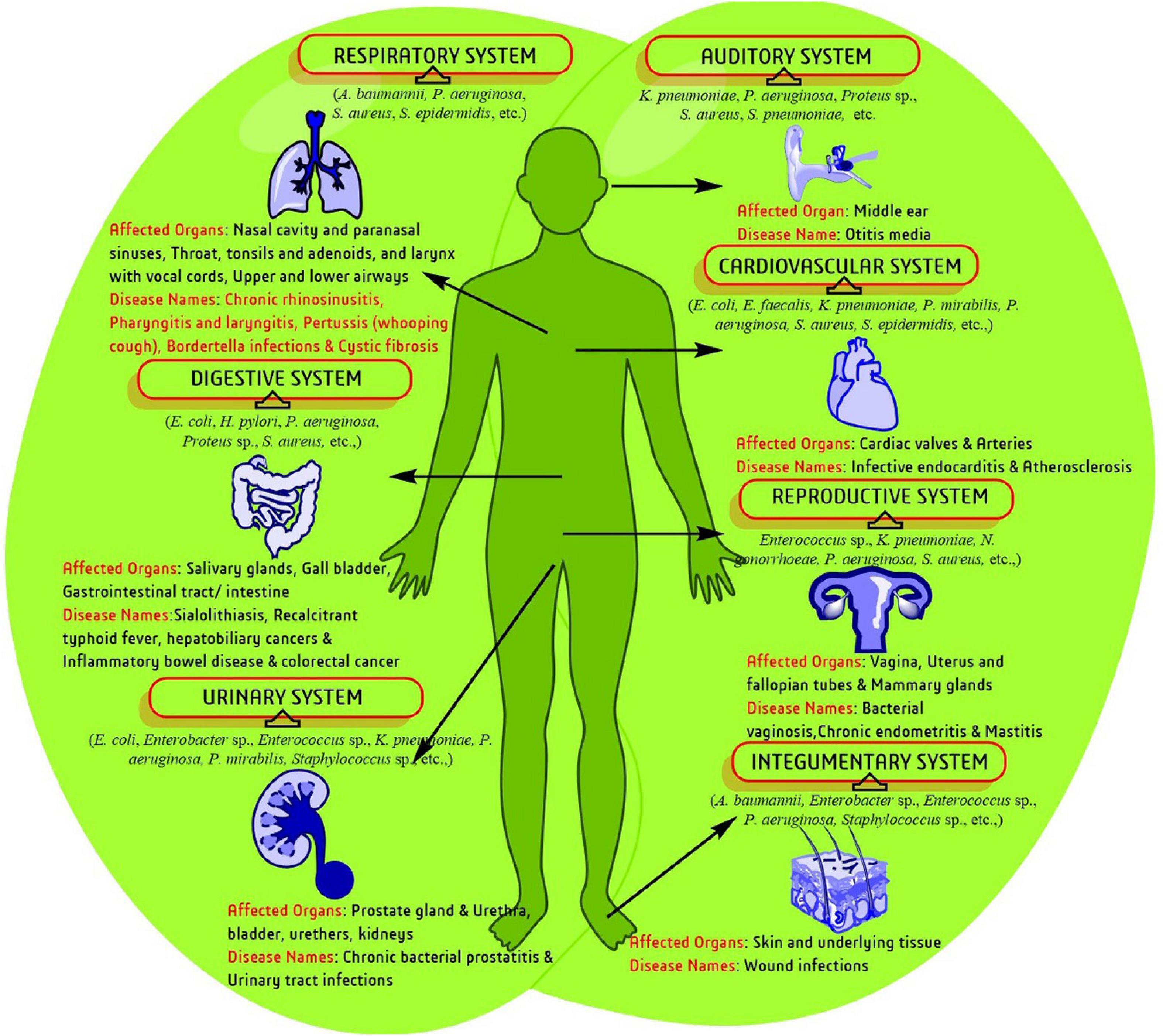
Figure 1. A schematic representation of biofilm-associated human diseases and affected biological systems/organs.
Bacteria are continuously presented in urinary system disorders like bacterial prostatitis (BP) and UTI. Acute BP infection harbors several pathogenic bacteria such as, Escherichia coli, Proteus mirabilis, P. aeruginosa, Klebsiella sp., Enterobacteriaceae and E. faecalis which are associated with infertility, bladder infection, prostatic abscesses, urosepsis, kidney damage, bacteremia, septicemia, and death. Biofilm producing E. coli also frequently occur in BP and UTI and enhance the complications by reducing the potency of antibiotics administered (Flores-Mireles et al., 2015). The human integumentary system is mostly associated with and effected by pathogenic bacteria. Damaged living tissue attract various bacterial species. Sometimes healing is disrupted, and wound tissues acquire severe infection by planktonic bacteria or biofilm producing bacteria. Wound infection in immuno-compromised, geriatric, and diabetic cases is more complicated (Bjarnsholt et al., 2011). Biofilm has been assumed to be the underlying cause for transformation of wounds into chronic stages. Several studies have confirmed that biofilm or biofilm producing pathogens are directly associated with the failure of antibiotics and delay in optimal wound healing/management (Zhao et al., 2013).
Bacterial vaginosis (BV) is the most common genital tract infection in women throughout reproductive years during which several anaerobic bacteria such as Gardnerella vaginalis, and Atopobium vagnae decreases the number of protective lactobacilli. Generally, the vaginal population of G. vaginalis of a healthy woman has less chance of causing BV. However, several transcriptome studies proposed that, G. vaginalis can result different phenotypes of the pathogen through large changes in gene expression (Kumar et al., 2011). Simultaneously, environmental pressure and ecological disturbances in the vaginal niche produces biofilms influenced by other bacterial species leading to pathogenesis. Similarly, chronic endometritis complications are also affected by biofilm producing E. faecalis, E. coli, G. vaginalis, K. pneumoniae, P. aeruginosa, Staphylococcus sp., and Streptococcus sp. Acute otitis media (OM), one of the most common pediatric inflammation agents in the middle ear cavity of <5-year aged children, is also caused by biofilm development that leads to high-risk pathogenesis through colonization of the otopathogenic bacteria, S. pneumoniae.
Typhoid fever, the most acute food borne illness generally caused by Salmonella enterica, is associated with several complications and even death if untreated. Several studies in a murine gall stone model with S. typhimurium identified biofilm formation in the gall bladder in chronic typhoid carrier states (Gonzalez-Escobedo et al., 2011). Antibiotics, those generally effective against the acute infection, are ineffective against the biofilm associated chronic colonization of the gall bladder. Other digestive system-associated disorders like ulcerative colitis and Crohn’s disease, an inflammatory bowel disease (IBD) lead to chronic inflammation of the digestive tract with some common symptoms like pain, diarrhea, weight loss, fatigue, etc. Bacteriodes fragilis, Enterobacteriaceae, and E. coli produce biofilms in epithelium site which stimulate an inflammatory response over failure of maintaining the integrity of the mucosal barrier resulting in a reduced ability to clear the infection.
Other specific clinical observations have confirmed that bacterial biofilms were pledged in every patient with clinical post-operative infections (∼75%) and recurrent sialadenitis (a digestive disorder that forms stones in salivary gland)/pus drainage. The presence of bacterial biofilms may enhance the severity of sialadenitis. Isolated microorganism (staphylococci, streptococci, enterococci, etc.) from infective endocarditis (a cardiovascular disorder) also produce biofilms, which is involved in physical disruption of valve function and overcome antibiotic therapy by bloodstream infection. Recently, biofilm producing bacteria have also reported in atherosclerotic arteries through fluorescence microscopy and fluorescence in situ hybridization (FISH) (Vestby et al., 2020).
Current Therapeutic Practices Against Biofilm Producing Bacteria
Biofilm is the bacterial community’s self-motived mechanism for pathogenesis and, alternatively, associated with antibiotic/antibacterial resistance/tolerance. Recently, several methods have been examined to tackle biofilm or biofilm-producing bacteria through innovative techniques such as non-coating, surface coating or individual/synergistic antibiotics, anticancer drugs, natural products, and peptides in application-specific manner (Pletzer and Hancock, 2016; Li et al., 2018; Reen et al., 2018; Verderosa et al., 2019b). Concomitantly, several experiments with different model systems to prevent biofilm formation are going on. The overall strategy may be divided into three parts as described below:
Biofilm Inhibition Strategy
Several target specific strategies have been used to control/inhibit bacterial biofilm development. As bacterial adhesion promotes mature biofilm formation, preventing bacterial attachment or bacterial adhesion is an ideal strategy. Development of medical devices, biomaterials or coating could alter the surface morphology of target tissues to avoid an extension unfavorable to bacteria (Bazaka et al., 2012; Li et al., 2017). Arciola et al. (2012) have shown an effective method for preventing biofilm-related infections/complications associated with orthopedic implants. Overall, this method is more suitable for large-scale surface modification to prevent biofilm formation (Bazaka et al., 2012; Campoccia et al., 2013). Similarly, using a small therapeutic inhibitor/agent is another approach used to prevent the formation of biofilm. The biofilm inhibitors are often employed to passivate the medical biomaterial/devise (Boase et al., 2018). A wide variety of biofilm inhibitors of bromo-pyrrole, furanone, imidazole, indole, phenol, etc., class of compounds have been reported (Simões et al., 2010; Worthington et al., 2012; Rabin et al., 2015).
Biofilm Dispersal Strategy
Biofilm dispersal agents mostly target the biofilm activated/regulated bacterial biochemical pathways such as QS, c-di-GMP, and sRNAs pathways. Inhibiting the function of enzymes involved in biofilm matrix formation is one such approach (Kaplan, 2010; McDougald et al., 2012; Fleming and Rumbaugh, 2017). These disperser cells are more suitable in antimicrobial treatment than biofilm-residing cells and currently, this approach becomes an intense area aiming to develop promising dispersal agents (Fleming and Rumbaugh, 2017; Roy et al., 2018). Briefly, alginate lyase of P. aeruginosa, a surface-protein-releasing enzyme of S. mutans, thermonuclease of S. aureus, LapG protease of P. putida, hemagglutinin protease of Vibrio cholerae, endo-β-1,4-mannanase of Xanthomonas campestris are some well-characterized target enzymes for biofilm dispersal strategy (Kaplan, 2010). However, this treatment method becomes problematic if the disperser cells are not treated or translocated into new areas, which may spread the infection like the initial stage. Therefore, in most cases, a potent dispersal agent is a co-administrated/synergistic approach with an antimicrobial agent to get a promising result (Marvasi et al., 2014; Reffuveille et al., 2015; Roizman et al., 2017).
Biofilm Eradication Strategy
Currently, the development of novel antimicrobial agents to eradicate biofilm is an emerging area of research. Till date, several promising agents have already been developed/proposed including antimicrobial peptides or AMPs like LL-37, oritavancin, novispirin G10, etc., quaternary ammonium compounds or QACs like tris-QAC-10, XF-70, XF-73, etc. (Jennings et al., 2014; Forman et al., 2016); antimicrobial lipids like glycerol monolaurate, docosahexaenoic acid, etc. (Schlievert and Peterson, 2012; Sun et al., 2016); nitric oxide releasing antibiotics or nitro-oxide functionalized antibiotics, like cephalosporin-3’-diazeniumdiolate, ciprofloxacin-nitroxides-27, poly (amidoamine) dendrimer, etc. (Barraud et al., 2009; Worley et al., 2015; Verderosa et al., 2016, 2019a); redox-active secondary metabolites or phenazines and quinolones (phenazine-14, bromophenazine-8, halogenated quinoline-3, etc.) (Cezairliyan et al., 2013; Garrison et al., 2015; Basak et al., 2016; Huigens et al., 2019). Many of them were proved against biofilm producing ESKAPE pathogens. Overall, the use of AMPs as an alternative to antibiotics and particularly biofilm has received significant attention over a couple of years.
Pathways and Molecular Mechanisms Involved in Biofilm Formation
The development of a biofilm occurs in four distinct stages: (a) attachment to a surface, (b) binding to the surface through adhesins followed by both a reversible and irreversible process of extension, (c) development of micro-colonies, and (d) maturation of biofilm architecture (Figure 2). Bacterial biofilm formation is a highly regulated process that occurs through bacterial quorum sensing (QS), bis-(3′-5′)-cyclic di-guanosine monophosphate (c-di-GMP) and small RNAs (sRNAs) pathways.
Quorum sensing is a unique signaling pathway in the bacterial community that maintains communication through a small “autoinducer” molecule that leads to biofilm formation. When a surficial density of bacteria is present and the autoinducers’ concentration reaches a threshold level, bacteria start to activate certain target genes. Overall, QS controls 10% of the bacterial genome and plays a crucial role in the formation and dispersal of biofilms through the proposed signaling. The QS-system is not involved in the biofilm’s initial attachment or growth stage but is the main pathway for biofilm dispersal.
The second primary biofilm regulator pathway is the c-di-GMP network the most complex secondary signaling bacterial system that varies between species. After binding to various cellular receptors, c-di-GMP regulates bacterial transcription, enzyme activity, and larger cellular structures via QS signaling. Overall, c-di-GMP plays a decisive role in the switch between planktonic and biofilm formation, as well as biofilm structure development through the synthesis of exopolysaccharides, adhesive pili, secretion of extracellular DNA, along with regulating cell death and survival. Lastly, the non-coding sRNA molecules actively participate in bacterial post-transcriptional gene regulation, metabolic processing, stress adaptation and microbial pathogenesis (Mandin and Gottesman, 2010; Michaux et al., 2014). Overall, sRNA plays an influential role in the biofilm life cycle.
Tan et al. (2018), published a review interpreting the “molecular mechanisms underlying agr quorum sensing and the regulation of agr expression.” They, deemed that agr system is as an attractive therapeutic target for controlling S. aureus by blocking or interfering with the agr system. For detail pathways, see Figure 3.
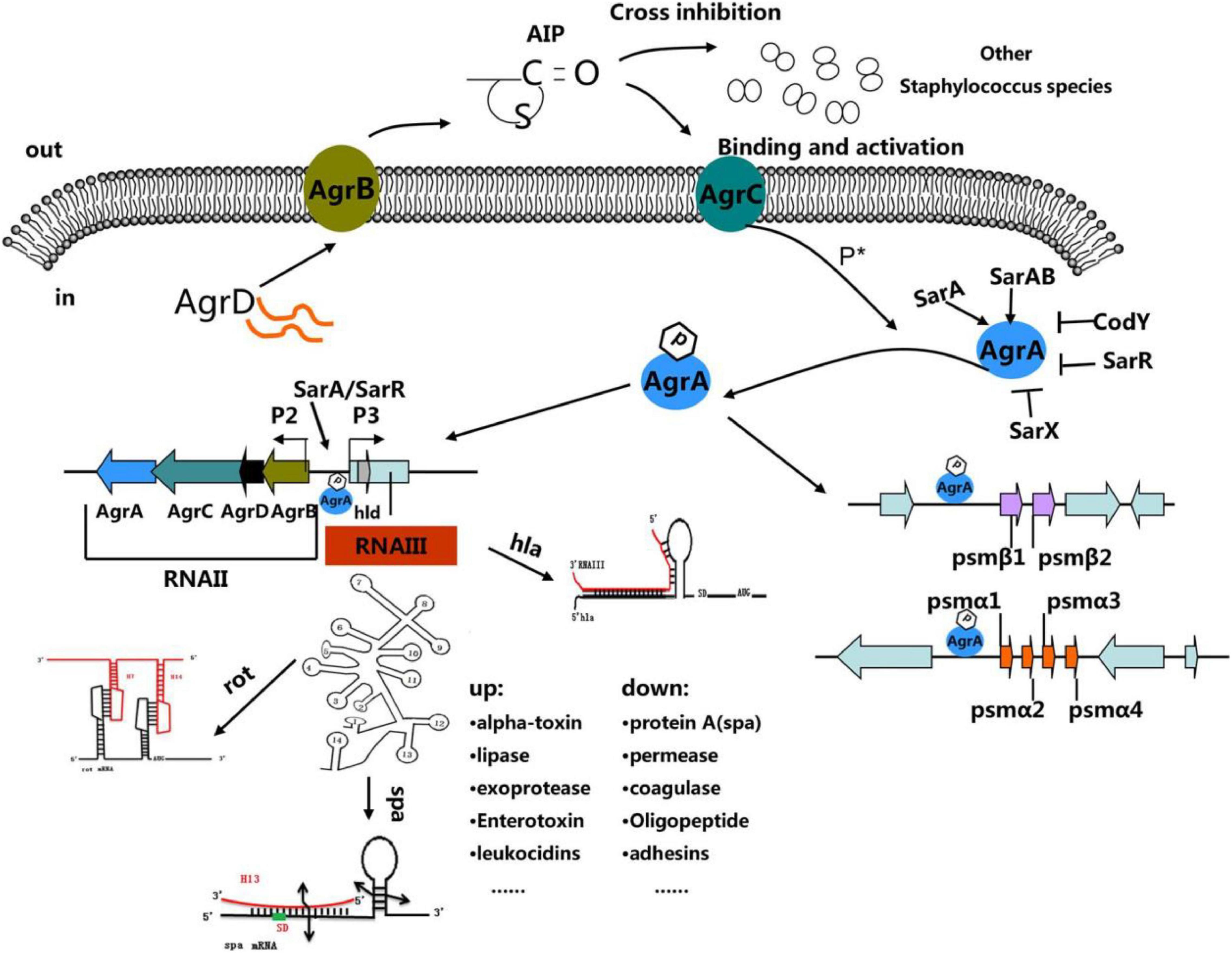
Figure 3. “The Staphylococcus quorum-sensing system. The agr locus is composed of divergent transcripts designated RNAII and RNAIII, driven by promoters P2 and P3, respectively. The AIP signal is produced from the AgrD precursor, while the membrane-localized enzyme AgrB participates in the maturation and export of the AIP. At a critical threshold concentration, AIP activates the two-component signal transduction system, AgrC–AgrA, and causes the phosphorylation of AgrA. Once phosphorylated, AgrA binds to the P2 and P3 promoter regions, as well as promoters PSM-a and PSM-b, resulting in agr system transcription. RNAIII encodes the delta-toxin encoding gene hld, and 14 stem-loop motifs. These domains regulate the expression of numerous virulence factors. Other regulators (such as SarA, SrrAB, SarR, and SarX) can enhance or inhibit agr activity (Tan et al., 2018).”
Recently, Yan and Wu (2019) reviewed the transcriptomic data and validated the possibility to reverse the biofilm formation in P. aeruginosa through QS. The QS system of P. aeruginosa constitutes 3 systems including its own specific QS system (PQS). For more details, see Figure 2, Yan and Wu (2019). Besides, Qvortrup et al. (2019), in a recent review, described the “anti-biofilm agents developed on the basis of mechanistic understanding of biofilm formation” and highlighted the process of biofilm formation and the molecular mechanism used as targets for the development of anti-biofilm chemicals with special reference to A. baumannii, E. coli, and P. aeruginosa.
Biofilm formation by P. aeruginosa can be initiated via the adhesive action of a number of components such as “flagella (O’Toole and Kolter, 1998), type IV pili (O’Toole and Kolter, 1998; Déziel et al., 2001; Chiang and Burrows, 2003), cup fimbria (Vallet et al., 2001), extracellular DNA (Whitchurch et al., 2002), and Psl polysaccharide (Ma et al., 2006).” Most of the biofilm matrix components such as polysaccharide, exopolysaccharide, alginate, CdrA, type IV pili, and Cup fimbriae are positively controlled by c-di-GMP (Fazli et al., 2014), which is a negative controller of motility of P. aeruginosa (Simm et al., 2004). It is witnessed that biofilm infection with P. aeruginosa can be treated by a reduction of the bacterial c-di-GMP content (Christensen et al., 2013). Quorum sensing (QS) plays an important role in the formation and persistence of P. aeruginosa biofilms. Although several compounds are controlled by QS, extracellular DNA that contributes toward antimicrobial resistance and the stability of biofilms is an important factor (Allesen-Holm et al., 2006; Chiang et al., 2013). The other responsible molecules is the rhamnolipid in the development of biofilm formation by phagocytizing immune cells (Pamp and Tolker-Nielsen, 2007).
In the case of E. coli, formation of biofilm is regulated by several adhesins and extracellular matrix components. The proteinaceous curli fibers are a major component required for the initial attachment to the host cells for biofilm formation (Olsén et al., 1989; Nasr et al., 1996; Prigent-Combaret et al., 2000; Chapman et al., 2002; Serra et al., 2013). The other important components are type 1 and P pili (Pratt and Kolter, 1998; Schembri and Klemm, 2001; Niba et al., 2008). It is evidenced that absence of FimH molecules significantly reduces adhesion capability both in vitro and in vivo (Langermann et al., 1997; Mulvey et al., 1998). The other factor is P pili (contain PapA subunits), that anchors the adhesin PapG (Gong and Makowski, 1992; Bullitt and Makowski, 1995), enables to bind E. coli to host epithelial cells (Busch et al., 2015). Beside the proteinaceous component, it is also evidenced that the exopolysaccharides, poly-GlcNAc (PGA), and colanic acid (Danese et al., 2000; Wang et al., 2004; Serra et al., 2013; Subashchandrabose et al., 2013; Besharova et al., 2016) and extracellular DNA are the key components of E. coli biofilm formation (Devaraj et al., 2015). In E. coli. Like P. aeruginosa, PGA and curli fimbria are also positively regulated by c-di-GMP.
The mechanisms of biofilm formation in A. baumannii is not well studied. Nevertheless, a few adhesins and extracellular components are detected for their role in biofilm formation. Csu pili and the OmpA are the outer membrane protein (Dorsey et al., 2002; Tomaras et al., 2003; Gaddy et al., 2009) which binds to epithelial cells to form the biofilm. BAP, another important protein, is responsible for cell-cell adhesion and in maintaining the structure of mature biofilms (Loehfelm et al., 2008). Furthermore, this bacterium can produce the exopolysaccharides alginate and poly-β-1,6-N-acetylglucosamine (PNAG) which can serve as an important constituent of the biofilm matrix (Lee et al., 2008; Choi et al., 2009). So far, the role of c-di-GMP signaling in A. baumannii, biofilm formation has not been documented. While QS has been proven to regulate the formation of biofilm (Anbazhagan et al., 2012), A. baumannii have an “AHL-based QS system with AbaI functioning as the AHL synthase and AbaR functioning and the AHL receptor.” Niu et al. (2008) observed that an AbaI mutant (could not produce AHL), have imperfections in the late stage of biofilm formation. Addition of AHL developed an increased expression of Csu pili, as well as stimulation of biofilm formation (Luo et al., 2015).
Antimicrobial Peptides (AMPs)
Antimicrobial peptides are a widespread feature of the innate immune systems, a principal defense system, present in almost all living organisms ranging from fungi to higher plants and animals. So, both eukaryotic and prokaryotic cells produce AMPs naturally as part of their immune/immune system (Rossi et al., 2008). Generally, a peptide’s primary role is to kill the invading pathogens (bacterial, fungal, viral, parasitic, etc.) through modulating the innate immune response of the host. However, the activities vary in different host systems depending on the organism and its location in that organism. Bacteria were among the first source of AMPs (called bacteriocins), which could be a new therapeutic source for various human ailments. Bacterial AMPs do not defend against infection by other species of bacteria; they kill other species (target) of bacteria as a source of nutrients or to decrease competition for nutrients. Some AMPs are narrow spectrum, most of the broad-spectrum activities target various bacterial enzymes, pathways, or structures (like lipid bilayers).
Distinctive characteristics of most AMPs are their small size containing 15 to 30 amino acids along with positively charged ones, and that they target the cell membrane (Rossi et al., 2008; Melo et al., 2009). As a result, positively charged peptides are attracted to cell membranes compared to poorly (negatively) charged ones by bacteria and biofilm sites. AMPs lead to increased antimicrobial activity in bacteria that are active and slow-growing in biofilms (Ma et al., 2012; Xu et al., 2014; Tiwari et al., 2015) and easily kill them (Jorge et al., 2012). However, in low-intensity environments, AMPs can be bacteriostatic (Beloin et al., 2014). AMPs are also classified depending on their secondary structure in liquid media (Dalton and March, 1998; Stoica et al., 2017). Some are mainly beta-pleated-sheet structures whereas others are primarily alpha-helical. In both the cases, cysteines form an intramolecular disulphide bridge, that stabilizes the structure, and helps AMPs cross the cell membrane (Whittaker et al., 1996; Garrett et al., 2008). Cell adhesion is favored due to hydrophobic interactions of AMPs (Costerton et al., 1999; Reffuveille et al., 2014; De La Fuente-Núñez et al., 2016). The antibacterial activity depends on the balance between the charge density, the hydrophobic character, and the length of the polypeptide chain (Stewart, 1996; Mah and O’Toole, 2001; Arciola et al., 2012). Increasing the number of basic amino acids or altering their configuration in the peptide chain can affect the secondary structure of AMPs, and thus their antibacterial activity. Insects are known to be highly resistant to bacterial infections. They can produce many proteins and peptides as the first line of defense against microbial infection (De La Fuente-Núñez et al., 2016).
Antibacterial Peptides From Insect Source
Since the time AMPs were first discovered forty years back, researchers have attempted to relate amino acid sequences for antibacterial activities in order to get better peptides (Figure 4). Bioassay-guided purification seems to be the best strategy. However, information on bioassay-guided purification is rare. Figure 4 provides a schematic presentation of a systematic approach for identification and characterization of bioactive peptides for developing potent antimicrobial peptides. The regular methodology is to substitute amino acids in the arrangement to control cationic charge and hydrophobicity and obtaining about 5-10 peptides that are tasted for antimicrobial activity. For the parent sequencing, the derived peptides record moderate function against antimicrobial activity (Fjell et al., 2012; Haney et al., 2012; Kim H. et al., 2016; Agrawal et al., 2018), or may be having lower toxic effect, and ∼1–4 of these peptides may be concentrated further to determine their mechanism of action. A large number of sequences are registered in the databases. For example, >3000 sequence are present in AMP Database (Wang et al., 2016) and >17000 sequences in DRAMP database (Fan et al., 2016).
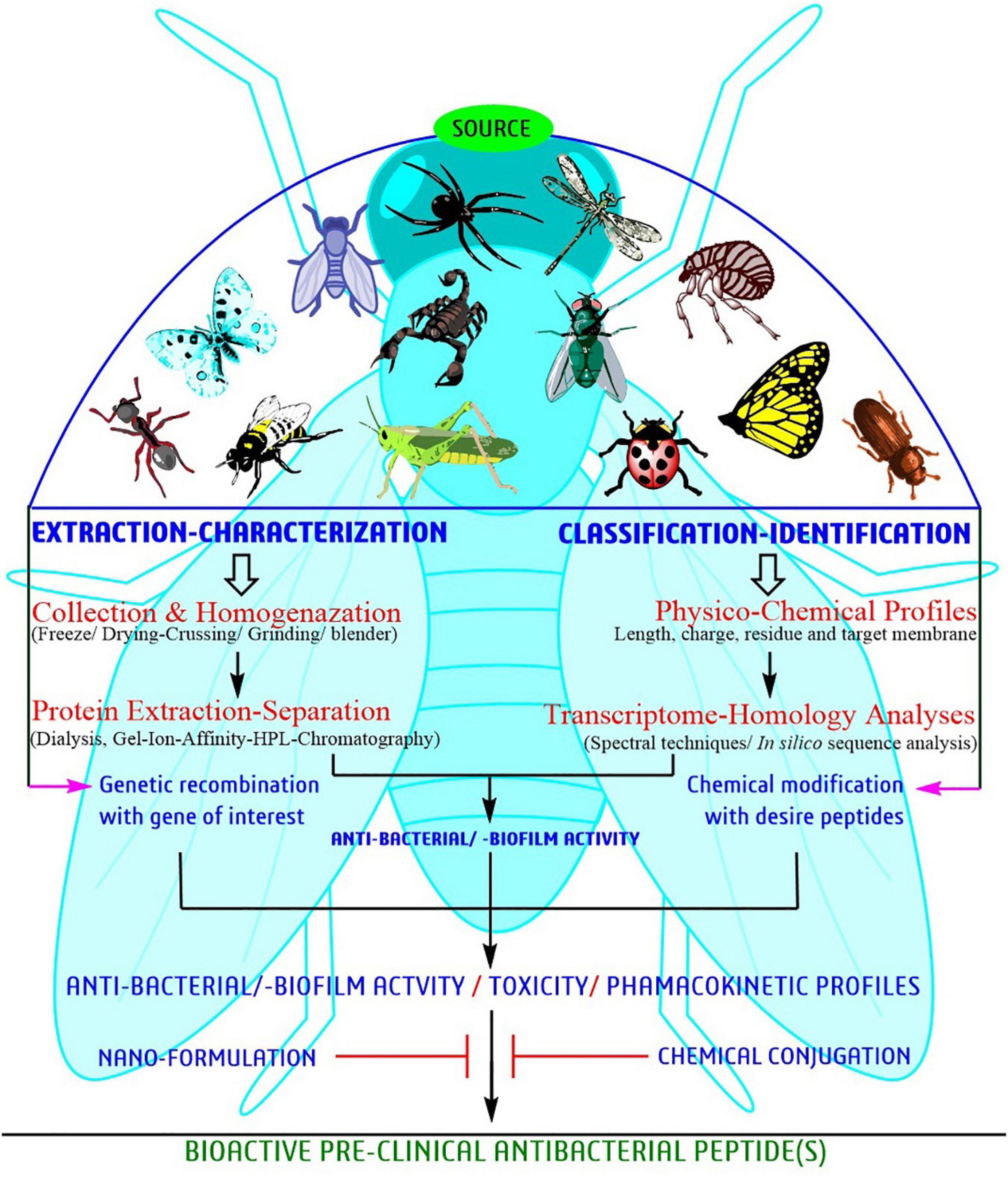
Figure 4. A systematic approach of bioactive peptide identification and characterization toward development of potent AMPs against biofilm-producing pathogens.
The development of high-throughput techniques, like next-generation sequencing and transcriptome analysis, is helping to find new antimicrobial peptides in different organisms. The process involves total RNA extraction, construction of RNA library, sequencing, and de novo assembling of transcriptome (Haas and Zody, 2010). In addition, different strategies of in silico analysis are being used to identify peptides of therapeutic interest, predict their three dimensional structures and enrich the search for antimicrobial peptides based on physicochemical properties and nucleotide sequence similarity (Lata et al., 2007; Amaral et al., 2012; Slavokhotova et al., 2015). Chemical synthesis of the identified/predicted AMPs can be carried out through solid-phase chemical synthesis and purified via RP-HPLC and obtained with differential purity levels. Mass spectroscopic technique is useful to determine the molecular mass of the molecules. Such molecules can be tested against selected microbes for antimicrobial activity. Before being finalized to be used as a drug, the molecules can be tested for their cytotoxic and human erythrocyte hemolytic activity. However, the molecular mechanism of these peptides is largely not known till date.
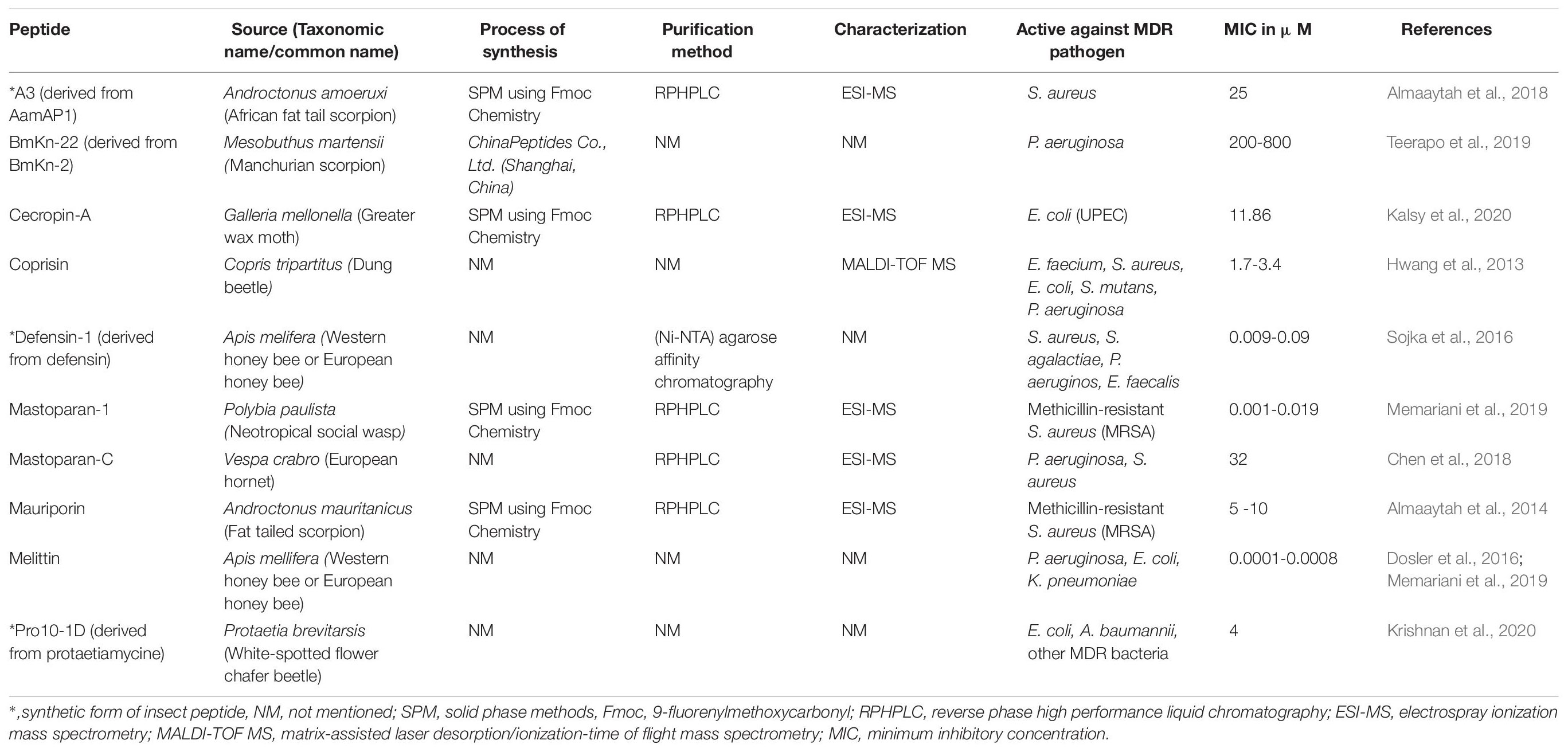
About 103 antimicrobial peptides isolated from insect sources are listed in Supplementary Table 1. The majority of AMPs are derived from insects like Acalolepta luxuriosa, Apis mellifera, Bombyx mori, Galleria mellonella, Heterometrus spinifer, Holotrichia diomphalia, Hyalophora cecropia, Oxysternon conspicillatum, Pandinus imperator, and Sarcophaga peregrine. These antimicrobial compounds are found to be effective against a wide range of bacteria both from Gm -ve and Gm +ve, including MDR strain (Supplementary Table 1). The MIC in most insect peptides is quite potent (0.02 to 20 μg/mL). Many of them are known to prevent the formation of biofilms in ESKAPE bacteria (Table 1). Sojka et al. (2016) isolated defencin-1 (derived from defensin) from A. melifera with strong antibacterial activity against both planktonic and biofilm cells of the bacterial pathogens such as S. aureus, S. agalactiae, P. aeruginosa and E. faecalis (MIC = 0.009-0.09 μM). Another broad-spectrum antimicrobial peptide coprisin isolated from Copris tripartitus has antibiofilm property against a wide range of pathogenic bacteria such as E. faecium, S. aureus, E. coli, S. mutans, P. aeruginosa (MIC = 1.7-3.4 μM) (Hwang et al., 2009). Melittin, isolated from A. mellifera has broad spectrum activity against Gm -ve, P. aeruginosa, E. coli, and K. pneumoniae (MIC = 0.0001-0.0008 μM) (Dosler et al., 2016; Memariani et al., 2019). Mastoparan-1 is a narrow spectrum AMP isolated from Polybia paulista and effective against methicillin-resistant S. aureus (MRSA) (Memariani et al., 2018). Table 1 provides information on a list of potent AMPs reported to inhibit biofilm.
Oxysterlins 1, 2, and 3, isolated from the dung beetles O. conspicillatum (Toro Segovia et al., 2017), similar to Cecropin A, Aedesin, Lucilin and HKABF, were found selective for Gm –ve bacteria and efficiently kill multidrug resistant (MDR) strains, including E. coli ESBL, E. cloacae, S. typhi, and E. coli with MIC values between 3.12 and 50 μg/mL. Cecropins were first isolated from the silk moth H. cecropia. Insect cecropins also have other names such as bactericidin, lepidopteran, sarcotoxin etc. “Cecropins can lyse bacterial cellular membranes and can also inhibit proline uptake as well as cause leaky membranes (Moore et al., 1996; Bechinger and Lohner, 2006).” Cecropin B is found to have the strongest antibacterial activity (Srisailam et al., 2000) and proved to decrease the load of E. coli in a rat model (Giacometti et al., 2001). A recombinant cecropin D was found to be active against both Gm + and Gm -ve bacteria (Guo et al., 2012). Most probably the C-terminal lysine residue is responsible to increase the antibacterial activity due to activated phosphorylation (Park et al., 2013).
Mechanisms of Antibiofilm Peptides With Special Reference to ESKAPE Pathogens
Insects are capable to produce variety of antimicrobial proteins and peptides most of which are smaller in size and contain cationic groups. Generally, antimicrobial peptides are categorized according to their structure such as α-helical, β-sheet, loop, or extended constructions; however, some does not fit into any specific class due to the presence of both α-helical and β-sheet domains. Thus, the peptide structures can be observed through membrane interactions (Jenssen et al., 2006). For example, indolicidin like neutrophil peptides from bovine is unstructured in an aqueous medium but become boat-like after interaction with the membrane mimicry surface of sodium dodecyl sulfate and dodecyl phosphocholine. The antibacterial mechanism of each peptide depends on its character/composition and physiological conditions. A peptide contains many positively charged residues that enable them to interact electrostatically with negatively charged cell surface molecules of surface peptidoglycan. Overall, isolated short cationic amphiphilic host-defense peptides are responsible for antibacterial activity through direct cell killing and immunomodulatory action (Zasloff, 2019; Mukhopadhyay et al., 2020). In general, peptides are highly effective against Gm -ve bacteria than Gm +ve bacteria due to differences in cell wall composition. Additionally, non-specific inhibition mechanisms target membrane, intracellular biomolecules, and oxidative pathways (Wimley and Hristova, 2011; Band and Weiss, 2014). The therapeutic potential of peptide antibiotic drugs lies in their ability to kill bacterial cells effectively without exhibiting significant cytotoxicity toward mammalian cells. Overall, the activity and mode of peptides are variable due to their structure like β-hairpin or loops, β-sheet and amphipathic α-helical constituents (Gomes et al., 2018; Hollmann et al., 2018).
Several models have been projected to elucidate the bacterial cell membrane disruption out of which, the Barrel-stave, Carpet model and Toroidal-pore are the most accepted models (Zasloff, 2019). Mechanically, peptides disrupt/kill/inhibit the bacterial growth through some essential steps such as attraction, attachment, insertion, and inhibition of bacterial biofilm formation. In the attachment stage, peptides penetrate the entire depth of bacterial surface polysaccharide and further join with the lipopolysaccharides in Gm -ve bacteria and teichoic/lipoteichoic acid in Gm +ve bacteria (Figure 4, left side). The right side of Figure 4 describe a computational model with a molecular docking approach graphically targeting LuxR genetic factor of S. aureus with insect-derived antibiofilm peptide Pro10-1D. Several computational methods like molecular docking and dynamic simulation are the most widely used, cost-effective programs to elucidate several unknown features, mechanisms, and potency targeting individual biofilm-associated targets.
Some Exclusive Examples of Antibiofilm Peptides Derived From Insects
Basically, four groups of AMPs are found in insects based on their structure and amino acid composition. They include proline rich peptides (e.g., drosocin, apidaecin, and lebocin), α-helical peptides (e.g., moricin and cecropin), cysteine rich peptides (e.g., defensin and drosomycin), and glycine-rich proteins (e.g., attacin and gloverin) (Otvos, 2000; Bulet and Stocklin, 2005). The major components of innate immunity in numerous groups of organisms including insects are cysteine-rich peptides (Pushpanathan et al., 2013; Slavokhotova et al., 2017) and are known for their molecular action to inhibit biofilms. In addition to these several new class of insect peptides is still isolated from several insects; diptericins, drosomycin metchnikowin and ponericins are most investigated antibacterial insect peptides. On the other hand, most insect peptides proved antibacterial activity in planktonic compared to biofilm. Overall, the glycine and proline-rich peptides are significantly active against Gm -ve bacteria (Wu et al., 2018).
Defensins are very small (<4 kDa) antibacterial peptides containing three intramolecular disulphide bridges by the help of six cysteine residue and are found in all living organisms. They are classified in to three families based on their structural characteristics, such as classical defensins, β-defensins and defensins of insects (Ganz and Lehrer, 1994). Vertebrate defensins in innate immunity have attracted many workers (Ding et al., 2009; Lehrer and Lu, 2012; Jarczak et al., 2013; Zhu et al., 2013; Wilson et al., 2013; Zhao and Lu, 2014). Insect defensins contain a cationic group of 34–51 peptide residues with six conserved cysteines. It basically inhibits Gm +ve bacteria, including S. aureus. Some insect defensins also perform their biological activity (Maget-Dana and Ptak, 1997). The AMPs lucifensin isolated from Lucilia sericata contain 44 amino acid residue (Čeřovský et al., 2010). However, the molecular mechanism of cysteine rich peptides is not well established against biofilms.
The other important classes of insect peptides are α-helical peptides (e.g., moricin and cecropin). Cecropins were first isolated from the pupae of the cecropia moth H. cecropia, whence the term cecropin was derived. Basically, different groups of cecropins are found such as cecropins A, B, C, D, E, and F among which Cecropin D with 37 amino acids is the major cecropin. They function against both Gm -ve and Gm +ve bacteria as well as fungi (Kockum et al., 1984; Tryselius et al., 1992; Moore et al., 1996; DeLucca et al., 1997; Cavallarin et al., 1998; Ekengren and Hultmark, 1999; Vizioli et al., 2000). Most cases of cecropins at the C-terminal end are altered to amide, and amidation is essential for the interaction of cecropins with liposomes, and hence the antimicrobial property (Li et al., 1988). The basic molecular mechanism of cecropins is described recently by Mukherjee on E. coli biofilm (Figure 5; Mukherjee, 2020).
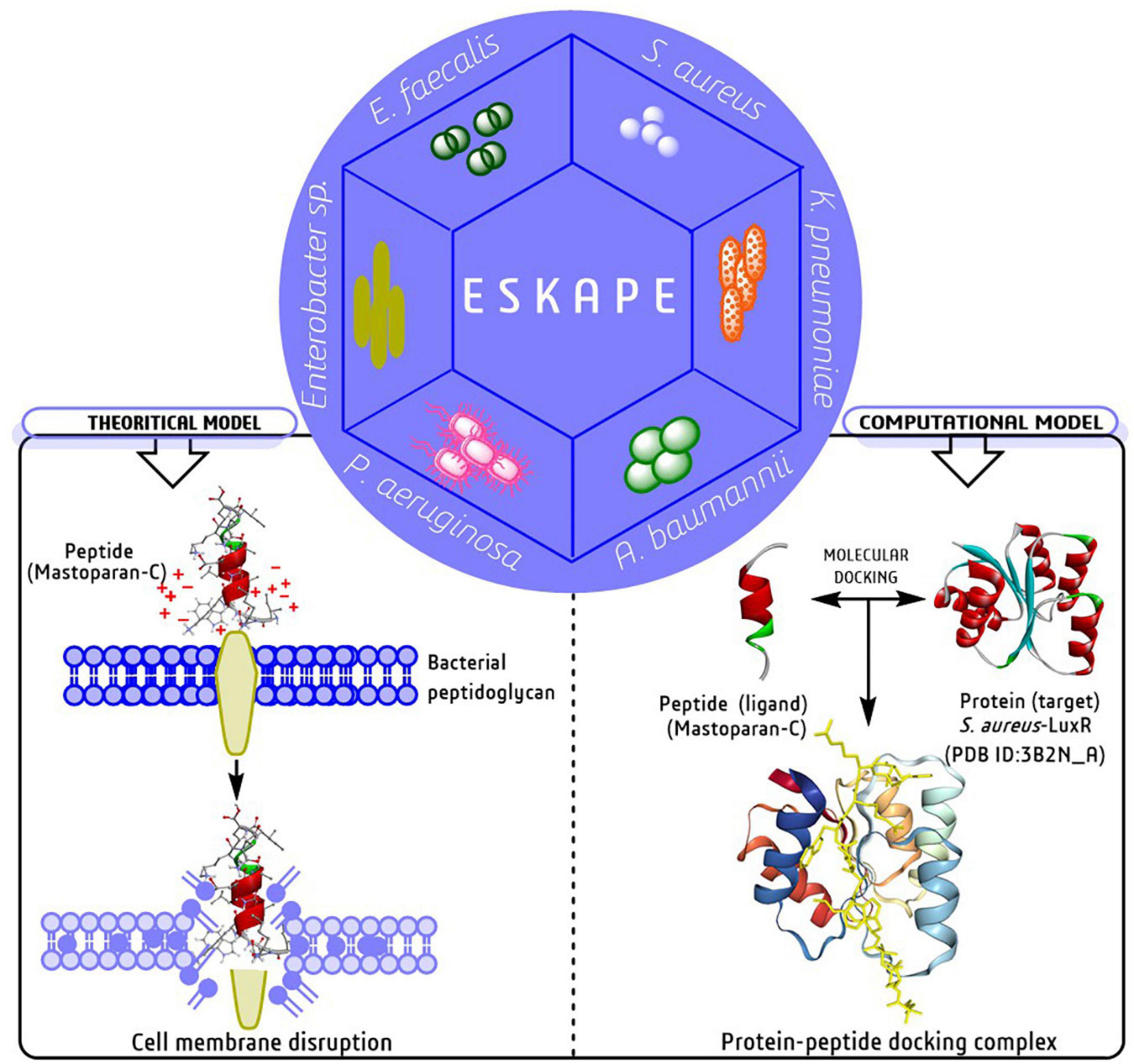
Figure 5. Graphical representation of membrane disruption mechanism of peptides against ESKAPE pathogens in both theoretical and computational models (Chen et al., 2018; Silveira et al., 2021).
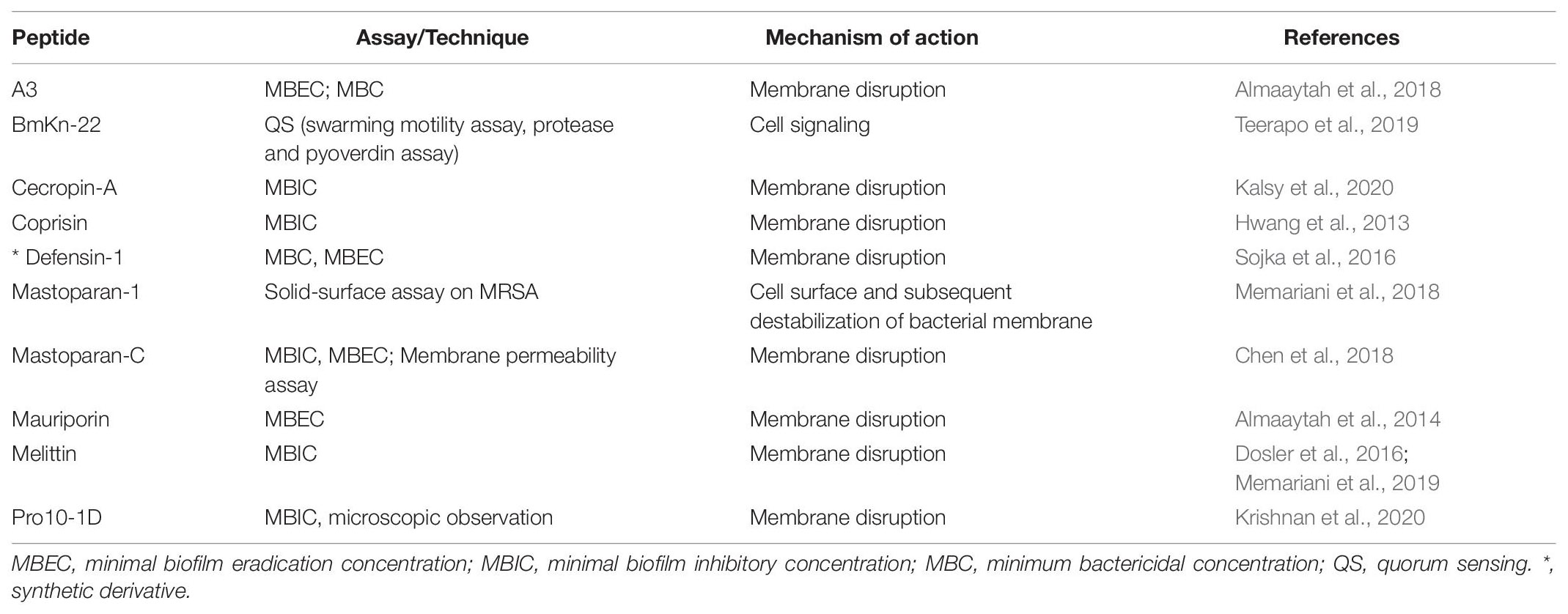
Glycine-rich proteins e.g., attacin and gloverin are the other important insect-derived peptides. Attacin (MW-20-23 kDa), isolated and purified from the hemolymph of the immunized bacteria H. cecropia and its isoforms, can be divided into two groups: the basic attacins (A–D) and acidic attacins (E and F) (Hultmark et al., 1983). Both are similar in amino acid sequence but acidic attacin contains higher proportion of Asp residues compared to basic attacin. Besides, both are encoded by two different genes (Kockum et al., 1984; Sun et al., 1991). Attacins are synthesized as inactive pre-proproteins with a signal peptide, a pro-peptide (P domain), and an N-terminal attacin domain followed by two glycine rich domains (G1 and G2 domains) (Sun et al., 1991; Hedengren et al., 2000). Attacin-coleoptericin is a hybrid protein with greater antibacterial activity against E. coli, Burkholderia glumae, and B. subtilis related to either attacin or coleoptericin alone (Lee et al., 2013). But leucine-rich attacins do not exhibit antimicrobial activity. Attacins mostly act by blocking the synthesis of the major outer membrane proteins of Gm -ve bacteria, as a result disturbing the integrity of the cell wall and causing the bacteria to grow in long chains (Carlsson et al., 1998). Attacin causes increased permeability of outer membrane of E. coli and inhibits synthesis of outer-membrane proteins at the transcriptional level without entering the inner membrane or cytoplasm (Carlsson et al., 1998). Biofilm-associated bacteria are more resistant to antibiotics than live (planktonic) cells. Despite the availability of several reports on the antibacterial property against Gm –ve bacteria with isolation of attacin from different insect species, no such data is available on biofilm which should attract priority. The antibiofilm mechanism of action, assay and techniques associated with antimicrobial peptides from insect sources are summarized in Table 2.
At present, most insect peptides can be synthesized in laboratory conditions and identified the potency through computational screening at an early stage. Indeed, the advanced in silico next-generation sequencing, transcriptome profiling, de novo assembly directed toward locating signature bioactive peptides (Kim I.-W. et al., 2016; Prajapati et al., 2020). Briefly, in silico analysis, able to find out the vital function with their encoded genetic factors from the large sequence (Toro Segovia et al., 2017). On the other hand, the modification of natural peptides in laboratory conditions also provides massive success in achieving the desired goal of improving activity and pharmacokinetics profiles. For example, several glycosylated and unglycosylated analog of the insect peptide apidaecin and drosocin were proposed by Gobbo et al. (2002) where synthesized unglycosylated containing intrachain disulfide bond have more potential activity than glycosylated derivatives (Gobbo et al., 2002). Similarly, A3 (AamAP1), BmKn-22 (BmKn-2), defensin-1, (defensin), Pro10-1D (protaetiamycine), several other synthetic derivatives, namely, LL1037, LL7-31 (LL-37), 1018 (bactenecin), AS10 (CRAMP), battacin (lipopeptides), BMAP27-melittin (melittin), CAMA (cecropin A and melittin A), NRC-16 (pleurocidin), P10 (P60.4Ac), P318 (CRAMP), are some improvised insect peptides toward control of biofilm (De La Fuente-Núñez et al., 2016; Almaaytah et al., 2018; Krishnan et al., 2020).
Antimicrobial Peptides: Synergy Studies With Antibiotics
Biologically, the synergistic approach of non-homologous insect peptide(s) and conventional antibiotics/antibacterial agents showed potential against resistance pathogens. Overall, the interaction of peptide and membrane is directly promotional to antibiofilm/antibacterial activity (for overall inhibition mechanisms refer Figure 4). Mechanically, a peptide kills bacteria through different routes, including disruption by electrostatic or hydrophobic interaction, interference of bacterial metabolism, targeting cytoplasmic components, etc. Currently, four models such as, barrel-stave, carpet, toroidal-pore, and disordered toroidal-pore are used to predict the modes of action of peptides and all models required a threshold concentration to conduct antibacterial activity (Hollmann et al., 2018; Wu et al., 2018; Manniello et al., 2021; Silveira et al., 2021). In general, the activity and mode of peptides are variable due to their structure (Gomes et al., 2018; Hollmann et al., 2018). For example, the potential cecropin A or CecA peptide isolated from G. mellonella recorded an antibiofilm activity; but, when applied synergistically with nalidixic acid, showed significant antibiofilm activity at lower concentration against uropathogenic-cum-biofilm producing E. coli (Kalsy et al., 2020).
De La Fuente-Núñez et al. (2012) have shown that AMP 1037 stimulates cell proliferation of P. aeruginosa PA2204 but does not affect the motility and biofilm formation (De La Fuente-Núñez et al., 2012). Antimicrobial peptides NA-CATH: ATRA1-ATRA1, a synthetic cathelicidin, inhibited S. aureus biofilm form, and peptide LL-37 regulated P. aeruginosa biofilm formation when used at levels below MIC (Overhage et al., 2008; Dean et al., 2011). These AMPs inhibit the expression of coded proteins in the genes involved in the formation of biofilm. In P. aeruginosa, genes contain type IV pili code, rhamnolipid synthesis, quorum sensing, and flagella assembly (Overhage et al., 2008). However, a few AMPs have specific antimicrobial properties; for example, lactoferrin chelates iron and inhibits biofilm formation by P. aeruginosa. Binding AMPs to extracellular DNA may improve the detection of biofilms (Das et al., 2010).
The S. aureus biofilm formation regulatory system (GraRS) perform a crucial role in resisting microorganisms in AMPs (Herbert et al., 2007). This resistance was inverted when AMPs were added in mixtures with other antimicrobial compounds. AMPs from a variety of sources, when combined with standard prescribed antibiotics, effectively prevent the formation of biofilm by P. aeruginosa (Eckert et al., 2006; Minardi et al., 2007; Herrmann et al., 2010; Hirt and Gorr, 2013; Dosler and Karaaslan, 2014). Combination of A3 (isolated from Androctonus amoerux) with conventional antibiotics performed a synergistic mode of action when compared with the natural scorpion venom peptide. The combination of A3 with conventional antibiotics may be used as a possible new treatment strategy against MDR and biofilm forming bacteria (Almaaytah et al., 2018). Combination of BmKn–22 peptide with azithromycin also reduces P. aeruginosa biofilms (Teerapo et al., 2019). Kalsy et al. (2020) have synergistically reported that insect antimicrobial peptide cecropin A (CecA) can reduce planktonic and sessile biofilm-forming UPEC cells, either alone or in combination with the antibiotic nalidixic acid (NAL) (Figure 6). Coprisin also performed antibiofilm activity alone and in mixtures with antibiotics (Hwang et al., 2013).
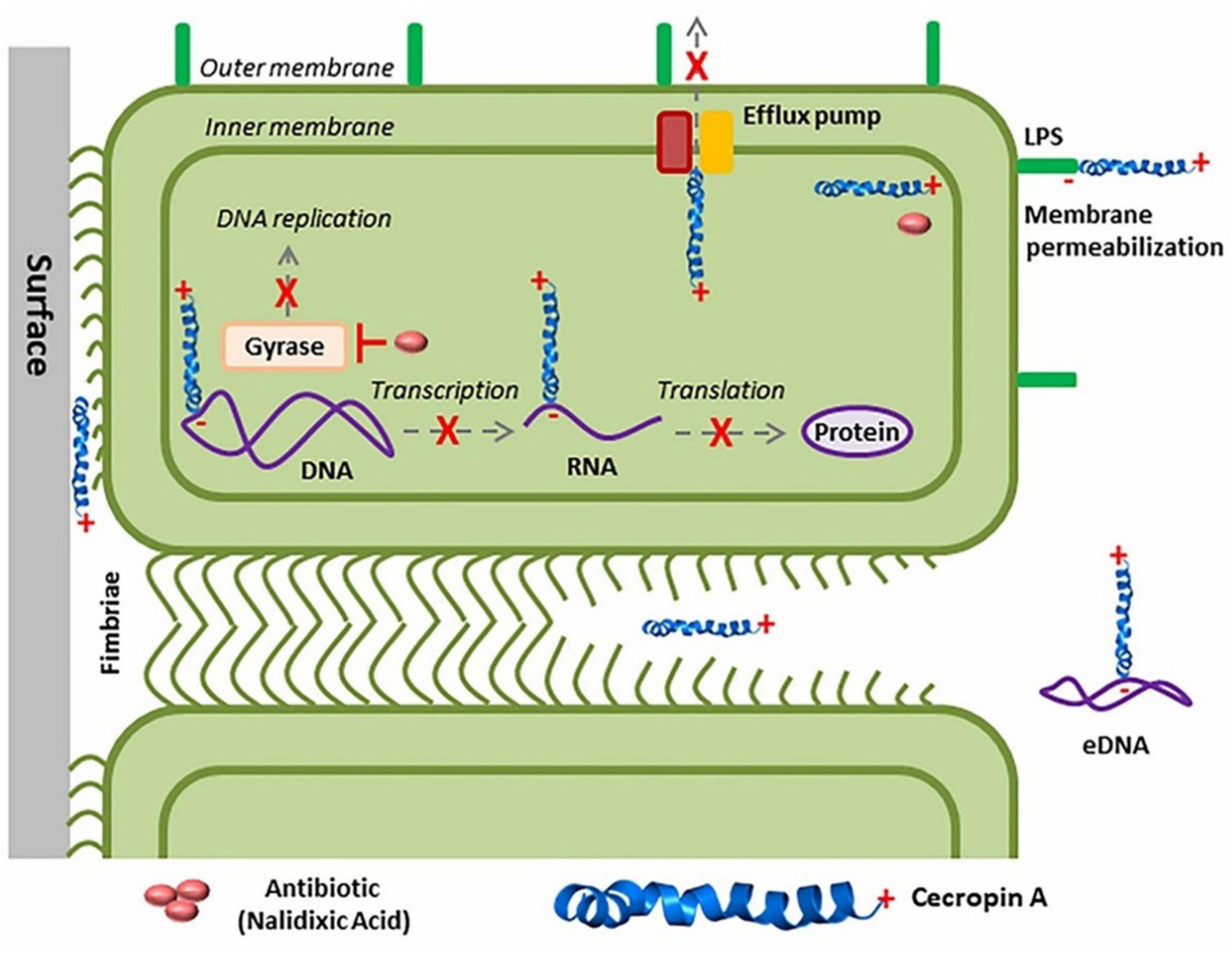
Figure 6. Multi-target mechanism of cecropin A (CecA) action in uropathogenic Escherichia coli (UPEC) biofilms. CecA interacts with LPS to permeabilize bacterial membrane enhancing the diffusion of nalidixic acid (NAL) into the cell. CecA and NLA then bind to different molecular targets in bacteria to disrupt UPEC biofilms (Adopted from https://naturemicrobiologycommunity.nature.com/posts/59576-in-search-of-new-anti-biofilm-agents-from-insects, Mukherjee, 2020).
Akbari et al. (2019) observed interesting synergistic effects of peptide melittin when combined with existing antibiotic doripenem against A. baumannii (61.5 fold reduction on MIC) as well as against P. aeruginosa (31.5 fold reduction on MIC). Same group also demonstrated two new melittin-derived peptides MDP1 and MDP2 with potent antibacterial activity against MDR S. aureus and E. coli (Akbari et al., 2018). Melittin also inhibits MRSA strains (Choi et al., 2015) and later proved to eradicate MRSA-infected mice (Moreno and Giralt, 2015). Another recent finding with AMP against MDR S. aureus is clavaspirin (Lee et al., 2018). Indeed, few synergistic effects of AMPs along with conventional antibiotics were demonstrated (Akbari et al., 2019) but quantitative methods are rarely used, or in vivo validation is not performed. Another synergetic study is by Wu et al. (2017) with AMP DP7 on MDRs (S. aureus, E. coli) using quantitative polymerase chain reaction.
In vivo Studies Related to Antimicrobial Peptide Research and Insect
In vivo studies has shown the effect of interactions on the biofilms of MRSA in which nisin was mixed with daptomycin/ciprofloxacin; indolicidin and teicoplanin, cecropin (1-7), melittin A (2-9), and amide (CAMA) with ciprofloxacin (Mataraci and Dosler, 2012; Dosler and Mataraci, 2013). The combination of cationic peptide IB-367 and LZD in antibiotic lock technology eliminated S. aureus biofilms in CVCs (Ghiselli et al., 2007). Significant reduction of biofilm-associated S. aureus in vascular grafts was observed when sub-MIC levels of vancomycin was combined with the lipopeptides Pal-Lys-Lys-NH2 and Pal-Lys-Lys (54). Some AMPs have a wide range of antibiotic activity, such as peptide 1018, which is blocked or degraded by guanosine [pentaphosphate (p) ppGpp] and is essential for biofilm formation. In the lower extremities, peptide 1018 inhibited the formation of biofilm but eliminated pre-packaged biofilms when applied at high concentrations (De La Fuente-Núñez et al., 2014). AMPs combined with generic antibiotics may be a better alternative than antibiotics alone. The interaction of AMPs with antibiotics against biofilm viruses should draw the attention of researchers to study the mechanical properties of these compounds.
Clinical Trials Related to Insect Derived Antimicrobial Peptides
Out of the 208 drugs approved during the last five-years (2015-2019) (FDA-drug approval records), 15 (approximately 7%) are peptide-based drugs (De La Torre and Albericio, 2020). As a result, the peptide-based drug market/business crossed one billion USD. Newer strategy and agents are urgently needed to counter-attack the biofilm-producing and multidrug-resistant pathogenic bacteria. At the same time, the potency, non-specific antimicrobial inhibition mechanism and other advantages over conventional antibiotics are rapidly gaining attention as an intellectual source toward the development of peptide-based antibacterial drugs (Browne et al., 2020; Magana et al., 2020). Bacitracin, daptomycin, colistin, gramicidin D, oritavancin, polymyxin B, teicoplanin, telavancin, vancomycin, etc., are some well-known peptide based-successive and revolutionized antibacterial (Browne et al., 2020). Similarly, many antimicrobial peptides are entering clinical trials with several positive responses and withdrawn from different clinical trial stages due to toxicity, lack of in vivo stability pharmacokinetic properties (Butler et al., 2013; Koo and Seo, 2019). Herein, relevant clinical status reports of ten antibacterial peptide based drug information such as origin, mechanism of action, route of administration, resisted clinical trials number are tabulated, particularly those currently presented in clinical-II onward (Koo and Seo, 2019; Browne et al., 2020).
Briefly, as per potency, several drugs are now in a different stage of clinical validation (Table 3). As we know, clinical proof is a long-term process that focuses on the lead candidates’ dose and adverse effects. MU1140, D2A21, Arenicin (AP139), AP138, Novarifyn, hLF1-11, Wap-8294A2 (10, lotilibcin) like peptide-based antibacterial drug candidates are entering in the clinical trial-1 study. Similarly, EA-230, CZEN-002, Delmitide, Ghrelin, C16G2, DPK-060, PAC113, LTX-109, OP-145, LL-37, Novexatin like candidates in trail-II and D2A21, SGX942, PXL01, POL7080, POL7080, Ramoplanin are in trail-III stage. Overall, most of the drugs failed in phase-II and III due to their inability to reach the required clinical endpoints. For example, omiganan, pexiganan and surotomycin like lead drugs are withdrawn in clinical trial-III due to lesser potency. Similarly, iseganian, talactoferrin (for mortality) and murepavadin (significantly for higher renal toxicity), were stopped/to be withdrawn due to serious adverse effects in comparison with control (Divyashree et al., 2019; Koo and Seo, 2019; Theuretzbacher et al., 2020). Additionally, NVB-302, XMP-629, Neuprex, and Friulimicin B like candidate drugs failed to achieve the required clinical features. Thus, clinical acceptance is always the last part of drug development where isolation/identification of such potential peptide candidates plays a significant role in initiating that drug development strategy.
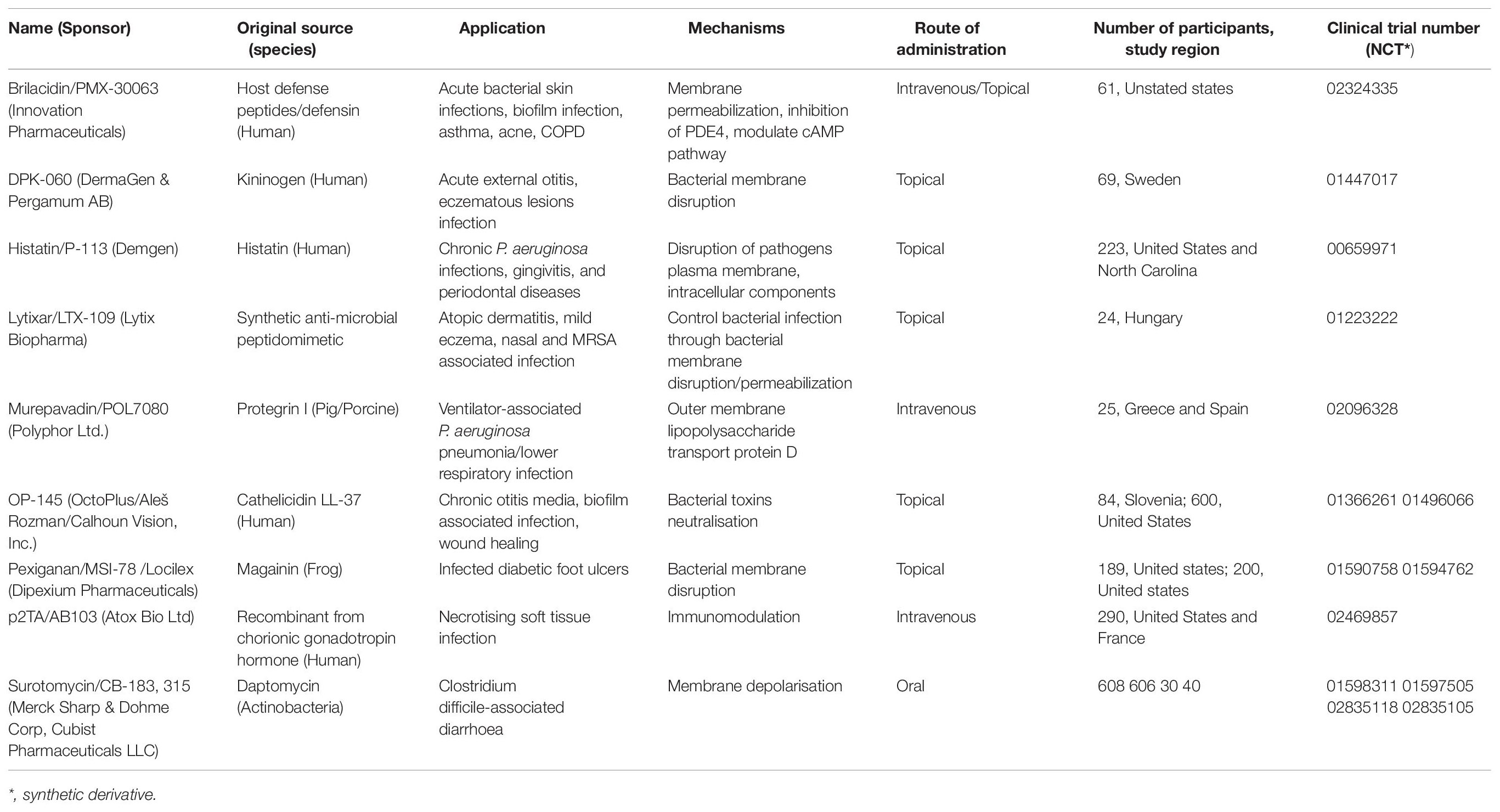
Table 3. Some potent exclusive antibacterial/anti-infective peptide-based therapeutical regimens (which are exited in clinical trials in phase II onward/not terminated in the clinical phase).
Strategically, several specific chemical modifications/synthetic conjugations are also available to boost the clinical success rate with withdrawn lead candidates and improved antibiofilm activity against different pathogenic bacteria (De La Fuente-Núñez et al., 2016). A3 (AamAP1), BmKn-22 (BmKn-2), defensin-1, (defensin), Pro10-1D (protaetiamycine), several other synthetic derivatives namely, LL1037, LL7-31 (LL-37), 1018 (bactenecin), AS10 (CRAMP), battacin (lipopeptides), BMAP27-melittin (melittin), CAMA (cecropin A and melittin A), NRC-16 (pleurocidin), P10 (P60.4Ac), P318 (CRAMP), are some chemical modification attempts toward improving the activity against biofilm than their parental form (De La Fuente-Núñez et al., 2016; Almaaytah et al., 2018; Krishnan et al., 2020). Similarly, nano-synthesis techniques like nanoparticle, nano-formulation, liposome, PLGA nanoparticle drug delivery also significantly help overcome the lack of physiochemical properties and pharmacokinetic profiles of peptides (Biswaro et al., 2018; Dostert et al., 2019; Galdiero et al., 2019). No such combination is available for the insect. Documentation of more scientific literature, critical discussion, and assessment will help peptide-based antibacterial/antibiofilm future drug development strategy.
Bioinformatics Tools to Accelerate the Peptide-Based Drug Discovery
Currently, in silico drug discovery approach or computational intelligence platform-based drug discovery plays a crucial role both in academia as well as in industry by locating possible active molecules from a bunch of desired molecules, predicts the structure and function of the active molecule with homologous/similar candidates information, able to predict toxicity-pharmacokinetic profiles based on chemical composition, mechanism of inhibition targeting any particular disease associated targets/enzyme, etc., (Malathi and Ramaiah, 2018; Duarte et al., 2019; Swain et al., 2020). Similarly, various tools, databases and software’s of bioinformatics can also help speed up the peptide-based research through analysis within a limited time and resources (Gupta et al., 2016; Agrawal et al., 2018; Câmara et al., 2020).
The universal National Center for Biotechnology Information (NCBI)1, and Universal Protein Resource or UniPort2, protein data bank or PDB3, with some specific sources such as data repository of antimicrobial peptides or DRAMP4, update linking antimicrobial peptides or LAMP25. Yet another database of antimicrobial peptides or YADAMP6, database of antimicrobial activity and structure prediction of DBAASP7, collection of antimicrobial peptides or CAMPR38, classification of antimicrobial peptides or ClassAMP9, structure database of bioactive peptides or StraPep10, ligand-protein binding database or BioLip11 are the most important open access resources able to guide any peptide associated research from prediction to submission. To date >200 peptide base databases are available for any kind of reference and study.
From the history of antibacterial drug discovery, validation of any kind of potential antibacterial agents (herein peptides) are facing a complex validation procedure or filtrate through different ideal drug parameters. Physiochemical properties play a crucial role in selecting the most potent antibacterial-cum-antibiofilm peptides at the first stage of clinical trials. On the other hand, the revolution in bioinformatics tools toward predicting ideal drug candidates directly reduces the time, resource, and cost in current drug discovery. Briefly, in the preclinical trial, most of the compounds showed the most potent activity during in vitro testing; however, most candidates do not contain physiochemical and are withdrawn later in different clinical trial stages. For example, the predicted hydropathicity index or GRAVY represents the hydrophobicity of the respect insect peptide. According to ExPASy, positive GRAVY values indicate hydrophobic and negative values indicated the hydrophilic value as a crucial parameter for clinical validation. The isoelectric point (pI)/IEP is the value at which the overall net surface charge is widely used in proteomics and peptide-based drug discovery. For a peptide, pI values mostly depend on the acid dissociation constant (pKa) of the ionizable groups of charged amino acids glutamate (δ-carboxyl group), aspartate (ß-carboxyl group), cysteine (thiol group), tyrosine (phenol group), histidine (imidazole side chains), lysine (ε-ammonium group), and arginine (guanidinium group). Overall, a peptide’s net charge is strongly related to pH and greatly influenced in druggable candidate selection. Similarly, molecular weight, half-life period, etc., are smaller parameters with significant influence in most potential peptide selection. Bioinformatics tools predict the possible values without any expensive experiments through an extensive training dataset. Thus, different bioinformatics not recommended but guided toward peptide-based drug discovery.
Similarly, the universal bioinformatics tools such as, protein homology/analogy recognition engine2.0 or Phyre212, protein secondary structure prediction or PSIPRED13, iterative threading assembly refinement or I-TASSER14, basic local alignment search tool or BLAST15, physicochemical analysis tool, ProtParam16, pharmacokinetic-toxicity property prediction tool, SwissADME17 with some specific tools namely, improved prediction of antimicrobial peptide or iAMPpre18, biofilm-active peptides or BaAMPs19, collection of antimicrobial peptides signature or CAMPSign20, the de novo approach peptide structure prediction tool, PEP-FOLD21 and for protein-peptide/protein-protein docking tools, HPEPDOCK22, HDOCK23, ClusPro24, and PeptiDock25 helps significantly in peptide research. Advanced artificial intelligence techniques and huge biological information always provide ideal opportunities to make some desired/objective-oriented tools for guidance at the early stage of peptide drug discovery by predicting several anonymous druggable information.
Physicochemical Property and Homology/Phylogenetic Analysis
Bioactive peptide identification in current antibacterial drug discovery era is one of the most interesting research area (Jakubczyk et al., 2020). The amino acid composition of a peptide directly influences drug-likeliness or drug suitability characteristics, including absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties. Moreover, an ideal isoelectric point, hydrophobicity, lipophilicity, half-life period, the amino acid composition, mass, etc., collectively known as physicochemical properties, is an essential feature for a lead bioactive therapeutic peptide (Boone et al., 2018; Zafar et al., 2020). The above properties are always estimated by chromatographic techniques; however, this technique is expensive and time-consuming to calculate protein fragments one by one in a considerable amount of data set. On the other hand, most bioactive peptides are withdrawn from the clinical study due to unfavorable physicochemical properties. Several computational based tools have been developed taking a massive number of training data sets as a possible solution; thus, anyone can check/predict the above properties freely before synthesis/derivatization or an exponential study. For example, the universal tool, ProtParam was used to estimate the physicochemical properties for selected antibiofilm insect-derived peptides (Table 4) and this information guides/useful for peptide-based antibacterial/antibiofilm drug-development research.
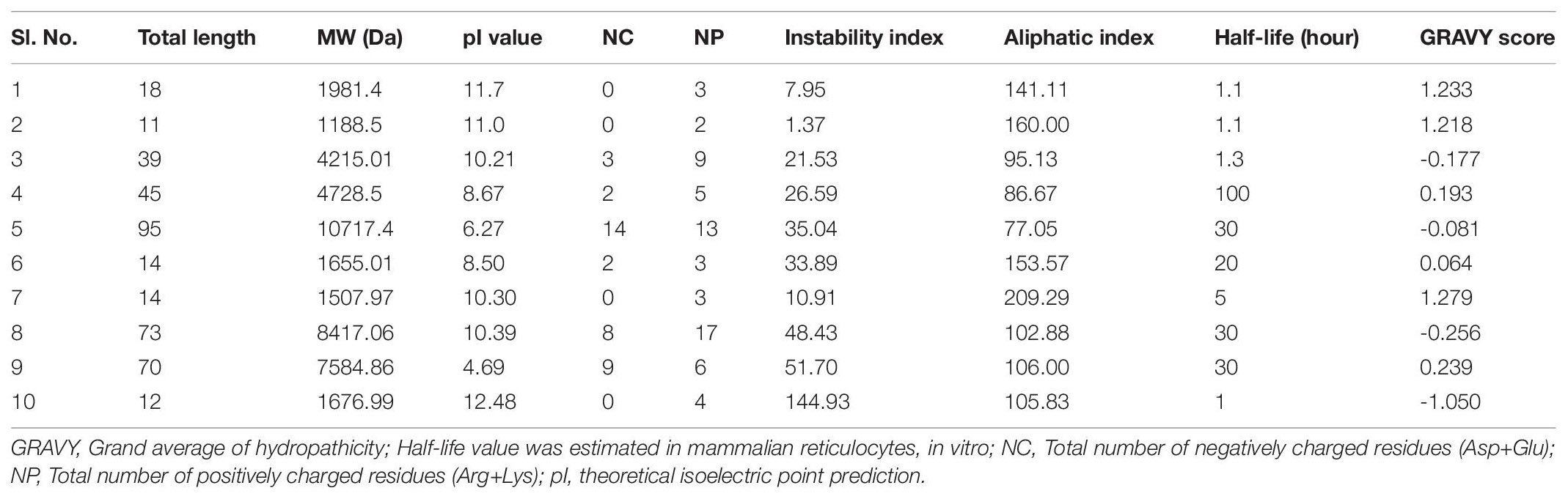
Table 4. Predicted physico-chemical properties for selective antibiofilm insect peptides using bioinformatics tool ProtParam.
Similarly, PepCalc.com26, PepDraw27, peptide property calculator28, protein descriptor calculator29, peptide analyzing tool30, etc., are several freely accessible tools that are available to predict the physicochemical properties of any desired peptide.
Homologous/structural similarity compound search is another high-through approach in ongoing drug discovery toward discovering more bioactive compounds in a minimum time and resource (Kanduc, 2012; Zheng et al., 2019). For example, if someone finds an active peptide from an insect or any other source and validated it to have potent antibiofilm property, then next homologous search is more appropriate to search structurally related/identical peptide from different databases than comparison with any physical documentation. The well-known protein-BLAST or BLASTp31, Phyre2, PSIPRED, SPIDER32, HHpred33, etc., are different bioinformatics tools that give homologous information to accelerate the peptide-based drug discovery. On the other hand, phylogenetic search helps give the proper identification or even source of an unknown peptide. Generally, a homologous peptide having a common origin is always present in the same clade or branch of a phylogenetic tree. The clustal omega34 and the molecular evolutionary genetics analysis or MEGA35 are the most widely used open-source bioinformatics tools for phylogenetic tree analysis. Overall, this homologous-cum-phylogenetic search is commonly used in genomics and proteomics analysis. Thus, bioinformatics is always trying to produce more relevant tools/software to reduce computational and experimental peptide research gaps.
Molecular Modeling and Docking Analysis
Molecular modeling is a technique used to generate a three-dimensional (3D) structure of any protein, peptide or DNA structure using two different approaches, namely homology modeling with known template and de novo/physical design without any template (Nikolaev et al., 2018). The 3D structure of any peptide or protein always gives some relevant information on the active amino acid geometry/active site of a disease associated enzyme for drug development modules.
Protein data bank is one of the largest and open-source 3D-structural database of peptides, proteins, DNAs, and nucleic acids generated through NMR, X-ray or cryo-EM. However, this technique is costly and limiting. At the same time, bioinformatics tools can generate the 3D structure followed by cost-effective homology and de nove modelling approach to overcome the structural complexity in drug discovery. Notably, the homology approach tools, SWISS-MODEL36, CPHmodels 3.237, and de novo approach tools, Phyre2, I-TASSER, PEP-FOLD338 are most prevalent for 3D protein/peptide structure prediction. For example, the 3D structure of the ten selected antibiofilm insect-derived peptides were generated with the PEP-FOLD3 tool and visualized using the software BIOVIA-Discovery Studio Visualizer (Figure 7). Thus, molecular modeling an essential tool in a systematic and cost-effective platform for peptides. Similarly, molecular docking is another essential tool for computer-aided drug discovery platforms (Swain et al., 2018).
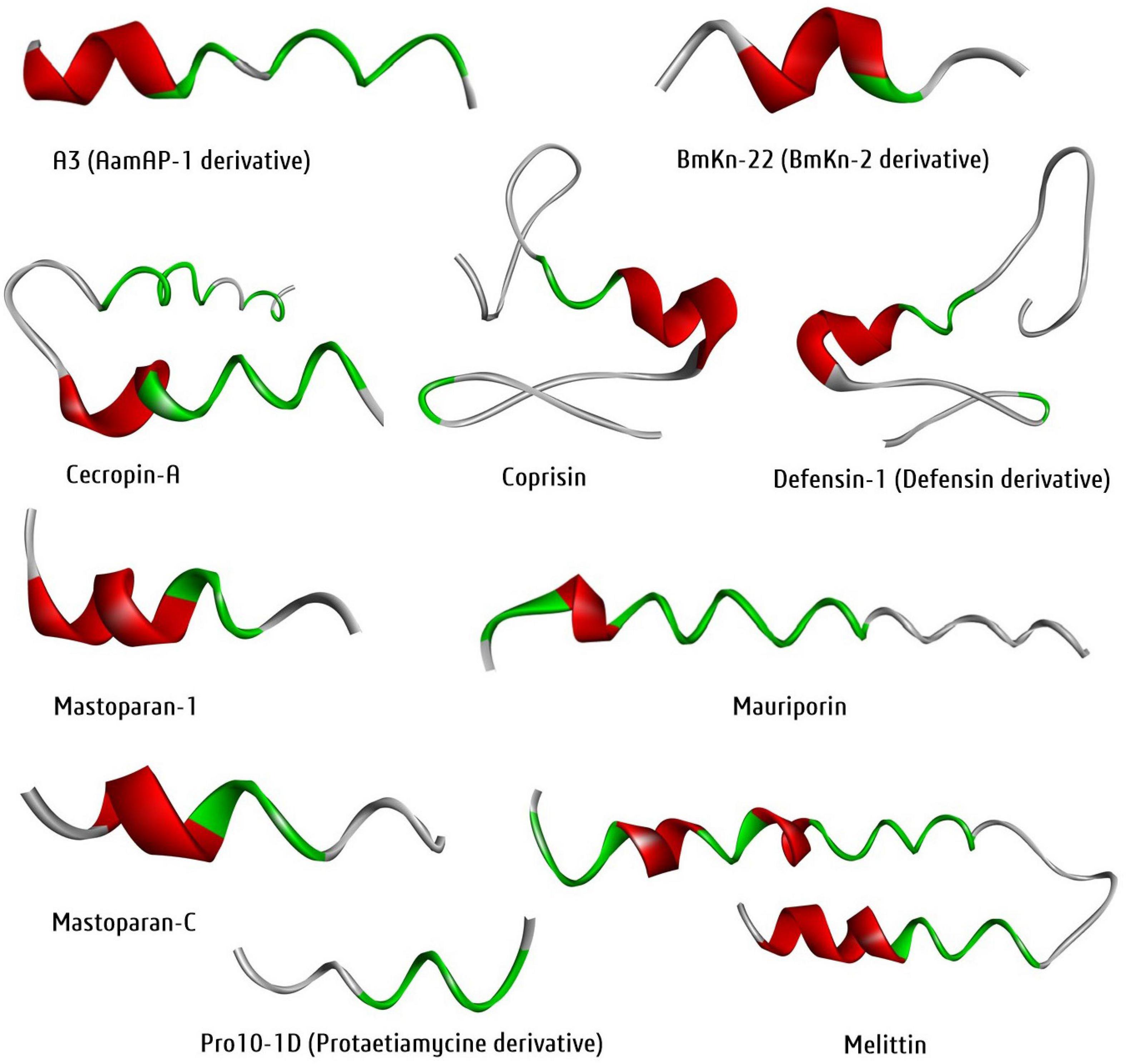
Figure 7. Three-dimensional structure of newly generated ten active antibiofilm peptide derivatives using the PEP-FOLD3 tool. The BIOVIA-Discovery studio visualizer software was used for 3D-visualization in secondary structure format with clean geometry. The backbone protein portion is more fat than other regions, where red color indicated helices regions, gray color indicated coil regions and green color indicated the turn regions of the protein structure.
Molecular docking is a technique used to estimate the binding affinity between a target and a ligand, which means any proposed therapeutic agents biding affinity/interaction with a desired target protein of interest. In peptide-based antibiofilm research, molecular docking can predict the binding affinity of any desired peptide against any putative biofilm-associated target enzyme. As we know, bacterial Lux operon encoded genetic factors, LuxR and LuxL are two well-known QS-regulator genes during biofilm formation; thus, as an example, the selective antibiofilm peptides were docked against the protein structure of S. aureus-LuxR (PDB ID: 3B2N) and recorded the binding affinity using HPEPDOCK tool with proper interaction (Figure 8). Among all, Pro-10-1D was the most effective peptide against S. aureus-LuxR based on the recorded docking score of -220.938 kcal/mol (some docking tool/software represent the docking score in negative sign and some in positive sign based on their algorithms). Similarly, several protein-peptide docking tools, namely, HDOCK, ClusPro PeptiDock, MDockPeP39 are also available for molecular docking analysis. Overall, molecular docking is the most widely used tool for potential peptides, drug-able agents and repurposing therapeutic lead section.
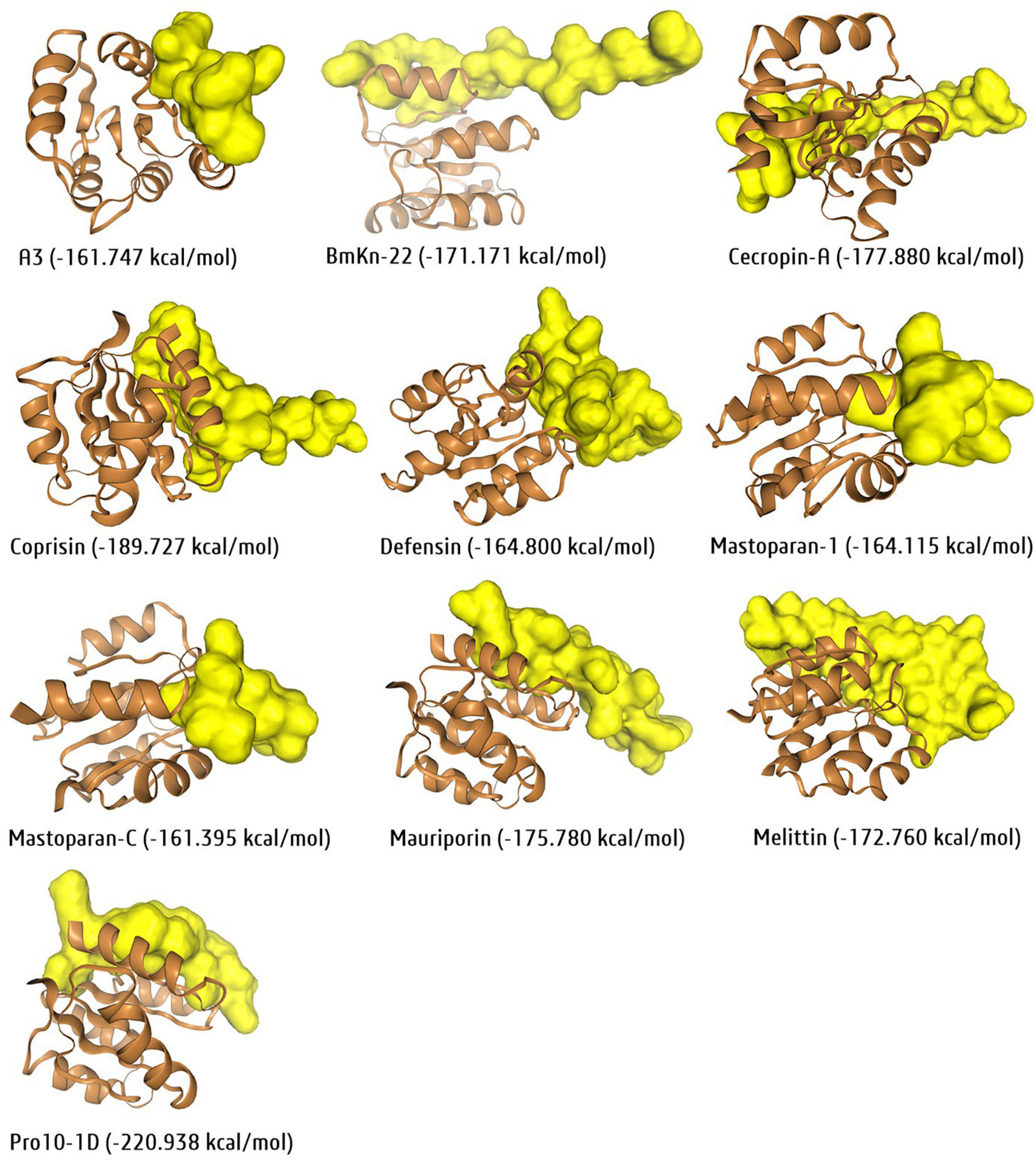
Figure 8. Molecular docking interaction of ten exclusive antibiofilm insect-peptides against the putative biofilm target DNA-binding response regulator/transcriptional factor, LuxR (PDB ID:3B2N) of S. aureus by the peptide docking server, HPEPDOCK. Docking score presented in parentheses with a negative sign and more in negative docking score more in potency according to docking tool.
Currently, bioinformatics tools can mimicry the whole natural biological system through advanced computational coding and artificial intelligence techniques. Different types of bioinformatics tools are available for individual parameter analysis, like peptide identification to peptide validation to clinical trial information (Lee et al., 2019; Lin et al., 2020). Thus, molecular docking, pharmacophore modeling, and molecular dynamic simulation are the most widely used bioinformatics tools to minimize resource, time, and cost while selecting potential candidates by different pharmaceutical industries and drug developers (Figure 9). Simultaneously, the prediction of such possible biological, physiological, toxicity profiles directly reduce the most expensive experiments and removes the unwanted/unfavorable candidates at the early stage of drug discovery or computer-aided drug discovery (CADD) or machine-learning approach platform (Lin et al., 2020; Mulligan, 2020). However, for every platform and approach, there is a limitation. The CADD platform also contains some demerits. Most tools and software are developed in different algorithms and hypotheses; thus, the same analysis can vary in decision-making. Particularly, there are several challenges such as, solubility, permeability, delivery associated with success of peptide-based drug discovery (Farhadi and Hashemian, 2018; Wagner et al., 2018; Lee et al., 2019). Computational programming can be used to some extent and always not a hundred percent similar to natural biological processes. Thus, bioinformatics tools are most suitable as a guidance/reference for selecting and validating drug candidates, but do not recommend any drug candidates for human consumption. In conclusion, bioinformatics could be a cost-effective essential tool in 21st century for any research analysis and mainly for drug discovery.
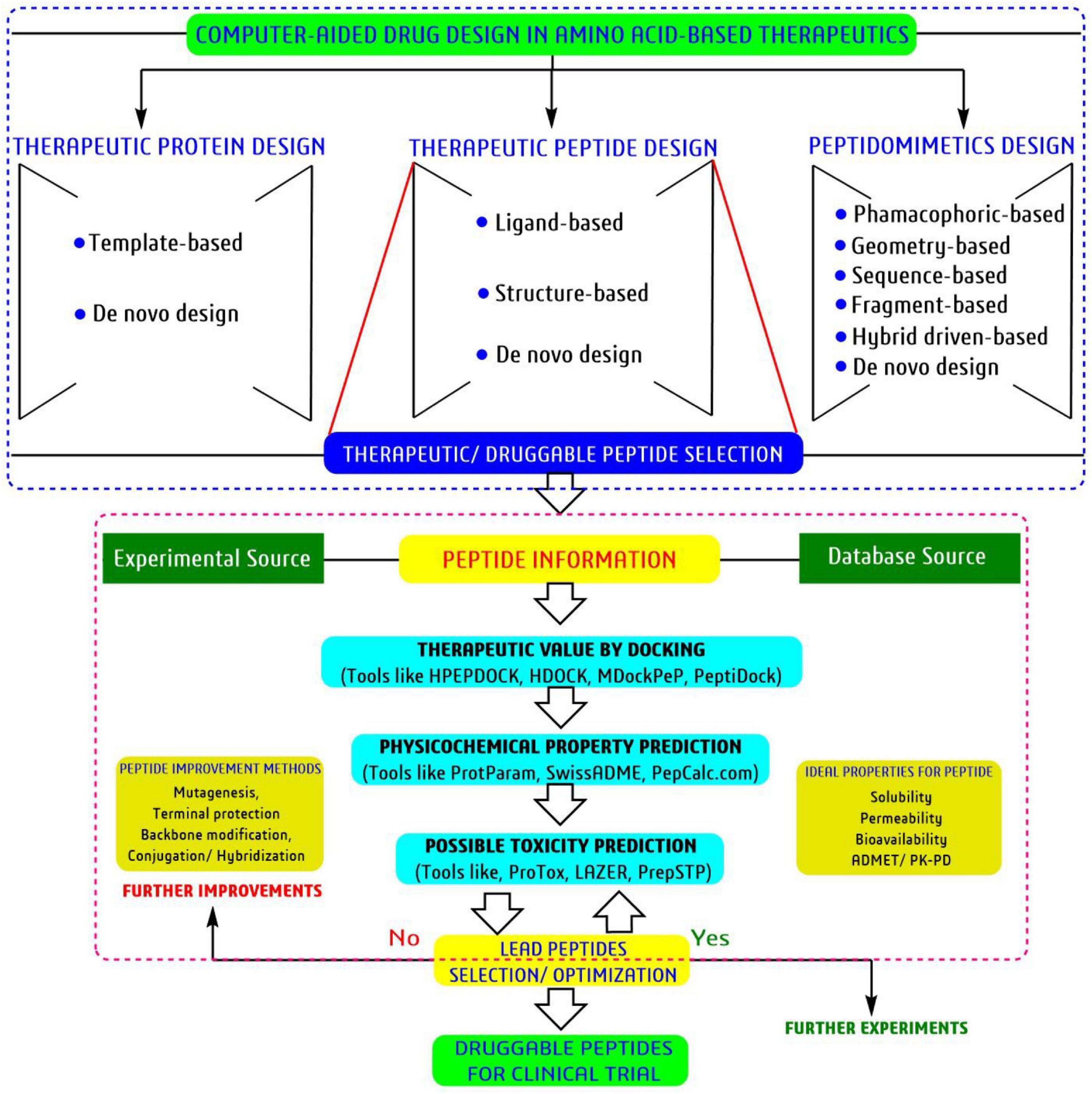
Figure 9. Schematic diagram toward selection and validation of therapeutic peptides using tools of bioinformatics.
Future Perspective and Concluding Remarks
It is now well evident that the main immune effector molecules of Insects are AMPs. Diversity of insects are huge and there is growing evidence that the natural system of insects is fairly dynamic. At the same time, due to the conserved biological evolution, bioactive molecules and signaling pathways within the natural system of insects exhibit much more similarity with vertebrates (including humans). Researchers worldwide are in search of novel bioactive molecule(s) with novel mechanism of action as antimicrobials. Nevertheless, it is convincing that AMPs could be an innovative antimicrobial candidate. Several factors such as sequence, the charge, the helicity, the amphipathicity and the overall hydrophobicity of AMPs are crucial in considering them as effective antimicrobial agents. However, a few studies have been carried out on insect derived peptides as antimicrobial agents that can prevent formation of biofilm/eradicate the biofilm. It is also obvious that no AMP from insects have been progressed to the pre- clinical and clinical stages of development as therapeutics.
It is evident that peptides show potential antimicrobial activity with several non-specific inhibition mechanisms (Dosler and Karaaslan, 2014; Wu et al., 2018; Raheem and Straus, 2019). Insect-derived antibacterial agents show potential activities against planktonic as well as some biofilm producing pathogenic specific bacterial stains such as uropathogenic E. coli, methicillin-resistant S. aureus, vancomycin-resistant S. aureus, carbapenem-resistant A. baumannii and others (Dosler and Karaaslan, 2014; Wu et al., 2018). The present review includes 103 peptides from insect sources with antimicrobial property in planktonic cells but, of these, only seven isolated and three derived peptides were documented against biofilm. However, screening all these AMPs in biofilm models are yet to be investigated. Thus, this review encourages researchers to screen insect derived peptides on different biofilm models. Moreover, more research should be directed toward finding novel antimicrobial peptides from insects.
Author Contributions
AB, AS, SS, and SP designed the hypothesis. AS and SS performed the computational work. SS analyzed the computationally generated data. SP prepared the draft manuscript. GS and PM commented on the final manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
SP was financially supported by a postdoc fellowship from RUSA 2.0. AS was supported by inhouse doctoral scholarship of Siksha O Anusandhan University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to the authorities of IMS and SUM Hospital, Siksha O Anusandhan and Utkal University, Bhubaneswar, India, for their inspiration to develop this manuscript. AS is thankful to the President, Siksha O Anusandhan University for providing inhouse PhD Scholarship. GS, PM, and SP are thankful to the RUSA 2.0 for supporting Centre of Excellence in Environment, Climate Change and Public Health (ECCPH). AB and SS are largely self-supported.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.661195/full#supplementary-material
Abbreviations
ADMET, absorption, distribution, metabolism, excretion, and toxicity; AMPs, antimicrobial peptides; Bap, biofilm associated protein; BP, bacterial prostatitis; BPs, bioactive peptides; BV, bacterial vaginosis; CecA, cecropin A; c-di-GMP, bis-(3′-5′)-cyclic di-guanosine monophosphate; CF, cystic fibrosis; COPD, chronic obstructive pulmonary disease; cylA, cytolysin A; ebp, endocarditis biofilm-associated protein; EPS, exopolysaccharide; ESKAPE, Enterococcus faecalis, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter; Esp, enterococcal surface protein; fib, fibrinogen−binding protein; FISH, fluorescence microscopy and fluorescence in situ hybridization; GP, Gram-positive; IBD, crohn’s disease; MIC, minimum inhibitory concentration; MRSA, methicillin-resistant S. aureus; NAL, nalidixic acid; NCBI, National Center for Biotechnology Information; OM, otitis media; PDB, protein data bank; PIA, intercellular adhesin; PNAG, polymeric N-acetyl-glucosamine; QS, quorum sensing; sprE, serine protease; sRNAs, small RNAs; UPEC, uropathogenic E. coli; UTI, urinary tract infection.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.uniprot.org/
- ^ https://www.rcsb.org/
- ^ http://dramp.cpu-bioinfor.org/
- ^ http://biotechlab.fudan.edu.cn/database/lamp./
- ^ http://yadamp.unisa.it/about.aspx
- ^ https://dbaasp.org/home
- ^ http://www.camp.bicnirrh.res.in/
- ^ http://www.bicnirrh.res.in/classamp/
- ^ http://isyslab.info/StraPep/
- ^ https://zhanglab.ccmb.med.umich.edu/BioLiP/
- ^ http://www.sbg.bio.ic.ac.uk/∼phyre2/
- ^ http://bioinf.cs.ucl.ac.uk/psipred/
- ^ https://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://web.expasy.org/protparam/
- ^ http://www.swissadme.ch/index.php
- ^ http://cabgrid.res.in:8080/amppred/index.html
- ^ http://www.baamps.it/
- ^ http://www.campsign.bicnirrh.res.in/
- ^ https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/
- ^ http://huanglab.phys.hust.edu.cn/hpepdock/
- ^ http://hdock.phys.hust.edu.cn/
- ^ https://cluspro.org/home.php
- ^ https://cluspro.org/peptide/index.php
- ^ https://pepcalc.com/
- ^ http://www.tulane.edu/∼biochem/WW/PepDraw/
- ^ http://biotools.nubic.northwestern.edu/proteincalc.html
- ^ http://biotriangle.scbdd.com/protein/index/
- ^ https://www.thermofisher.com/in/en/home/life-science/protein-biology/peptides-proteins/custom-peptide-synthesis-services/peptide-analyzing-tool.html
- ^ https://www.bioinfor.com/peptide-mutations-homology-searching/
- ^ https://toolkit.tuebingen.mpg.de/tools/hhpred
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://www.megasoftware.net/
- ^ https://swissmodel.expasy.org/
- ^ http://www.cbs.dtu.dk/services/CPHmodels/
- ^ https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/
- ^ https://zougrouptoolkit.missouri.edu/mdockpep/index.html
References
Agrawal, P., Bhalla, S., Chaudhary, K., Kumar, R., Sharma, M., and Raghava, G. P. S. (2018). In silico approach for prediction of antifungal peptides. Front. Microbiol. 9:323. doi: 10.3389/fmicb.2018.00323
Akbari, R., Hakemi Vala, M., Hashemi, A., Aghazadeh, H., Sabatier, J. M., and Pooshang Bagheri, K. (2018). Action mechanism of melittin-derived antimicrobial peptides, MDP1 and MDP2, de novo designed against multidrug resistant bacteria. Amino Acids 50, 1231–1243. doi: 10.1007/s00726-018-2596-5
Akbari, R., Hakemi-Vala, M., Pashaie, F., Bevalian, P., Hashemi, A., and Bagheri, K. P. (2019). Highly synergistic effects of melittin with conventional antibiotics against multidrug-resistant isolates of Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 25, 193–202. doi: 10.1089/mdr.2018.0016
Allesen-Holm, M., Barken, K. B., Yang, L., Klausen, M., Webb, J. S., Kjelleberg, S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128. doi: 10.1111/J.1365-2958.2005.05008.X
Almaaytah, A., Farajallah, A., Abualhaijaa, A., and Al-balas, Q. (2018). A3, a Scorpion venom derived peptide analogue with potent antimicrobial and antibiofilm activity against clinical isolates of multi-drug resistant gram positive bacteria. Molecules 23:1603. doi: 10.20944/PREPRINTS201805.0356.V1
Almaaytah, A., Tarazi, S., Alsheyab, F., Al-Balas, Q., and Mukattash, T. (2014). Antimicrobial and antibiofilm activity of mauriporin, a multifunctional scorpion venom peptide. Int. J. Pept. Res. Ther. 20, 397–408. doi: 10.1007/s10989-014-9405-0
Amala Reena, A., Subramaniyan, A., and Kanungo, R. (2017). Biofilm formation as a virulence factor of Acinetobacter bauhamannii: An emerging pathogen in critical care units. J. Curr. Res. Sci. Med. 3:74. doi: 10.4103/jcrsm.jcrsm_66_17
Amaral, A. C., Silva, O. N., Mundim, N. C. C. R., De Carvalho, M. J. A., Migliolo, L., Leite, J. R. S. A., et al. (2012). Predicting antimicrobial peptides from eukaryotic genomes: In silico strategies to develop antibiotics. Peptides 37, 301–308. doi: 10.1016/j.peptides.2012.07.021
Anbazhagan, D., Mansor, M., Yan, G. O. S., Md Yusof, M. Y., Hassan, H., and Sekaran, S. D. (2012). Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS One 7:e36696. doi: 10.1371/journal.pone.0036696
Andreu, D., and Rivas, L. (1998). Animal antimicrobial peptides: an overview. Biopolymers 47, 415–433. doi: 10.1002/(sici)1097-0282(1998)47:6<415::aid-bip2>3.0.co;2-d
Anwar, H., Dasgupta, M. K., and Costerton, J. W. (1990). Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob. Agents Chemother. 34, 2043–2046. doi: 10.1128/AAC.34.11.2043
Arciola, C. R., Campoccia, D., Speziale, P., Montanaro, L., and Costerton, J. W. (2012). Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33, 5967–5982. doi: 10.1016/j.biomaterials.2012.05.031
Armbruster, C. R., and Parsek, M. R. (2018). New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 115, 4317–4319. doi: 10.1073/pnas.1804084115
Atshan, S. S., Nor Shamsudin, M., Sekawi, Z., Lung, L. T. T., Hamat, R. A., Karunanidhi, A., et al. (2012). Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. J. Biomed. Biotechnol. 2012:976972. doi: 10.1155/2012/976972
Azara, E., Longheu, C., Sanna, G., and Tola, S. (2017). Biofilm formation and virulence factor analysis of Staphylococcus aureus isolates collected from ovine mastitis. J. Appl. Microbiol. 123, 372–379. doi: 10.1111/jam.13502
Band, V., and Weiss, D. (2014). Mechanisms of antimicrobial peptide resistance in gram-negative bacteria. Antibiotics 4, 18–41. doi: 10.3390/antibiotics4010018
Barbu, E. M., Mackenzie, C., Foster, T. J., and Höök, M. (2014). SdrC induces staphylococcal biofilm formation through a homophilic interaction. Mol. Microbiol. 94, 172–185. doi: 10.1111/mmi.12750
Barraud, N., Storey, M. V., Moore, Z. P., Webb, J. S., Rice, S. A., and Kjelleberg, S. (2009). Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2, 370–378. doi: 10.1111/j.1751-7915.2009.00098.x
Basak, A., Abouelhassan, Y., Norwood, V. M., Bai, F., Nguyen, M. T., Jin, S., et al. (2016). Synthetically tuning the 2-position of halogenated quinolines: optimizing antibacterial and biofilm eradication activities via alkylation and reductive amination pathways. Chemistry 22, 9181–9189. doi: 10.1002/chem.201600926
Bazaka, K., Jacob, M. V., Crawford, R. J., and Ivanova, E. P. (2012). Efficient surface modification of biomaterial to prevent biofilm formation and the attachment of microorganisms. Appl. Microbiol. Biotechnol. 95, 299–311. doi: 10.1007/s00253-012-4144-7
Bechinger, B., and Lohner, K. (2006). Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 1758, 1529–1539. doi: 10.1016/j.bbamem.2006.07.001
Beloin, C., Renard, S., Ghigo, J. M., and Lebeaux, D. (2014). Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 18, 61–68. doi: 10.1016/j.coph.2014.09.005
Besharova, O., Suchanek, V. M., Hartmann, R., Drescher, K., and Sourjik, V. (2016). Diversification of gene expression during formation of static submerged biofilms by Escherichia coli. Front. Microbiol. 7:1568. doi: 10.3389/fmicb.2016.01568
Biswaro, L. S., Sousa, M. G. D. C., Rezende, T. M. B., Dias, S. C., and Franco, O. L. (2018). Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 9:855. doi: 10.3389/fmicb.2018.00855
Bjarnsholt, T. (2013). The role of bacterial biofilms in chronic infections. APMIS. Suppl. 121, 1–51. doi: 10.1111/apm.12099
Bjarnsholt, T., Moser, C., and Jensen, P. Ø, and Høiby, N. (2011). Biofilm Infections. New York, NY: Springer, doi: 10.1007/978-1-4419-6084-9
Boase, N. R. B., Torres, M. D. T., Fletcher, N. L., De La Fuente-Nunez, C., and Fairfull-Smith, K. E. (2018). Polynitroxide copolymers to reduce biofilm fouling on surfaces. Polym. Chem. 9, 5308–5318. doi: 10.1039/c8py01101j
Boles, B. R., Thoende, M., Roth, A. J., and Horswill, A. R. (2010). Identification of genes involved in polysaccharide- independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146
Bongers, J., and Heimer, E. P. (1994). Recent applications of enzymatic peptide synthesis. Peptides 15, 183–193. doi: 10.1016/0196-9781(94)90189-9
Boone, K., Camarda, K., Spencer, P., and Tamerler, C. (2018). Antimicrobial peptide similarity and classification through rough set theory using physicochemical boundaries. BMC Bioinformatics 19:469. doi: 10.1186/s12859-018-2514-6
Brooks, J. L., and Jefferson, K. K. (2014). Phase variation of poly-N-Acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog. 10:e1004292. doi: 10.1371/journal.ppat.1004292
Browne, K., Chakraborty, S., Chen, R., Willcox, M. D., Black, D. S., Walsh, W. R., et al. (2020). A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 21:7047. doi: 10.3390/ijms21197047
Bulet, P., and Stocklin, R. (2005). Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 12, 3–11. doi: 10.2174/0929866053406011
Bullitt, E., and Makowski, L. (1995). Structural polymorphism of bacterial adhesion pili. Nature 373, 164–167. doi: 10.1038/373164a0
Busch, A., Phan, G., and Waksman, G. (2015). Molecular mechanism of bacterial type 1 and P pili assembly. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 373:20130153. doi: 10.1098/rsta.2013.0153
Butler, M. S., Blaskovich, M. A., and Cooper, M. A. (2013). Antibiotics in the clinical pipeline in 2013. J. Antibiot. (Tokyo) 66, 571–591. doi: 10.1038/ja.2013.86
Câmara, G. A., Nishiyama-Jr, M. Y., Kitano, E. S., Oliveira, U. C., da Silva, P. I., Junqueira-de-Azevedo, I. L., et al. (2020). A multiomics approach unravels new toxins with possible in silico antimicrobial, antiviral, and antitumoral activities in the venom of Acanthoscurria rondoniae. Front. Pharmacol. 11:1075. doi: 10.3389/fphar.2020.01075
Campoccia, D., Montanaro, L., and Arciola, C. R. (2013). A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 34, 8533–8554. doi: 10.1016/j.biomaterials.2013.07.089
Campoccia, D., Speziale, P., Ravaioli, S., Cangini, I., Rindi, S., Pirini, V., et al. (2009). The presence of both bone sialoprotein-binding protein gene and collagen adhesin gene as a typical virulence trait of the major epidemic cluster in isolates from orthopedic implant infections. Biomaterials 30, 6621–6628. doi: 10.1016/j.biomaterials.2009.08.032
Candan, E. D., and Aksöz, N. (2015). Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 62, 867–874. doi: 10.18388/abp.2015_1148
Carlsson, A., Nyström, T., De Cock, H., and Bennich, H. (1998). Attacin – an insect immune protein - binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 144, 2179–2188. doi: 10.1099/00221287-144-8-2179
Cavallarin, L., Andreu, D., and San Segundo, B. (1998). Cecropin A-derived peptides are potent inhibitors of fungal plant pathogens. Mol. Plant Microbe Interact. 11, 218–227. doi: 10.1094/MPMI.1998.11.3.218
Čeřovský, V., Žd’árek, J., Fučík, V., Monincová, L., Voburka, Z., and Bém, R. (2010). Lucifensin, the long-sought antimicrobial factor of medicinal maggots of the blowfly Lucilia sericata. Cell. Mol. Life Sci. 67, 455–466. doi: 10.1007/s00018-009-0194-0
Cezairliyan, B., Vinayavekhin, N., Grenfell-Lee, D., Yuen, G. J., Saghatelian, A., and Ausubel, F. M. (2013). Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 9:e1003101. doi: 10.1371/journal.ppat.1003101
Chapartegui-González, I., Lázaro-Díez, M., Bravo, Z., Navas, J., Icardo, J. M., and Ramos-Vivas, J. (2018). Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS One 13:e0201961. doi: 10.1371/journal.pone.0201961
Chapman, M. R., Robinson, L. S., Pinkner, J. S., Roth, R., Heuser, J., Hammar, M., et al. (2002). Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855. doi: 10.1126/science.1067484
Chauang, W. U., Pan, B. S., and Tsai, J. S. (2000). Inhibition of lipoxygenase and blood thinning effects of mackerel protein hydrolysate. J. Food Biochem. 24, 333–343. doi: 10.1111/j.1745-4514.2000.tb00704.x
Chen, X., Zhang, L., Wu, Y., Wang, L., Ma, C., Xi, X., et al. (2018). Evaluation of the bioactivity of a mastoparan peptide from wasp venom and of its analogues designed through targeted engineering. Int. J. Biol. Sci. 14, 599–607. doi: 10.7150/ijbs.23419
Chiang, P., and Burrows, L. L. (2003). Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185, 2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003
Chiang, W. C., Nilsson, M., Jensen, P. Ø, Høiby, N., Nielsen, T. E., Givskov, M., et al. (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361. doi: 10.1128/AAC.00001-13
Choi, A. H. K., Slamti, L., Avci, F. Y., Pier, G. B., and Maira-Litrán, T. (2009). The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963. doi: 10.1128/JB.00647-09
Choi, J. H., Jang, A. Y., Lin, S., Lim, S., Kim, D., Park, K., et al. (2015). Melittin, a honeybee venom-derived antimicrobial peptide, may target methicillin-resistant Staphylococcus aureus. Mol. Med. Rep. 12, 6483–6490. doi: 10.3892/mmr.2015.4275
Christensen, L. D., van Gennip, M., Rybtke, M. T., Wu, H., Chiang, W. C., Alhede, M., et al. (2013). Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-gmp level in the bacteria. Infect. Immun. 81, 2705–2713. doi: 10.1128/IAI.00332-13
Cortés, M. E., Consuegra Bonilla, J., and Ruben, D. S. (2011). Biofilm formation, control and novel strategies for eradication. Sci. Against Microbial. Pathog. Commun. Curr. Res. Technol. Adv. 2, 896–905.
Costerton, J. W., Cheng, K. J., Geesey, G. G., Ladd, T. I., Nickel, J. C., Dasgupta, M., et al. (1987). Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464. doi: 10.1146/annurev.mi.41.100187.002251
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Cucarella, C., Solano, C., Valle, J., Amorena, B., and Lasa, Í, and Penadés, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Dahdouh, E., Gómez-Gil, R., Pacho, S., Mingorance, J., Daoud, Z., and Suárez, M. (2017). Clonality, virulence determinants, and profiles of resistance of clinical Acinetobacter baumannii isolates obtained from a Spanish hospital. PLoS One 12:e0176824. doi: 10.1371/journal.pone.0176824
Dalton, H. M., and March, P. E. (1998). Molecular genetics of bacterial attachment and biofouling. Curr. Opin. Biotechnol. 9, 252–255. doi: 10.1016/S0958-1669(98)80055-4
Danese, P. N., Pratt, L. A., Dove, S. L., and Kolter, R. (2000). The outer membrane protein, Antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37, 424–432. doi: 10.1046/j.1365-2958.2000.02008.x
Das, T., Sharma, P. K., Busscher, H. J., Van Der Mei, H. C., and Krom, B. P. (2010). Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76, 3405–3408. doi: 10.1128/AEM.03119-09
De La Fuente-Núñez, C., Cardoso, M. H., De Souza Cândido, E., Franco, O. L., and Hancock, R. E. W. (2016). Synthetic antibiofilm peptides. Biochim. Biophys. Acta Biomembr. 1858, 1061–1069. doi: 10.1016/j.bbamem.2015.12.015
De La Fuente-Núñez, C., Korolik, V., Bains, M., Nguyen, U., Breidenstein, E. B. M., Horsman, S., et al. (2012). Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704. doi: 10.1128/AAC.00064-12
De la Fuente-Núñez, C., Reffuveille, F., Fernández, L., and Hancock, R. E. W. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
De La Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2014). Broad-Spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
De La Torre, B. G., and Albericio, F. (2020). Peptide therapeutics 2.0. Molecules 25:2293. doi: 10.3390/molecules25102293
Dean, S. N., Bishop, B. M., and van Hoek, M. L. (2011). Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 11:114. doi: 10.1186/1471-2180-11-114
DeLucca, A. J., Bland, J. M., Jacks, T. J., Grimm, C., Cleveland, T. E., and Walsh, T. J. (1997). Fungicidal activity of cecropin A. Antimicrob. Agents Chemother. 41, 481–483. doi: 10.1128/aac.41.2.481
Devaraj, A., Justice, S. S., Bakaletz, L. O., and Goodman, S. D. (2015). DNABII proteins play a central role in UPEC biofilm structure. Mol. Microbiol. 96, 1119–1135. doi: 10.1111/mmi.12994
Déziel, E., Comeau, Y., and Villemur, R. (2001). Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183, 1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001
Ding, J., Chou, Y. Y., and Chang, T. L. (2009). Defensins in viral infections. J. Innate Immun. 1, 413–420. doi: 10.1159/000226256
Divyashree, M., Mani, M. K., Reddy, D., Kumavath, R., Ghosh, P., Azevedo, V., et al. (2019). Clinical applications of antimicrobial peptides (AMPs): where do we stand now? Protein Pept. Lett. 27, 120–134. doi: 10.2174/0929866526666190925152957
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
Dorsey, C. W., Tomaras, A. P., and Actis, L. A. (2002). Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68, 6353–6360. doi: 10.1128/AEM.68.12.6353-6360.2002
Dosler, S., and Karaaslan, E. (2014). Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 62, 32–37. doi: 10.1016/j.peptides.2014.09.021
Dosler, S., Karaaslan, E., and Alev Gerceker, A. (2016). Antibacterial and anti-biofilm activities of melittin and colistin, alone and in combination with antibiotics against Gram-negative bacteria. J. Chemother. 28, 95–103. doi: 10.1179/1973947815Y.0000000004
Dosler, S., and Mataraci, E. (2013). In vitro pharmacokinetics of antimicrobial cationic peptides alone and in combination with antibiotics against methicillin resistant Staphylococcus aureus biofilms. Peptides 49, 53–58. doi: 10.1016/j.peptides.2013.08.008
Dostert, M., Belanger, C. R., and Hancock, R. E. W. (2019). Design and assessment of anti-biofilm peptides: steps toward clinical application. J. Innate Immun. 11, 193–204. doi: 10.1159/000491497
Duarte, Y., Márquez−Miranda, V., Miossec, M. J., and González−Nilo, F. (2019). Integration of target discovery, drug discovery and drug delivery: a review on computational strategies. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11:e1554. doi: 10.1002/wnan.1554
Eckert, R., Brady, K. M., Greenberg, E. P., Qi, F., Yarbrough, D. K., He, J., et al. (2006). Enhancement of antimicrobial activity against Pseudomonas aeruginosa by coadministration of G10KHc and tobramycin. Antimicrob. Agents Chemother. 50, 3833–3838. doi: 10.1128/AAC.00509-06
Ekengren, S., and Hultmark, D. (1999). Drosophila cecropin as an antifungal agent. Insect Biochem. Mol. Biol. 29, 965–972. doi: 10.1016/S0965-1748(99)00071-5
Evans, B. A., Hamouda, A., and Amyes, S. G. B. (2012). The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 19, 223–238. doi: 10.2174/138161213804070285
Fan, L., Sun, J., Zhou, M., Zhou, J., Lao, X., Zheng, H., et al. (2016). DRAMP: a comprehensive data repository of antimicrobial peptides. Sci. Rep. 6:24482.
Farhadi, T., and Hashemian, S. M. (2018). Computer-aided design of amino acid-based therapeutics: a review. Drug Des. Devel. Ther. 12, 1239–1254. doi: 10.2147/DDDT.S159767
Fazli, M., Almblad, H., Rybtke, M. L., Givskov, M., Eberl, L., and Tolker-Nielsen, T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981. doi: 10.1111/1462-2920.12448
Fjell, C. D., Hiss, J. A., Hancock, R. E. W., and Schneider, G. (2012). Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51. doi: 10.1038/nrd3591
Fleming, D., and Rumbaugh, K. (2017). Approaches to dispersing medical biofilms. Microorganisms 5:15. doi: 10.3390/microorganisms5020015
Flores-Mireles, A. L., Walker, J. N., Caparon, M., and Hultgren, S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284. doi: 10.1038/nrmicro3432
Forman, M. E., Jennings, M. C., Wuest, W. M., and Minbiole, K. P. C. (2016). Building a better quaternary ammonium compound (QAC): branched tetracationic antiseptic amphiphiles. ChemMedChem 11, 1401–1405. doi: 10.1002/cmdc.201600176
Frank, D. W. (1997). The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26, 621–629. doi: 10.1046/j.1365-2958.1997.6251991.x
Gaddy, J. A., Tomaras, A. P., and Actis, L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160. doi: 10.1128/IAI.00096-09
Galdiero, E., Lombardi, L., Falanga, A., Libralato, G., Guida, M., and Carotenuto, R. (2019). Biofilms: novel strategies based on antimicrobial peptides. Pharmaceutics 11:322. doi: 10.3390/pharmaceutics11070322
Ganz, T., and Lehrer, R. I. (1994). Defensins. Curr. Opin. Immunol. 6, 584–589. doi: 10.1016/0952-7915(94)90145-7
Garrett, T. R., Bhakoo, M., and Zhang, Z. (2008). Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 18, 1049–1056. doi: 10.1016/j.pnsc.2008.04.001
Garrison, A. T., Abouelhassan, Y., Kallifidas, D., Bai, F., Ukhanova, M., Mai, V., et al. (2015). Halogenated phenazines that potently eradicate biofilms, MRSA persister cells in non-biofilm cultures, and mycobacterium tuberculosis. Angew. Chem. 54, 14819–14823. doi: 10.1002/anie.201508155
Ghasemi, E., Ghalavand, Z., Goudarzi, H., Yeganeh, F., Hashemi, A., Dabiri, H., et al. (2018). Phenotypic and genotypic investigation of biofilm formation in clinical and environmental isolates of Acinetobacter baumannii. Arch. Clin. Infect. Dis. 13:14. doi: 10.5812/archcid.12914
Ghiselli, R., Giacometti, A., Cirioni, O., Mocchegiani, F., Silvestri, C., Orlando, F., et al. (2007). Pretreatment with the protegrin ib-367 affects gram-positive biofilm and enhances the therapeutic efficacy of linezolid in animal models of central venous catheter infection. J. Parenter. Enter. Nutr. 31, 463–468. doi: 10.1177/0148607107031006463
Giacometti, A., Cirioni, O., Ghiselli, R., Viticchi, C., Mocchegiani, F., Riva, A., et al. (2001). Effect of mono-dose intraperitoneal cecropins in experimental septic shock. Crit. Care Med. 29, 1666–1669. doi: 10.1097/00003246-200109000-00002
Gobbo, M., Biondi, L., Filira, F., Gennaro, R., Benincasa, M., Scolaro, B., et al. (2002). Antimicrobial peptides: synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1b analogues. J. Med. Chem. 45, 4494–4504. doi: 10.1021/jm020861d
Gomes, B., Augusto, M. T., Felício, M. R., Hollmann, A., Franco, O. L., Gonçalves, S., et al. (2018). Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 36, 415–429. doi: 10.1016/j.biotechadv.2018.01.004
Gong, M., and Makowski, L. (1992). Helical structure of P pili from Escherichia coli. Evidence from X-ray fiber diffraction and scanning transmission electron microscopy. J. Mol. Biol. 228, 735–742. doi: 10.1016/0022-2836(92)90860-M
Gonzalez-Escobedo, G., Marshall, J. M., and Gunn, J. S. (2011). Chronic and acute infection of the gall bladder by Salmonella typhi: understanding the carrier state. Nat. Rev. Microbiol. 9, 9–14. doi: 10.1038/nrmicro2490
Guo, C., Huang, Y., Zheng, H., Tang, L., He, J., Xiang, L., et al. (2012). Secretion and activity of antimicrobial peptide cecropin D expressed in Pichia pastoris. Exp. Ther. Med. 4, 1063–1068. doi: 10.3892/etm.2012.719
Gupta, S., Sharma, A. K., Jaiswal, S. K., and Sharma, V. K. (2016). Prediction of biofilm inhibiting peptides: an in silico approach. Front. Microbiol. 7:949. doi: 10.3389/fmicb.2016.00949
Haas, B. J., and Zody, M. C. (2010). Advancing RNA-Seq analysis. Nat. Biotechnol. 28, 421–423. doi: 10.1038/nbt0510-421
Hall-Stoodley, L., and Stoodley, P. (2009). Evolving concepts in biofilm infections. Cell Microbiol. 11, 1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x
Hall-Stoodley, L., Stoodley, P., Kathju, S., Høiby, N., Moser, C., Costerton, J., et al. (2012). Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65, 127–145. doi: 10.1111/j.1574-695X.2012.00968.x
Haney, E. F., Nguyen, L. T., Schibli, D. J., and Vogel, H. J. (2012). Design of a novel tryptophan-rich membrane-active antimicrobial peptide from the membrane-proximal region of the HIV glycoprotein, gp41. Beilstein J. Org. Chem 8, 1172–1184. doi: 10.3762/bjoc.8.130
Hashem, Y. A., Amin, H. M., Essam, T. M., Yassin, A. S., and Aziz, R. K. (2017). Biofilm formation in enterococci: genotype-phenotype correlations and inhibition by vancomycin. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-05901-0
Hedengren, M., Borge, K., and Hultmark, D. (2000). Expression and evolution of the Drosophila Attacin/Diptericin gene family. Biochem. Biophys. Res. Commun. 279, 574–581. doi: 10.1006/bbrc.2000.3988
Herbert, S., Bera, A., Nerz, C., Kraus, D., Peschel, A., Goerke, C., et al. (2007). Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in Staphylococci. PLoS Pathog. 3:e102. doi: 10.1371/journal.ppat.0030102
Herrmann, G., Yang, L., Wu, H., Song, Z., Wang, H., Høiby, N., et al. (2010). Colistin−tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202, 1585–1592. doi: 10.1086/656788
Hirt, H., and Gorr, S. U. (2013). Antimicrobial peptide GL13K is effective in reducing biofilms of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 4903–4910. doi: 10.1128/AAC.00311-13
Høiby, N. (2017). A short history of microbial biofilms and biofilm infections. APMIS 125, 272–275. doi: 10.1111/apm.12686
Hollmann, A., Martinez, M., Maturana, P., Semorile, L. C., and Maffia, P. C. (2018). Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 6:204. doi: 10.3389/fchem.2018.00204
Huigens, R. W., Abouelhassan, Y., and Yang, H. (2019). Phenazine antibiotic-inspired discovery of bacterial biofilm-eradicating agents. ChemBioChem 20, 2885–2902. doi: 10.1002/cbic.201900116
Hultmark, D., Engström, A., Andersson, K., Steiner, H., Bennich, H., and Boman, H. G. (1983). Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 2, 571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x
Hwang, I. S., Hwang, J. S., Hwang, J. H., Choi, H., Lee, E., Kim, Y., et al. (2013). Synergistic effect and antibiofilm activity between the antimicrobial peptide coprisin and conventional antibiotics against opportunistic bacteria. Curr. Microbiol. 66, 56–60. doi: 10.1007/s00284-012-0239-8
Hwang, J.-S., Lee, J., Kim, Y.-J., Bang, H.-S., Yun, E.-Y., Kim, S.-R., et al. (2009). Isolation and characterization of a defensin-like peptide (Coprisin) from the dung beetle, Copris tripartitus. Int. J. Pept. 2009, 1–5. doi: 10.1155/2009/136284
Jakubczyk, A., Karaś, M., Rybczyńska-Tkaczyk, K., Zielińska, E., and Zieliński, D. (2020). Current trends of bioactive peptides—new sources and therapeutic effect. Foods 9:846. doi: 10.3390/foods9070846
Jarczak, J., Kościuczuk, E. M., Lisowski, P., Strzałkowska, N., Jóźwik, A., Horbańczuk, J., et al. (2013). Defensins: natural component of human innate immunity. Hum. Immunol. 74, 1069–1079. doi: 10.1016/j.humimm.2013.05.008
Jennings, M. C., Ator, L. E., Paniak, T. J., Minbiole, K. P. C., and Wuest, W. M. (2014). Biofilm-eradicating properties of quaternary ammonium amphiphiles: simple mimics of antimicrobial peptides. ChemBioChem 15, 2211–2215. doi: 10.1002/cbic.201402254
Jenssen, H., Hamill, P., and Hancock, R. E. W. (2006). Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511. doi: 10.1128/CMR.00056-05
Jorge, P., Lourenço, A., and Pereira, M. O. (2012). New trends in peptide-based anti-biofilm strategies: a review of recent achievements and bioinformatic approaches. Biofouling 28, 1033–1061. doi: 10.1080/08927014.2012.728210
Kalsy, M., Tonk, M., Hardt, M., Dobrindt, U., Zdybicka-Barabas, A., Cytrynska, M., et al. (2020). The insect antimicrobial peptide cecropin A disrupts uropathogenic Escherichia coli biofilms. NPJ Biofilms Microbiomes 6:6. doi: 10.1038/s41522-020-0116-3
Kanduc, D. (2012). Homology, similarity, and identity in peptide epitope immunodefinition. J. Pept. Sci. 18, 487–494. doi: 10.1002/psc.2419
Kaplan, J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218. doi: 10.1177/0022034509359403
Kim, H., Jang, J., Kim, S., and Cho, J. H. (2016). Enhancement of the antimicrobial activity and selectivity of GNU7 against Gram-negative bacteria by fusion with LPS-targeting peptide. Peptides 82, 60–66.
Kim, I.-W., Lee, J. H., Subramaniyam, S., Yun, E.-Y., Kim, I., Park, J., et al. (2016). De Novo transcriptome analysis and detection of antimicrobial peptides of the american cockroach Periplaneta americana (Linnaeus). PLoS One 11:e0155304. doi: 10.1371/journal.pone.0155304
Kockum, K., Faye, I., Hofsten, P. V., Lee, J.-Y., Xanthopoulos, K. G., and Boman, H. G. (1984). Insect Immunity. Isolation and Sequence of Two cDNA Clones Corresponding to Acidic and Basic Attacins from Hyalophora cecropia. Heidelberg: European Molecular Biology Organization.
Kongthai, P., and Sitthisak, S. (2016). Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates molecular characterization of genotypes, virulence factors, gene transfer and copper transport system of Acinetobacter baumanni. Int. Microbiol. 19, 121–129. doi: 10.2436/20.1501.01.270
Koo, H. B., and Seo, J. (2019). Antimicrobial peptides under clinical investigation. Pept. Sci. 111:e24122. doi: 10.1002/pep2.24122
Krishnan, M., Choi, J., Jang, A., and Kim, Y. (2020). A novel peptide antibiotic, Pro10-1D, designed from insect defensin shows antibacterial and anti-inflammatory activities in sepsis models. Int. J. Mol. Sci. 21:6216. doi: 10.3390/ijms21176216
Kumar, N., Behera, B., Sagiri, S. S., Pal, K., Ray, S. S., and Roy, S. (2011). Bacterial vaginosis: etiology and modalities of treatment – A brief note. J. Pharm. Bioallied Sci. 3, 496–503. doi: 10.4103/0975-7406.90102
Langermann, S., Palaszynski, S., Barnhart, M., Auguste, G., Pinkner, J. S., Burlein, J., et al. (1997). Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276, 607–611. doi: 10.1126/science.276.5312.607
Lata, S., Sharma, B. K., and Raghava, G. P. S. (2007). Analysis and prediction of antibacterial peptides. BMC Bioinformatics 8:263. doi: 10.1186/1471-2105-8-263
Lauderdale, K. J., Boles, B. R., Cheung, A. L., and Horswill, A. R. (2009). Interconnections between sigma b, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation ∇. Infect. Immun. 77, 1623–1635. doi: 10.1128/IAI.01036-08
Lee, A. C.-L., Harris, J. L., Khanna, K. K., and Hong, J.-H. (2019). A comprehensive review on current advances in peptide drug development and design. Int. J. Mol. Sci. 20:2383. doi: 10.3390/ijms20102383
Lee, H. W., Koh, Y. M., Kim, J., Lee, J. C., Lee, Y. C., Seol, S. Y., et al. (2008). Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14, 49–54. doi: 10.1111/j.1469-0691.2007.01842.x
Lee, J. K., Luchian, T., and Park, Y. (2018). New antimicrobial peptide kills drug-resistant pathogens without detectable resistance. Oncotarget 9, 15616–15634. doi: 10.18632/oncotarget.24582
Lee, M., Bang, K., Kwon, H., and Cho, S. (2013). Enhanced antibacterial activity of an attacin-coleoptericin hybrid protein fused with a helical linker. Mol. Biol. Rep. 40, 3953–3960. doi: 10.1007/s11033-012-2472-4
Lehrer, R. I., and Lu, W. (2012). α-Defensins in human innate immunity. Immunol. Rev. 245, 84–112. doi: 10.1111/j.1600-065X.2011.01082.x
Li, X., Wu, B., Chen, H., Nan, K., Jin, Y., Sun, L., et al. (2018). Recent developments in smart antibacterial surfaces to inhibit biofilm formation and bacterial infections. J. Mater. Chem. B 6, 4274–4292. doi: 10.1039/C8TB01245H
Li, Y., Yang, L., Fu, J., Yan, M., Chen, D., and Zhang, L. (2017). The novel loop-mediated isothermal amplification based confirmation methodology on the bacteria in Viable but Non-Culturable (VBNC) state. Microb. Pathog. 111, 280–284. doi: 10.1016/j.micpath.2017.09.007
Li, Z. Q., Merrifield, R. B., Boman, I. A., and Boman, H. G. (1988). Effects on electrophoretic mobility and antibacterial spectrum of removal of two residues from synthetic sarcotoxin IA and addition of the same residues to cecropin B. FEBS Lett. 231, 299–302. doi: 10.1016/0014-5793(88)80837-8
Lin, X., Li, X., and Lin, X. (2020). A review on applications of computational methods in drug screening and design. Molecules 25:1375. doi: 10.3390/molecules25061375
Loehfelm, T. W., Luke, N. R., and Campagnari, A. A. (2008). Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 190, 1036–1044. doi: 10.1128/JB.01416-07
Luo, L. M., Wu, L. J., Xiao, Y. L., Zhao, D., Chen, Z. X., Kang, M., et al. (2015). Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 15:62. doi: 10.1186/s12866-015-0397-5
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222. doi: 10.1128/CMR.15.2.194-222.2002
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/JB.01202-06
Ma, L., Liu, X., Liang, H., Che, Y., Chen, C., Dai, H., et al. (2012). Effects of 14-alpha-lipoyl andrographolide on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 6088–6094. doi: 10.1128/AAC.01119-12
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20, e216–e230. doi: 10.1016/S1473-3099(20)30327-3
Maget-Dana, R., and Ptak, M. (1997). Penetration of the insect defensin A into phospholipid monolayers and formation of defensin A-lipid complexes. Biophys. J. 73, 2527–2533. doi: 10.1016/S0006-3495(97)78281-X
Magiorakos, A. P., Burns, K., Rodríguez Baño, J., Borg, M., Daikos, G., Dumpis, U., et al. (2017). Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European centre for disease prevention and control. Antimicrob. Resist. Infect. Control 6, 1–17. doi: 10.1186/s13756-017-0259-z
Mah, T. F. C., and O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/S0966-842X(00)01913-2
Malathi, K., and Ramaiah, S. (2018). Bioinformatics approaches for new drug discovery: a review. Biotechnol. Genet. Eng. Rev. 34, 243–260. doi: 10.1080/02648725.2018.1502984
Mandin, P., and Gottesman, S. (2010). Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29, 3094–3107. doi: 10.1038/emboj.2010.179
Manniello, M. D., Moretta, A., Salvia, R., Scieuzo, C., Lucchetti, D., Vogel, H., et al. (2021). Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 1:3. doi: 10.1007/s00018-021-03784-z
Marvasi, M., Chen, C., Carrazana, M., Durie, I. A., and Teplitski, M. (2014). Systematic analysis of the ability of Nitric Oxide donors to dislodge biofilms formed by Salmonella enterica and Escherichia coli O157:H7. AMB Express 4, 1–11. doi: 10.1186/s13568-014-0042-y
Mataraci, E., and Dosler, S. (2012). In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 56, 6366–6371. doi: 10.1128/AAC.01180-12
Matz, C., Bergfeld, T., Rice, S. A., and Kjelleberg, S. (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6, 218–226. doi: 10.1111/j.1462-2920.2004.00556.x
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D., and Kjelleberg, S. (2012). Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Melo, M. N., Ferre, R., and Castanho, M. A. R. B. (2009). Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 7, 245–250. doi: 10.1038/nrmicro2095
Memariani, H., Memariani, M., and Pourmand, M. R. (2018). Venom-derived peptide Mastoparan-1 eradicates planktonic and biofilm-embedded methicillin-resistant Staphylococcus aureus isolates. Microb. Pathog. 119, 72–80. doi: 10.1016/j.micpath.2018.04.008
Memariani, H., Memariani, M., Shahidi-Dadras, M., Nasiri, S., Akhavan, M. M., and Moravvej, H. (2019). Melittin: from honeybees to superbugs. Appl. Microbiol. Biotechnol. 103, 3265–3276. doi: 10.1007/s00253-019-09698-y
Michaux, C., Verneuil, N., Hartke, A., and Giard, J. C. (2014). Physiological roles of small RNA molecules. Microbiology 160, 1007–1019. doi: 10.1099/mic.0.076208-0
Minardi, D., Ghiselli, R., Cirioni, O., Giacometti, A., Kamysz, W., Orlando, F., et al. (2007). The antimicrobial peptide Tachyplesin III coated alone and in combination with intraperitoneal piperacillin-tazobactam prevents ureteral stent Pseudomonas infection in a rat subcutaneous pouch model. Peptides 28, 2293–2298. doi: 10.1016/j.peptides.2007.10.001
Mohamed, J. A., and Huang, D. B. (2007). Biofilm formation by enterococci. J. Med. Microbiol. 56, 1581–1588. doi: 10.1099/jmm.0.47331-0
Moore, A. J., Beazley, W. D., Bibby, M. C., and Devine, D. A. (1996). Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 37, 1077–1089. doi: 10.1093/jac/37.6.1077
Moreno, M., and Giralt, E. (2015). Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: Melittin, apamin and mastoparan. Toxins (Basel) 7, 1126–1150. doi: 10.3390/toxins7041126
Mottola, C., Semedo-Lemsaddek, T., Mendes, J. J., Melo-Cristino, J., Tavares, L., Cavaco-Silva, P., et al. (2016). Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J. Biomed. Sci. 23, 1–10. doi: 10.1186/s12929-016-0250-7
Mukherjee, K. (2020). In Search of New Anti-biofilm Agents from Insects | Nature Research Microbiology Community. Available online at: https://naturemicrobiologycommunity.nature.com/posts/59576-in-search-of-new-anti-biofilm-agents-from-insects (accessed December 26, 2020).
Mukhopadhyay, S., Bharath Prasad, A. S., Mehta, C. H., and Nayak, U. Y. (2020). Antimicrobial peptide polymers: no escape to ESKAPE pathogens–a review. World J. Microbiol. Biotechnol. 36, 131–132. doi: 10.1007/s11274-020-02907-1
Mulligan, V. K. (2020). The emerging role of computational design in peptide macrocycle drug discovery. Expert Opin. Drug Discov. 15, 833–852. doi: 10.1080/17460441.2020.1751117
Mulvey, M. A., Lopez-Boado, Y. S., Wilson, C. L., Roth, R., Parks, W. C., Heuser, J., et al. (1998). Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497. doi: 10.1126/science.282.5393.1494
Munita, J. M., and Arias, C. A. (2016). “Mechanisms of antibiotic resistance,” in Virulence Mechanisms of Bacterial Pathogens, eds I. T. Kudva, N. A. Cornick, P. J. Plummer, Q. Zhang, T. L. Nicholson, J. P. Bannantine, et al. (Washington, DC: ASM Press), 481–511. doi: 10.1128/9781555819286.ch17
Naorem, R. S., Urban, P., Goswami, G., and Fekete, C. (2020). Characterization of methicillin-resistant Staphylococcus aureus through genomics approach. 3 Biotech 10:401. doi: 10.1007/s13205-020-02387-y
Nasr, A., Olsén, A., Sjöbring, U., Müller-Esterl, W., and Björck, L. (1996). Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20, 927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x
Nehete, J., Bhambar, R., Narkhede, M., and Gawali, S. (2013). Natural proteins: sources, isolation, characterization and applications. Pharmacogn. Rev. 7, 107–116. doi: 10.4103/0973-7847.120508
Niba, E. T. E., Naka, Y., Nagase, M., Mori, H., and Kitakawa, M. (2008). A Genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 14, 237–246. doi: 10.1093/dnares/dsm024
Nikolaev, D. M., Shtyrov, A. A., Panov, M. S., Jamal, A., Chakchir, O. B., Kochemirovsky, V. A., et al. (2018). A comparative study of modern homology modeling algorithms for rhodopsin structure prediction. ACS Omega 3, 7555–7566. doi: 10.1021/acsomega.8b00721
Niu, C., Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2008). Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190, 3386–3392. doi: 10.1128/JB.01929-07
O’Brien, M. M., Walsh, E. J., Massey, R. C., Peacock, S. J., and Foster, T. J. (2002). Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. 4, 759–770. doi: 10.1046/j.1462-5822.2002.00231.x
Olsén, A., Jonsson, A., and Normark, S. (1989). Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655. doi: 10.1038/338652a0
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Otvos, L. (2000). Antibacterial peptides isolated from insects. J. Pept. Sci. 6, 497–511. doi: 10.1002/1099-1387(200010)6:10<497::aid-psc277>3.0.co;2-w
Overhage, J., Campisano, A., Bains, M., Torfs, E. C. W., Rehm, B. H. A., and Hancock, R. E. W. (2008). Human Host defense peptide ll-37 prevents bacterial biofilm formation †. Infect. Immun. 76, 4176–4182. doi: 10.1128/IAI.00318-08
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/mmbr.00078-15
Paganelli, F. L., Willems, R. J. L. W., Jansen, P., Hendrickx, A., Zhang, X., Bonten, M. J. M. B., et al. (2013). Enterococcus faecium biofilm formation: identification of major autolysin AtlAefm, associated acm surface localization, and AtlAefm-independent extracellular DNA release. mBio 4:e00154-13. doi: 10.1128/mBio.00154-13
Pamp, S. J., and Tolker-Nielsen, T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189, 2531–2539. doi: 10.1128/JB.01515-06
Park, S.-I., An, H. S., Chang, B. S., and Yoe, S. M. (2013). Expression, cDNA cloning, and characterization of the antibacterial peptide cecropin D from Agrius convolvuli. Animal Cells Syst. 17, 23–30. doi: 10.1080/19768354.2013.769465
Passador, L., and Iglewski, W. (1994). ADP-ribosylating toxins. Methods Enzymol. 235, 617–631. doi: 10.1016/0076-6879(94)35175-9
Peleg, A. Y., and Hooper, D. C. (2010). Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 362, 1804–1813. doi: 10.1056/nejmra0904124
Piperaki, E.-T., Syrogiannopoulos, G. A., Tzouvelekis, L. S., and Daikos, G. L. (2017). Klebsiella pneumoniae: virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Dis. J. 36, 1002–1005. doi: 10.1097/INF.0000000000001675
Pletzer, D., and Hancock, R. E. W. (2016). Antibiofilm peptides: potential as broadspectrum agents. J. Bacteriol. 198, 2572–2578. doi: 10.1128/JB.00017-16
Prajapati, V. K., Varma, M., and Vadassery, J. (2020). In silico identification of effector proteins from generalist herbivore Spodoptera litura. BMC Genomics 21:819. doi: 10.1186/s12864-020-07196-4
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Prigent-Combaret, C., Prensier, G., Le Thi, T. T., Vidal, O., Lejeune, P., and Dorel, C. (2000). Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2, 450–464. doi: 10.1046/j.1462-2920.2000.00128.x
Punsalang, A. P., and Sawyer, W. D. (1973). Role of Pili in the virulence of Neisseria gonorrhoeae. Infect. Immun. 8, 255–263. doi: 10.1128/iai.8.2.255-263.1973
Pushpanathan, M., Gunasekaran, P., and Rajendhran, J. (2013). Antimicrobial peptides: versatile biological properties. Int. J. Pept. 2013:675391. doi: 10.1155/2013/675391
Qvortrup, K., Hultqvist, L. D., Nilsson, M., Jakobsen, T. H., Jansen, C. U., Uhd, J., et al. (2019). Small Molecule anti-biofilm agents developed on the basis of mechanistic understanding of biofilm formation. Front. Chem. 7:742. doi: 10.3389/fchem.2019.00742
Rabin, N., Zheng, Y., Opoku-Temeng, C., Du, Y., Bonsu, E., and Sintim, H. O. (2015). Agents that inhibit bacterial biofilm formation. Future Med. Chem. 7, 647–671. doi: 10.4155/fmc.15.7
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Raheem, N., and Straus, S. K. (2019). Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front. Microbiol. 10:2866. doi: 10.3389/fmicb.2019.02866
Reen, F. J., Gutiérrez-Barranquero, J. A., Parages, M. L., and O’Gara, F. (2018). Coumarin: a novel player in microbial quorum sensing and biofilm formation inhibition. Appl. Microbiol. Biotechnol. 102, 2063–2073. doi: 10.1007/s00253-018-8787-x
Reffuveille, F., de la Fuente-Núñez, C., Fairfull-Smith, K. E., and Hancock, R. E. W. (2015). Potentiation of ciprofloxacin action against Gram-negative bacterial biofilms by a nitroxide. Pathog. Dis. 73:16. doi: 10.1093/femspd/ftv016
Reffuveille, F., De La Fuente-Núñez, C., Mansour, S., and Hancock, R. E. W. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. doi: 10.1128/AAC.03163-14
Rohde, H., Burandt, E. C., Siemssen, N., Frommelt, L., Burdelski, C., Wurster, S., et al. (2007). Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720. doi: 10.1016/j.biomaterials.2006.11.046
Roizman, D., Vidaillac, C., Givskov, M., and Yang, L. (2017). In vitro evaluation of biofilm dispersal as a therapeutic strategy to restore antimicrobial efficacy. Antimicrob. Agents Chemother. 61, e1028–e1017. doi: 10.1128/AAC.01088-17
Rossi, E., Cimdins, A., Lüthje, P., Brauner, A., Sjöling, Å, Landini, P., et al. (2018). “It’s a gut feeling”–Escherichia coli biofilm formation in the gastrointestinal tract environment. Crit. Rev. Microbiol. 44, 1–30. doi: 10.1080/1040841X.2017.1303660
Rossi, L. M., Rangasamy, P., Zhang, J., Qiu, X. Q., and Wu, G. Y. (2008). Research advances in the development of peptide antibiotics. J. Pharm. Sci. 97, 1060–1070. doi: 10.1002/jps.21053
Roy, R., Tiwari, M., Donelli, G., and Tiwari, V. (2018). Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9, 522–554. doi: 10.1080/21505594.2017.1313372
Schembri, M. A., and Klemm, P. (2001). Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20, 3074–3081. doi: 10.1093/emboj/20.12.3074
Schlievert, P. M., and Peterson, M. L. (2012). Glycerol monolaurate antibacterial activity in broth and biofilm cultures. PLoS One 7:e40350. doi: 10.1371/journal.pone.0040350
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Seno, Y., Kariyama, R., Mitsuhata, R., Monden, K., and Kumon, H. (2005). Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med. Okayama 59, 79–87. doi: 10.18926/AMO/31979
Serra, D. O., Richter, A. M., and Hengge, R. (2013). Cellulose as an architectural element in spatially structured escherichia coli biofilms. J. Bacteriol. 195, 5540–5554. doi: 10.1128/JB.00946-13
Serray, B., Oufrid, S., Hannaoui, I., Bourjilate, F., Soraa, N., Mliji, M., et al. (2016). Genes encoding adhesion factors and biofilm formation in methicillinresistant Staphylococcus aureus in Morocco. J. Infect. Dev. Ctries. 10, 863–869. doi: 10.3855/jidc.8361
Shannon, O., and Flock, J. I. (2004). Extracellular fibrinogen binding protein, Efb, from Staphylococcus aureus binds to platelets and inhibits platelet aggregation. Thromb. Haemost. 91, 779–789. doi: 10.1160/th03-05-0287
Shirmohammadlou, N., Zeighami, H., Haghi, F., and Kashefieh, M. (2018). Resistance pattern and distribution of carbapenemase and antiseptic resistance genes among multidrug-resistant Acinetobacter baumannii isolated from intensive care unit patients. J. Med. Microbiol. 67, 1467–1473. doi: 10.1099/jmm.0.000826
Shirtliff, M. E., Peters, B. M., and Jabra-Rizk, M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299, 1–8. doi: 10.1111/j.1574-6968.2009.01668.x
Silveira, G. G. O. S., Torres, M. D. T., Ribeiro, C. F. A., Meneguetti, B. T., Carvalho, C. M. E., De La Fuente-Nunez, C., et al. (2021). Antibiofilm peptides: relevant preclinical animal infection models and translational potential. ACS Pharmacol. Transl. Sci. 4, 55–73. doi: 10.1021/acsptsci.0c00191
Simm, R., Morr, M., Kader, A., Nimtz, M., and Römling, U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x
Simões, M., Simões, L. C., and Vieira, M. J. (2010). A review of current and emergent biofilm control strategies. LWT Food Sci. Technol. 43, 573–583. doi: 10.1016/j.lwt.2009.12.008
Slavokhotova, A. A., Shelenkov, A. A., Korostyleva, T. V., Rogozhin, E. A., Melnikova, N. V., Kudryavtseva, A. V., et al. (2017). Defense peptide repertoire of Stellaria media predicted by high throughput next generation sequencing. Biochimie 135, 15–27. doi: 10.1016/j.biochi.2016.12.017
Slavokhotova, A. A., Shelenkov, A. A., and Odintsova, T. I. (2015). Prediction of Leymus arenarius (L.) antimicrobial peptides based on de novo transcriptome assembly. Plant Mol. Biol. 89, 203–214. doi: 10.1007/s11103-015-0346-6
Sojka, M., Valachova, I., Bucekova, M., and Majtan, J. (2016). Antibiofilm efficacy of honey and bee-derived defensin-1 on multispecies wound biofilm. J. Med. Microbiol. 65, 337–344. doi: 10.1099/jmm.0.000227
Srisailam, S., Arunkumar, A. I., Wang, W., Yu, C., and Chen, H. M. (2000). Conformational study of a custom antibacterial peptide cecropin B1: implications of the lytic activity. Biochim. Biophys. Acta 1479, 275–285. doi: 10.1016/S0167-4838(00)00008-X
Stewart, P. S. (1996). Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40, 2517–2522. doi: 10.1128/aac.40.11.2517
Stoica, P., Chifiriuc, M. C., Rapa, M., and Lazãr, V. (2017). “Overview of biofilm-related problems in medical devices,” in Biofilms and Implantable Medical Devices: Infection and Control, eds Y. Deng and W. Lv (Amsterdam: Elsevier Inc), 3–23. doi: 10.1016/B978-0-08-100382-4.00001-0
Stoodley, P., Sauer, K., Davies, D. G., and Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Subashchandrabose, S., Smith, S. N., Spurbeck, R. R., Kole, M. M., and Mobley, H. L. T. (2013). Genome-wide detection of fitness genes in uropathogenic escherichia coli during systemic infection. PLoS Pathog. 9:e1003788. doi: 10.1371/journal.ppat.1003788
Sun, M., Zhou, Z., Dong, J., Zhang, J., Xia, Y., and Shu, R. (2016). Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microb. Pathog. 99, 196–203. doi: 10.1016/j.micpath.2016.08.025
Sun, S. C., Lindstrom, I., Lee, J. −Y., and Faye, I. (1991). Structure and expression of the attacin genes in Hyalophora cecropia. Eur. J. Biochem. 196, 247–254. doi: 10.1111/j.1432-1033.1991.tb15811.x
Sung, J. Y. (2018). Molecular Characterization and antimicrobial susceptibility of biofilm-forming acinetobacter baumannii clinical isolates from Daejeon, Korea. Korean J. Clin. Lab. Sci. 50, 100–109. doi: 10.15324/kjcls.2018.50.2.100
Swain, S. S., Paidesetty, S. K., Dehury, B., Das, M., Vedithi, S. C., and Padhy, R. N. (2020). Computer-aided synthesis of dapsone-phytochemical conjugates against dapsone-resistant Mycobacterium leprae. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-63913-9
Swain, S. S., Paidesetty, S. K., Dehury, B., Sahoo, J., Vedithi, S. C., Mahapatra, N., et al. (2018). Molecular docking and simulation study for synthesis of alternative dapsone derivative as a newer antileprosy drug in multidrug therapy. J. Cell. Biochem. 119, 9838–9852. doi: 10.1002/jcb.27304
Tan, L., Li, S. R., Jiang, B., Hu, X. M., and Li, S. (2018). Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 9:25. doi: 10.3389/fmicb.2018.00055
Tang, J., Chen, J., Li, H., Zeng, P., and Li, J. (2013). Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among Staphylococcus aureus strains isolated from different sources. Foodborne Pathog. Dis. 10, 757–763. doi: 10.1089/fpd.2012.1474
Tavares, W. (2000). Problem gram-positive bacteria: Resistance in staphylococci, enterococci, and pneumococi to antimicrobial drugs. Rev. Soc. Bras. Med. Trop. 33, 281–301. doi: 10.1590/s0037-86822000000300008
Teerapo, K., Roytrakul, S., Sistayanarain, A., and Kunthalert, D. (2019). A scorpion venom peptide derivative BmKn?22 with potent antibiofilm activity against Pseudomonas aeruginosa. PLoS One 14:e0218479. doi: 10.1371/journal.pone.0218479
Theuretzbacher, U., Outterson, K., Engel, A., and Karlén, A. (2020). The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 18, 275–285. doi: 10.1038/s41579-019-0288-0
Thomas, V. C., Thurlow, L. R., Boyle, D., and Hancock, L. E. (2008). Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190, 5690–5698. doi: 10.1128/JB.00314-08
Tiwari, S. K., Noll, K. S., Cavera, V. L., and Chikindas, M. L. (2015). Improved antimicrobial activities of synthetic-hybrid bacteriocins designed from enterocin E50-52 and pediocin PA-1. Appl. Environ. Microbiol. 81, 1661–1667. doi: 10.1128/AEM.03477-14
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., and Actis, L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484. doi: 10.1099/mic.0.26541-0
Toro Segovia, L. J., Téllez Ramírez, G. A., Henao Arias, D. C., Rivera Duran, J. D., Bedoya, J. P., and Castaño Osorio, J. C. (2017). Identification and characterization of novel cecropins from the Oxysternon conspicillatum neotropic dung beetle. PLoS One 12:e0187914. doi: 10.1371/journal.pone.0187914
Tryselius, Y., Samakovlis, C., Kimbrell, D. A., and Hultmark, D. (1992). CecC, a cecropin gene expressed during metamorphosis in Drosophila pupae. Eur. J. Biochem. 204, 395–399. doi: 10.1111/j.1432-1033.1992.tb16648.x
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A., and Filloux, A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 98, 6911–6916. doi: 10.1073/pnas.111551898
Verderosa, A. D., Harris, J., Dhouib, R., Totsika, M., and Fairfull-Smith, K. E. (2019a). Eradicating uropathogenic Escherichia coli biofilms with a ciprofloxacin-dinitroxide conjugate. Medchemcomm 10, 699–711. doi: 10.1039/c9md00062c
Verderosa, A. D., Mansour, S. C., De La Fuente-Núñez, C., Hancock, R. E. W., and Fairfull-Smith, K. E. (2016). Synthesis and evaluation of ciprofloxacin-nitroxide conjugates as anti-biofilm agents. Molecules 21:841. doi: 10.3390/molecules21070841
Verderosa, A. D., Totsika, M., and Fairfull-Smith, K. E. (2019b). Bacterial biofilm eradication agents: a current review. Front. Chem. 7:824. doi: 10.3389/fchem.2019.00824
Vestby, L. K., Grønseth, T., Simm, R., and Nesse, L. L. (2020). Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel) 9:59. doi: 10.3390/antibiotics9020059
Vizioli, J., Bulet, P., Charlet, M., Lowenberger, C., Blass, C., Müller, H. M., et al. (2000). Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 9, 75–84. doi: 10.1046/j.1365-2583.2000.00164.x
Wagner, A. M., Gran, M. P., and Peppas, N. A. (2018). Designing the new generation of intelligent biocompatible carriers for protein and peptide delivery. Acta Pharm. Sin. B 8, 147–164. doi: 10.1016/j.apsb.2018.01.013
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093.
Wang, X., Preston, J. F., and Romeo, T. (2004). The pgaABCD Locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487
Whittaker, C. J., Klier, C. M., and Kolenbrander, P. E. (1996). Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50, 513–552. doi: 10.1146/annurev.micro.50.1.513
Wilson, S. S., Wiens, M. E., and Smith, J. G. (2013). Antiviral mechanisms of human defensins. J. Mol. Biol. 425, 4965–4980. doi: 10.1016/j.jmb.2013.09.038
Wimley, W. C., and Hristova, K. (2011). Antimicrobial peptides: Successes, challenges and unanswered questions. J. Membr. Biol. 239, 27–34. doi: 10.1007/s00232-011-9343-0
Worley, B. V., Schilly, K. M., and Schoenfisch, M. H. (2015). Anti-biofilm efficacy of dual-action nitric oxide-releasing alkyl chain modified poly(amidoamine) dendrimers. Mol. Pharm. 12, 1573–1583. doi: 10.1021/acs.molpharmaceut.5b00006
Worthington, R. J., Richards, J. J., and Melander, C. (2012). Small molecule control of bacterial biofilms. Org. Biomol. Chem. 10, 7457–7474. doi: 10.1039/c2ob25835h
Wu, Q., Patočka, J., and Kuča, K. (2018). Insect antimicrobial peptides, a mini review. Toxins (Basel). 10:461. doi: 10.3390/toxins10110461
Wu, X., Li, Z., Li, X., Tian, Y., Fan, Y., Yu, C., et al. (2017). Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Devel. Ther. 11, 939–946. doi: 10.2147/DDDT.S107195
Xu, W., Zhu, X., Tan, T., Li, W., and Shan, A. (2014). Design of embedded-hybrid antimicrobial peptides with enhanced cell selectivity and anti-biofilm activity. PLoS One 9:e98935. doi: 10.1371/journal.pone.0098935
Yan, S., and Wu, G. (2019). Can biofilm be reversed through quorum sensing in Pseudomonas aeruginosa? Front. Microbiol. 10:23. doi: 10.3389/fmicb.2019.01582
Zafar, F., Gupta, A., Thangavel, K., Khatana, K., Sani, A. A., Ghosal, A., et al. (2020). Physicochemical and pharmacokinetic analysis of anacardic acid derivatives. ACS Omega 5, 6021–6030. doi: 10.1021/acsomega.9b04398
Zasloff, M. (2019). Antimicrobial peptides of multicellular organisms: my perspective. Adv. Exp. Med. Biol. 1117, 3–6. doi: 10.1007/978-981-13-3588-4_1
Zhao, G., Usui, M. L., Lippman, S. I., James, G. A., Stewart, P. S., Fleckman, P., et al. (2013). Biofilms and inflammation in chronic wounds. Adv. Wound Care 2, 389–399. doi: 10.1089/wound.2012.0381
Zhao, L., and Lu, W. (2014). Defensins in innate immunity. Curr. Opin. Hematol. 21, 37–42. doi: 10.1097/MOH.0000000000000005
Zheng, J. X., Lin, Z. W., Chen, C., Chen, Z., Lin, F. J., Wu, Y., et al. (2018). Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front. Cell. Infect. Microbiol. 8:21. doi: 10.3389/fcimb.2018.00021
Zheng, W., Wuyun, Q., Li, Y., Mortuza, S. M., Zhang, C., Pearce, R., et al. (2019). Detecting distant-homology protein structures by aligning deep neural-network based contact maps. PLoS Comput. Biol. 15:e1007411. doi: 10.1371/journal.pcbi.1007411
Keywords: antimicrobial peptide, biofilms, ESKAPE, insect, multidrug-resistant bacteria, molecular docking, anti-biofilm mechanism of action, therapeutic and prophylactic strategies
Citation: Sahoo A, Swain SS, Behera A, Sahoo G, Mahapatra PK and Panda SK (2021) Antimicrobial Peptides Derived From Insects Offer a Novel Therapeutic Option to Combat Biofilm: A Review. Front. Microbiol. 12:661195. doi: 10.3389/fmicb.2021.661195
Received: 02 February 2021; Accepted: 12 April 2021;
Published: 10 June 2021.
Edited by:
Ester Boix, Universitat Autònoma de Barcelona, SpainReviewed by:
Jhon Carlos Castaño, University of Quindío, ColombiaCarolina Muñoz-Camargo, University of Los Andes, Colombia
Copyright © 2021 Sahoo, Swain, Behera, Sahoo, Mahapatra and Panda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sujogya Kumar Panda, sujogyapanda@gmail.com
 Alaka Sahoo
Alaka Sahoo Shasank Sekhar Swain
Shasank Sekhar Swain Ayusman Behera
Ayusman Behera Gunanidhi Sahoo4
Gunanidhi Sahoo4 Sujogya Kumar Panda
Sujogya Kumar Panda