- 1Medfuture Research Center for Advanced Medicine, Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
- 2Research Center for Functional Genomics, Biomedicine and Translational Medicine, Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
- 3African Organisation for Research and Training in Cancer, Cape Town, South Africa
- 4Surgical Department, Municipal Hospital, Cluj-Napoca, Romania
- 5Department of Surgery, Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
- 6Department of Functional Genomics and Experimental Pathology, The Oncology Institute “Prof. Dr. I. Chiricuta”, Cluj-Napoca, Romania
The small nucleolar RNA host genes (SNHGs) are a group of long non-coding RNAs, which are reported in many studies as being overexpressed in various cancers. With very few exceptions, the SNHGs (SNHG1, SNHG3, SNHG5, SNHG6, SNHG7, SNHG12, SNHG15, SNHG16, SNHG20) are recognized as inducing increased proliferation, cell cycle progression, invasion, and metastasis of cancer cells, which makes this class of transcripts a viable biomarker for cancer development and aggressiveness. Through our literature research, we also found that silencing of SNHGs through small interfering RNAs or short hairpin RNAs is very effective in both in vitro and in vivo experiments by lowering the aggressiveness of solid cancers. The knockdown of SNHG as a new cancer therapeutic option should be investigated more in the future.
Introduction
Worldwide, in 91 developed countries, cancer is the first or second leading cause of mortality with 18.1 million new cases and 9.5 million deaths estimated in 2018 (1). According to Siegel et al., in the United States, between 2000 and 2014, the most frequently diagnosed types of cancer in men are: prostate, lung, and colorectal cancer; while in women, these are: breast, lung, and colorectal cancer (2). These statistics show that the malignant disease has a high impact over the socioeconomical welfare of developed countries; thus, each new discovery of its molecular context should be carefully analyzed and evaluated for its potential as biomarker and/or therapeutic target.
In malignant diseases, the imbalance between overactivation of proto-oncogenes and inhibition of tumor suppressor genes is usually the consequence of a faulty regulatory system, which includes, among others, the non-coding RNAs (3–5). Most of these transcripts do not have the ability to interact with the ribosome and are not translated into proteins (6–11); however, they interact with other coding or non-coding RNAs (ncRNAs), being part of complex regulatory networks (12). The ncRNAs are divided into two classes: the small ncRNAs (with <200 nt) and the long ncRNAs (lncRNAs), with >200 nt (13). The short ncRNAs are generated through multistep processing of the primary transcript, while the lncRNAs remain highly similar to their primary transcripts (14–17). The short ncRNAs are divided into: small interfering RNAs (siRNAs), microRNAs (miRNA), piwi-interacting RNAs (piRNAs), and others (18). The lncRNAs, even if very heterogeneous in terms of biogenesis, structure, mechanism of action, and biological functions, are still mentioned under the general term of lncRNAs (6, 19, 20).
The snoRNAs (small nucleolar RNAs) were discovered in the late 1980s (21). However, only recently these transcripts were considered as potential biomarkers of cancer (22). The snoRNAs have the general characteristics of small ncRNAs, with the processed transcripts having between 65 and 300 nt. The snoRNAs do not have a poly-A tail and are 5′Cap, which means that they are not exported from the nucleus. They are transcribed from clusters of protein-coding genes or genes coding for other ncRNAs, and a small franction of snoRNAs are originated in the intergenetic regions. Only three snoRNAs (U3, U8, and U13] and telomerase snoRNA are transcribed and processed as single units. The polycistronic transcription of snoRNAs has a phylogenetic progression, which means that in higher organisms, more snoRNAs originating from clusters of genes are found (23).
There are two types of snoRNAs: C/D box snoRNA and H/ACA snoRNA. The C/D box snoRNAs have a C box (RUGAUGA, R = A/G) and two D boxes (CUGA). The H/ACA snoRNAs contain the H box (ANANNA, N = A/T/C/G) and the ACA sequence (24). The C/D box snoRNAs are involved in the 2-O′-methylation of rRNA, and the H/ACA snoRNA cause the pseudouridylation of rRNAs. Some snoRNAs are further processed into microRNAs, which repress the translation of mRNA in the cytoplsm (23) or into piwi-interacting RNAs, essential in the regulation of transposomes mobility in the nucleus (25).
The snoRNA host genes are mainly located in the introns of protein-coding genes, with ancestral positionally conserved (APC) snoRNA sequences constituting only 2% of host genes (26). However, the intronic sequences of snoRNAs show a high degree of conservation across species (27) in comparioson with intergenetic regions (26). The generation of snoRNAs across species may be a result of mobility of the nested snoRNAs in protein-coding genes, since orthologous introns can encode for non-orthologous snoRNAs in different species. However, as the new element of non-coding sequence moves in a protein-coding gene, it results in loss-of-function genetic variants (28).
Some snoRNA genes, without any capacity to code for proteins, still contain both introns and exons in their sequences However, snoRNAs are generated only from introns. If the full-length transcript, including exons, is kept, it will function as a type of lncRNA, named small nucleolar RNA host gene (SNHG) (29). A pan-cancer analysis of 31 cancer types based on “omics” data retrieved from TCGA revealed that the snoRNA host genes are enriched in non-protein coding genomic regions and that these have higher expression (30). There seems to be a positive correlation between SNHGs and their corresponding snoRNAs. More precisely, the intracellular level of snoRNAs is influenced by their structure and the availability of SNHGs (23). In murine genome, Zfas1 is an antisense transcript, which shares a bidirectional promoter with the protein coding gene Znfx1 (zinc finger NFX-1-type containing), their expression in different tissues being positively correlated. Zfas is also an lncRNA from an snoRNA gene, and it can be processed into three types of intronic snoRNAs: SNORD12, SNORD12b, and SNORD12c. SNORD12b has a higher expression than SNORD12 and SNORD12c, in cells with a lower degree of differentiation, due to a higher stability of the processed structure, which contains an additional short hairpin structure. The snoRNAs are located only in the nucleus, whereas Zfas is present in the cytoplasm and nucleus as well. The siRNA knockdown of Zfas only slightly affected the expression of snoRNAs (31). In addition, SNHG12 knockdown in hepatocellular carcinoma did not statistically significantly affect the expression of its snoRNAs (SNORA44, SNORA61, SNORA16A, and SNORD99] (32). However, the expression of some SNHGs, such as SNHG17, is regulated by the copy number variations (CNV) of their host gene, this being also correlated with the aggressiveness of non-small cell lung cancer and squamous cell lung cancer. A change in SNHG intracellular level can generate a significant downstream effect, such as siRNA inhibition of SNHG17 that caused 637 protein coding RNAs to be up-regulated and 581 to be down-regulated (33).
The proteins that target the SNHGs can also target their corresponding snoRNAs. A recent article reported that p53 activation repressed the intracellular level of SNHG1 that also reduces the level of snoRNAs: SNORD22, SNORD25, SNORD26, SNORD27, SNORD28, and SNORD75. The SNORD28 is further processed into an miRNA, named sno-miR-28. This miRNA repressed the expression of TAF9B, a coactivator of p53. As follows, a positive feedback loop forms between sno-miR-28-p53-SNHG1-SNRD28 (34).
The cellular location differs between snoRNAs and SNHGs. While the snoRNAs remain in the nucleus (35), SNHGs are present in the nucleus as well as in the cytoplasm (29) (Figure 1).
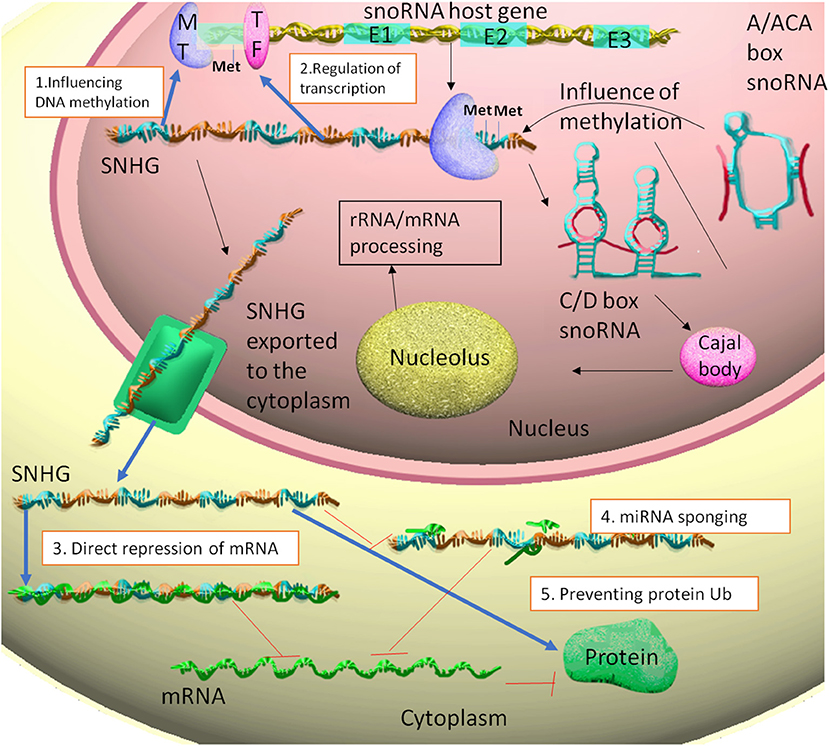
Figure 1. The snoRNA host gene are transcribed to SNHG. Some of these transcripts generate from their introns C/D box snoRNAs or A/ACA box snoRNAs. The snoRNAs can influence the methylation of their SNHG of origin. The SNHGs are then associated with Cajal bodies and nucleolus, thus participating in rRNA and mRNA primary processing. The SNHGs in the nucleus fulfill the following functions: 1. Influencing DNA methylation through interaction with methyltransferases (MT), such as EZH2. 2. Regulating transcription through interaction with transcription factors (TF), such as E2F1. The SNHGs are then exported into the cytoplasm, where: 3. Can directly interact and repress mRNA translation; 4. act as ceRNA for miRNAs (miRNA sponging) and indirectly up-regulating the translation of miRNA targets. 5. Stabilizing proteins, by preventing protein ubiquitination.
Through our research of literature on SNHGs, we have identified five main types of molecular mechanism of action, with different cellular localizations.
Nuclear:
1. Influencing DNA methylation through modulation of methylation enzymes
2. Interaction with transcription factors and repression of gene transcription.
Cytoplasm:
3. MiRNA sponging and the releasing of miRNA targets
4. Direct binding to the mRNA and repression of translation
5. Prevention of protein ubiquitination through single protein interaction or multi-protein complex.
In the nucleus, SNHG1 binds to the Mediator complex and facilitates the interaction of the enhancer and promoter DNA region corresponding to SLC3A2 gene (36). SLC3A2 is a well-known cancer-promoting gene (37, 38). The Mediator complex is composed of multiple proteins and non-coding RNAs that cooperate with RNA polymerase II during transcription initiation (39). SNHG1 competitively binds to the Far Upstream Element Binding Protein 1 (FUBP1), and it prevents FUBP1 interaction with its repressor, followed by overstimulated transcription of c-MYC gene (39).
In hepatocellular carcinoma, SNHG6 lowers DNA methylation through inhibition of S-adenosylmethionine and down-regulation of MAT1A (methionine adenosyltransferase 1A). SNHG3 binds to enhancer of zeste homolog 2 (EZH2) and causes the methylation of several genes (40, 41).
In the cytoplasm, SNHGs fulfill the role of competing endogenous RNAs (ceRNAs) for miRNAs (42). ceRNA is a term used for the non-coding RNA species that share common miRNA binding sites with mRNAs. The process is also known under the name of miRNA sponging (43). The SNHGs can also interact directly to the mRNA and cause their overexpression, such as in the case of SNHG1 and p53 in colorectal cancer (44).
The SNHGs can also post-translationally interact with proteins and prevent their ubiquitination. This is the case of SNHG15 and SLUG in colorectal cancer (45).
The SNHGs have an oncogenic role in cancer through the versatility of their interactions at DNA–RNA–protein level. The correlation between SNHGs and cancer still lacks important information on the general activity of SNHGs, mainly because the data are still fractioned and focused on each SNHG. In current article, we reviewed over 200 articles on the role of SNHG in different malignancies. We first briefly consider the oncogenic activity of each SNHG separately, and in the final section of the article, we draw some general lines of consideration on the realistic potential of SNHGs to reach clinical applications, in the context of a comprehensive overview of their biological influence. To our knowledge, this is the first time that a comprehensive clarification of the general mechanism of action of SNHGs in cancer is given.
Small Nucleolar RNA Host Gene 1 (SNHG1)
SNHG1 generates, through alternative splicing, eight snoRNAs: SNORD22 (46), SNORD25, SNORD26, SNORD27, SNORD28, SNORD29, SNORD30, and SNORD31 (47).
In colorectal cancer, SNHG1 leads to poor prognosis (44, 48, 49). It promotes cancer cell viability, proliferation, cell cycle progression, tumor growth, and increased invasion/migration capacity (44, 48–50). Its mechanism of action consists of targeting the p53 gene (44) and overstimulating the Wnt/β-catenin signaling pathway (48, 50]. It also acts as a ceRNA for miR-145 (50), a suppressor of a colorectal cancer invasion and migration (51).
In ovarian cancer, SNHG1 promotes cell proliferation, invasion, and new colony formation, while impairing cell apoptosis (52). In prostate cancer, SNHG1 acts as an miR-199a sponge, thus increasing the level of CDK7 (cyclin-dependent kinase 7), which stimulates cell division (53), and HIF-1α (hypoxia-inducible factor 1-alpha), which leads to enhanced angiogenesis (54). HIF-1α is a transcription factor (TF) activated in case of hypoxic microenvironment present at the tumor site. This TF stimulates the expression of VEGFα, an important promoter of tumor neoangiogenesis. This process is essential for tumor survival (provision of nutrients) and spreading to new sites of the body (connection with systemic blood circulation) (55).
In esophageal cancer, SNGH1 sponges miR-338 (42) miR-338 is involved in stimulating the radiotherapy-induced apoptosis of esophageal cancer cells, by targeting Survivin gene (56). SNHG1 in esophageal cancer promotes cell proliferation and EMT (epithelial to mesenchymal transition)-mediated invasion (56), by down-regulation E-CAD (E-cadherin) and up-regulation VIM (Vimentin) and N-CAD, as well as activation of Notch signaling pathway (57). EMT is the process through which epithelial cells lose their tight junctions and cell polarity, while acquiring a higher degree of mobility and ability of invasion through basement membrane. There are three types of EMTs: (1) during embryogenesis, this process is necessary for the transformation of epiblast into mesinchyme; (2) during wound healing and chronic inflammation when an additional supply of fibroblasts are locally needed; (3) during cancer progression, when the cells from the in situ primary tumor invade the local lymph nodes or enter into systemic blood circulation in order to from distant metastasis (58). E-cadherin is a cell adhesion molecule (CAM) specific for the formation of adherent junctions between epithelial cells. It is also considered as a tumor suppressor through DNA methylation of oncogenes (59). N-CAD suppresses the activity of E-CAD and stimulates the motility of cells. During embryonic development, it supports gastrulation and the formation of neuronal crest; however, N-CAD can be reactivated during malignant development to support EMT progression (60). VIM is a component of the intermediate filaments from cytoskeleton that keep the organelles in place, while also allowing to some degree their mobility. VIM is a highly flexible component found in all cells, but its overexpression is specific for mesenchymal cells. In cancer, VIM is a major stimulator of EMT and inhibitor of autophagy through its repression of Beclin 1 and 14-3-3 (61).
In hepatocellular carcinoma, SNHG1 was associated with advanced stages, larger tumor size, and poor differentiation due to its interaction with the tumor suppressor p53 (62). SNHG1 sponges miR-195 (63), a miRNA that inhibits metastasis and angiogenesis in hepatocellular carcinoma by targeting FGF2 (fibroblast growth factor) and VEGFA (vascular endothelial growth factor alpha) (62, 64). Similarly, SNHG1 overexpression results in worse prognosis in glioma because it increases proliferation and invasion while inhibiting apoptosis (65).
In lung cancer, SNHG1 acts as a tumor-promoter by sponging miR-145 (66) and miR-101 (67) and enhancing the Wnt/β-catenin signaling pathway (67). miR-145 acts as a tumor suppressor in lung cancer by impairing cell migration and invasion (68, 69). In lung cancer, miR-101 underexpression is associated with advanced stages and lymph node metastasis and it induces cell apoptosis by targeting MCL-1 (70).
In osteosarcoma, SNHG1 promotes cell proliferation, tumor growth, invasion, and EMT by sponging miR-326, resulting in overexpression of NOB1 [NIN1 (RPN12) Binding Protein 1 Homolog] (70). miR-326 suppresses the antiapoptotic gene BCL-2 in osteosarcoma and acts as a tumor suppressor (71). BCL-2 is a major antiapoptotic marker that inhibits the liberation of cytochrome C from mitochondria (72), thus impairing the cleavage of the effector caspases CASP3 and CASP7.
SNHG1 also represses miR-577 and activates WNT/β-catenin pathway (73). WNT2B, a member of WNT signaling pathway, plays a significant oncogenic role in osteosarcoma (73). Further research and details regarding the latest studies on SNHG1 oncogenic function in malignant disease are presented in Supplementary Table 1.
Small Nucleolar RNA Host Gene 3 (SNHG3)
SNHG3 is an oncogenic lncRNA that generates SNORD17 (74).
In osteosarcoma, SNHG3 overexpression facilitates cell invasion and migration in vitro, through the inhibition of miR-151a-3p (75) and miR-196a-5p (76).
In glioma, SNHG3 induces a more aggressive phenotype through increased cell proliferation and apoptosis resistance of malignant cells. SNHG3 interacts with the protein EZH2, which determines the methylation of KLF2 (Krüppel-like Factor 2), and p21 gene promoters, which inhibit their transcription through an epigenetic mechanism (40). KLF2 is a zinc-finger transcription factor that activates CD4+ T cells (77). As a tumor suppressor, KLF2 also impairs invasion and stimulates apoptosis through down-regulation of MMP9 and BCL-2 (78). P21 is a major tumor suppressor activated by p53 in cancer. During cell cycle, p21 activates CDK4 (cyclin-dependent kinase 4) and CDK6 (cyclin-dependent kinase 6), resulting in transition of cells from G1 to S phase. During S phase, however, it impairs the interaction between CDK2 (cyclin-dependent kinase 2) and cyclin E, hence stopping the transition of cells from S to G2 phase. Moreover, in G2, P21 also functions as a disruptor of CDK1 (cyclin- dependent kinase 1) and Cyclin B1 interaction, thus preventing mitosis initiation (79). In gastric cancer, SNHG3 binds to EZH2 and epigenetically silence MED18 (Mediator Complex Subunit 18) expression (41).
Furthermore, SNHG3 causes proliferation of colorectal cancer cells by sponging miR-182 and allowing the overexpression of the tumor promoting transcription factor c-MYC (80). In hepatocellular carcinoma, SNHG3 is positively associated with larger tumor size and relapse (81), in addition to Sorafenib resistance, as a consequence of ceRNA activity on miR-128. Further research and details on the role of SNHG3 in malignant diseases are presented in Supplementary Table 2.
Small Nucleolar RNA Host Gene 5 (SNHG5)
SNHG5 is processed into SNORD 50 and SNORD50′ (82).
In SNHG5, overexpression increases the ability of breast cancer cells to proliferate and go through cell cycle, by releasing PCNA (proliferating cell nuclear antigen) from the inhibition of miR-154-5p (83).
In bladder cancer, SNHG5 induces p27 silencing, followed by enhanced proliferation rate and cell cycle progression, associated with apoptosis inhibition (84). In chronic myelogenous leukemia (CML), SNHG5 up-regulation stimulates imatinib resistance, through down-regulation of miR-205 and up-regulation of ABCC2 (ATP binding cassette subfamily C member 2) (85). In CML patients with rare BCR-ABL variants, this microRNA was found to be down-regulated in imatinib resistant patients compared with imatinib responsive patients (86).
In gastric cancer, there are controversial results. One study reported SNHG5 as an oncogene that stimulates cell proliferation and migration, by sponging miR-32 and up-regulation of KLF4 (Krüppel-like factor 4) (87), while another study reported that SNHG5 is a tumor suppressor, which interacts with MTA2 (metastasis associated 1 family member 2), and it impairs the MTA2 translocation from the cytoplasm to the nucleus. The overexpression of SNHG5 was also associated with a lower level of MMP9 (metalloproteinase 9), MMP1 (metalloproteinase 1), MMP13 (metalloproteinase 13), and EGFR (epithelial growth factor receptor) (88). MTA2 is a major repressor of E-CAD; thus, it stimulates EMT (89). MMPs are a class of metalloproteinases that degrade non-cellular components of extracellular matrix, such as collagen, gelatin, lamins, and others. The secretion of MMPs helps malignant cells to degrade the basement membrane from their site of origin and to get access to the local blood vessels and lymphatic system (90).
In colorectal cancer, SNHG5 interacts with the SPATS2 (spermatogenesis associated serine rich 2) and increases its stability by blocking the mRNA degradation in the cytoplasm, caused by STATS2 mRNA interaction with STAU1 (staufen double-stranded RNA binding protein 1) (82).
Through miR-26 sponging and up-regulation of TRPC3 (transient receptor potential channel 3), SNHG5 stimulates melanoma cell growth. The exogenous knockdown of SNHG5 in melanoma cells in vitro decreases their proliferation rate and invasive capacity, while it stimulated apoptosis (91). Further research and details on the role of SNHG5 in malignant diseases are presented in Supplementary Table 3.
Small Nucleolar RNA Host Gene 6 (SNHG6)
SNHG6 is the lncRNA of origin for U87 SNORD (92) and U88 small Cajal bodies (93).
SNHG6 has an oncogenic role in gastric cancer by promoting cell growth, migration, and EMT-mediated invasion. These effects are a consequence of miR-101 (94) and miR-26a sponging (95). In colorectal cancer, the shRNA inhibition of SNHG6 causes increased level of miR-181a-5p and decreased level of E2F5 (E2F transcription factor 5), a transcription factor, which further decreases the number of colonies and invasive cells, as well as cell cycle arrest during in vitro studies (96). SNHG6 sponging of miR-26a/b and miR-214 causes an increased level of EZH2 (enhancer of zeste homolog 2), which further affects the epigenetic landscape of the colorectal cells (97).
SNHG6 sponges miR-101 causing the EMT through up-regulation of N-CAD and VIM and down-regulation of E-CAD and β-catenin (94). By suppressing SOCS2 (suppressor of cytokine signaling 2), miR-101 inhibits Helicobacter pylori–induced gastric cancer tumorigenesis (98) and tumor growth (99).
SNHG6 level has a positive correlation with disease progression and formation of local lymph node metastasis in the case of esophageal carcinoma (100).
In hepatocellular carcinoma, SNHG6 promotes cell cycle progression and apoptosis evasion. miR-101 is targeted by SNHG6, which further stimulates ZEB1 (zinc finger E-box-binding homeobox 1) expression (101).
In osteosarcoma, SNHG6 exogenous suppression impairs cell autophagy, apoptosis, colony formation, and invasion through miR-26a-5p sponging, which results in ATF3 (activating transcription factor 3) up-regulation. ULK1 (autophagy activating kinase) is a positive up-stream regulator of SNHG6 (102). Further research and details on the role of SNHG6 in malignant diseases are presented in Supplementary Table 4.
Small Nucleolar RNA Host Gene 7 (SNHG7)
SNHG7 is the origin for SNORA17 and SNORA43 (103).
SNHG7 is up-regulated in high grade bladder cancer. siRNA-mediated inhibition of SNHG7 leads to decreased wound closure speed in scratch assay, lower number of invasive cells in transwell assay, and an increased intracellular level of the proapoptotic marker BAX (Bcl-2-associated X protein), cell cycle inhibitor p21, and anti-invasive adhesion molecule, E-cadherin (104), while the expression of mesenchymal promoting proteins, namely, N-cadherin, VIM, and SNAIL are increased (105). SNHG7 activity in bladder cancer also extends to the activation of WNT/β-catenin pathway (106). BAX is an important proapoptotic marker. Its expression and activation are stimulated by p53 during apoptosis, when BAX is released from nucleus into the cytoplasm, where it binds to mitochondrial membrane. This is followed by mitochondrial pore formation necessary for the release of cytochrome C. BAX also directly interacts and represses the antiapoptotic protein BCL-2 (107). During WNT/β-catenin pathway activation, WNT protein binds to Frizzled receptor and stimulates its interaction with the LRP6 (lipoprotein receptor related protein 6). This is followed by accumulation and nuclear translocation of β-catenin. In the nucleus, β-catenin disrupts the interaction between T cell factor and lymphoid enhancer factor (TCF/LEF), thus stimulating the transcription of genes involved in cell self-renewal and proliferation capacity, necessary for tumorigenesis. In cancer, different components of this pathway acquire gain-of-function mutations (108).
In breast cancer, when comparing exogenous silencing of SNHG7 with controls, the following was observed: decreased self-renewal capacity of malignant cells, lower number of invasive cells during transwell assay, and smaller volume of xenograft tumors. At the molecular level, SNHG7 silencing lowered VIM and SNAIL level while increasing E-cadherin expression. SNHG7 overexpression leads to up-regulation of the antiapoptotic gene Survivin and the cell cycle promoting gene, Cyclin D, through the activation of Notch signaling pathway (109). In breast cancer, SNHG7 acts as ceRNA for several tumor suppressor miRNAs: miR-34a (109), miR-186 (110), and miR-381 (111).
Overexpression of SNHG7 stimulates the in vitro invasion capacity of colorectal cancer cell lines. The siRNA mediated silencing of SNHG7 increases cleaved PARP and cleaved Caspase 3 levels leading to apoptosis initiation. In vivo, colon cancer cells transfected with SNHG7 siRNA form smaller tumors, in comparison with negative control (112), while the overexpression of SNHG7 causes increased liver metastasis from primary colon tumors (113). In colon cancer, SNHG7 also targets miR-34a (112) and miR-216b (113).
In both esophageal (114) and gastric cancer, this lncRNA represses expression of p15 and p16, two tumor suppressors. Consequently, it stimulates proliferation and cell cycle progression and it inhibits apoptosis (114, 115).
In glioblastoma, SNHG7 induces proliferation, migration, and invasion of malignant cells, by inhibiting miR-5095 and by activating the WNT/β-catenin signaling pathway (116).
In hepatocellular carcinoma, inhibition of SNHG7 decreases the invasion capacity of malignant cells. The protein expression of RBM5 (RNA binding motif protein 5) was increased after SNHG7 silencing (117).
In melanoma, SNHG7 is overexpressed in malignant tissue vs. normal tissue. It increases cell invasion and migration capacity through positive correlation with SOX4 (SRY-box transcription factor 4) (118). miR-503, in prostate cancer, targets RNF31 and inhibits proliferation and metastasis (119). In prostate cancer, this lncRNA represses miR-503 and leads to tumor cell proliferation, cell cycle progression, and xenograft tumor growth (120).
In lung cancer, SNHG7 stimulates proliferation, migration, and invasion while inhibiting apoptosis through up-regulation of FAIM2 (Fas apoptotic inhibitory molecule 2) (121) and miR-193b sponging (122).
In neuroblastoma, it forms a positive feedback loop with miR-653-5p and STAT2 (signal transducer and activator of transcription 2), which sustains an aggressive phenotype of malignant cells (123). Further research and details on the role of SNHG7 in malignant diseases are presented in Supplementary Table 5.
Small Nucleolar RNA Host Gene 12 (SNHG12)
From the introns of SNHG12, four different types of snoRNAs are processed: SNORA44, SNORA61, SNORA16A, and SNORD99 (32). Latest studies show the SNHG12 up-regulation, and the inhibition of tumor suppressor miRNAs, in many cancer types (Supplementary Table 6).
SNHG12 has various biological functions through targeting of cell proliferation, invasion, migration, and apoptosis. SNHG12 is involved in different types of malignant diseases by targeting the following microRNAs: miR-424-5p in cervical cancer (124), miR-320 in gastric cancer (125), miR-199a-5p in renal cell carcinoma (126), miR-199a/b in hepatocellular carcinoma (32), miR-199a/b in gastric cancer (127), miR-138 in lung cancer (32), miR-181a in lung cancer (128), miR-16-5p in thyroid carcinoma (129), miR-129-5p in laryngeal squamous cell carcinoma (130), miR-199-5p in renal carcinoma (126), and miR-195-5p in osteosarcoma (131).
SNHG12 stimulates EMT in lung cancer by ceRNA targeting of miR-218 (132), and mediates doxorubicin resistance via miR-320a repression and MCL1 (MCL1 apoptosis regulator, BCL2 family member) up-regulation in osteosarcoma (133).
In glioma, SNHG12 targeting of miR-101-2 leads to enhanced cell growth (134), malignant progression via TDP43 (TAR DNA-binding protein 43) (135), and enhanced proliferation/migration capacity due to the association with the Hu antigen R (136).
In cervical cancer, SNHG12 promotes proliferation, invasion, and migration, by targeting miR-424-5p (124). This microRNA acts as a tumor suppressor in cervical cancer, by down-regulation of CHK1 (checkpoint kinase 1) gene (137) and up-regulation of aprataxin, which leads to radiosensitivity (138).
SNHG12 activation of PI3K/AKT signaling pathway in gastric cancer promoted cell proliferation, cell cycle progression, and inhibition of apoptosis (139). SNHG12 stimulation of PI3K/AKT pathway in colorectal cancer leads to cell growth (140). PI3K/AKT is a signaling pathway that becomes activated in response to growth factor stimulation. This signaling pathway is a master regulator of cell activity, and it acts by inhibiting p21, p53, and BAX, while activating the MDM2 oncogene (141, 142).
SNHG12 inhibits miR-320 (125), which acts as a tumor suppressor in gastric cancer, by lowering the expression of FoxM1 and P27KIP1 (143).
In hepatocellular carcinoma, SNHG12 induces miR-199a/b-5p underexpression, which leads to the overexpression of MLK3 (mixed-lineage kinase 3) and the stimulation of the NF-κB pathway (32).
In the non-small cell lung cancer, SNHG12 acts as a ceRNA for miR-138 (144) and miR-181a (128), causing increased proliferation and colony-formation capabilities, as well as impaired apoptosis (128, 144). MiR-138 is an inhibitor of proliferation, autophagy, and metastasis (145). Its down-regulation also leads to chemoresistance (146). MiR-181a hinders lung cancer cell proliferation and migration by targeting CDK1 (cyclin dependent kinase 1) (147) and KRAS (148). According to a recent meta-analysis, miR-181a is majorly linked to lung cancer patient's survival rate (149).
In osteosarcoma, SNHG12 causes increased cell proliferation and migration as well as stimulated angiogenesis, through up-regulation of AMOT (angiomotin) (150). In triple negative breast cancer, it also leads to increased cell proliferation, migration, and inhibited apoptosis, through the stimulation of MMP13 and SNHG12 genes activated by c-Myc (151).
Small Nucleolar RNA Host Gene 15 (SNHG15)
The SNHG15 is the origin for SNORA 9 (152).
The main molecular activity of SNHG15 is related to its targeting of miRNAs, as follows: miR-141-3p in hepatocellular carcinoma (153), miR-338-3p in prostate cancer (154), miR-211-3p in lung cancer (155, 156), and miR-211-3p in breast cancer (157). SNHG15 also leads to activation of NF-KB pathway in renal cell carcinoma (158).
SNHG15 is involved in colorectal cancer cell proliferation and migration via miR-141 (159) and miR-338-3p sponging (160), along with up-regulation of AIF (Allograft Inflammatory Factor 1) (152), and activation of transcription factor SLUG (45). In thyroid cancer, SNHG15 is a ceRNA for miR-510-5p (161) and miR-200a-3p (162).
In osteosarcoma, SNHG15 promotes cell proliferation, autophagy, and migration by acting as a ceRNA for miR-141. The involvement of SNHG15 was also responsible for the proliferation, autophagy, and invasion of osteosarcoma cells (163). MiR-141 is a tumor suppressor in osteosarcoma, inhibiting proliferation and activating apoptosis (164). In pancreatic cancer, SNHG15 was found to be up-regulated and to suppress the transcription of P15 and KLF2 (Kruppel Like Factor 2) by binding to EZH2 and the subsequent methylation at the promoter region of the histone 3 (H3K27me3) (165). This lncRNA is also involved in stimulating the proliferation of endothelial cells from glioma. SNHG15 silencing leads to down-regulation of VEGFA (vascular endothelial growth factor A) and CDC42 (cell division cycle 42), both being proangiogenic genes (166). In gastric cancer, this SNHG15 functions as a tumor promoter by up-regulating MMP2 and MMP9, followed by stimulation of proliferation and invasion (167).
In colorectal cancer, a new mechanism for SNHG15 was found: this lncRNA binds to the zinc-finger domain of SLUG and prevents its ubiquitination. The synchronous coexpression of SNHG and SLUG leads to increased colon cancer cell migration and tumorigenesis capacity (45). Further research and details on the role of SNHG15 in malignant diseases are presented in Supplementary Table 7.
Small Nucleolar RNA Host Gene 16 (SNHG16)
From the introns of SNHG16, the snoRD1A, snoRD1B, and snoRD1C are generated (168). SNHG16 sponges miR-4518 and activates the PI3K/Akt pathway in glioma, thus causing increased tumor cell proliferation and migration (169). In esophageal cancer, SNHG16 maintains cancer cell viability, impairs apoptosis, and enhances cell migration, by means of miR-140 entrapment, which further results in ZEB1 up-regulation (170).
In cervical cancer, the lncRNA SNHG16 is involved in cell proliferation, apoptosis, and migration by down-regulating miR-216 and up-regulating ZEB1 (171).
In breast cancer, SNHG16 is positively associated with increased proliferation rate, apoptosis evasion, and cell cycle progression through ceRNA binding of miR-98, followed by E2F5 up-regulation (172). Moreover, this lncRNA also has the potential to become a biomarker of early-stage pulmonary malignancy, which is very difficult to diagnose with currently available methods (173). In ovarian cancer, SNHG16 increases malignant cell migration capacity through phosphorylation-mediated activation of AKT (AKT serine/threonine kinase 1) and overexpression of MMP9 (174). Further research and details on the role of SNHG16 in malignant disease are presented in Supplementary Table 8.
Small Nucleolar RNA Host Gene 20 (SNHG20)
The SNHG20 has 2183 nt (175), and its introns encode for SCARNA16 (176). The SNHG20 is up-regulated in colorectal cancer, hepatocellular cancer, lung cancer, ovarian cancer, and breast cancer. This lncRNA was proven in many studies to interact with p21 (Supplementary Table 9).
SNHG20 down-regulates p21 and E-cadherin in ovarian cancer (177), activates STAT6 in hepatocellular carcinoma (178), represses miR140 in laryngeal squamous cell carcinoma (179), modulates ATM (ataxia telangiectasia mutated)–JAK (janus kinase 2)–PD-L1 (programmed death-ligand 1) pathway in esophageal carcinoma (180), up-regulates TGF-B1 in nasopharyngeal carcinoma (181), and enhances EMT and apoptosis via miR-139 -RUNX2 (runt-related transcription factor 2) in osteosarcoma (182, 183). In breast cancer, SNHG20 causes miR-495 sponging (184), while in oral cancer, it induces cell proliferation via down-regulating PCNA and Ki67 (185) and via targeting miR-197/LIN28 axis (186).
SNHG20, like all SNHG transcripts, is involved in many biological processes, such as: cell proliferation, cell cycle progression, tumorigenesis, apoptosis evasion, and resistance to therapy.
In gastric cancer, it inhibits p21 expression (187), it can interact with miR140-5p to induce resistance to therapy (188), or it can sponge miR-495-3p in order to enhance cell proliferation (189). In ovarian cancer, SNHG20 can activate WNT/β-catenin signaling pathway, which promotes cell proliferation (190).
SNHG20 supports glioma cell survival by sponging miR-4486 and up-regulating MDM2-p53 pathway (191), and silencing p21 (192). This lncRNA is also involved in vasculogenic mimicry, a process of pseudo-vascular formation in highly aggressive cancers, via enhancement of ZRANB2/SNHG20/FOX1 axis (193).
SNHG20 promotes gastric cancer by stimulating cancer cell proliferation, cell cycle progression, invasion, and migration. This is done by down-regulating the expression of the p21 and by activating GSK-3β/β-catenin signaling pathway (187). In ovarian cancer, the overexpression of this lncRNAs also leads to enhanced proliferation and invasion through the activation of the WNT/β-catenin signaling pathway (194).
A number of proinvasion proteins such as ZEB1, ZEB2, N-CAD, and VIM are positively correlated with SNHG20, thus leading to stimulated cell cycle progression, proliferation rate, and migration capacity hepatocellular carcinoma (195) and breast cancer (184).
In lung cancer, SNHG20 overexpression causes increased cell proliferation, invasion, and migration capacity, by changing the DNA methylation pattern, after interaction with EZH2 and epigenetic repression of p21 (175). p21 is suppressed by SNHG20 in colorectal cancer, where it stimulates cyclin A1, leading to intensification of proliferation and migration (196) P21 is classically considered as a tumor suppressor, being involved in the downstream pathway of the p53 cell cycle arrest and in the assembly of cyclin D-CDK4/CDK6. However, when p53 is dysregulated or when p21 acquires mutations, it becomes an oncogene (197). Further research and details on the role of SNHG20 in malignant diseases are presented in Supplementary Table 9.
SNHGs are overexpressed from the malignant transformation of a normal cell to the installment of metastasis. They sustain replicative immortality, speeding up of proliferation rate, resistance to programmed cell death, tumor growth, neoangiogenesis (and in some instances vascular mimicry), local invasion, migration, and formation of metastasis. The overall representation of different SNHGs' involvement in cancer initiation and progression is found in Figure 2.
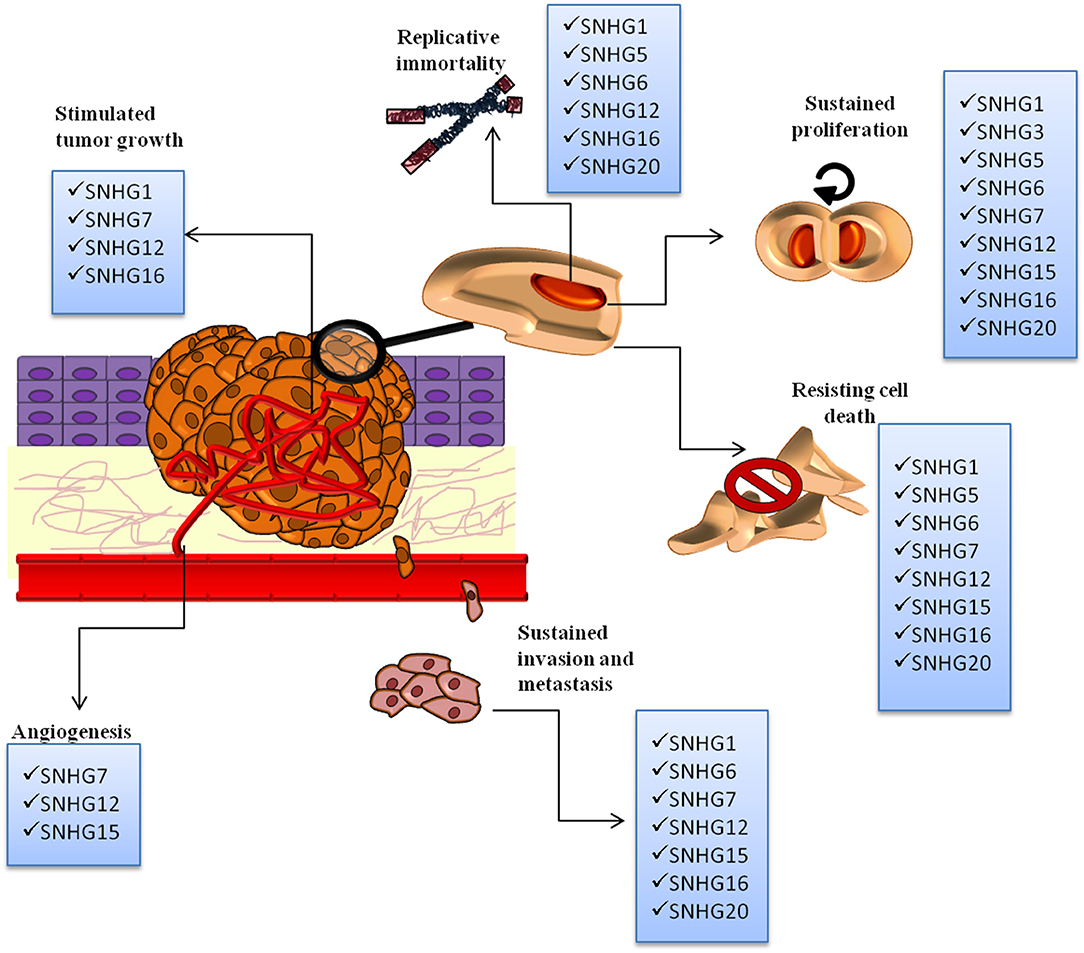
Figure 2. The SNHGs' involvement in the following malignant processes: replicative immortality, sustained proliferation, angiogenesis, resisting cell death, invasion, and metastasis and tumor growth. All SNHGs promote tumor development and progression.
Future Applications Based on Current Knowledge-Discussions
Through our research of literature, we identified that the general activity of SNHGs in malignant disease is related to stimulation of the following malignant processes: EMT, invasion, proliferation, cell cycle progression, and apoptosis evasion. The most common signaling pathways activated by SNHGs are: WNT/β-catenin (67, 198) and mTOR/PI3K/AKT (199–201). SNHGs also influence the NF-kB (202) and Hippo signaling pathways (203); however, more data are needed to include these two pathways in the general mechanisms of SNHG activity (Figure 3).
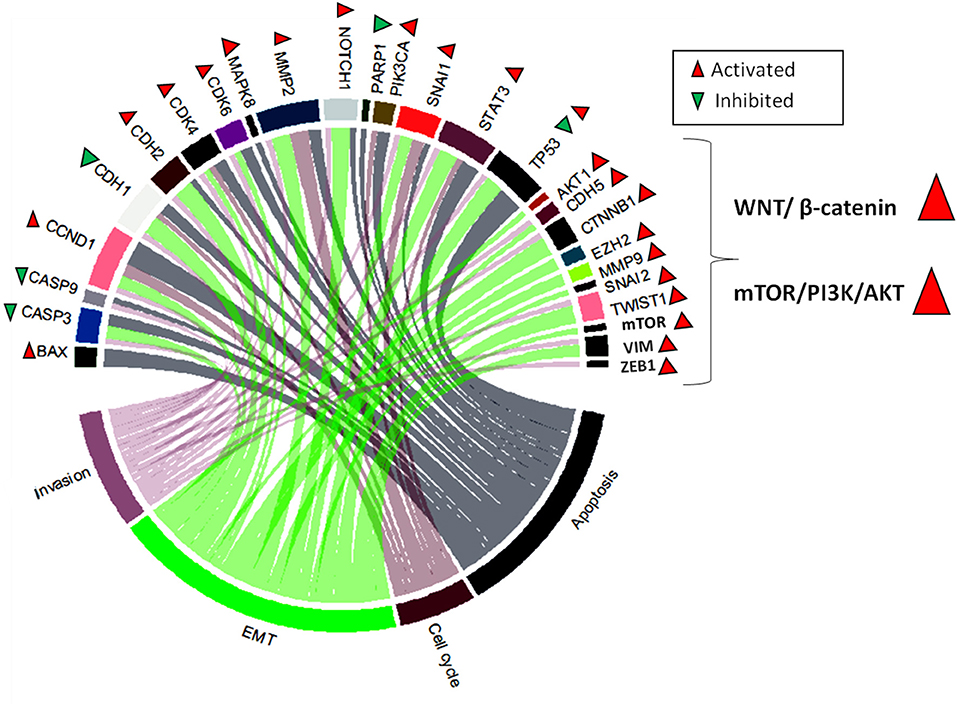
Figure 3. Chord plot representing the most frequently modulated genes by SNHGs. These are: BAX (Bcl-2-associated X protein), CASP3 (caspase 3), CASP9 (caspase 9), CCND1 (cyclin-D1), CDH1 (E-cadherin), CDH2 (N-cadherin), CDK4 (cyclin-dependent kinase 4), CDK6 (cyclin-dependent kinase 6), MAPK8 (mitogen-activated protein kinase), MMP2 (metalloproteinase 2), NOTCH1 (Notch homolog 1), PARP1 (poly [ADP-ribose] polymerase 1), PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha), SNAI1, STAT3 (signal transducer and activator of transcription 3), TP53, AKT1 (AKT serine/threonine kinase 1), CDH5 (vascular endothelial cadherin), CTNNB1 (beta-catenin), EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit), MMP9 (metalloproteinase 9), SNAI2 (snail family transcriptional repressor 2), TWIST1, mTOR, VIM (vimentin), and ZEB1 (zinc finger E-box binding homeobox 1). These are associate in a different degree to the most frequently dysregulated cellular processes in cancer: invasion, EMT, cell cycle, and apoptosis. SNHGs also act by activating two major signaling pathways: WNT/β-catenin and mTOR/PI3K/AKT.
The oncogenic role of SNHGs in malignancies is supported by solid scientific data. These ncRNAs act on multiple levels to induce a more aggressive phenotype of cancer cell, and their overexpression is correlated with lower overall and progression-free survival. There are a number of meta-analyses that offer additional support in favor of bringing the research of SNHGs into clinical trials. SNHG16 is associated with advanced clinicopathological features and worse overall survival in the case of malignancies located in the lung, ovaries, cervix, bladder, and esophagus (204). SNHG1 has a progressive increase in expression correlated with disease progression in the case of the following tumor localizations: lung, esophagus, bone, brain, stomach, liver, and colon. A high level of SNHG1 is associated with poor overall and disease-free survival in all of these malignancies (205–207). SNHG6 leads to worse prognosis and a propensity of distant metastasis formation (208).
The oncogenic activity of SNHGs can be impaired by the temporary silencing of SNHGs at RNA level with the help of RNA interference (194, 209) or the permanent deletion of these lncRNAs in cancer cells through genome editing techniques (152, 210).
A careful evaluation of the SNHGs inhibition in normal cells would be extremely useful in evaluating the potential druggability of SNHGs. From what we know so far, the downstream effect of SNHG inhibition on their corresponding snoRNAs-miRNAs/piRNAs will be dependent on the type of change: knock out or knockdown. SNHGs are targeted through permanent changes at the DNA level through gene therapy. This is a desired effect if the therapy is carefully monitored to have high specificity for tumor cells and to avoid normal cells. In normal cells, snoRNAs have essential physiological functions in rRNA and mRNA processing. A recent study has already evaluated the ability of the CRISPR/Cas9 to target protospacer adjacent motifs (PAMs) located in the structure of snoRNAs from the same SNHG, GAS5. The induced mutations affected specifically the ability to form secondary Kink-turn structure in each SNORD (SNORD74, SNORD77, and SNORD80). Moreover, the editing of PAM located in the D' box of SNORD75 affected the alternative splicing of SNHG, thus showing that snoRNAs can modulate the alternative splicing of their SNHG of origin by affecting the m6A methyltransferase complex (211). A better alternative to genome editing is the temporal inhibition of SNHGs through siRNAs. This is also the main therapeutic strategy evaluated across various studies. For instance, in renal cell carcinoma, the RNAi silencing of SNHG15, in vitro, led to decreased proliferation, invasion, and migration capacity (158). The silencing of SNHG1 in colon cancer cells lowered their malignant potential (212). The knockdown of SNHG6 in glioma causes a reduced growth rate of treated cells (213). The SNHG inhibition can reverse chemotherapeutic resistance, as in the case of sorafenib and SNHG1 (200). However, because of their ability of acting as ceRNAs for miRNAs (214) or being the primary transcript for various snoRNAs, piRNAs, and miRNAs, the down-stream effect of SNHG therapeutic modulation would be very difficult to predict and control, especially in case of non-specific targeting of tumor cells. For instance, we still don't know whether siRNA silencing of SNHGs affects the downstream levels of snoRNAs-piRNAs/miRNAs originated from the targeted SNHG.
Conclusion
Because of the versatility and function in the majority of cancers, SNHGs are becoming increasingly important for molecular research in cancer. The data on these non-coding RNAs are abundant, and the validation of their role in the progression and severity of malignant diseases has been made clear during the last year. However, a general overlook on these lncRNAs is still lacking as well as a better understanding of their influence at the molecular level. The majority of SNHGs (SNHG1, SNHG12, SNHG20, SNHG15, SNHG16, SNHG3, SNHG5, SNHG6, SNHG7) function by sponging tumor- suppressing microRNAs, allowing the oncogene transcripts to be expressed. The SNHGs can also cause epigenetic alterations in the genome, transcription initiation through interaction with transcription factors, direct binding and up-regulation of mRNAs and extension of protein life through prevention of ubiquitination. SNHGs also activate the signaling pathways commonly involved in cancer development and progression, such as Wnt/β-catenin and mTOR/PI3K/AKT. The SNHGs have the potential to act as cancer biomarkers or even therapeutic targets because of their ability to retain the tumor suppressor microRNAs. The studies on SNHGs increase by the day, and these classes of non-coding RNAs might constitute the next miRNAs in the field of molecular oncology. However, intrapopulation heterogeneity is still a matter of concern in the analysis of every transcript as biomarker and more data are needed considering especially that there are some reports, even if scarce, on the role of these lncRNAs acting as tumor suppressors. More analysis should also be done on regulatory mechanism at DNA and protein level and SNHGs stability maintenance in the disadvantage of snoRNAs, although data present a positive correlation between SNHGs and their corresponding snoRNAs.
Author Contributions
A-AZ: first co-author and PhD student specializing in molecular biology who wrote the majority of the paper. AT: PhD student in cellular and molecular biology who majorly contributed to the literature review regarding functional in vitro assays and tables 1–9 writing. CB: researcher with experience in molecular biology who contributed to the design of figures, general concept of the article, and text writing. CS: MD, PhD expert in global oncology, researcher with multiple specializations in pedriatic oncology, gynecology, and public health who wrote about the clinical applications of SNHGs. CI: surgeon and physician scientist with expertise in surgical oncology, ncRNAs, and molecular profiling of cancers who critically revised the manuscript and wrote part of the introduction. IB-N: Professor of Medical Biotechnology who coordinated the article topic, analysis, and interpretation and was responsible for the final version of the article, including proofreading, writing, and reorganizing the article.
Funding
This work was supported by Competitivity Operational Program, 2014–2020, entitled Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance—CANTEMIR, no. 35/01.09.2016, MySMIS 103375, project PNCDI III 2015-2020 entitled Increasing the performance of scientific research and technology transfer in translational medicine through the formation of a new generation of young researchers—ECHITAS, no. 29PFE/18.10.2018 and by PN-III-P1-1.2-PCCDI2017-0737, grant 35/2018 entitled Genomic population mapping of radioactive and heavy metals in order to increase national security—ARTEMIS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00389/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
3. Braicu C, Pileczki V, Irimie A, Berindan-Neagoe I. p53siRNA therapy reduces cell proliferation, migration and induces apoptosis in triple negative breast cancer cells. Mol Cell Biochem. (2013) 381:61–8. doi: 10.1007/s11010-013-1688-5
4. Pop-Bica C, Gulei D, Cojocneanu-Petric R, Braicu C, Petrut B, Berindan-Neagoe I. Understanding the role of non-coding RNAs in bladder cancer: from dark matter to valuable therapeutic targets. Int J Mol Sci. (2017) 18:E1514. doi: 10.3390/ijms18071514
5. Rigoutsos I, Lee SK, Nam SY, Anfossi S, Pasculli B, Pichler M, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. (2017) 18:98. doi: 10.1186/s13059-017-1224-0
6. Braicu C, Catana C, Calin GA, Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr Pharm Des. (2014) 20:6565–74. doi: 10.2174/1381612820666140826153529
7. Catana CS, Calin GA, Berindan-Neagoe I. Inflamma-miRs in aging and breast cancer: are they reliable players? Front Med. (2015) 2:85. doi: 10.3389/fmed.2015.00085
8. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. (2018) 24:257–77. doi: 10.1016/j.molmed.2018.01.001
9. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. (2018) 419:152–66. doi: 10.1016/j.canlet.2018.01.053
10. Irimie AI, Zimta AA, Ciocan C, Mehterov N, Dudea D, Braicu C, et al. The unforeseen non-coding RNAs in head and neck cancer. Genes. (2018) 9:134. doi: 10.3390/genes9030134
11. Chen D, Lu T, Tan J, Li H, Wang Q, Wei L. Long non-coding RNAs as communicators and mediators between the tumor microenvironment and cancer cells. Front Oncol. (2019) 9:739. doi: 10.3389/fonc.2019.00739
12. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. (2006) 15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046
13. Catana CS, Pichler M, Giannelli G, Mader RM, Berindan-Neagoe I. Non-coding RNAs, the Trojan horse in two-way communication between tumor and stroma in colorectal and hepatocellular carcinoma. Oncotarget. (2017) 8:29519–34. doi: 10.18632/oncotarget.15706
14. Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. (2009) 21:416–25. doi: 10.1016/j.ceb.2009.04.001
15. Braicu C, Calin GA, Berindan-Neagoe I. MicroRNAs and cancer therapy - from bystanders to major players. Curr Med Chem. (2013) 20:3561–73. doi: 10.2174/0929867311320290002
16. Jurj A, Braicu C, Pop LA, Tomuleasa C, Gherman CD, Berindan-Neagoe I. The new era of nanotechnology, an alternative to change cancer treatment. Drug Des Devel Ther. (2017) 11:2871–90. doi: 10.2147/DDDT.S142337
17. Sonea L, Buse M, Gulei D, Onaciu A, Simon I, Braicu C, et al. Decoding the emerging patterns exhibited in non-coding RNAs characteristic of lung cancer with regard to their clinical significance. Curr Genomics. (2018) 19:258–78. doi: 10.2174/1389202918666171005100124
18. Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. (2013) 14:16010–39. doi: 10.3390/ijms140816010
19. Berindan-Neagoe I, Braicu C, Gulei D, Tomuleasa C, Calin GA. Noncoding RNAs in lung cancer angiogenesis. In: Simionescu D, Simionescu A, editors. Physiologic and Pathologic Angiogenesis - Signaling Mechanisms and Targeted Therapy. Rijeka: InTech (2017). p. 14.
20. Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. (2017) 33:540–52. doi: 10.1016/j.tig.2017.05.004
21. Scott MS, Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. (2011) 93:1987–92. doi: 10.1016/j.biochi.2011.05.026
22. Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. (2012) 1826:121–8. doi: 10.1016/j.bbcan.2012.03.005
23. Kufel J, Grzechnik P. Small nucleolar RNAs tell a different tale. Trends Genet. (2019) 35:104–17. doi: 10.1016/j.tig.2018.11.005
24. Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays. (2013) 35:46–54. doi: 10.1002/bies.201200117
25. Halic M, Moazed D. Transposon silencing by piRNAs. Cell. (2009) 138:1058–60. doi: 10.1016/j.cell.2009.08.030
26. Hoeppner MP, White S, Jeffares DC, Poole AM. Evolutionarily stable association of intronic snoRNAs and microRNAs with their host genes. Genome Biol Evol. (2009) 1:420–8. doi: 10.1093/gbe/evp045
27. Tanaka-Fujita R, Soeno Y, Satoh H, Nakamura Y, Mori S. Human and mouse protein-noncoding snoRNA host genes with dissimilar nucleotide sequences show chromosomal synteny. RNA. (2007) 13:811–6. doi: 10.1261/rna.209707
28. Boivin V, Deschamps-Francoeur G, Scott MS. Protein coding genes as hosts for noncoding RNA expression. Semin Cell Dev Biol. (2018) 75:3–12. doi: 10.1016/j.semcdb.2017.08.016
29. Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. (2012) 12:84–8. doi: 10.1038/nrc3195
30. Gong J, Li Y, Liu C-J, Xiang Y, Li C, Ye Y, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. (2017) 21:1968–81. doi: 10.1016/j.celrep.2017.10.070
31. Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. (2011) 17:878–91. doi: 10.1261/rna.2528811
32. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. (2017) 36:11. doi: 10.1186/s13046-016-0486-9
33. Xu T, Yan S, Jiang L, Yu S, Lei T, Yang D, et al. Gene amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer. Mol Ther Nucleic Acids. (2019) 17:405–13. doi: 10.1016/j.omtn.2019.06.008
34. Yu F, Bracken CP, Pillman KA, Lawrence DM, Goodall GJ, Callen DF, et al. p53 represses the oncogenic Sno-MiR-28 derived from a SnoRNA. PLoS ONE. (2015) 10:e0129190. doi: 10.1371/journal.pone.0129190
35. Peculis BA. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA. (2001) 7:207–19. doi: 10.1017/S1355838201001625
36. Sun Y, Wei G, Luo H, Wu W, Skogerbo G, Luo J, et al. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene. (2017) 36:6774–83. doi: 10.1038/onc.2017.286
37. Yang Y, Toy W, Choong LY, Hou P, Ashktorab H, Smoot DT, et al. Discovery of SLC3A2 cell membrane protein as a potential gastric cancer biomarker: implications in molecular imaging. J Proteome Res. (2012) 11:5736–47. doi: 10.1021/pr300555y
38. Zhu B, Cheng D, Hou L, Zhou S, Ying T, Yang Q. SLC3A2 is upregulated in human osteosarcoma and promotes tumor growth through the PI3K/Akt signaling pathway. Oncol Rep. (2017) 37:2575–82. doi: 10.3892/or.2017.5530
39. Poss ZC, Ebmeier CC, Taatjes DJ. The mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. (2013) 48:575–608. doi: 10.3109/10409238.2013.840259
40. Fei F, He Y, He S, He Z, Wang Y, Wu G, et al. LncRNA SNHG3 enhances the malignant progress of glioma through silencing KLF2 and p21. Biosci Rep. (2018) 38:BSR20180420. doi: 10.1042/BSR20180420
41. Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death dis. (2019) 10:694. doi: 10.1038/s41419-019-1940-3
42. Yan Y, Fan Q, Wang L, Zhou Y, Li J, Zhou K. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget. (2017) 8:35750–60. doi: 10.18632/oncotarget.16189
43. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. (2016) 17:272–83. doi: 10.1038/nrg.2016.20
44. Zhao Y, Qin ZS, Feng Y, Tang XJ, Zhang T, Yang L. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53. Eur Rev Med Pharmacol Sci. (2018) 22:976–84. doi: 10.26355/eurrev_201802_14379
45. Jiang H, Li T, Qu Y, Wang X, Li B, Song J, et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Letters. 425:78–87 doi: 10.1016/j.canlet.2018.03.038
46. Frey MR, Wu W, Dunn JM, Matera AG. The U22 host gene (UHG): chromosomal localization of UHG and distribution of U22 small nucleolar RNA. Histochem Cell Biol. (1997) 108:365–70. doi: 10.1007/s004180050177
47. Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. (1996) 379:464–6. doi: 10.1038/379464a0
48. Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, et al. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget. (2017) 8:111715–27. doi: 10.18632/oncotarget.22903
49. Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. (2018) 9:2128–39. doi: 10.18632/oncotarget.23255
50. Qi H, Wang J, Wang F, Ma H. Long non-coding RNA SNHG1 promotes cell proliferation and tumorigenesis in colorectal cancer via Wnt/beta-catenin signaling. Pharmazie. (2017) 72:395–401. doi: 10.1691/ph.2017.7463
51. Li S, Wu X, Xu Y, Wu S, Li Z, Chen R, et al. miR-145 suppresses colorectal cancer cell migration and invasion by targeting an ETS-related gene. Oncol Rep. (2016) 36:1917–26. doi: 10.3892/or.2016.5042
52. Ge J, Wu XM, Yang XT, Gao JM, Wang F, Ye KF. Role of long non-coding RNA SNHG1 in occurrence and progression of ovarian carcinoma. Eur Rev Med Pharmacol Sci. (2018) 22:329–35. doi: 10.26355/eurrev_201801_14176
53. Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. (2017) 487:146–52. doi: 10.1016/j.bbrc.2017.03.169
54. Zhong J, Huang R, Su Z, Zhang M, Xu M, Gong J, et al. Downregulation of miR-199a-5p promotes prostate adeno-carcinoma progression through loss of its inhibition of HIF-1alpha. Oncotarget. (2017) 8:83523–38. doi: 10.18632/oncotarget.18315
55. Shi Y-H, Fang W-G. Hypoxia-inducible factor-1 in tumour angiogenesis. World J Gastroenterol. (2004) 10:1082–7. doi: 10.3748/wjg.v10.i8.1082
56. Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. MiR-338-5p enhances the radiosensitivity of esophageal squamous cell carcinoma by inducing apoptosis through targeting survivin. Sci Rep. (2017) 7:10932. doi: 10.1038/s41598-017-10977-9
57. Zhang Y, Jin X, Wang Z, Zhang X, Liu S, Liu G. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomarkers. (2017) 21:1–8. doi: 10.3233/CBM-170286
58. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. (2009) 119:1420–8. doi: 10.1172/JCI39104
59. Van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. (2008) 65:3756–88. doi: 10.1007/s00018-008-8281-1
60. Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. (2004) 48:463–76. doi: 10.1387/ijdb.041793ld
61. Kidd ME, Shumaker DK, Ridge KM. The role of vimentin intermediate filaments in the progression of lung cancer. Am J Respir Cell Mol Biol. (2014) 50:1–6. doi: 10.1165/rcmb.2013-0314TR
62. Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. (2016) 80:73–9. doi: 10.1016/j.biopha.2016.02.036
63. Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, et al. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. (2016) 22:4820–9. doi: 10.12659/MSM.898574
64. Wang M, Zhang J, Tong L, Ma X, Qiu X. MiR-195 is a key negative regulator of hepatocellular carcinoma metastasis by targeting FGF2 and VEGFA. Int J Clin Exp Pathol. (2015) 8:14110–20.
65. Wang Q, Li Q, Zhou P, Deng D, Xue L, Shao N, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. (2017) 91:906–11. doi: 10.1016/j.biopha.2017.05.014
66. Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. (2018) 32:3957–67. doi: 10.1096/fj.201701237RR
67. Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. (2017) 8:17785–94. doi: 10.18632/oncotarget.14854
68. Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med. (2016) 8:29. doi: 10.1186/s13073-016-0284-1
69. Li Y, Li Y, Liu J, Fan Y, Li X, Dong M, et al. Expression levels of microRNA-145 and microRNA-10b are associated with metastasis in non-small cell lung cancer. Cancer Biol Ther. (2016) 17:272–9. doi: 10.1080/15384047.2016.1139242
70. Luo L, Zhang T, Liu H, Lv T, Yuan D, Yao Y, et al. MiR-101 and Mcl-1 in non-small-cell lung cancer: expression profile and clinical significance. Med Oncol. (2012) 29:1681–6. doi: 10.1007/s12032-011-0085-8
71. Cao L, Wang J, Wang PQ. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed Pharmacother. (2016) 84:828–35. doi: 10.1016/j.biopha.2016.10.008
72. Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. (1997) 275:1129–32. doi: 10.1126/science.275.5303.1129
73. Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/beta-catenin pathway. Biochem Biophys Res Commun. (2018) 495:238–45. doi: 10.1016/j.bbrc.2017.11.012
74. Pelczar P, Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5'-terminal oligopyrimidine gene family. Mol Cell Biol. (1998) 18:4509–18. doi: 10.1128/MCB.18.8.4509
75. Zheng S, Jiang F, Ge D, Tang J, Chen H, Yang J, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother. (2019) 112:108695. doi: 10.1016/j.biopha.2019.108695
76. Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. (2019) 33:2058738418820743. doi: 10.1177/2058738418820743
77. Wu J, Lingrel JB. Kruppel-like factor 2, a novel immediate-early transcriptional factor, regulates IL-2 expression in T lymphocyte activation. J Immunol. (2005) 175:3060–6. doi: 10.4049/jimmunol.175.5.3060
78. Mao QQ, Chen JJ, Dong L, Zhong L, Sun X. Kruppel-like factor 2 suppresses growth and invasion of gastric cancer cells in vitro and in vivo. J Biol Regul Homeost Agents. (2016) 30:703–12.
79. Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. (2009) 9:400–14. doi: 10.1038/nrc2657
80. Huang W, Tian Y, Dong S, Cha Y, Li J, Guo X, et al. The long non-coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep. (2017) 38:1402–10. doi: 10.3892/or.2017.5837
81. Zhang T, Cao C, Wu D, Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biol. (2016) 37:2379–85. doi: 10.1007/s13277-015-4052-4
82. Damas ND, Marcatti M, Come C, Christensen LL, Nielsen MM, Baumgartner R, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. (2016) 7:13875. doi: 10.1038/ncomms13875
83. Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. (2019) 17:138–49. doi: 10.1016/j.omtn.2019.05.013
84. Turgeon M-O, Perry NJS, Poulogiannis G. DNA damage, repair, and cancer metabolism. Front Oncol. (2018) 8:15. doi: 10.3389/fonc.2018.00015
85. He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. (2017) 7:1704–13.
86. Jurkovicova D, Lukackova R, Magyerkova M, Kulcsar L, Krivjanska M, Krivjansky V, et al. microRNA expression profiling as supportive diagnostic and therapy prediction tool in chronic myeloid leukemia. Neoplasma. (2015) 62:949–58. doi: 10.4149/neo_2015_115
87. Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. (2017) 31:893–903. doi: 10.1096/fj.201600994R
88. Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. (2016) 35:5770–80. doi: 10.1038/onc.2016.110
89. Zhang B, Zhang H, Shen G. Metastasis-associated protein 2 (MTA2) promotes the metastasis of non-small-cell lung cancer through the inhibition of the cell adhesion molecule Ep-CAM and E-cadherin. Jpn J Clin Oncol. (2015) 45:755–66. doi: 10.1093/jjco/hyv062
90. Jabłonska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhibition Med Chem. (2016) 31:177–83. doi: 10.3109/14756366.2016.1161620
91. Gao J, Zeng K, Liu Y, Gao L, Liu L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. (2019) 12:169–79. doi: 10.2147/OTT.S184078
92. Makarova JA, Kramerov DA. Analysis of C/D box snoRNA genes in vertebrates: the number of copies decreases in placental mammals. Genomics. (2009) 94:11–9. doi: 10.1016/j.ygeno.2009.02.003
93. Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. (2013) 14:9099–110. doi: 10.3390/ijms14059099
94. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through Epigenetically Silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. (2017) 42:999–1012. doi: 10.1159/000478682
95. Zhang M, Duan W, Sun W. LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis. Onco Targets Ther. (2019) 12:3349–60. doi: 10.2147/OTT.S197433
96. Yu C, Sun J, Leng X, Yang J. Long noncoding RNA SNHG6 functions as a competing endogenous RNA by sponging miR-181a-5p to regulate E2F5 expression in colorectal cancer. Cancer Manag Res. (2019) 11:611–24. doi: 10.2147/CMAR.S182719
97. Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. (2019) 12:3. doi: 10.1186/s13045-018-0690-5
98. Zhou X, Xia Y, Li L, Zhang G. MiR-101 inhibits cell growth and tumorigenesis of Helicobacter pylori related gastric cancer by repression of SOCS2. Cancer Biol Ther. (2015) 16:160–9. doi: 10.4161/15384047.2014.987523
99. He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, et al. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. (2012) 279:4201–12. doi: 10.1111/febs.12013
100. Zhang Y, Li R, Ding X, Zhang K, Qin W. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell malignancy and its diagnostic value. Am J Transl Res. (2019) 11:1084–91.
101. Chang L, Yuan Y, Li C, Guo T, Qi H, Xiao Y, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. (2016) 383:183–94. doi: 10.1016/j.canlet.2016.09.034
102. Zhu X, Yang G, Xu J, Zhang C. Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int. (2019) 19:82. doi: 10.1186/s12935-019-0794-1
103. Boone D, Warburton A, Som S, Lee A. A negative feedback loop between Insulin-like Growth Factor signaling and the lncRNA SNHG7 tightly regulates transcript levels and proliferation. (2019). doi: 10.1101/709352
104. Xu C, Zhou J, Wang Y, Wang A, Su L, Liu S, et al. Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7. J Cancer. (2019) 10:539–46. doi: 10.7150/jca.25507
105. Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J Buon. (2018) 23:776–81.
106. Chen Y, Peng Y, Xu Z, Ge B, Xiang X, Zhang T, et al. Knockdown of lncRNA SNHG7 inhibited cell proliferation and migration in bladder cancer through activating Wnt/beta-catenin pathway. Pathol Res Pract. (2019) 215:302–7. doi: 10.1016/j.prp.2018.11.015
107. Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. (2008) 18:157–64. doi: 10.1016/j.tcb.2008.01.007
108. Macdonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. (2009) 17:9–26. doi: 10.1016/j.devcel.2009.06.016
109. Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur J Pharmacol. (2019) 856:172407. doi: 10.1016/j.ejphar.2019.172407
110. Luo X, Song Y, Tang L, Sun DH, Ji DG. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. (2018) 22:7788–97. doi: 10.26355/eurrev_201811_16403
111. Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci. (2019) 23:6588–95. doi: 10.26355/eurrev_201908_18545
112. Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. (2018) 11:89. doi: 10.1186/s13045-018-0632-2
113. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. (2018) 9:1–13. doi: 10.1038/s41419-018-0759-7
114. Xu LJ, Yu XJ, Wei B, Hui HX, Sun Y, Dai J, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. (2018) 22:2653–61. doi: 10.26355/eurrev_201805_14961
115. Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. (2017) 21:4613–22.
116. Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. (2018) 496:712–8. doi: 10.1016/j.bbrc.2018.01.109
117. Sun BZ, Ji DG, Feng ZX, Wang Y. Long noncoding RNA SNHG7 represses the expression of RBM5 to strengthen metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. (2019) 23:5699–704. doi: 10.26355/eurrev_201907_18307
118. Zhang C, Zhu B, Li XB, Cao YQ, Yang JC, Li X, et al. Long non-coding RNA SNHG7 promotes migration and invasion of melanoma via upregulating SOX4. Eur Rev Med Pharmacol Sci. (2019) 23:4828–34. doi: 10.26355/eurrev_201906_18069
119. Guo J, Liu X, Wang M. miR-503 suppresses tumor cell proliferation and metastasis by directly targeting RNF31 in prostate cancer. Biochem Biophys Res Commun. (2015) 464:1302–8. doi: 10.1016/j.bbrc.2015.07.127
120. Qi H, Wen B, Wu Q, Cheng W, Lou J, Wei J, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. (2018) 102:326–32. doi: 10.1016/j.biopha.2018.03.011
121. She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. (2016) 36:2673–80. doi: 10.3892/or.2016.5105
122. She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. (2018) 51:e12406. doi: 10.1111/cpr.12406
123. Chi R, Chen X, Liu M, Zhang H, Li F, Fan X, et al. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol. (2019) 234:13403–12. doi: 10.1002/jcp.28017
124. Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem. (2018) 45:2086–94. doi: 10.1159/000488045
125. Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. (2018) 17:2743–9. doi: 10.3892/mmr.2017.8143
126. Chen Q, Zhou W, Du SQ, Gong DX, Li J, Bi JB, et al. Overexpression of SNHG12 regulates the viability and invasion of renal cell carcinoma cells through modulation of HIF1alpha. Cancer Cell Int. (2019) 19:128. doi: 10.1186/s12935-019-0782-5
127. Yang BF, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. (2018) 22:1297–306. doi: 10.26355/eurrev_201803_14471
128. Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. (2017) 8:84086–101. doi: 10.18632/oncotarget.20475
129. Feng X, Dong X, Wu D, Zhao H, Xu C, Li H. Long noncoding RNA small nucleolar RNA host gene 12 promotes papillary thyroid carcinoma cell growth and invasion by targeting miR-16-5p. Histol Histopathol. (2019) 35:217–24. doi: 10.14670/HH-18-155
130. Li J, Sun S, Chen W, Yuan K. Small nucleolar RNA host gene 12 (SNHG12) promotes proliferation and invasion of laryngeal cancer cells via sponging miR-129-5p and potentiating WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) expression. Med Sci Monit. (2019) 25:5552–60. doi: 10.12659/MSM.917088
131. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. (2018) 495:1822–32. doi: 10.1016/j.bbrc.2017.12.047
132. Wang Y, Liang S, Yu Y, Shi Y, Zheng H. Knockdown of SNHG12 suppresses tumor metastasis and epithelial-mesenchymal transition via the Slug/ZEB2 signaling pathway by targeting miR-218 in NSCLC. Oncol Lett. (2019) 17:2356–64. doi: 10.3892/ol.2018.9880
133. Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. (2018) 106:850–7. doi: 10.1016/j.biopha.2018.07.003
134. Sun Y, Liu J, Chu L, Yang W, Liu H, Li C, et al. Long noncoding RNA SNHG12 facilitates the tumorigenesis of glioma through miR-101-3p/FOXP1 axis. Gene. (2018) 676:315–21. doi: 10.1016/j.gene.2018.08.034
135. Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z, et al. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol Ther Nucleic Acids. (2018) 10:142–58. doi: 10.1016/j.omtn.2017.12.001
136. Lei W, Wang ZL, Feng HJ, Lin XD, Li CZ, Fan D. Long non-coding RNA SNHG12promotes the proliferation and migration of glioma cells by binding to HuR. Int J Oncol. (2018) 53:1374–84. doi: 10.3892/ijo.2018.4478
137. Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. (2013) 32:976–87. doi: 10.1038/onc.2012.121
138. Wang X, Li Q, Jin H, Zou H, Xia W, Dai N, et al. miR-424 acts as a tumor radiosensitizer by targeting aprataxin in cervical cancer. Oncotarget. (2016) 7:77508–15. doi: 10.18632/oncotarget.12716
139. Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. (2017) 50:e6079. doi: 10.1590/1414-431x20176079
140. Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. (2015) 1855:104–21. doi: 10.1016/j.bbcan.2014.09.008
141. Brazil DP, Hemmings BA. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci. (2001) 26:657–64. doi: 10.1016/S0968-0004(01)01958-2
142. Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. (2002) 277:21843–50. doi: 10.1074/jbc.M109745200
143. Wang Y, Zeng J, Pan J, Geng X, Li L, Wu J, et al. MiR-320a inhibits gastric carcinoma by targeting activity in the FoxM1-P27KIP1 axis. Oncotarget. (2016) 7:29275–86. doi: 10.18632/oncotarget.8676
144. Wang X, Qi G, Zhang J, Wu J, Zhou N, Li L, et al. Knockdown of long noncoding RNA small nucleolar RNA host gene 12 inhibits cell growth and induces apoptosis by upregulating miR-138 in nonsmall cell lung cancer. DNA Cell Biol. (2017) 36:892–900. doi: 10.1089/dna.2017.3830
145. Ye Z, Fang B, Pan J, Zhang N, Huang J, Xie C, et al. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep. (2017) 37:3244–52. doi: 10.3892/or.2017.5619
146. Jin Z, Guan L, Song Y, Xiang GM, Chen SX, Gao B. MicroRNA-138 regulates chemoresistance in human non-small cell lung cancer via epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci. (2016) 20:1080–6.
147. Al-Hebshi NN, Alharbi FA, Mahri M, Chen T. Differences in the bacteriome of smokeless tobacco products with different oral carcinogenicity: compositional and predicted functional analysis. Genes. (2017) 8:106. doi: 10.3390/genes8040106
148. Ma Z, Qiu X, Wang D, Li Y, Zhang B, Yuan T, et al. MiR-181a-5p inhibits cell proliferation and migration by targeting Kras in non-small cell lung cancer A549 cells. Acta Biochim Biophys Sin. (2015) 47:630–8. doi: 10.1093/abbs/gmv054
149. Pop-Bica C, Pintea S, Cojocneanu-Petric R, Del Sal G, Piazza S, Wu ZH, et al. MiR-181 family-specific behavior in different cancers: a meta-analysis view. Cancer Metastasis Rev. (2018) 37:17–32. doi: 10.1007/s10555-017-9714-9
150. Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. (2016) 37:4065–73. doi: 10.1007/s13277-015-4256-7
151. Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. (2017) 9:533–45.
152. Saeinasab M, Bahrami AR, González J, Marchese FP, Martinez D, Mowla SJ, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. (2019) 38:172. doi: 10.1186/s13046-019-1169-0
153. Ye J, Tan L, Fu Y, Xu H, Wen L, Deng Y, et al. LncRNA SNHG15 promotes hepatocellular carcinoma progression by sponging miR-141-3p. J Cell Biochem. (2019) 120:19775–83. doi: 10.1002/jcb.29283
154. Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. LncRNA SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. (2019) 705:44–50. doi: 10.1016/j.gene.2019.04.033
155. Cui HX, Zhang MY, Liu K, Liu J, Zhang ZL, Fu L. LncRNA SNHG15 promotes proliferation and migration of lung cancer via targeting microRNA-211-3p. Eur Rev Med Pharmacol Sci. (2018) 22:6838–44. doi: 10.26355/eurrev_201810_16152
156. Ma XR, Xu YL, Qian J, Wang Y. Long non-coding RNA SNHG15 accelerates the progression of non-small cell lung cancer by absorbing miR-211-3p. Eur Rev Med Pharmacol Sci. (2019) 23:1536–44. doi: 10.26355/eurrev_201902_17112
157. Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. (2018) 495:1594–600. doi: 10.1016/j.bbrc.2017.12.013
158. Du Y, Kong C, Zhu Y, Yu M, Li Z, Bi J, et al. Knockdown of SNHG15 suppresses renal cell carcinoma proliferation and EMT by regulating the NF-kappaB signaling pathway. Int J Oncol. (2018) 53:384–94. doi: 10.3892/ijo.2018.4395
159. Sun X, Bai Y, Yang C, Hu S, Hou Z, Wang G. Long noncoding RNA SNHG15 enhances the development of colorectal carcinoma via functioning as a ceRNA through miR-141/SIRT1/Wnt/beta-catenin axis. Artif Cells Nanomed Biotechnol. (2019) 47:2536–44. doi: 10.1080/21691401.2019.1621328
160. Li M, Bian Z, Jin G, Zhang J, Yao S, Feng Y, et al. LncRNA-SNHG15 enhances cell proliferation in colorectal cancer by inhibiting miR-338-3p. Cancer Med. (2019) 8:2404–13. doi: 10.1002/cam4.2105
161. Liu Y, Li J, Li M, Li F, Shao Y, Wu L. microRNA-510-5p promotes thyroid cancer cell proliferation, migration, and invasion through suppressing SNHG15. J Cell Biochem. (2019). doi: 10.1002/jcb.28454. [Epub ahead of print].
162. Wu DM, Wang S, Wen X, Han XR, Wang YJ, Shen M, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. (2018) 9:947. doi: 10.1038/s41419-018-0975-1
163. Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. (2017) 24:46. doi: 10.1186/s12929-017-0353-9
164. Xu H, Mei Q, Xiong C, Zhao J. Tumor-suppressing effects of miR-141 in human osteosarcoma. Cell Biochem Biophys. (2014) 69:319–25. doi: 10.1007/s12013-013-9801-7
165. Ma Z, Huang H, Wang J, Zhou Y, Pu F, Zhao Q, et al. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget. (2017) 8:84153–67. doi: 10.18632/oncotarget.20359
166. Ma Y, Xue Y, Liu X, Qu C, Cai H, Wang P, et al. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol Rep. (2017) 38:3265–77. doi: 10.3892/or.2017.5985
167. Chen SX, Yin JF, Lin BC, Su HF, Zheng Z, Xie CY, et al. Upregulated expression of long noncoding RNA SNHG15 promotes cell proliferation and invasion through regulates MMP2/MMP9 in patients with GC. Tumour Biol. (2016) 37:6801–12. doi: 10.1007/s13277-015-4404-0
168. Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. (2016) 10:1266–82. doi: 10.1016/j.molonc.2016.06.003
169. Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, et al. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. (2018) 45:1975–85. doi: 10.1159/000487974
170. Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. (2018) 9:1028–40. doi: 10.18632/oncotarget.23178
171. Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. (2018) 637:1–8. doi: 10.1016/j.abb.2017.11.003
172. Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. (2017) 485:272–8. doi: 10.1016/j.bbrc.2017.02.094
173. Zhu H, Zhang L, Yan S, Li W, Cui J, Zhu M, et al. LncRNA16 is a potential biomarker for diagnosis of early-stage lung cancer that promotes cell proliferation by regulating the cell cycle. Oncotarget. (2017) 8:7867–77. doi: 10.18632/oncotarget.13980
174. Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. (2018) 22:616–22. doi: 10.26355/eurrev_201802_14284
175. Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. (2017) 8:e3092. doi: 10.1038/cddis.2017.484
176. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. (2018) 75:467–84. doi: 10.1007/s00018-017-2626-6
177. Wang D, Dai J, Hou S, Qian Y. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci Rep. (2019) 39:BSR20182186. doi: 10.1042/BSR20182186
178. Wang B, Li X, Hu W, Zhou Y, Din Y. Silencing of lncRNA SNHG20 delays the progression of nonalcoholic fatty liver disease to hepatocellular carcinoma via regulating liver Kupffer cells polarization. IUBMB Life. (2019) 71:1952–61. doi: 10.1002/iub.2137
179. Li Y, Xu J, Guo YN, Yang BB. LncRNA SNHG20 promotes the development of laryngeal squamous cell carcinoma by regulating miR-140. Eur Rev Med Pharmacol Sci. (2019) 23:3401–9. doi: 10.26355/eurrev_201904_17704
180. Zhang C, Jiang F, Su C, Xie P, Xu L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem. (2019) 120. doi: 10.1002/jcb.28444
181. Sun C, Sun Y, Zhang E. Long non-coding RNA SNHG20 promotes nasopharyngeal carcinoma cell migration and invasion by upregulating TGF-beta1. Exp Ther Med. (2018) 16:4967–74. doi: 10.3892/etm.2018.6849
182. Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H, et al. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. (2018) 503:1927–33. doi: 10.1016/j.bbrc.2018.07.137
183. Zhang J, Ju C, Zhang W, Xie L. LncRNA SNHG20 is associated with clinical progression and enhances cell migration and invasion in osteosarcoma. IUBMB Life. (2018) 70:1115–21. doi: 10.1002/iub.1922
184. Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu SZ, Zhang YL. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem. (2018) 119:7971–81. doi: 10.1002/jcb.26588
185. Gao P, Fan R, Ge T. SNHG20 serves as a predictor for prognosis and promotes cell growth in oral squamous cell carcinoma. Oncol Lett. (2019) 17:951–7. doi: 10.3892/ol.2018.9709
186. Wu J, Zhao W, Wang Z, Xiang X, Zhang S, Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. (2019) 23:680–8. doi: 10.1111/jcmm.13987
187. Liu J, Liu L, Wan JX, Song Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3beta/ beta-catenin signaling pathway. Oncotarget. (2017) 8:80700–8. doi: 10.18632/oncotarget.20959
188. Yu J, Shen J, Qiao X, Cao L, Yang Z, Ye H, et al. SNHG20/miR-140-5p/NDRG3 axis contributes to 5-fluorouracil resistance in gastric cancer. Oncol Lett. (2019) 18:1337–43. doi: 10.3892/ol.2019.10439
189. Cui N, Liu J, Xia H, Xu D. LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer. J Cell Biochem. (2019) 120:3114–23. doi: 10.1002/jcb.27539
190. Zhao Q, Gao S, Du Q, Liu Y. Long non-coding RNA SNHG20 promotes bladder cancer via activating the Wnt/beta-catenin signalling pathway. Int J Mol Med. (2018) 42:2839–48. doi: 10.3892/ijmm.2018.3819
191. Liu J, Cheng LG, Li HG. LncRNA SNHG20 promoted the proliferation of glioma cells via sponging miR-4486 to regulate the MDM2-p53 pathway. Eur Rev Med Pharmacol Sci. (2019) 23:5323–31. doi: 10.26355/eurrev_201906_18199
192. Li XS, Shen FZ, Huang LY, Hui L, Liu RH, Ma YJ, et al. lncRNA small nucleolar RNA host gene 20 predicts poor prognosis in glioma and promotes cell proliferation by silencing P21. Onco Targets Ther. (2019) 12:805–14. doi: 10.2147/OTT.S192641
193. Li X, Xue Y, Liu X, Zheng J, Shen S, Yang C, et al. ZRANB2/SNHG20/FOXK1 Axis regulates Vasculogenic mimicry formation in glioma. J Exp Clin Cancer Res. (2019) 38:68. doi: 10.1186/s13046-019-1073-7
194. He S, Zhao Y, Wang X, Deng Y, Wan Z, Yao S, et al. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/beta-catenin signaling. Biosci Rep. (2018) 38:BSR20170681. doi: 10.1042/BSR20170681
195. Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. (2017) 89:857–63. doi: 10.1016/j.biopha.2017.01.011
196. Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. (2016) 16:655. doi: 10.1186/s12885-016-2719-x
197. Gartel AL. Is p21 an oncogene? Mol Cancer Ther. (2006) 5:1385–6. doi: 10.1158/1535-7163.MCT-06-0163
198. Liu Z, Tao H. Small nucleolar RNA host gene 3 facilitates cell proliferation and migration in oral squamous cell carcinoma via targeting nuclear transcription factor Y subunit gamma. J Cell Biochem. (2020) 121:2150–8. doi: 10.1002/jcb.29421
199. Zhang Y, Zhang R, Luo G, Ai K. Long noncoding RNA SNHG1 promotes cell proliferation through PI3K/AKT signaling pathway in pancreatic ductal adenocarcinoma. J Cancer. (2018) 9:2713–22. doi: 10.7150/jca.26207
200. Li W, Dong X, He C, Tan G, Li Z, Zhai B, et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. (2019) 38:183. doi: 10.1186/s13046-019-1177-0
201. Zhang R, Liu Y, Liu H, Chen W, Fan H-N, Zhang J, et al. The long non-coding RNA SNHG12 promotes gastric cancer by activating the phosphatidylinositol 3-kinase/AKT pathway. Aging. (2019) 11:10902–22. doi: 10.18632/aging.102493
202. Li Z, Li X, Du X, Zhang H, Wu Z, Ren K, et al. The interaction between lncRNA SNHG1 and miR-140 in regulating growth and tumorigenesis via the TLR4/NF-kappaB pathway in cholangiocarcinoma. Oncol Res. (2019) 27:663–72. doi: 10.3727/096504018X15420741307616
203. Gao L, Cao H, Cheng X. A positive feedback regulation between long noncoding RNA SNHG1 and YAP1 modulates growth and metastasis in laryngeal squamous cell carcinoma. Am J Cancer Res. (2018) 8:1712–24.
204. Zhang C, Ren X, He J, Wang W, Tu C, Li Z. The prognostic value of long noncoding RNA SNHG16 on clinical outcomes in human cancers: a systematic review and meta-analysis. Cancer Cell Int. (2019) 19:261. doi: 10.1186/s12935-019-0971-2
205. Dong B, Chen X, Zhang Y, Zhu C, Dong Q. The prognostic value of lncRNA SNHG1 in cancer patients: a meta-analysis. BMC Cancer. (2019) 19:780. doi: 10.1186/s12885-019-5987-4
206. Yu J, Yan Y, Hua C, Ming L. Upregulation of lncRNA SNHG1 is associated with metastasis and poor prognosis in cancers: a meta-analysis. Medicine. (2019) 98:e15196. doi: 10.1097/MD.0000000000015196
207. Yu Y, Yang J, Yang S, Li Q, Zhang M, Wang L, et al. Expression level and clinical significance of SNHG1 in human cancers: a meta-analysis. Onco Targets Ther. (2019) 12:3119–27. doi: 10.2147/OTT.S184803
208. Wu Y, Zhang Y, Zhu X, Liu H. The association of long non-coding RNA SNHG6 expression with clinicopathological features and prognosis in cancer. Clin Lab. (2019) 65. doi: 10.7754/Clin.Lab.2019.190346
209. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. (2018) 118:24–30. doi: 10.1016/j.ijbiomac.2018.06.033
210. Zhen S, Li X. Application of CRISPR/Cas9 for long non-coding RNA genes in cancer research. Hum Gene Ther. (2018) 30:3–9. doi: 10.1089/hum.2018.063
211. Filippova JA, Matveeva AM, Zhuravlev ES, Balakhonova EA, Prokhorova DV, Malanin SJ, et al. Are small nucleolar RNAs “CRISPRable”? A report on box C/D small nucleolar RNA editing in human cells. Front Pharmacol. (2019) 10:1246. doi: 10.3389/fphar.2019.01246
212. Yang H, Wang S, Kang YJ, Wang C, Xu Y, Zhang Y, et al. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol Rep. (2018) 40:261–71. doi: 10.3892/or.2018.6412
213. Cai G, Zhu Q, Yuan L, Lan Q. LncRNA SNHG6 acts as a prognostic factor to regulate cell proliferation in glioma through targeting p21. Biomed Pharmacother. (2018) 102:452–7. doi: 10.1016/j.biopha.2018.03.083
Keywords: lncRNAs, SNHG, snoRNA, cancer, malignant disease, proliferation, invasion
Citation: Zimta A-A, Tigu AB, Braicu C, Stefan C, Ionescu C and Berindan-Neagoe I (2020) An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 10:389. doi: 10.3389/fonc.2020.00389
Received: 02 December 2019; Accepted: 04 March 2020;
Published: 07 April 2020.
Edited by:
Ondrej Slaby, Brno University of Technology, CzechiaReviewed by:
Sarah D. Diermeier, University of Otago, New ZealandMingjun Bi, The University of Texas Health Science Center at San Antonio, United States
Copyright © 2020 Zimta, Tigu, Braicu, Stefan, Ionescu and Berindan-Neagoe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Calin Ionescu, calin.ionescu@umfcluj.ro; Ioana Berindan-Neagoe, ioananeagoe29@gmail.com
 Alina-Andreea Zimta
Alina-Andreea Zimta Adrian Bogdan Tigu
Adrian Bogdan Tigu Cornelia Braicu
Cornelia Braicu Cristina Stefan3
Cristina Stefan3 Ioana Berindan-Neagoe
Ioana Berindan-Neagoe