- 1Key Laboratory of Earth and Planetary Physics, Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing, China
- 2France-China Joint Laboratory for Evolution and Development of Magnetotactic Multicellular Organisms, Chinese Academy of Sciences, Beijing, China
- 3College of Earth and Planetary Sciences, University of Chinese Academy of Sciences, Beijing, China
- 4State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Magnetotactic bacteria (MTB) are a group of microbes that biomineralize membrane-bound, nanosized magnetite (Fe3O4), and/or greigite (Fe3S4) crystals in intracellular magnetic organelle magnetosomes. MTB belonging to the Nitrospirae phylum can form up to several hundreds of Fe3O4 magnetosome crystals and dozens of sulfur globules in a single cell. These MTB are widespread in aquatic environments and sometimes account for a significant proportion of microbial biomass near the oxycline, linking these lineages to the key steps of global iron and sulfur cycling. Despite their ecological and biogeochemical importance, our understanding of the diversity and ecophysiology of magnetotactic Nitrospirae is still very limited because this group of MTB remains unculturable. Here, we identify and characterize two previously unknown MTB populations within the Nitrospirae phylum through a combination of 16S rRNA gene-based and genome-resolved metagenomic analyses. These two MTB populations represent distinct morphotypes (rod-shaped and coccoid, designated as XYR, and XYC, respectively), and both form more than 100 bullet-shaped magnetosomal crystals per cell. High-quality draft genomes of XYR and XYC have been reconstructed, and they represent a novel species and a novel genus, respectively, according to their average amino-acid identity values with respect to available genomes. Accordingly, the names Candidatus Magnetobacterium cryptolimnobacter and Candidatus Magnetomicrobium cryptolimnococcus for XYR and XYC, respectively, were proposed. Further comparative genomic analyses of XYR, XYC, and previously reported magnetotactic Nitrospirae reveal the general metabolic potential of this MTB group in distinct microenvironments, including CO2 fixation, dissimilatory sulfate reduction, sulfide oxidation, nitrogen fixation, or denitrification processes. A remarkably conserved magnetosome gene cluster has been identified across Nitrospirae MTB genomes, indicating its putative important adaptive roles in these bacteria. Taken together, the present study provides novel insights into the phylogenomic diversity and ecophysiology of this intriguing, yet poorly understood MTB group.
Introduction
A diverse group of bacteria in nature can sense and swim along the geomagnetic field, a behavior known as magnetotaxis or microbial magnetoreception (Blakemore, 1975; Lefèvre and Wu, 2013; Lin et al., 2020a). These unique microorganisms, referred to as magnetotactic bacteria (MTB), can actively uptake a large amount of Fe(II) and/or Fe(III) from environments and accumulate it within the cell during the biomineralization of nanosized, membrane-bound Fe3O4, and/or Fe3S4 magnetosomes, which are usually arranged in well-ordered, chain-like structures that are responsible for magnetotaxis. Consequently, the intracellular iron content of MTB is much higher (ca. 100- to 1,000-fold) than that in other microorganisms (Lin et al., 2014a; Amor et al., 2020a,c). MTB have been discovered across various environments from freshwater to marine ecosystems, and they make important contributions to the global cycling of iron and other elements (such as sulfur, phosphorus, nitrogen, and carbon; Cox et al., 2002; Rivas-Lamelo et al., 2017; Schulz-Vogt et al., 2019; Monteil and Lefèvre, 2020). The determination of the genetic basis for magnetosome biogenesis reveals a complex sequence of steps controlled by dozens of genes clustered at one genomic locus, known as a magnetosome gene cluster (MGC; Grünberg et al., 2001; Murat et al., 2010; Uebe and Schüler, 2016; Lin et al., 2017a; McCausland and Komeili, 2020). For many years, the taxonomic distribution of MTB has been considered to be restricted to a few bacterial phyla (Amann et al., 2006). However, over the past decade, our knowledge of MTB diversity has been significantly expanded through the applications of cultivation-dependent and -independent approaches (Kolinko et al., 2012; Bazylinski et al., 2013; Lefèvre and Bazylinski, 2013; Lin et al., 2017a; Amor et al., 2020b). MTB have thus far been identified in at least 16 phylum-level lineages (Lin et al., 2018, 2020b; Uzun et al., 2020), suggesting unexpected taxonomic diversity of magnetosome biogenesis and magnetotaxis across the domain Bacteria.
Magnetotactic bacteria phylogenetically affiliated within the Nitrospirae phylum are of great interest because of their biomineralization of up to several hundreds of Fe3O4 magnetosomal crystals and formation of dozens of sulfur globules in a single cell, linking these microorganisms to the key steps of iron and sulfur cycling in aquatic ecosystems (Spring et al., 1993; Jogler et al., 2010; Lin et al., 2014b; Li et al., 2020). Historically, Nitrospirae MTB have only been discovered in freshwater environments (Spring et al., 1993; Flies et al., 2005; Amann et al., 2006); however, recent studies have revealed that these MTB are globally distributed in a wider range of environments than anticipated previously, including hot springs (Lefèvre et al., 2010; Uzun et al., 2020), estuary and marine ecosystems (Lin et al., 2017a; Qian et al., 2019), and acidic peatlands (Lin et al., 2020b). Due to the lack of cultivated representatives, much of our understanding of Nitrospirae MTB was obtained through 16S rRNA gene-based methods. Recently, since the first report of a nearly complete draft genome of Candidatus (Ca.) Magnetobacterium casensis through a targeted metagenomic approach (Lin et al., 2014b), a growing number of Nitrospirae MTB genomes have been reconstructed from distinct ecosystems through applications of genome-resolved metagenomics (Lin et al., 2017b, 2018, 2020b; Koziaeva et al., 2020; Uzun et al., 2020) and single-cell genomics (Kolinko et al., 2016). Genomic analyses of Ca. Magnetobacterium casensis (Lin et al., 2014b) and Ca. Magnetobacterium bavaricum (Kolinko et al., 2016) indicate an autotrophic lifestyle with capacity of CO2 fixation via the reductive acetyl-CoA (Wood-Ljungdahl or WL) pathway or reductive TCA (rTCA) pathway. Both species are predicted to conduct denitrification, sulfur oxidation, and/or sulfate reduction in different microenvironments across anoxic and microoxic layers near the oxic-anoxic transition zone (Lin et al., 2014b; Kolinko et al., 2016).
Despite these glimpses into the diversity and metabolic potential of magnetotactic Nitrospirae, the vast majority of MTB in this group are underexplored, and a comprehensive phylogenomic and comparative genomic analysis spanning a broad representation of the Nitrospirae MTB remains lacking. Here, we report the identification and characterization of two novel Nitrospirae MTB populations and the reconstruction of their high-quality draft genome sequences. The newly recovered genomes were further compared with the reported Nitrospirae MTB genomes. Findings of this study extend our understanding of the phylogenomic diversity and metabolic potential of this intriguing, yet poorly understood MTB group.
Materials and Methods
Observation and Magnetic Enrichment of Magnetotactic Bacteria
Surface sediments (0–10 cm) were collected from freshwater Lake Xianyang (34.33°N 108.72°E) in Xianyang City, Shaanxi Province, China, transferred to 600-ml plastic flasks, covered with approximately 100 ml of lake water and transported to the laboratory. The presence of MTB in sediments was examined using the hanging-drop method (Greenberg et al., 2005) under an Olympus BX51 microscope (Olympus, Tokyo, Japan). MTB cells were magnetically collected using a double-ended open magnetic separation apparatus (referred to as MTB trap) as previously described (Jogler et al., 2009). Briefly, approximately 300 ml of surface sediments were transferred to the MTB trap, and a homogeneous magnetic field was applied for MTB cell enrichment for 4 h. The retrieved MTB cells were then washed with double-distilled water.
Fluorescence in situ Hybridization and Transmission Electron Microscopy
Two distinct morphotypes of MTB have been observed, designated as XYR and XYC. XYR and XYC are abbreviations for Xianyang Lake rods and Xianyang Lake cocci, respectively. 16S rRNA genes were directly amplified from magnetically enriched MTB cells, using universal bacterial primers 27F (5′- AGAGTTTGATCCTGGCTCAG -3′) and 1492R (5′- GGTTACCTTGTTACGACTT -3′). The PCR protocol included denaturation at 94∘C for 3 min; 35 cycles of 94∘C for 45 s, 50∘C for 1 min, and 72∘C for 1.5 min; followed by 10 min at 72∘C and a holding temperature of 4∘C. Sequencing was conducted on 30 randomly selected clones. To identify the newly discovered MTB cells, two oligonucleotide probes specific to 16S rRNA gene sequences of XYR (5′-TCCCTTGCGAGAGTCGTTAT-3′) and XYC (5′- TGCCGAAACACCAGTCGTCC -3′) were designed and applied to in situ hybridization of the enriched MTB cells. The candidate probes were manually designed with district regions and further checked using SILVA TestProbe (Quast et al., 2012). The Ca. Magnetobacterium bavaricum-specific probe (5′-GCCATCCCCTCGCTTACT-3′, named BaP in this study; Spring et al., 1993) that is specific for most known Nitrospirae MTB (Lin et al., 2011) and the 4′,6-diamidino-2′-phenylindole were used as control. Probes for XYR and XYC were synthesized and fluorescently labeled with sulfoindocyanine dye Cy3 at the 5′ ends, and the probe BaP was labeled with fluorescein phosphoramidite FAM at the 5′ end. Fluorescence in situ hybridization (FISH) was then performed as previously described (Spring et al., 1993; Pernthaler et al., 2001). Briefly, 2 μl of cell mixtures were dropped onto gelatin-coated glass slides, allowed to dry, and dehydrated through an ethanol gradient (50, 80, and 100% for 3 min each). In situ hybridization was performed at 46∘C for 3 h in hybridization buffer with 35% (vol/vol) formamide. A fluorescence microscope Olympus BX51 (Olympus, Tokyo, Japan) was used to record optical sections. For transmission electron microscope (TEM) observation, a 10-μl drop of magnetic enrichment was deposited on a Formvar-carbon-coated copper grid and was allowed to air dry in a laminar flow hood. MTB cells were imaged using a JEM-2100 HR TEM (JEOL, Tokyo, Japan) at 200 kV.
Metagenomic Sequencing, Assembly, and Genome Binning
Metagenomic sequencing, assembly, and MTB genome binning were performed as previously described (Zhang et al., 2020). Briefly, metagenomic DNA was directly amplified from magnetically enriched cells using the Genomiphi V2 DNA amplification kit (GE Healthcare, United States) according to the manufacturer’s protocol. Shotgun sequencing was performed with an Illumina HiSeq 2500 using the pair-end 150 × 150 library with a 270-bp insert size. The metagenomic data set was assembled de novo using metaSPAdes (version 3.13.0; Nurk et al., 2017) with default parameters. Initial assembled scaffolds were binned separately using MetaBAT2 (version 2.12.1; Kang et al., 2019), MaxBin2 (version 2.2.4; Wu et al., 2016), CONCOCT (Alneberg et al., 2014), and MyCC (Lin and Liao, 2016), and the high-scoring, non-redundant set of bins were dereplicated and selected using DASTool (Sieber et al., 2018). The completeness and contamination of the acquired genomes were assessed with CheckM (Parks et al., 2015) using the “lineage_wf” workflow. Statistics for each genome were obtained using QUAST (version 4.2; Gurevich et al., 2013). Genomes were annotated using Prokka (version 1.13.3; Seemann, 2014). Reconstructed genomes were checked manually for the presence of magnetosome genes according to the known magnetosome genes from previously reported Nitrospirae MTB, including Ca. Magnetobacterium casensis (Lin et al., 2014b), Ca. Magnetobacterium bavaricum (Kolinko et al., 2016), and Ca. Magnetominusculus xianensis (Lin et al., 2017b). The average amino acid identity (AAI) and average nucleotide identity (ANI) values were calculated using enveomics (Rodriguez-R and Konstantinidis, 2016). The percentage of conserved proteins (POCP) was calculated using BLASTP according to Qin et al. (2014).
Phylogenetic Analyses
For genome-based phylogenetic analyses, all available magnetotactic Nitrospirae genomes (n = 38) and non-MTB Nitrospirae genomes (a total of 138 GTDB species representatives) as of October 2020 were downloaded from the NCBI (Agarwala et al., 2018) and GTDB (Parks et al., 2018) databases. Those non-MTB genomes were further dereplicated using dRep (Olm et al., 2017) with “-sa 0.99” for dereplication at 99% ANI. The phylogenomic tree was inferred from 120 concatenated bacterial single-copy marker proteins as described previously (Parks et al., 2017) and was calculated using IQ-TREE (version 1.6.9; Nguyen et al., 2015) under the “TEST” option for the best-fit substitution model selection (LG + F + I + G4) with 1,000 ultrafast bootstraps. The genome tree was rooted with Thermodesulfobacterium geofontis OPF15 and Thermodesulfobacterium commune DSM 2178.
For a 16S rRNA gene-based tree, 16S rRNA gene sequences retrieved from those two genomes reconstructed in this study were searched against NCBI’s nucleotide collection (nr/nt) using BLASTn. The hits with percentage identity ranging from 100 to 90% and query coverage from 100 to 95% were downloaded and aligned by SINA (Pruesse et al., 2012). trimAL (Capella-Gutiérrez et al., 2009; -gappyout) was performed to eliminate poorly aligned portions of the alignment. A phylogenetic tree was subsequently constructed using IQ-TREE (Nguyen et al., 2015) under the TEST option for best model selection (TIM3 + F + I + G4) with 1,000 ultrafast bootstraps. The tree was rooted with Thermodesulfobacterium geofontis OPF15 and Thermodesulfobacterium commune DSM 2178.
Comparative Genomic Analysis and Metabolic Predictions
For comparative genomic analysis, only MTB genomes from the Nitrospirae phylum with >90% completeness and <5% contamination were considered. These genomes were further dereplicated using dRep (Olm et al., 2017) with “-sa 0.99” for dereplication at 99% ANI. Finally, a total of 13 high-quality representative Nitrospirae MTB genomes were selected (including XYR and XYC reconstructed in the present study). These genomes were annotated against the KEGG database using the KofamKOALA server (Aramaki et al., 2020). An overview of the completeness of general metabolic pathways was estimated and visualized using KEGG-Decoder (Graham et al., 2018), and a heat map was generated using “static” visualization mode. Putative magnetosome genes of these genomes were checked using NCBI PSI-BLAST (Altschul et al., 1997) and further manually inspected. Comparison of MGCs were performed with clinker (Gilchrist and Chooi, 2021).
Data Availability
Genome sequences of Ca. Magnetobacterium cryptolimnobacter strain XYR and Ca. Magnetomicrobium cryptolimnococcus strain XYC have been deposited in GenBank under the accession numbers JAGYWH000000000 and JAGYWI000000000 (BioProject number PRJNA400260).
Results and Discussion
Two Novel Magnetotactic Bacteria Populations Affiliated Within the Nitrospirae Phylum
Light-microscope observation of surface sediments from freshwater Lake Xianyang showed two distinct morphotypes of MTB (rod-shaped and coccoid, Figure 1A). TEM observation reveals that cells of both rods and cocci form intracellular bullet-shaped magnetosome crystals organized into several bundles of chains (Figures 1C,D). Analysis of retrieved 16S rRNA gene sequences revealed only two populations (defined by a 97% identity threshold) in magnetically enriched MTB cells and FISH results turned out that they were either rods or cocci (Figure 1). To reconstruct their draft genomes, cells of MTB in the sediments were magnetically enriched using the “MTB trap,” and their metagenome was sequenced using Illumina techniques. Assembly and binning resulted in two near-complete genome bins (designated XYR and XYC), which consist of 195 and 91 scaffolds with average GC contents of 48.61 and 37.75%, respectively (Table 1). Genomes of XYR and XYC were 4.23 and 3.58 Mbp and were estimated to be 96.97% complete with 2.73% contamination and 99.94% complete with 1.82% contamination, respectively. Each genome contained a complement of 23S, 16S, and 5S rRNA genes and >18 tRNAs, thus exceeding the high-quality level of the minimum information about a metagenome-assembled genome standard (Bowers et al., 2017). 16S rRNA gene identities (91.98%) indicate that XYR and XYC belong to different genus (Yarza et al., 2014). The average AAI value between XYR and XYC was 53.73%, indicating that they represent organisms from two distinct genera according to Konstantinidis et al. (2017). The ANI value between XYR and XYC was too low (only ∼27%) to effectively define their genetic relationship. The pairwise value of the POCP between XYR and XYC was 50.32%, which is near the proposed genus threshold of 50% (Qin et al., 2014). Taking together, XYR and XYC represent two MTB populations from different genus. BLASTn analysis of 16S rRNA gene sequences of XYR and XYC revealed their best hits to an uncultured Nitrospirae bacterium clone OTU7 (GenBank accession number MT138725, 99.66% identity) and to Ca. Magnetoovum mohavensis strain LO-1 (GU979422, 98.60% identity; Lefèvre et al., 2011), respectively.
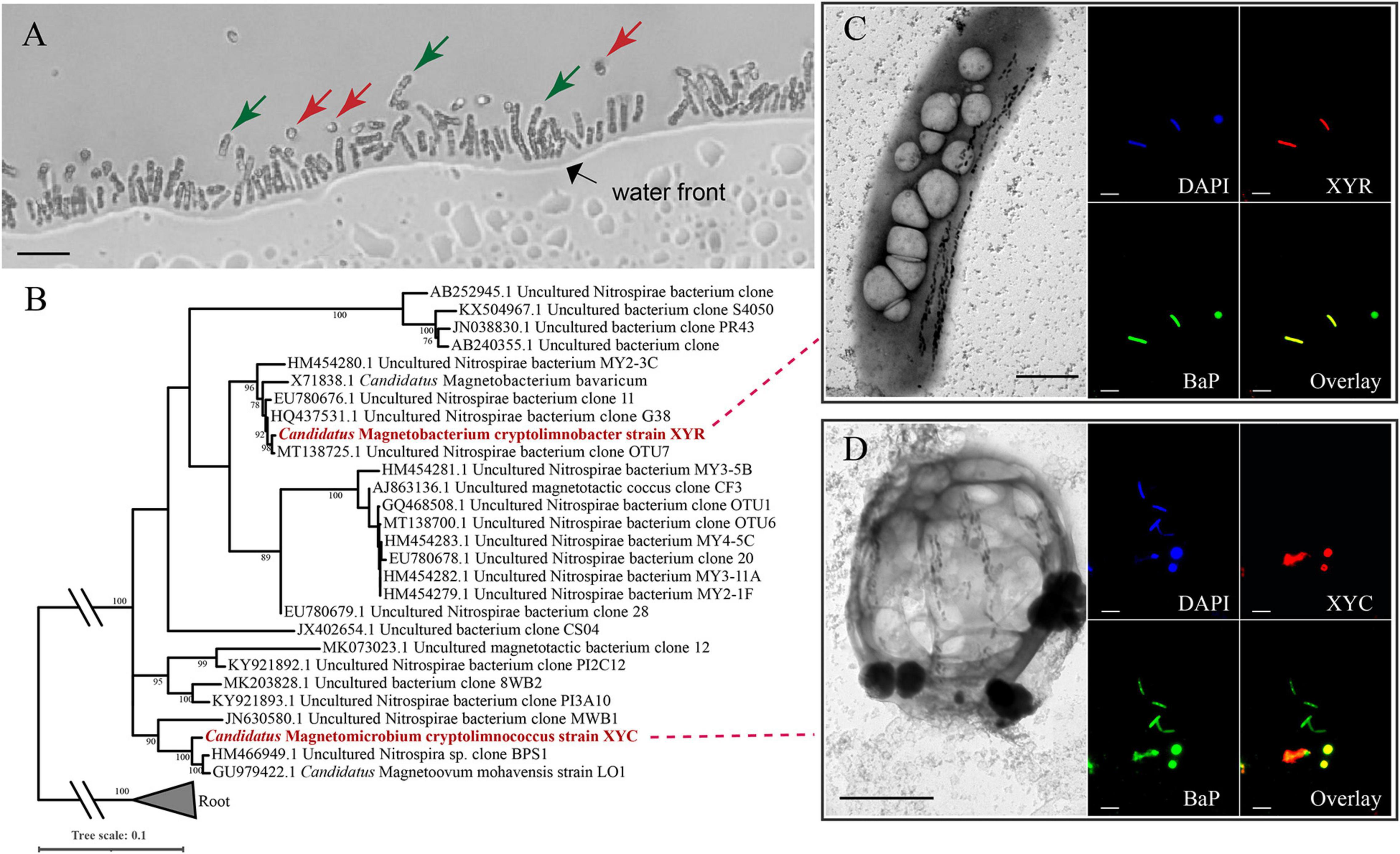
Figure 1. Phylogenetic and morphological identification of two novel Nitrospirae MTB. (A) Morphologies of Ca. Magnetobacterium cryptolimnobacter strain XYR cells (green arrows) and Ca. Magnetomicrobium cryptolimnococcus strain XYC cells (red arrows) as revealed by light microscopy (bar = 10 μm). (B) Phylogenetic tree based on comparative sequence analysis of 16S rRNA genes. Numbers at nodes are bootstrap support values (n = 1,000, only bootstrap values greater than 75% are shown). (C,D) Transmission electron microscopy pictures (bar = 1 μm) and fluorescence in situ hybridization results (bar = 10 μm) of cells of XYR and XYC, respectively. BaP is the probe specific for most Nitrospirae MTB (Spring et al., 1993; Lin et al., 2011).
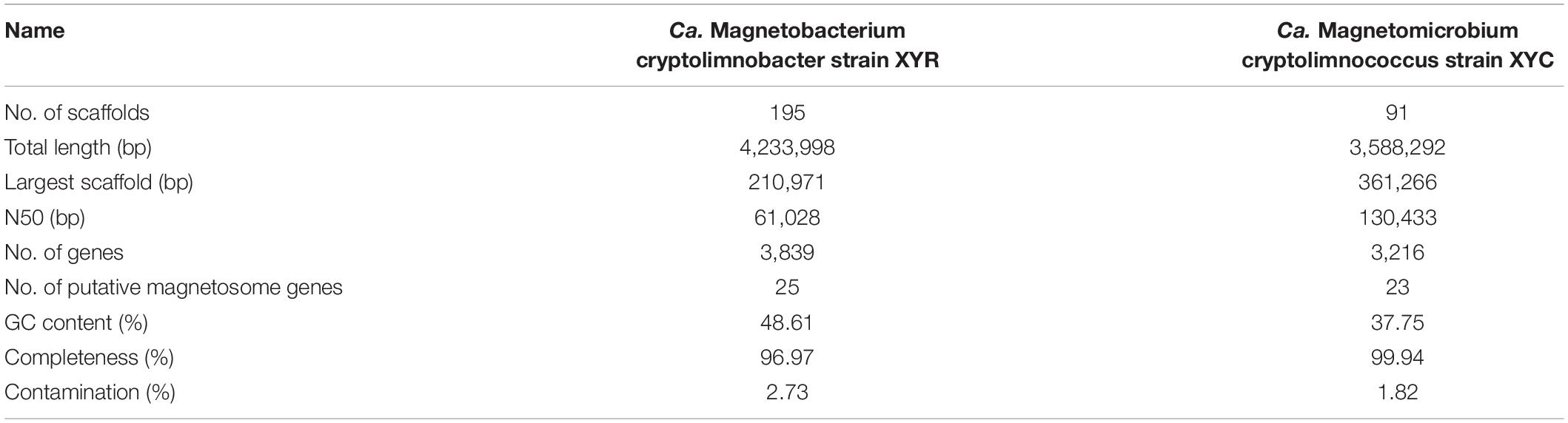
Table 1. Genome statistics of Ca. Magnetobacterium cryptolimnobacter strain XYR and Ca. Magnetomicrobium cryptolimnococcus strain XYC.
According to FISH results, cells of XYR were a rod-shaped morphotype that was similar to previously characterized Ca. Magnetobacterium bavaricum (Spring et al., 1993; Jogler et al., 2010) and Ca. Magnetobacterium casensis (Lin et al., 2014b), and the morphology of XYC cells represented coccoid and was similar to those of Ca. Magnetoovum mohavensis strain LO-1 (Lefèvre et al., 2011) and MWB-1 (Lin et al., 2012). According to TEM observation, cells of XYR are approximately 1–2 μm in width and 5–7 μm in length and contain up to 150 magnetosome crystals organized into two to three bundles of chains. The size of XYC cells ranges from 2 to 4 μm, containing approximately 50–145 magnetosome crystals that formed four to eight bundles of chains. The sizes of magnetosome crystals of XYR and XYC vary from 30 to 130 nm and 45 to 135 nm, respectively.
Phylogenomic Analysis of Magnetotactic Nitrospirae
Phylogenomic analysis of XYR and XYC and the available Nitrospirae MTB genomes was performed based on 120 bacterial single-copy concatenated protein sequence alignment (Figure 2). The genome tree includes 36 previously reported draft genomes of Nitrospirae MTB as of October 2020 and 130 representative non-MTB Nitrospirae genomes. The majority of Nitrospirae MTB genomes are from freshwater lakes (Lin et al., 2014b, 2017b, 2018, 2020b; Kolinko et al., 2016; Koziaeva et al., 2020; Uzun et al., 2020) and acidic peatland soils (Lin et al., 2020b) with one genome (Nitrospirae bacterium SG8_35_4) from a sulfate-rich zone estuary (Lin et al., 2017a) and one (Nitrospirae bacterium MAG_10313_ntr_31) from a hot spring (Uzun et al., 2020). The average estimated completeness and contamination of available Nitrospirae MTB genomes was 87.35 ± 12.69% and 3.08 ± 6.15%, respectively. Their genome sizes ranged from 1.91 to 6.08 Mbp (average 3.36 ± 0.82 Mbp) with a genomic GC content from 35.15 to 49.89% (average 45.13 ± 4.19%).
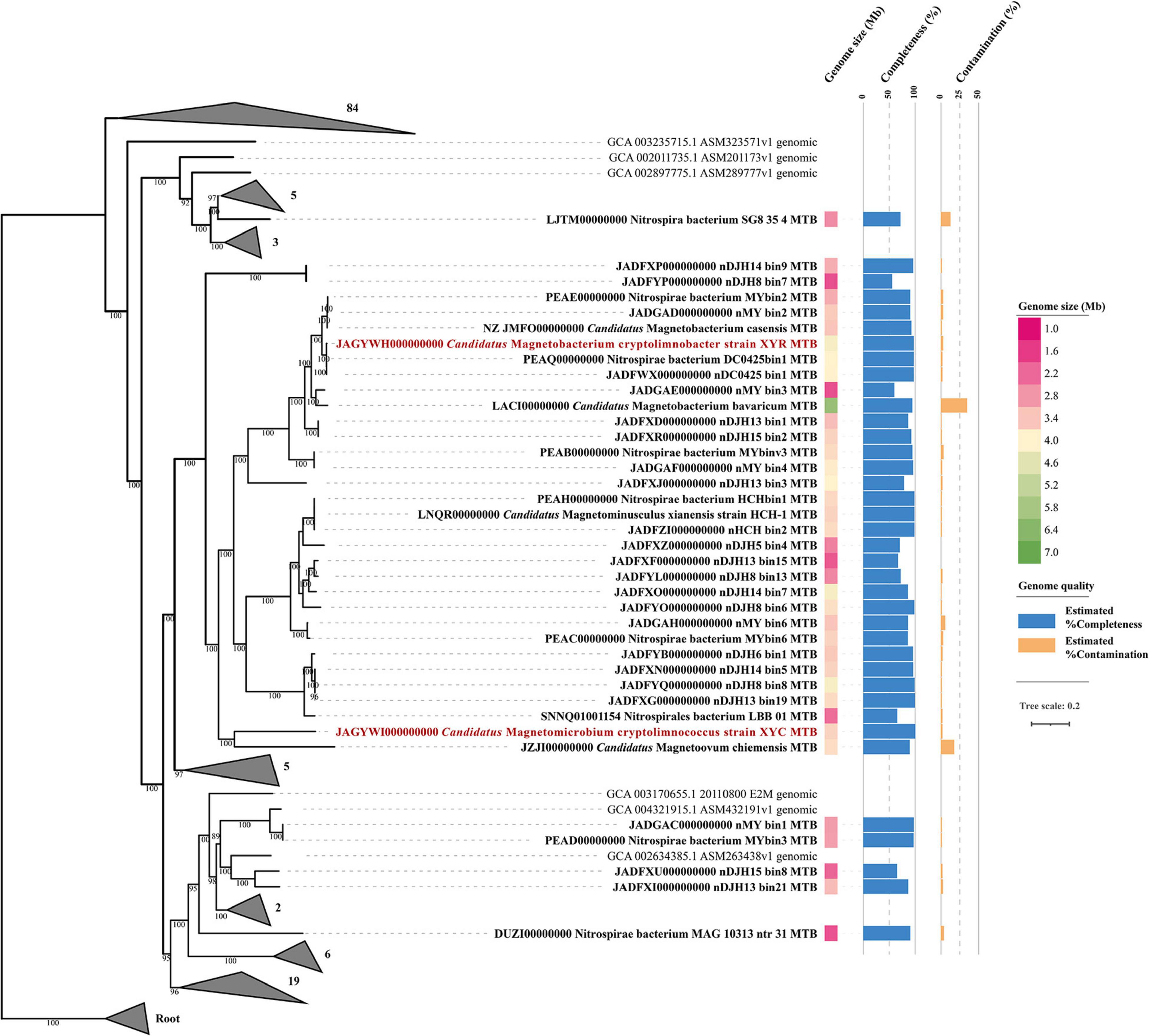
Figure 2. Phylogenomic analysis of 138 Nitrospirae genomes including 38 MTB genomes. A maximum likelihood tree was constructed using a concatenated alignment of 120 conserved bacterial markers with GTDB-Tk (120 conserved bacterial marker genes are provided by GTDB). Numbers at nodes are bootstrap support values (n = 1,000, only bootstrap values greater than 75% are shown). The genome size and quality of each MTB genome were shown.
The genome tree placed XYR as a close sister group of Ca. Magnetobacterium casensis (Lin et al., 2014b) with an average AAI value of 91.38% and an ANI value of 91.84%, indicating that XYR and Ca. Magnetobacterium casensis belong to the same genus but to two different species. Therefore, XYR was provisionally named Ca. Magnetobacterium cryptolimnobacter (Ma.gne.to.bac.te′ri.um. Gr. n. magnes, -etos a magnet; N.L. pref. magneto- pertaining to a magnet; N.L. neut. n. bacterium a rod; Cryp.to.lim.no.bac′ter. Gr. adj. kryptos hidden; Gr. fem. n. limne lake; N.L. masc. n. bacter a rod) while XYC represents a separated phylogenetic lineage of the genus level, sharing 55.48% AAI and 50.30% POCP with Ca. Magnetoovum chiemensis (Kolinko et al., 2016). XYC was accordingly designated Ca. Magnetomicrobium cryptolimnococcus (Ma.gne.to.mi.cro′bi.um. Gr. n. magnes, -etos a magnet; N.L. pref. magneto- pertaining to a magnet; N.L. neut. n. microbium a microbe; Cryp.to.lim.no.coc′cus. Gr. adj. kryptos hidden; Gr. fem. n. limne lake; N.L. masc. n. coccus coccus).
All available Nitrospirae MTB genomes were affiliated within the order Thermodesulfovibrionales except for Nitrospirae bacterium SG8_35_4 (belonging to the order UBA6902) based on phylogenomic analysis and GTDB taxonomy analysis (Figure 2). These genomes were classified into four family-level groups with a majority of genomes (n = 32) belonging to the family Magnetobacteriaceae. Interestingly, the Magnetobacteriaceae family exclusively consists of MTB, including previously well-characterized Ca. Magnetobacterium casensis (Lin et al., 2014b) and Ca. Magnetobacterium bavaricum (Kolinko et al., 2016). Considering that multiple instances of MGC loss may occur across the bacterial tree of life, the Magnetobacteriaceae would be interesting and useful to investigate MTB favorable physiological background and mechanisms to maintain magnetosome formation and magnetotaxis during the evolution. However, those possibilities that Magnetobacteriaceae may diverge very recently leading to little opportunity to lose MGCs or that non-MTB members in this family have so far not been sequenced yet cannot be fully ruled out.
Metabolic Potential of Magnetotactic Nitrospirae
Thirteen high-quality, representative Nitrospirae MTB genomes were annotated and compared in this study, which revealed that members of Nitrospirae MTB generally encoded similar sets of functional genes (Figure 3). Similar to previous studies (Lin et al., 2014b; Kolinko et al., 2016), all these microorganisms encode autotrophic lifestyles with potential capacity in fixation of CO2 via the Wood–Ljungdahl pathway. Although RubisCO-encoding genes were identified in four of 13 genomes (including XYR and Ca. Magnetobacterium casensis), the lack of carboxylase and oxygenase activity of these genes as previously suggested (Jogler et al., 2010) indicate that Nitrospirae MTB might not operate the Calvin–Benson–Bassham (CBB) cycle for CO2 fixation.
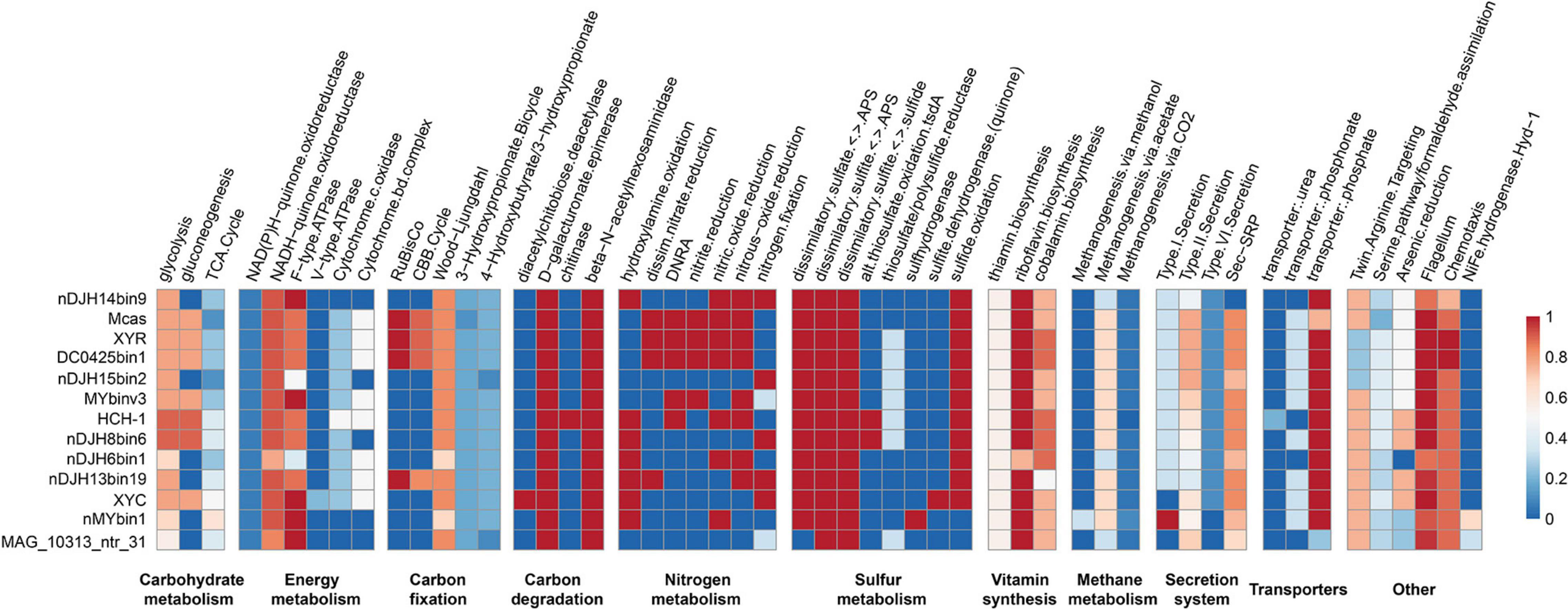
Figure 3. Completeness of metabolic pathways of representative Nitrospirae MTB genomes. The scale number represents the completeness of major metabolic pathways inferred from the presence or absence of genes as determined by the KEGG-Decoder. Dark red represents a complete or highly complete pathway, and dark blue represents a pathway that is absent or highly incomplete. nDJH14bin9, Nitrospirae bacterium nDJH14bin9; Mcas, Ca. Magnetobacterium casensis; HCH-1, Ca. Magnetominusculus xianensis strain HCH-1; XYR, Ca. Magnetobacterium cryptolimnobacter strain XYR; XYC, Ca. Magnetomicrobium cryptolimnococcus strain XYC; DC0425bin1, Nitrospirae bacterium DC0425bin1; nDJH15bin2, Nitrospirae bacterium nDJH15bin2; MYbinv3, Nitrospirae bacterium MYbinv3; nDJH8bin6, Nitrospirae bacterium nDJH8bin6; nDJH6bin1, Nitrospirae bacterium nDJH6bin1; nDJH13bin19, Nitrospirae bacterium nDJH13bin19; nMYbin1, Nitrospirae bacterium nMYbin1; and MAG_10313_ntr31, Nitrospirae bacterium MAG_10313_ntr31.
Genomes of Nitrospirae MTB encode proteins involved in the energy-producing dissimilatory sulfate-reduction pathway, suggesting that these microorganisms could reduce sulfate to sulfite and further to sulfide in anoxic microenvironments (Figure 3). In 11 of 13 genomes, we identified the potential for sulfide oxidation to elemental sulfur or sulfite either by sulfide:quinone oxidoreductase (Sqr) or by dissimilatory sulfite reductase complex (Dsr). Sulfite might be further oxidized to sulfate by adenosine 5′-phosphosulfate reductase (Apr) and 5′-triphosphate sulfurylase (Sat). Different from other Nitrospirae, the genome of XYC contains a small gene cluster encoding sulfite dehydrogenase (quinone) subunits SoeABC that may catalyze the oxidation of sulfite to sulfate in the cytoplasm (Dahl et al., 2013).
A nearly complete nitrogen fixation pathway has been identified in five Nitrospirae MTB genomes (including XYC), which indicates the potential of these MTB to reduce atmospheric molecular nitrogen to ammonia (Figure 3). Several genomes appeared to perform the dissimilatory nitrate reduction to ammonium pathway. Consistent with previous studies (Lin et al., 2014b; Kolinko et al., 2016), the majority of Nitrospirae MTB may conduct a denitrification process in which nitrate or nitrite is reduced as a terminal electron acceptor to produce gaseous nitrogen compounds under low-oxygen or anoxic conditions.
These bacteria also encode proteins involved in a series of B-vitamin biosynthesis, including thiamin (B1), riboflavin (B2), and cobalamin (B12). Not surprisingly, genes encoding components of the flagellar and chemotaxis machinery are present in all Nitrospirae MTB genomes because magnetotaxis is a flagellum-dependent motility that is generally recognized to be interconnected with chemotaxis (Frankel et al., 1997). Magnetotactic Nitrospirae may use the Sec-SRP pathway and the type II secretion system to secrete a variety of proteins and other molecules to the extracellular space. Although a few genes responsible for the type I and VI secretion systems have also been identified in their genomes, whether Nitrospirae MTB can perform these pathways needs further experimental confirmation.
A Remarkably Conserved Magnetosome Gene Cluster Across Magnetotactic Nitrospirae
Genes responsible for biosynthesis and organization of magnetosomes are clustered in all known MTB genomes, referred to as an MGC (Grünberg et al., 2001; Murat et al., 2010; Lin et al., 2017a). We have identified nearly complete MGCs in both genomes of XYR and XYC (Figure 4). Although XYR and XYC belong to different genera, the gene content and organization of MGCs between them are remarkably conserved, both of which contain genes homologous to magnetosome genes man1, mamK, man2, mad10, man3, mamP, mamM, mad31, mamQ-II, mamB, mad2, mamA, mamI, mamE, mamQ-I, man4, man5, man6, mamO, mad23, mad24, mad25, and mad26. This Man1-Mad26 gene cluster is across a region of approximately 19 kb in length. Two mad28 genes located upstream of man1 of XYR are not identified in the same region of the MGC of XYC (Figure 4).
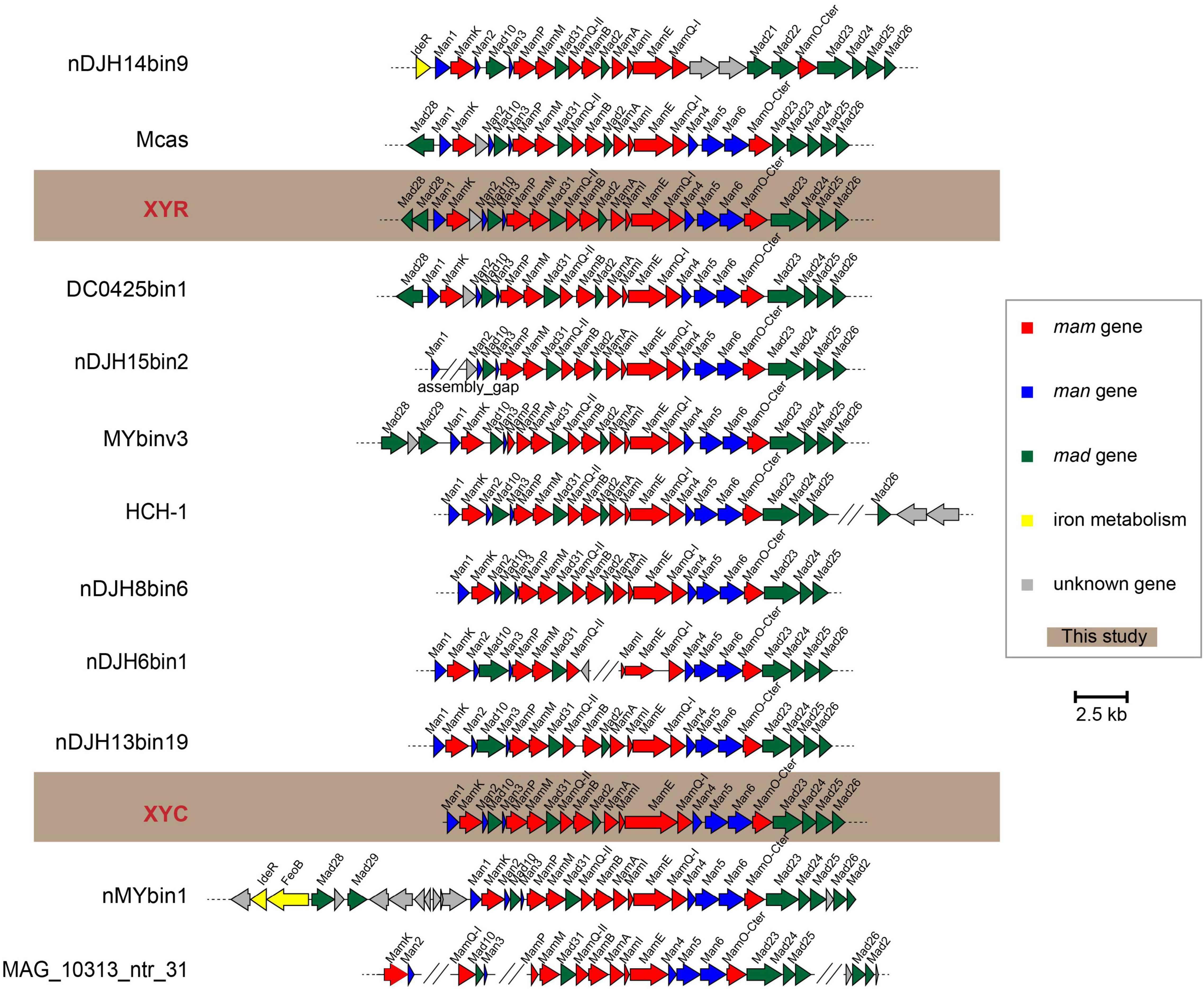
Figure 4. Comparison of MGCs recovered in this study with previously reported representative MGCs. Mcas, Ca. Magnetobacterium casensis; HCH-1, Ca. Magnetominusculus xianensis strain HCH-1; XYR, Ca. Magnetobacterium cryptolimnobacter strain XYR; XYC, Ca. Magnetomicrobium cryptolimnococcus strain XYC; DC0425bin1, Nitrospirae bacterium DC0425bin1; nDJH15bin2, Nitrospirae bacterium nDJH15bin2; MYbinv3, Nitrospirae bacterium MYbinv3; nDJH8bin6, Nitrospirae bacterium nDJH8bin6; nDJH6bin1, Nitrospirae bacterium nDJH6bin1; nDJH13bin19, Nitrospirae bacterium nDJH13bin19; nMYbin1, Nitrospirae bacterium nMYbin1; and MAG_10313_ntr31, Nitrospirae bacterium MAG_10313_ntr31.
A further manual check of 13 representative Nitrospirae MTB genomes revealed the high stability of the man1-mad26 gene cluster. A total of nine genomes have been identified to contain a complete man1-mad26 gene cluster in a single scaffold except for the lack of the mad26 gene in the genome of nDJH8bin6 (Figure 4). Both gene content and gene order of man1-mad26 gene clusters are extraordinarily conserved across the Nitrospirae MTB from different environmental conditions with the only exception being nDJH14bin9, which suggests that magnetosome biosynthesis and magnetotaxis play important adaptive roles in this MTB group. Alternatively, recent horizontal gene transfer(s) of MGCs into this group or unknown selective pressure(s) to maintain the content and order of magnetosome genes are also possible. A group of six magnetosome genes (mamABIKMQ) was identified previously to be shared by the majority of MTB genomes across different bacterial phyla (Lin et al., 2020b). All these six genes have been identified in the man1-mad26 gene cluster. Proteins encoded by these genes are identified to be either essential for magnetosomal biogenesis (mamBIMQ) or responsible for important accessory functions, including magnetosome membrane assembly (mamA) and magnetosome chain formation (mamK) in Magnetospirillum strains (Murat et al., 2010; Lohße et al., 2014; Uebe and Schüler, 2016). Similar to their counterparts in Magnetospirillum, Nitrospirae MamE and MamP contain the PDZ domain that is often involved in organizing signaling complexes by forming protein–protein interactions. MamE is involved in localization of magnetosome proteins and in magnetosomal crystal maturation (Murat et al., 2010; Siponen et al., 2012), and MamP is responsible for redox control of iron biomineralization (Jones et al., 2015) in Magnetospirillum. Nitrospirae MamO-Cter contains a predicted TauE-like transporter domain and its homolog MamO in Magnetospirillum, and homologs MamO and TauE in Desulfovibrio magneticus RS-1 has been speculated to be directly involved in nucleation of iron oxide particles (Quinlan et al., 2011; Rahn-Lee et al., 2015).
In addition to the abovementioned Mam proteins with homology to well-characterized magnetosome proteins, the man1-mad26 gene cluster contains additional genes (man and mad genes) with no apparent homology to MTB belonging to the Magnetospirillum (Figure 4). Among these genes, only mad2 has been functionally characterized previously. Mutant of mad2 in Desulfovibrio magneticus RS-1 resulted in cells containing rare, unusual-looking particles with no magnetic response, indicating the important role of Mad2 in magnetosome biomineralization in this magnetotactic bacterium (Rahn-Lee et al., 2015). Although functions of the other man and mad genes in the Nitrospirae have not been investigated so far, their remarkable conservation suggests that they should govern important functions in magnetosome formation and magnetotaxis in Nitrospirae MTB.
Despite the stability of man1-mad26 gene clusters, a few differences were noticed across Nitrospirae MTB. For example, genes of man4, man5, and man6 are absent from MGC of nDJH14bin9 but are present in those of other magnetotactic Nitrospirae, and MGC of nDJH14bin9 contains mad21 and mad22 genes that are absent from other Nitrospirae MGCs (Figure 4). A gene between mamK and man2 encoding a Uma2 family endonuclease is found in MGCs of XYR, Ca. Magnetobacterium casensis and DC0425bin1 but is absent from the rest of the genomes (Figure 4).
Conclusion
In summary, we have identified and characterized two novel MTB populations, Ca. Magnetobacterium cryptolimnobacter strain XYR and Ca. Magnetomicrobium cryptolimnococcus strain XYC, belonging to the Nitrospirae phylum, which expand the genomic diversity of this MTB lineage. Thoroughly comparative analyses of Nitrospirae MTB genomes reveal metabolic plasticity of this widely distributed MTB group and a remarkably conserved MGC. Results of this study deepen our understanding of the taxonomic diversity, genomic property, ecophysiology, and magnetosome biogenesis of MTB members within the Nitrospirae phylum. In the future, successful pure cultivation and a combination of metagenomics, metatranscriptomics, and other culture-independent analyses will further deepen our understanding of MTB belonging to the Nitrospirae phylum.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Genome sequences of Ca. Magnetobacterium cryptolimnobacter strain XYR and Ca. Magnetomicrobium cryptolimnococcus strain XYC have been deposited in GenBank under the accession numbers JAGYWH000000000 and JAGYWI000000000 (BioProject number PRJNA400260).
Author Contributions
WL and WZ conceived and designed the study. YW collected field samples. WZ conducted laboratory work. WZ and WL analyzed the data. WZ and WL wrote the manuscript with input from YW, LL, and YP. All the authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) grants 41822704 and 41621004, and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Lixin Gu and Xu Tang for their assistance in electron microscope observation.
References
Agarwala, R., Barrett, T., Beck, J., Benson, D. A., Bollin, C., Bolton, E., et al. (2018). Database resources of the national center for biotechnology information. Nucleic Acids Res. 46, D8–D13. doi: 10.1093/nar/gkx1095
Alneberg, J., Bjarnason, B. S., De Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Amann, R., Peplies, J., and Schüler, D. (2006). “Diversity and taxonomy of magnetotactic bacteria,” in Magnetoreception and Magnetosomes in Bacteria, ed. D. Schüler (Berlin: Springer), 25–36. doi: 10.1007/7171_037
Amor, M., Ceballos, A., Wan, J., Simon, C. P., Aron, A. T., Chang, C. J., et al. (2020a). Magnetotactic bacteria accumulate a large pool of iron distinct from their magnetite crystals. Appl. Environ. Microbiol. 86:e1278. doi: 10.1128/AEM.01278-20
Amor, M., Mathon, F. P., Monteil, C. L., Busigny, V., and Lefèvre, C. T. (2020b). Iron-biomineralizing organelle in magnetotactic bacteria: function, synthesis and preservation in ancient rock samples. Environ. Microbiol. 22, 3611–3632. doi: 10.1111/1462-2920.15098
Amor, M., Tharaud, M., Gélabert, A., and Komeili, A. (2020c). Single-cell determination of iron content in magnetotactic bacteria: implications for the iron biogeochemical cycle. Environ. Microbiol. 22, 823–831. doi: 10.1111/1462-2920.14708
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2020). KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Bazylinski, D. A., Lefèvre, C. T., and Schüler, D. (2013). “Magnetotactic bacteria,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 453–494.
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cox, B. L., Popa, R., Bazylinski, D. A., Lanoil, B., Douglas, S., Belz, A., et al. (2002). Organization and elemental analysis of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiol. J. 19, 387–406. doi: 10.1080/01490450290098504
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Flies, C. B., Peplies, J., and Schüler, D. (2005). Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71, 2723–2731. doi: 10.1128/AEM.71.5.2723-2731.2005
Frankel, R. B., Bazylinski, D. A., Johnson, M. S., and Taylor, B. L. (1997). Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73, 994–1000. doi: 10.1016/S0006-3495(97)78132-3
Gilchrist, C. L. M., and Chooi, Y.-H. (2021). Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 47:btab007. doi: 10.1093/bioinformatics/btab007
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2018). Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866. doi: 10.1038/s41396-018-0091-3
Greenberg, M., Canter, K., Mahler, I., and Tornheim, A. (2005). Observation of magnetoreceptive behavior in a multicellular magnetotactic prokaryote in higher than geomagnetic fields. Biophys. J. 88, 1496–1499. doi: 10.1529/biophysj.104.047068
Grünberg, K., Wawer, C., Tebo, B. M., and Schüler, D. (2001). A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67, 4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Jogler, C., Lin, W., Meyerdierks, A., Kube, M., Katzmann, E., Flies, C., et al. (2009). Toward cloning of the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75, 3972–3979. doi: 10.1128/AEM.02701-08
Jogler, C., Niebler, M., Lin, W., Kube, M., Wanner, G., Kolinko, S., et al. (2010). Cultivation-independent characterization of ‘Candidatus Magnetobacterium bavaricum’ via ultrastructural, geochemical, ecological and metagenomic methods. Environ. Microbiol. 12, 2466–2478. doi: 10.1111/j.1462-2920.2010.02220.x
Jones, S. R., Wilson, T. D., Brown, M. E., Rahn-Lee, L., Yu, Y., Fredriksen, L. L., et al. (2015). Genetic and biochemical investigations of the role of MamP in redox control of iron biomineralization in Magnetospirillum magneticum. Proc. Natl. Acad. Sci. U.S.A. 112, 3904–3909. doi: 10.1073/pnas.1417614112
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Kolinko, S., Jogler, C., Katzmann, E., Wanner, G., Peplies, J., and Schüler, D. (2012). Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 14, 1709–1721. doi: 10.1111/j.1462-2920.2011.02609.x
Kolinko, S., Richter, M., Glöckner, F. O., Brachmann, A., and Schüler, D. (2016). Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ. Microbiol. 18, 21–37. doi: 10.1111/1462-2920.12907
Koziaeva, V. V., Alekseeva, L. M., Uzun, M. M., Leão, P., Sukhacheva, M. V., Patutina, E. O., et al. (2020). Biodiversity of magnetotactic bacteria in the freshwater lake beloe bordukovskoe, Russia. Microbiology 89, 348–358. doi: 10.1134/S002626172003008X
Konstantinidis, K. T., Rosselló-Móra, R., and Amann, R. (2017). Uncultivated microbes in need of their own taxonomy. ISME J. 11, 2399–2406. doi: 10.1038/ismej.2017.113
Lefèvre, C. T., Abreu, F., Schmidt, M. L., Lins, U., Frankel, R. B., Hedlund, B. P., et al. (2010). Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl. Environ. Microbiol. 76, 3740–3743. doi: 10.1128/AEM.03018-09
Lefèvre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/mmbr.00021-13
Lefèvre, C. T., Frankel, R. B., Abreu, F., Lins, U., and Bazylinski, D. A. (2011). Culture-independent characterization of a novel, uncultivated magnetotactic member of the Nitrospirae phylum. Environ. Microbiol. 13, 538–549. doi: 10.1111/j.1462-2920.2010.02361.x
Lefèvre, C. T., and Wu, L.-F. (2013). Evolution of the bacterial organelle responsible for magnetotaxis. Trends Microbiol. 21, 534–543. doi: 10.1016/j.tim.2013.07.005
Li, J., Liu, P., Wang, J., Roberts, A. P., and Pan, Y. (2020). Magnetotaxis as an adaptation to enable bacterial shuttling of microbial sulfur and sulfur cycling across aquatic oxic-anoxic interfaces. J. Geophys. Res. Biogeosci. 125:e2020JG006012. doi: 10.1029/2020JG006012
Lin, H.-H., and Liao, Y.-C. (2016). Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci. Rep. 6:24175. doi: 10.1038/srep24175
Lin, W., Bazylinski, D. A., Xiao, T., Wu, L.-F., and Pan, Y. (2014a). Life with compass: diversity and biogeography of magnetotactic bacteria. Environ. Microbiol. 16, 2646–2658. doi: 10.1111/1462-2920.12313
Lin, W., Deng, A., Wang, Z., Li, Y., Wen, T., Wu, L. F., et al. (2014b). Genomic insights into the uncultured genus “Candidatus magnetobacterium” in the phylum Nitrospirae. ISME J. 8, 2463–2477. doi: 10.1038/ismej.2014.94
Lin, W., Jogler, C., Schüler, D., and Pan, Y. (2011). Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the phylum Nitrospirae. Appl. Environ. Microbiol. 77, 323–326. doi: 10.1128/AEM.01476-10
Lin, W., Kirschvink, J. L., Paterson, G. A., Bazylinski, D. A., and Pan, Y. (2020a). On the origin of microbial magnetoreception. Natl. Sci. Rev. 7, 472–479. doi: 10.1093/nsr/nwz065
Lin, W., Li, J., and Pan, Y. (2012). Newly isolated but uncultivated magnetotactic bacterium of the phylum Nitrospirae from Beijing, China. Appl. Environ. Microbiol. 78, 668–675. doi: 10.1128/AEM.06764-11
Lin, W., Pan, Y., and Bazylinski, D. A. (2017a). Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ. Microbiol. Rep. 9, 345–356. doi: 10.1111/1758-2229.12550
Lin, W., Paterson, G. A., Zhu, Q., Wang, Y., Kopylova, E., Li, Y., et al. (2017b). Origin of microbial biomineralization and magnetotaxis during the Archean. Proc. Natl. Acad. Sci. U.S.A. 114, 2171–2176. doi: 10.1073/pnas.1614654114
Lin, W., Zhang, W., Paterson, G. A., Zhu, Q., Zhao, X., Knight, R., et al. (2020b). Expanding magnetic organelle biogenesis in the domain Bacteria. Microbiome 8:152. doi: 10.1186/s40168-020-00931-9
Lin, W., Zhang, W., Zhao, X., Roberts, A. P., Paterson, G. A., Bazylinski, D. A., et al. (2018). Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J. 12, 1508–1519. doi: 10.1038/s41396-018-0098-9
Lohße, A., Borg, S., Raschdorf, O., Kolinko, I., Tompa, É, Pósfai, M., et al. (2014). Genetic dissection of the mamAB and mms6 operons reveals a gene set essential for magnetosome biogenesis in magnetospirillum gryphiswaldense. J. Bacteriol. 196, 2658–2669. doi: 10.1128/JB.01716-14
McCausland, H. C., and Komeili, A. (2020). Magnetic genes: studying the genetics of biomineralization in magnetotactic bacteria. PLoS Genet. 16:e1008499. doi: 10.1371/journal.pgen.1008499
Monteil, C. L., and Lefèvre, C. T. (2020). Magnetoreception in microorganisms. Trends Microbiol. 28, 266–275. doi: 10.1016/j.tim.2019.10.012
Murat, D., Quinlan, A., Vali, H., and Komeili, A. (2010). Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U.S.A. 107, 5593–5598. doi: 10.1073/pnas.0914439107
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Olm, M. R., Brown, C. T., Brooks, B., and Banfield, J. F. (2017). dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868. doi: 10.1038/ismej.2017.126
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Parks, D. H., Rinke, C., Chuvochina, M., Chaumeil, P. A., Woodcroft, B. J., Evans, P. N., et al. (2017). Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542. doi: 10.1038/s41564-017-0012-7
Pernthaler, J., Glöckner, F.-O., Schönhuber, W., and Amann, R. (2001). Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30, 207–226. doi: 10.1016/S0580-9517(01)30046-6
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Qian, X. X., Liu, J., Menguy, N., Li, J., Alberto, F., Teng, Z., et al. (2019). Identification of novel species of marine magnetotactic bacteria affiliated with Nitrospirae phylum. Environ. Microbiol. Rep. 11, 330–337. doi: 10.1111/1758-2229.12755
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quinlan, A., Murat, D., Vali, H., and Komeili, A. (2011). The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol. Microbiol. 80, 1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x
Rahn-Lee, L., Byrne, M. E., Zhang, M., Le Sage, D., Glenn, D. R., Milbourne, T., et al. (2015). A genetic strategy for probing the functional diversity of magnetosome formation. PLoS Genet. 11:e1004811. doi: 10.1371/journal.pgen.1004811
Rivas-Lamelo, S., Benzerara, K., Lefèvre, C. T., Monteil, C. L., Jézéquel, D., Menguy, N., et al. (2017). Magnetotactic bacteria as a new model for P sequestration in the ferruginous Lake Pavin. Geochem. Perspect. Lett. 5, 35–41. doi: 10.7185/geochemlet.1743
Rodriguez-R, L., and Konstantinidis, K. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 4:e1900v1. doi: 10.7287/peerj.preprints.1900
Schulz-Vogt, H. N., Pollehne, F., Jürgens, K., Arz, H. W., Beier, S., Bahlo, R., et al. (2019). Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J. 13, 1198–1208. doi: 10.1038/s41396-018-0315-6
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Siponen, M. I., Adryanczyk, G., Ginet, N., Arnoux, P., and Pignol, D. (2012). Magnetochrome: a c-type cytochrome domain specific to magnetotatic bacteria. Biochem. Soc. Trans. 40, 1319–1323. doi: 10.1042/BST20120104
Spring, S., Amann, R., Ludwig, W., Schleifer, K. H., Van Gemerden, H., and Petersen, N. (1993). Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59, 2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993
Uebe, R., and Schüler, D. (2016). Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 14, 621–637. doi: 10.1038/nrmicro.2016.99
Uzun, M., Alekseeva, L., Krutkina, M., Koziaeva, V., and Grouzdev, D. (2020). Unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. Sci. Data 7:252. doi: 10.1038/s41597-020-00593-0
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Keywords: magnetotactic bacteria, Nitrospirae, metagenomics, magnetotaxis, magnetosome gene cluster
Citation: Zhang W, Wang Y, Liu L, Pan Y and Lin W (2021) Identification and Genomic Characterization of Two Previously Unknown Magnetotactic Nitrospirae. Front. Microbiol. 12:690052. doi: 10.3389/fmicb.2021.690052
Received: 06 April 2021; Accepted: 29 June 2021;
Published: 27 July 2021.
Edited by:
Yohey Suzuki, The University of Tokyo, JapanReviewed by:
Lilah Rahn-Lee, William Jewell College, United StatesMaxim Rubin-Blum, National Institute of Oceanography, Israel Oceanographic and Limnological Research (IOLR), Israel
Copyright © 2021 Zhang, Wang, Liu, Pan and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Lin, weilin@mail.iggcas.ac.cn
 Wensi Zhang
Wensi Zhang Yinzhao Wang
Yinzhao Wang Li Liu1,2,3
Li Liu1,2,3 Yongxin Pan
Yongxin Pan Wei Lin
Wei Lin