- 1Department of Animal Sciences, The Ohio State University, Columbus, OH, United States
- 2Department of Inspection of Animal Products, Ministry of Agriculture, Livestock and Food Supply (MAPA), Brasília, Brazil
The successional changes in the early intestinal microbiota occur concomitantly with the development, expansion, and education of the mucosal immune system. Although great attention of researchers has been focused on understanding the linkage between microbiota and immune functions, many essential details of the symbiotic relationship between the intestinal pioneer microbiota and the avian immune system remain to be discovered. This study was conducted to understand the impact of different early life intestinal colonizers on innate and adaptive immune processes in chicks and further identify immune-associated proteins expressed in the intestinal tissue. To accomplish it, we performed an in ovo application of two apathogenic Enterobacteriaceae isolates and lactic acid bacteria (L) to determine their influences on the intestinal proteome profile of broilers at the day of hatch (DOH) and at 10 days old. The results indicated that there were predicted biological functions of L-treated chicks associated with the activation and balanced function of the innate and adaptive immune systems. At the same time, the Enterobacteriaceae-exposed birds presented dysregulated immunological mechanisms or downregulated processes related to immune development. Those findings suggested that a proper immune function was dependent on specific gut microbiota exposure, in which the prenatal probiotic application may have favored the fitting programming of immune functions in chicks.
Introduction
In recent years, attention has focused on understanding the linkage between microbiota and immune functions. Such an intimate relationship creates mechanisms for mutual benefits to both microbes and the host (Chow et al., 2010). At homeostasis, this mutualistic partnership enables the maintenance of microbial tolerance in the intestinal ecosystem, and, in turn, the proper microbiota colonization contributes to the development, maturation, and function of the immune system (Bar-Shira et al., 2003; Kelly et al., 2007; Brisbin et al., 2008; Chung et al., 2012).
Nevertheless, the cooperative arrangements between gut microbiota and host mucosal immunity are constantly threatened during the early life of chickens. Evidence has suggested that undesirable microbial colonization during the prenatal period may negatively influence the function and expansion of the immune system in broilers (Bar-Shira et al., 2003; Rodrigues et al., 2020a). Previous work has also demonstrated that perturbation of the intestinal pioneer microbiota with antibiotic exposure affects early immune programming and has been shown to boost negative antibody response in hens after cessation of antibiotic treatment (Simon et al., 2016). Besides, our previous reports indicated that pre-hatch colonization by Enterobacteriaceae promoted immune dysregulation and chronic inflammation (Rodrigues et al., 2020a,b; Wilson et al., 2020). Another example of dysregulated immune response triggered by early disturbance of enteric microbiota in chicks was shown by Schokker et al. (2010). It was revealed that early colonization by Salmonella enterica serotype Enteritidis delayed the morphological processes of the jejunum, thereby interrupting the spatial-temporal development of the immune system in chicks. On account of this fact, intestinal dysbiosis during the early post-hatch phase in chicks may have short- and long-term consequences on immune responses. Against this background, many essential details of the symbiotic relationship between the intestinal pioneer microbiota and the avian immune system remain to be discovered, and an important outstanding question was whether a proper early immune response depends on host-specific gut microbiota.
Recently, the emergence of in ovo techniques made it possible to manipulate the intestinal bacteria colonization before chicks have even been hatched or exposed to farm environments (Pedroso et al., 2016; Roto et al., 2016; Teague et al., 2017). Our lab has previously used the in ovo technique as an experimental model to address how early intestinal colonization shapes the microbiome composition in hatching chicks (Wilson et al., 2020). The technique of delivering various nutrients, supplements, immunostimulants, vaccines, and drugs via the in ovo route is gaining wide attention among researchers for boosting the production performance and immunity and for safeguarding the health of poultry (Saeed et al., 2019; Alagawany et al., 2021; Hassan et al., 2021).
In order to better understand the impact of different early life intestinal colonizers on innate and adaptive immune processes in chicks and further identify immune-associated proteins expressed in the intestinal tissue, we performed an in ovo application of two apathogenic Enterobacteriaceae isolates and lactic acid bacteria to determine their influences on the intestinal proteome profile of broilers at the day of hatch (DOH) and 10 days of age. Proteomics pathway enrichment and Gene Ontology (GO) function annotation analyses were performed to reveal the effect of different pioneer intestinal colonizers on innate and adaptive immune processes in chicks.
Materials and Methods
Study Design
The trial was performed on Ross 708 fertile eggs obtained from a local hatchery and incubated under standard conditions at the poultry research farm of the Ohio Agricultural Research and Development Center. All eggs were in the same incubator prior to inoculation. Once eggs were confirmed fertile, at embryonic day 18, inoculations containing one of the following: 0.9% sterile saline (S), ~102 cells of Citrobacter (CF), Citrobacter 2 (C2), or a lactic acid bacteria probiotic (L) were administered via in ovo injection into the amnion. After inoculation, the eggs were allocated by treatments into separate benchtop hatchers (Hova-Bator model 1602N, Savannah, GA, USA). Each inoculation treatment was separated into three hatchers, which contained up to 30 eggs. The chicks hatched between 48 and 72 h post-inoculation. The hatchability ranged from 86 to 100% among the in ovo treatments (Wilson et al., 2020). The bacterial inoculum was selected from our previous trial (Bielke et al., 2003), and the homology of strains was confirmed by next-generation sequencing. The L isolate was composed of a mixed culture of Lactobacillus salivarius and Pediococcus ssp. The CF strain was composed of Citrobacter freundii, and the C2 was identified as Citrobacter spp. Bacterial inoculum was prepared as described by Wilson et al. (2020). All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Ohio State University.
Sample Collection
Once all chicks were hatched, 10 chicks were randomly chosen from among the hatchers within each of the treatments (n = 40) and were immediately euthanized via cervical dislocation. The intestine was aseptically removed, from the duodenum to the cloaca. Tissues were placed in individual 2 ml tubes and flash-frozen in liquid nitrogen. Immediately post-hatch, the remaining 128 chicks were placed into treatment-separate brooder battery cages and had ad libitum access to a standard corn-soy diet and water (Nutrient Requirements of Nutrient Requirements of Poultry., 1994). At 10 days post-hatch, 12 chicks per treatment were randomly selected for ileal proteome analysis. The tissue of the region above the ileocecal junction, designated as lower ileum, was aseptically collected (n = 45). Since there were three mortalities in the CF, only nine birds were sampled for this treatment. Ileum tissue was also frozen and stored at −80°C until further use.
Once intestinal samples were thawed, a cumulative total of 0.1 g was individually placed into 5 ml of buffer (8 M urea/2 M thiourea, 2 mM DTT, 50 mM Tris, 5% SDS, pH 6.8). The extraction protocol is a modified version described previously by Iqbal et al. (2004) and Kong et al. (2016). In brief, samples were homogenized for 5 s (PRO250 Homogenizer; Pro Scientific, Oxford, CT, USA), and then, 500 μl of the solution was placed in 2 ml tubes containing 0.1 g stainless steel beads (SSB14B; Next Advance, Averill Park, NY, USA). Samples were homogenized for 3 min total in 30-s intervals (MiniBeadbeater-16, Model 607; BioSpec Products, Bartlesville, OK, USA) and centrifuged at 4°C at 14,000 g for 20 min. The supernatant was collected, aliquoted, and placed at −80°C until further use.
To ensure proper extraction, the concentration of total protein was quantified with the Bradford assay (Bradford reagent; VWR, Suwanee, GA, USA) and a standard bovine serum albumin curve (VWR, Suwanee, GA, USA) on a Synergy HTX multi-mode plate reader (BioTek Instruments, Winooski, VT, USA). Samples were mixed to create pooled samples from two birds per treatment/time and were sent to the Ohio State University Proteomics Core lab for performing in solution digestion and mass spectrometry via established methods.
Proteomics Analyses
Samples were precipitated with 25% (w/v) trichloroacetic acid (TCA) and then resuspended in 50 mM ammonium bicarbonate. A total of 5 ml of DTT (5 μg/μl in 50 mM ammonium bicarbonate) was added, and the samples were incubated at 56°C for 15 min. After incubation, 5 μl of iodoacetamide (15 mg/ml in 50 mM ammonium bicarbonate) was added, and the samples were kept in the dark at room temperature for 30 min. Sequencing grade-modified trypsin (Promega, Madison, WI, USA) prepared in 50 mM ammonium bicarbonate was added to each sample with an estimation of 1:20/1:100 enzyme-substrate ratio set at 37°C overnight. The reaction was quenched the following day by adding acetic acid for acidification. Once samples were quenched, the peptide concentration was measured by Nanodrop (Thermo Scientific Nanodrop 2000; Thermo Scientific, Waltham, MA, USA).
Capillary-liquid chromatography-nanospray tandem mass spectrometry (Capillary-LC/MS/MS) of global protein identification was performed on a Thermo Fisher Fusion mass spectrometer (Thermo Scientific, Waltham, MA, USA). Samples were separated on a Thermo Nano C18 column (UltiMate™ 3000 HPLC system; Thermo Scientific, Waltham, MA, USA). The MS/MS data sequences were scanned, and the scan sequence was based on the preview mode data-dependent TopSpeed™ method with CID and ETD as fragmentation methods. The raw data were searched on Sequest via Proteome Discoverer (Proteome Discoverer™ software; Thermo Scientific, Waltham, MA, USA). The data were searched against the most recent Uniprot Gallus gallus database for the identification of proteins. Only proteins with < 0.05 false discovery rate (FDR) were reported. Proteins with a Mascot score of 50 or higher with a minimum of two unique peptides from one protein having a -b or -y ion sequence tag of five residues or better were accepted. Any modifications or low score peptide/protein identifications were manually checked for validation.
BioInformatics and Statistical Analysis
Label-free quantitation was performed using the spectral count approach, in which the relative protein quantitation is measured by comparing the number of MS/MS spectra identified from the same protein in each of the multiple LC/MSMS datasets. Comparisons between in ovo bacterial treatments and S control group were performed in Scaffold (Scaffold 4.8.4; Proteome Software, Portland, OR, USA). The student's t-test (P < 0.05) was performed to identify significance across the fold-change values. Differentially expressed proteins (DEPs) were assigned at level P ≤ 0.1.
From the significant proteins were performed GO annotation enrichment terms analysis using the STRING v10.0 database (https://string-db.org). Pathway enrichment was assessed using upregulated DEPs based on Reactome databases. Then to evaluate the role of overexpressed proteins in the immune system, we searched for immune pathways based on biological processes using Reactome pathways (https://reactome.org/). P < 0.05 was set as the threshold value, and FDR was corrected using the Benjamini-Hochberg method. Immune-related DEPs were identified and their predicted functional partners were searched (STRING v10.0 database). Then, functional clusters in the protein-protein interaction (PPI) networks were determined. Active interaction sources, including experiments, databases, co-occurrence, and co-expression, as well as species limited to “Gallus gallus,” and an interaction score > 0.4 were applied to construct the PPI networks.
Results
Identification of DEPs
A total of 78, 107, and 39 proteins were identified as DEPs in the ileum of L, CF, and C2 treatments, based on the set threshold (P ≤ 0.1) at DOH. By 10 days of age, a total of 61, 44, and 63 proteins showed differential expression in L, CF, and C2, respectively, compared with the in ovo S control.
Functional Enrichment Analysis
To investigate whether different early colonizers in the gastrointestinal tract (GIT) could affect biological processes linked to the immune system, GO enrichment analyses were performed for overexpressed and underexpressed proteins in each dataset. The top 20 enriched GO terms, relative to the control, were exhibited for each in ovo treatment at two time points. Proteins associated with response to stimulus and immune annotation roles were displayed in Figure 1.
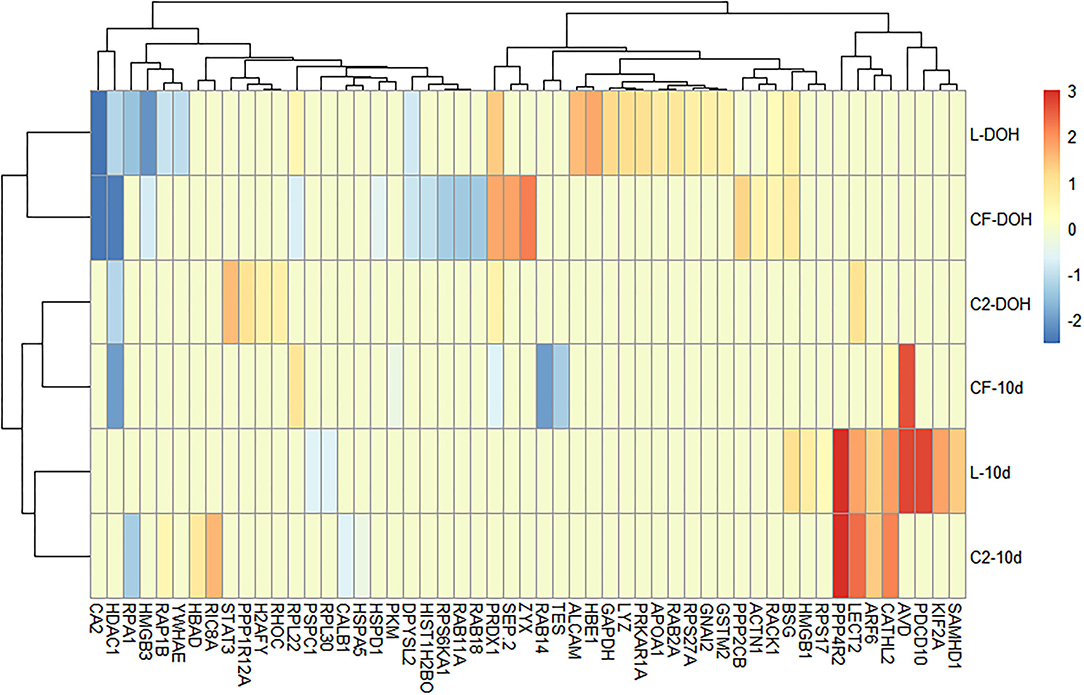
Figure 1. Differentially expressed proteins (DEPs) related to immune processes based on Gene Ontology analysis. Heatmap plot represents the log2-fold changes of proteins expressed intestine samples of broilers treated with lactic acid bacteria (L), Citrobacter freundii (CF) or Citrobacter spp. (C2) in relation to control treatment at the day of the hatch (DOH) and 10 days of age (10d). Blue shades represent downregulation, while red shades indicate upregulation of the particular protein. Light yellow cells indicate no expression.
The upregulated DEPs in L revealed that the major biological processes at DOH were related to protein and cellular metabolic process, cellular organization, and response to a stimulus (Blue bars in Figure 2A). On the 10th day, cellular differentiation and assembly, regulation of stress response, and immune system process were enriched. In terms of downregulated DEPs in the L dataset, the most enriched processes were biological regulation, cell communication, and response to stress at DOH. While by 10 days of age, proteolysis, cellular metabolic process, and immune system process were overrepresented (Red bars in Figure 2B).
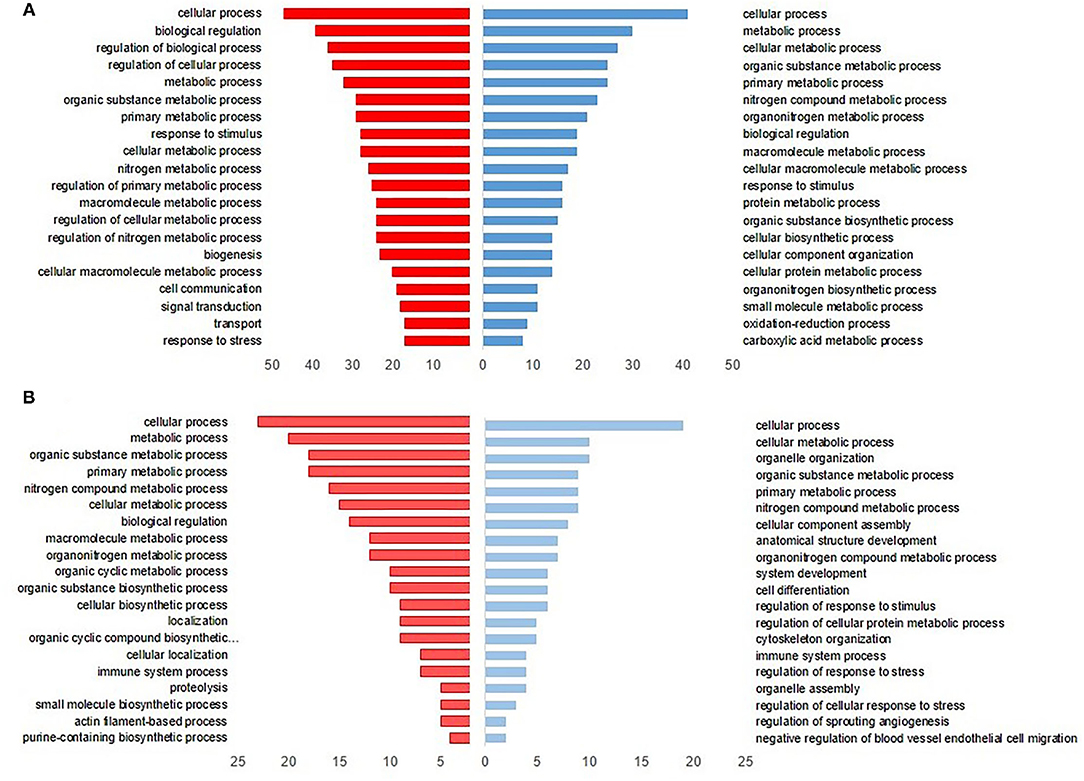
Figure 2. Gene Ontology (GO) enrichment analysis of intestinal differentially expressed proteins from broilers exposed to lactic acid bacteria (L) in ovo (A) at the day of hatch and (B) by 10 days of age. The blue bars to the right indicate the top 20 GO-enriched terms by upregulated proteins. The red bars to the left represent the top 20 GO terms enriched by downregulated proteins, while the axis at the bottom is the number of proteins in each biological process.
The upregulated DEGs in CF were involved in different GO terms, including cellular and metabolic processes, and protein folding at DOH (Figure 3A), whereas terms associated with protein stabilization and regulation of cellular were evident by 10 days of age (Figure 3B). Conversely, the downregulated DEPs in CF were related to transport, localization, and cellular organization component at DOH (Figure 3A). By 10 days of age, the GO terms were allied to the catabolic process and response to a stimulus (Figure 3B).
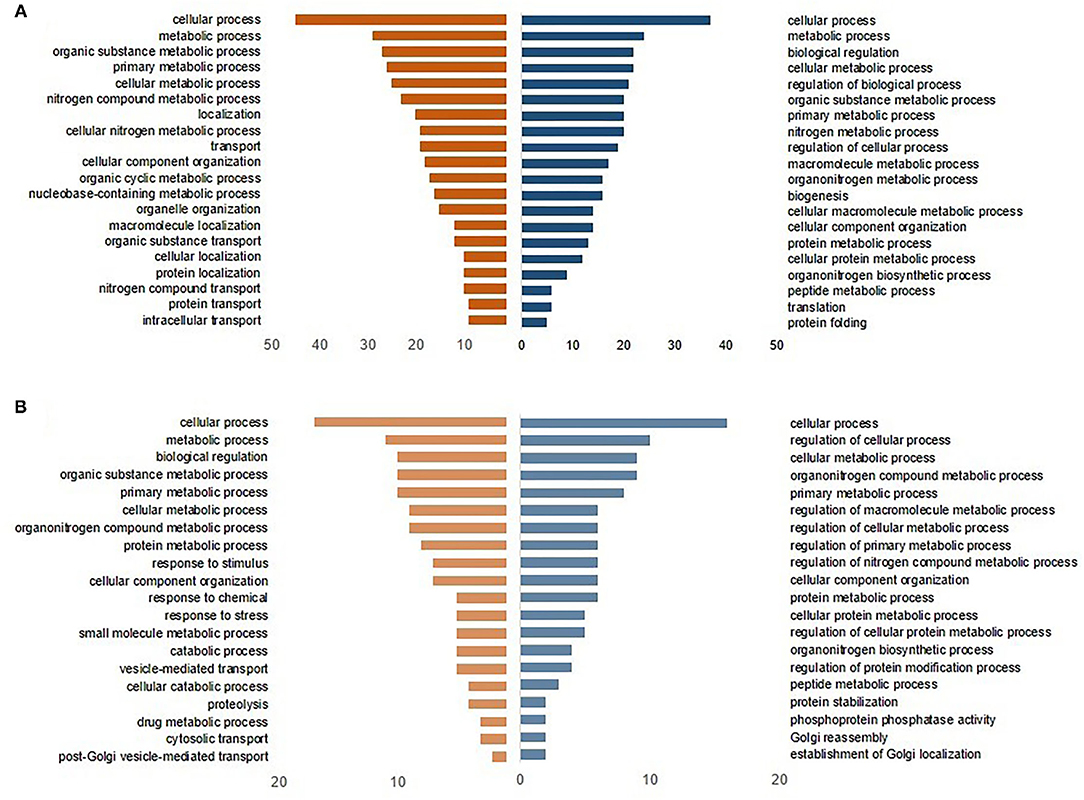
Figure 3. Gene Ontology (GO)-enriched terms of intestinal differentially expressed proteins from broilers exposed to Citrobacter freundii (CF) in ovo (A) at the day of hatch and (B) by 10 days of age. The blue bars to the right indicate the most GO-enriched terms by upregulated proteins. The orange bars to the left represent GO terms enriched by downregulated proteins, while the axis at the bottom is the number of proteins in each biological process.
The most significant GO annotation based on the overexpressed proteins in C2 was associated with a cellular process, biological regulation, and organelle organization at DOH (Figure 4A), whereas it was associated with cellular metabolic process, primary metabolic process, and response to a stimulus at 10 days of age (Figure 4B). The underrepresented proteins enhanced only a few terms at DOH, including cellular process and transport. Biological functions enriched by 10 days of age were related to protein metabolic, homeostatic, and cellular protein metabolic processes (Figure 4B).
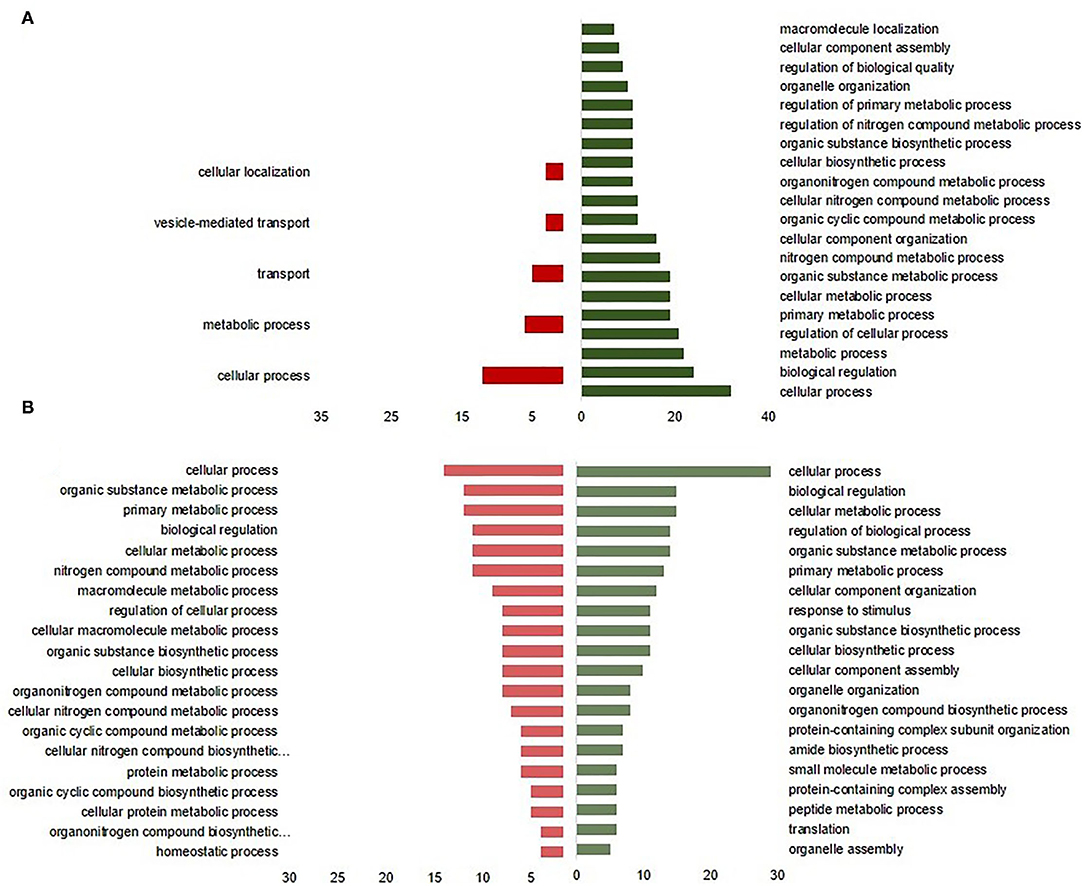
Figure 4. Enrichment of biological process of intestinal differentially expressed proteins from broilers exposed to Citrobacter spp. (C2) in ovo (A) at the day of hatch and (B) by 10 days of age. The green bars to the right indicate the most GO-enriched terms by upregulated proteins. The red bars to the right represent GO terms enriched by downregulated proteins, while the axis at the bottom is the number of proteins attributed to each GO term.
Pathways With DEPs
The five most significant pathways in L at DOH included amyloid fiber formation, glutathione conjugation, cellular responses to stress, HSF1 activation, and cellular responses to external stimuli (Table 1). The enriched pathways by 10 days of age were muscle contraction, smooth muscle contraction, kinesins, axon guidance, and COPI-dependent Golgi-to-ER retrograde traffic.
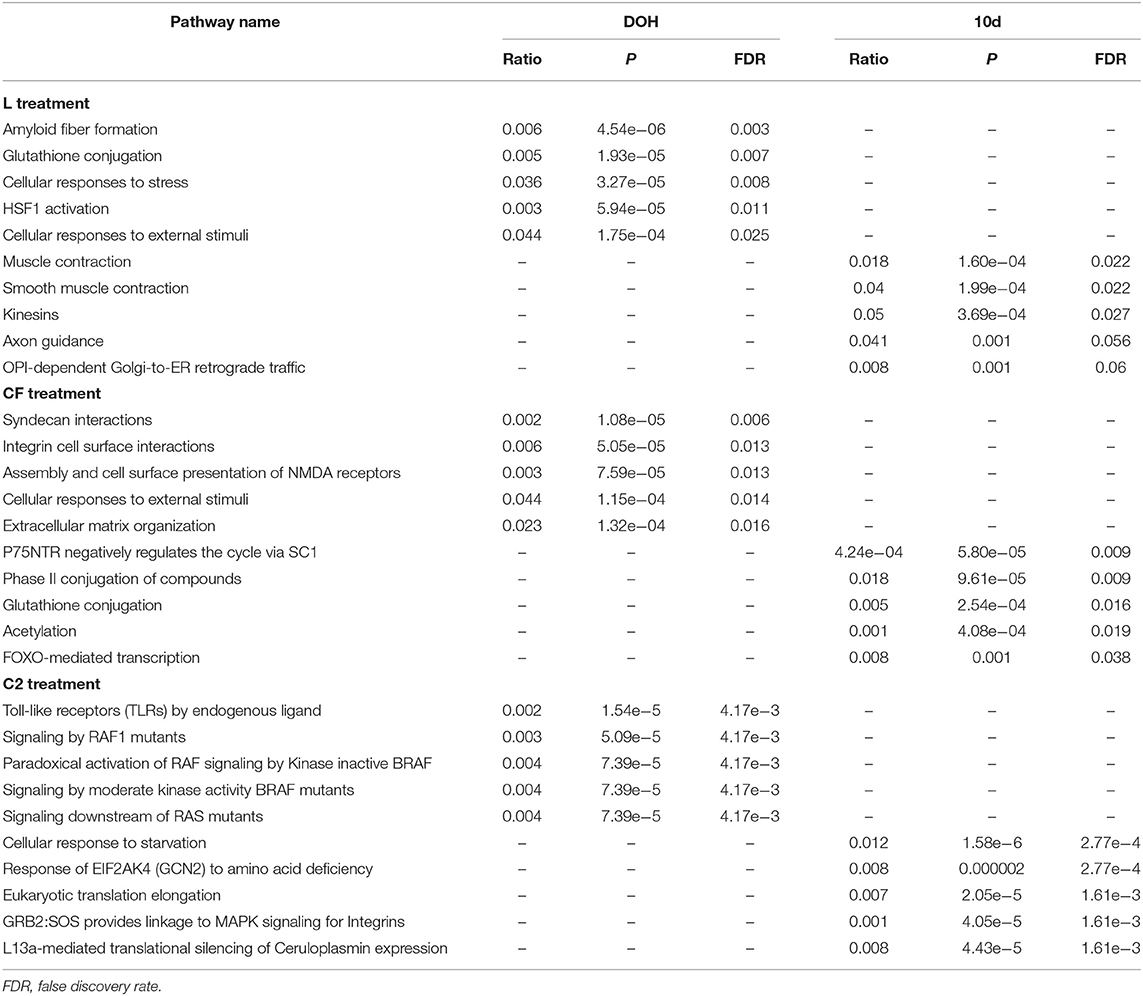
Table 1. The five most significant pathways based on the upregulated proteins expressed in intestinal samples of broilers treated with lactic acid bacteria (L), Citrobacter freundii (CF) or Citrobacter spp. (C2) in relation to control treatment at the day of the hatch (DOH) and 10 days of age (10d).
The most relevant predicted pathways in CF were syndecan interactions, integrin cell surface interactions, assembly and cell surface presentation of NMDA receptors, cellular responses to external stimuli, and extracellular matrix organization at DOH (Table 1). By 10 days of age, pathways associated with P75NTR negatively regulate the cycle via SC1, and phase II conjugation of compounds, glutathione conjugation, acetylation, and FOXO-mediated transcription were enriched.
The inoculation of C2 affected pathways related to the regulation of Toll-like receptors (TLRs) by endogenous ligand, signaling by RAF1 mutants, paradoxical activation of RAF signaling by kinase inactive BRAF, signaling by moderate kinase activity BRAF mutants, and signaling downstream of RAS mutants at DOH; whereas cellular response to starvation, response of EIF2AK4 (GCN2) to amino acid deficiency, eukaryotic translation elongation, GRB2:SOS linkage to MAPK signaling for integrins, and L13a-mediated translational silencing of ceruloplasmin expression was activated at 10 days of age (Table 1).
The presence of several immune genes prompted us to investigate whether the early exposure to different bacterial isolates could affect the immune biological processes in the GIT of broilers. Then, we performed the broad immune response pathways in Reactome for up- and downregulated proteins. Analyses showed substantially more upregulated DEPs in L treatment that accomplishes functions on immune signaling than the other in ovo treatment by DOH (Figure 5). Accordingly, an enrichment of heterophil degranulation (P = 0.0005; FDR = 0.034) and antimicrobial peptides pathways (P = 0.022; FDR = 0.129) at DOH was observed (Figure 5). By 10 days of age, MHC class II antigen presentation was overexpressed in L treatment (P = 0.003; FDR = 0.076). Using downregulated DEPs, Reactome analyses showed that Rap1 signaling was enriched in L at DOH (P = 0.004; FDR = 0.053; Figure 5), while the pathways heterophil degranulation (P = 0.018; FDR= 0.223) and Butyrophilin (BTN) family interactions (P = 0.037; FDR= 0.223) were significant at 10 days of age.
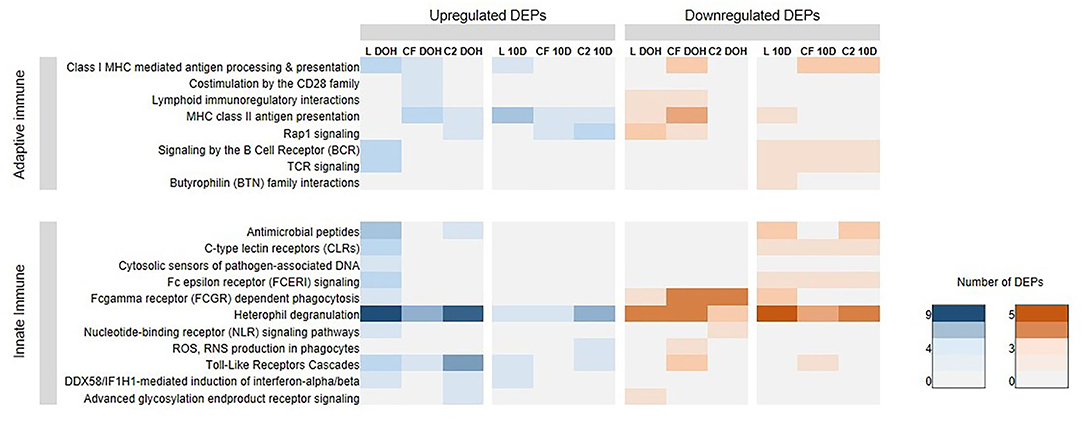
Figure 5. Reactome Pathways analyses, at the day of hatch (DOH) and 10 days of age (10d), related to immune response based on differentially expressed proteins (DEPs) in intestinal samples of broilers treated with lactic acid bacteria (L), Citrobacter freundii (CF) or Citrobacter spp. (C2).
While there was not any pathway enhanced with upregulated DEPs at DOH in CF, the pathway Fcgamma receptor (FCGR)-dependent phagocytosis (P = 0.025; FDR = 0.100) was overrepresented in downregulted DEPs. Regarding C2 treatment, within upregulated DEPs, there were enriched pathways related to heterophil degranulation (P = 0.001; FDR = 0.022) and TLRs cascades (P = 0.001; FDR = 0.023) at DOH. By 10 days of age, Rap1 pathway (P = 0.003; FDR = 0.019) was significant. Within the underexpressed DEPs profile, the FCGR-dependent phagocytosis was enriched at DOH (P = 0.0002; FDR = 0.007). By 10 days of age, antimicrobial peptides (P = 0.031; FDR = 0.130) and heterophil degranulation (P = 0.022; FDR= 0.130) were enhanced.
Identification of Immune-Related DEPs
Proteins related to immunity among the treatments were ranked based on a functional comparison involving the GO terms. The key immune-related DEPs included leukocyte cell-derived chemotaxin-2 (LECT2), avidin (AVD), high mobility group protein B1 (HMGB1), activated leukocyte cell adhesion molecule (ALCAM), cathelicidin-2 (CAMP), and lysozyme C (LYZ). The DEPs associated with immune function were obtained from the following categories: immune system process, immune response, response to stress, cellular response to stimulus, and related functions (Supplementary Data).
Then, a PPI network for each key protein was screened for a better understanding of their roles and underlying mechanisms involving immune processes. Figure 6 shows the interactome networks for LECT2, AVD, HMGB1, ALCAM, CAMP, and LYZ, in which the nodes represent proteins, while the edges are physical, biochemical, or functional interactions between them.
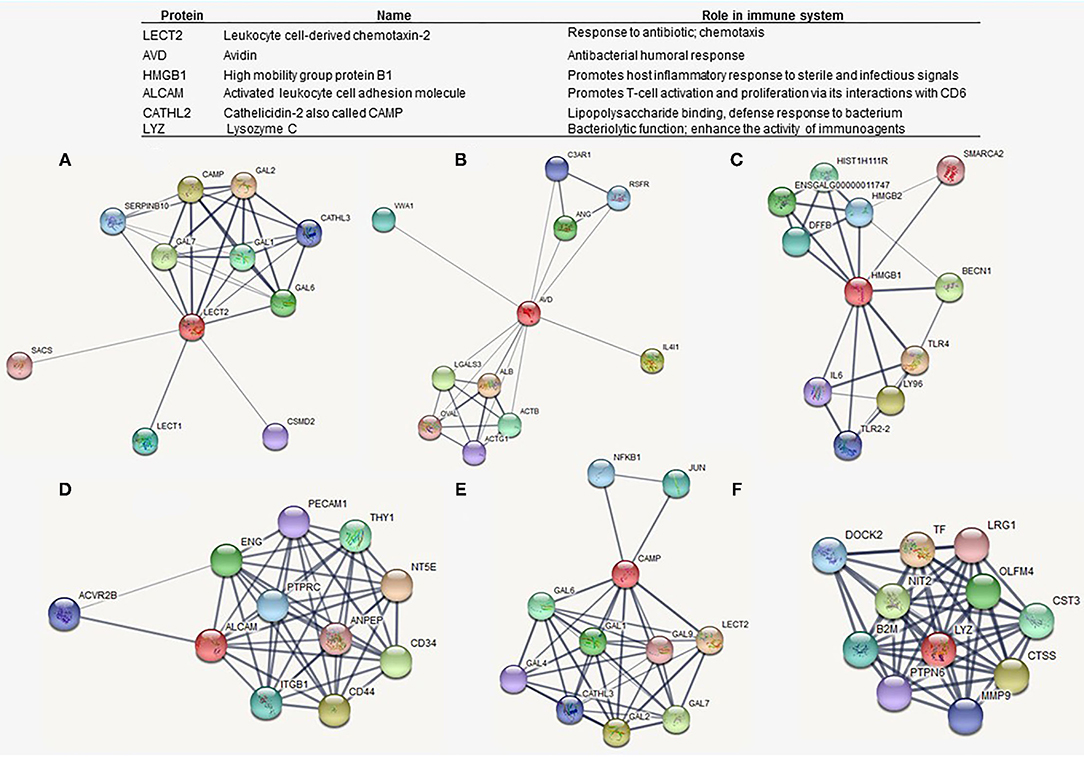
Figure 6. Protein-protein interaction (PPI) network analyses of selected differentially expressed proteins associated with immunomodulatory functions. Red nodes denote the selected immune-related protein, and edges show the biological relationship between nodes. (A) Leukocyte cell-derived chemotaxin-2 (LECT2). (B) Avidin (AVD). (C) High mobility group protein B1 (HMGB1). (D) Activated leukocyte cell adhesion molecule (ALCAM). (E) Cathelicidin-2 also (CAMP). (F) Lysozyme C (LYZ).
Discussion
The presence of distinct intestinal proteome profiles in chicks inoculated in ovo with Enterobacteriaceae isolates or L-probiotic poses the question of whether the expression of intestinal immune-related proteins is dependent on specific early microbiota exposure. Here, we conducted different proteomic approaches as pathway enrichment and GO function annotation analyses to understand the impact of different early life intestinal colonizers on innate and adaptive immune processes in chicks. It was also the focus of this research to identify candidate protein markers for monitoring immune biological functions in broilers. The results of this study provide a significant comprehension of how early exposure to beneficial bacteria may affect immune programming in broilers.
Our previous work also addressed the in ovo technique as an experimental model to study how early intestinal colonization can shape the development and persistence of microbiome in chicks (Rodrigues et al., 2020a; Wilson et al., 2020). In those studies, it was shown that different bacterial isolates provided in ovo resulted in distinct microbiome profiles on DOH and by 10 days of age. Notably, inoculation of L resulted in increased Lactobacilli populations at DOH, which may have influenced the establishment of butyrate-producing bacteria and segmented filamentous bacterium (SFB) in young broilers. The poultry GIT microbiota undergoes a period of heavy changes during the first days of life. Some reports have shown that the microbiota composition of post-hatched chicks is primarily Proteobacteria derived from opportunistic environmental communities (Ballou et al., 2016; Donaldson et al., 2017; Rodrigues et al., 2020a; Wilson et al., 2020). Concomitant with this process occurs the development, expansion, and education of the mucosal immune system (Chow et al., 2010; Gensollen et al., 2016; Zheng et al., 2020). The adaptive immune functions of newly hatched chicks develop only toward the end of the first week post-hatch (Barshira and Friedman, 2006). Therefore, maternal antibodies and the innate immune system are the main apparatus for dealing with any early pathogenic assault.
From that perspective, early exposures are significant determinants for programming innate immune functions. We found that L treatment enriched the key pathways of the innate immune signaling as heterophil degranulation and antimicrobial peptides at DOH (Figure 5). One of the mechanisms displayed by the innate immune system of the host in reaction to microbial stimulation or into the pathogen-containing phagosome is heterophil degranulation, which releases granule substances including antimicrobial peptides into the external environment (Kogut et al., 2001; Genovese et al., 2013). In agreement with our study, Farnell and Donoghue (2006) reported that oral administration of probiotics could stimulate heterophil oxidative bursting and degranulation in the poultry intestine. Likewise, the upregulated DEPs were predominantly associated with cellular response to stress and external stimuli pathways. Part of cellular responses to external stimuli is carried out by macroautophagy that is considered a cytoprotective host defense mechanism against damaged organelles, cytosolic proteins, and invasive microbes (Feng et al., 2014; Delorme-Axford and Klionsky, 2018). Macroautophagy assists both innate and adaptive immunity releasing lysosomes for the degradation of foreign substances, including pathogenic proteins within cells (Gannagé and Münz, 2009; Bel and Hooper, 2018). Following this process, the products of lysosomal degradation are presented for MHC class II molecules and recognized by CD4+ T cells (Schmid et al., 2007). Correspondingly, there was an enrichment of the MHC class II antigen presentation pathway in L by 10 days of age.
In contrast, these results revealed that the introduction of CF in ovo downregulated the MHC class II antigen presentation pathway. In addition, the downregulated DEPs in CF treatment were allied to transport, localization, and response to the stimulus at an early and later age, indicating that the functions associated with the trafficking of immune cells, detection, and response to a biotic and abiotic stimulus may have been impaired. Nedjic et al. (2008) have shown that a change in the MHC class II system by a genetic interference of autophagy in the thymus of mice resulted in severe colitis and multi-organ inflammation. Comparably, our recent research has indicated that pre-hatch Enterobacteriaceae colonization perturbed the initial microbial establishment and promoted intestinal proteomic changes accompanied by inflammation in chicks (Wilson et al., 2020). The failure of immune regulation suggested by these results might be connected with the onset of intestinal chronic inflammation signaling in CF treated broilers at 10 days of age (Rodrigues et al., 2020a,b).
Interestingly, we found that C2 overrepresented the regulation of TLRs by endogenous ligand and TLRs receptors cascade pathways at DOH. As feedback mechanisms during either infection or injury to the organism, pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) indicate a danger alert that activates TLRs (Piccinini and Midwood, 2010). Additionally, there was evidence of other innate mechanism activation by the enrichment of the heterophil degranulation pathway in C2 on DOH (Figure 5). Contrarily, the inoculation of C2 in ovo has been shown to downregulate the pathway FCGR-dependent phagocytosis at the same age. Phagocytosis is a significant event involving the recognition of invading foreign particles by specific types of phagocytic receptors and the subsequent internalization of the particles (Alberts et al., 2002; Henneke and Golenbock, 2004; Feng et al., 2014). These early events that are mediated by the innate immune system are critical to eliminating the invading infectious agents (Henneke and Golenbock, 2004). The clustering of FCGRs by IgG antibodies on the phagocyte initiates a variety of signals, which lead, through the reorganization of the actin cytoskeleton and membrane remodeling, to the formation of pseudopods and phagosomes (Joshi et al., 2006). As a result of this process, pathogen-derived molecules can be presented on the surface of the host cell, allowing the induction of pathogen-specific adaptive immunity (Alberts et al., 2002; Kumar et al., 2013). Collectively, the underexpression of DEPs on GO terms related to immune defense and the downregulation of pathways related to phagocytosis suggested that C2-treated chicks may have been able to activate the recognition of potentially harmful microorganisms, despite that their defense mechanisms may have failed in executing bacterial clearance. Although we found links between proteomic signatures and the immune system, this study did not assess the link with the host phenotypes. Future research is warranted to evaluate whether immune response-associated proteomic signature can affect body weight and gut health parameters.
We took our results one step further to identify DEPs directly relevant to biological functions associated with early-age immune response in poultry. The mass spec-based proteomics outcomes have been extensively used to discover potential biomarkers to predict or confirm health disorders in human medicine (Geyer et al., 2017). Despite the lack of information, the expansion of modern technologies is beginning to be approached for this purpose in poultry production (Arsenault et al., 2014; Kuttappan et al., 2017). Here, in this study, we suggest a panel of proteins for evaluating mucosal immune response in broilers (Figure 6).
Among the highlighted proteins, LECT2 plays an important role in the immune processes by increasing cytokine expression, inducing chemotaxis, and activating macrophages (Liu et al., 2014; Slowik and Apte, 2017; Jung et al., 2018). Previous work revealed a decreased expression of LECT2 on the proteome of heterophils from the spleen of the chicken in response to Salmonella Enteritidis infection. Birds vaccinated against Salmonella sp. succeeding a Salmonella challenge upregulated granular proteins as CATHL2, also called CAMP, and LECT2, suggesting that these peptides might be considered a positive marker of enhanced immune response to vaccination (Sekelova et al., 2017). Figure 6 demonstrates a co-expression, with a high score (0.976; see Supplementary Data), between LECT2 and CATHL2. Also, LECT2 is strongly related to the GAL1 and other β-defensins, which are a family of peptides with antimicrobial activity and immunomodulatory functions in chickens (Kalenik et al., 2018). In fact, CATHL2 was identified as upregulated in this study along with all treatments, while LECT2 was differentially expressed by L and C2 treatments.
Otherwise, ALCAM and HMGB1 were only upregulated by L treatment, which might have contributed to explaining the activation of immunostimulatory complexes by DOH and 10 days of age, respectively. HMGB1 is actively secreted by innate immune cells in response to PAMPs, where it mediates the activation of innate immune responses, including cytokine release (Yanai et al., 2011; Yang et al., 2013). HMGB1 can also act as a chemotactic mediator by transmitting signals to the cell interior via the activation of receptors that include TLR4 (Yang et al., 2013). In relevance of HMGB1 role in cellular and humoral immunity, this protein has been reported as a potential immunological adjuvant in poultry vaccines (Sawant et al., 2015; Yang et al., 2017; Vuong et al., 2020).
Avidin and LYZ are other proteins recently associated with innate antimicrobial activity by preventing direct access of bacteria to the intestinal epithelial surface in newly hatched chicks (Shira and Friedman, 2018). LYZ was found upregulated in L treatment at DOH. Given the potent function of LYZ in limiting the bacterial growth at mucosal surfaces, we speculate that early exposure to L might be a strategy to enhance the innate antimicrobial activity. Finally, further studies are recommended to validate this panel of proteins as potential biological markers of the enhanced immune response in broilers.
Our study indicated that there were predicted biological functions of L-treated chicks associated with the activation and balanced function of the innate and adaptive immune systems. At the same time, the Enterobacteriaceae exposed birds presented dysregulated immunological mechanisms or downregulated processes related to immune development. It is expected that the activated response at an early age may manifest the ability of the immune system to recognize, uptake, and destroy foreign microorganisms. Those findings highlighted that a proper immune function was dependent on specific GIT microbiota exposure, in which the prenatal probiotic application may have favored the fitting programming of immune functions in chicks.
Data Availability Statement
The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015504.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
Author Contributions
KW and DR carried out the project. DR performed the analyses, interpreted the results, and wrote the manuscript in consultation with LB. All authors contributed to the experimental design, discussed the results, and commented on the manuscript.
Funding
This research was supported by the OARDC Research Enhancement Competitive Grants Program (SEEDS) (Grant No. 2016035) and Arkansas Biosciences Institute (ABI: Little Rock, AR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Whitey Briggs, Kaylin Chasser, and Audrey Duff for their assistance. The authors would also like to thank the Proteomics Lab, Dr. Wilbur Ouma, and OARDC Poultry Research Farm for providing eggs and participating in the animal husbandry portion of the study. Lastly, the authors would like to thank the OARDC Research Enhancement Committee Grants Program (SEEDS) and Arkansas Biosciences Institute for providing the funding for this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.758183/full#supplementary-material
References
Alagawany, M., Elnesr, S. S., Farag, M. R., Abd El-Hack, M. E., Barkat, R. A., Gabr, A. A., et al. (2021). Potential role of important nutraceuticals in poultry performance and health—a comprehensive review. Res. Vet. Sci. 137, 9–29. doi: 10.1016/j.rvsc.2021.04.009
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). The Adaptive Immune System, 4th Edn. Mol. Biol. Cell. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK21070/ (accessed April 26, 2019).
Arsenault, R. J., Trost, B., and Kogut, M. H. (2014). A comparison of the chicken and turkey proteomes and phosphoproteomes in the development of poultry-specific immuno-metabolism kinome peptide arrays. Front. Vet. Sci. 1:22. doi: 10.3389/fvets.2014.00022
Ballou, A. L., Ali, R. A., Mendoza, M. A., Ellis, J. C., Hassan, H. M., Croom, W. J., et al. (2016). Development of the chick microbiome: how early exposure influences future microbial diversity. Front. Vet. Sci. 3:2. doi: 10.3389/fvets.2016.00002
Barshira, E., and Friedman, A. (2006). Development and adaptations of innate immunity in the gastrointestinal tract of the newly hatched chick. Dev. Comp. Immunol. 30, 930–941. doi: 10.1016/j.dci.2005.12.002
Bar-Shira, E., Sklan, D., and Friedman, A. (2003). Establishment of immune competence in the avian GALT during the immediate post-hatch period. Dev. Comp. Immunol. 27, 147–157. doi: 10.1016/S0145-305X(02)00076-9
Bel, S., and Hooper, L. V. (2018). Secretory autophagy of lysozyme in Paneth cells. Autophagy 14, 719–721. doi: 10.1080/15548627.2018.1430462
Bielke, L. R., Elwood, A. L., Donoghue, D. J., Donoghue, A. M., Newberry, L. A., and Neighbor, N. K. (2003). Approach for selection of individual enteric bacteria for competitive exclusion in turkey poults. Poult. Sci. 82, 1378–1382. doi: 10.1093/ps/82.9.1378
Brisbin, J. T., Gong, J., and Sharif, S. (2008). Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim. Health Res. Rev. 9, 101–110. doi: 10.1017/S146625230800145X
Chow, J., Lee, S. M., Shen, Y., Khosravi, A., and Mazmanian, S. K. (2010). Host–bacterial symbiosis in health and disease. Adv. Immunol. 107, 243–274. doi: 10.1016/B978-0-12-381300-8.00008-3
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific Microbiota. Cell 149, 1578–1593. doi: 10.1016/j.cell.2012.04.037
Delorme-Axford, E., and Klionsky, D. J. (2018). Secretory autophagy holds the key to lysozyme secretion during bacterial infection of the intestine. Autophagy 14, 365–367. doi: 10.1080/15548627.2017.1401425
Donaldson, E. E., Stanley, D., Hughes, R. J., and Moore, R. J. (2017). The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ 5:e3587. doi: 10.7717/peerj.3587
Farnell, M. B., and Donoghue, A. M. (2006). de los Santos FS, Blore PJ, Hargis BM, Tellez G. Upregulation of oxidative burst and degranulation in chicken heterophils stimulated with probiotic bacteria. Poult. Sci. 85, 1900–1906. doi: 10.1093/ps/85.11.1900
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi: 10.1038/cr.2013.168
Gannagé, M, and Münz, C. (2009). Macroautophagy in Immunity and Tolerance. Traffic 10, 615–620. doi: 10.1111/j.1600-0854.2009.00883.x
Genovese, K. J., He, H., Swaggerty, C. L., and Kogut, M. H. (2013). The avian heterophil. Dev. Comp. Immunol. 41, 334–340. doi: 10.1016/j.dci.2013.03.021
Gensollen, T., Iyer, S. S., Kasper, D. L., and Blumberg, R. S. (2016). How colonization by microbiota in early life shapes the immune system. Science. 352, 539–544. doi: 10.1126/science.aad9378
Geyer, P. E., Holdt, L. M., Teupser, D., and Mann, M. (2017). Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol.13:942. doi: 10.15252/msb.20156297
Hassan, H. A., Arafat, A. R., Farroh, K. Y., Bahnas, M. S., El-Wardany, I., and Elnesr, S. S. (2021). Effect of in ovo copper injection on body weight, immune response, blood biochemistry and carcass traits of broiler chicks at 35 days of age. Animal Biol. 2021, 1–8. doi: 10.1080/10495398.2021.1924763
Henneke, P., and Golenbock, D. T. (2004). Phagocytosis, innate immunity, and host–pathogen specificity. J. Exp. Med. 199, 1–4. doi: 10.1084/jem.20031256
Iqbal, M., Pumford, N. R., Tang, Z. X., Lassiter, K., Wing, T., Cooper, M., et al. (2004). Low feed efficient broilers within a single genetic line exhibit higher oxidative stress and protein expression in breast muscle with lower mitochondrial complex activity. Poult. Sci. 83, 474–484. doi: 10.1093/ps/83.3.474
Joshi, T., Butchar, J. P., and Tridandapani, S. (2006). Fcgamma receptor signaling in phagocytes. Int. J. Hematol. 84, 210–216. doi: 10.1532/IJH97.06140
Jung, T. W., Chung, Y. H., Kim, H. C., El-Aty, A. M. A., and Jeong, J. H. (2018). LECT2 promotes inflammation and insulin resistance in adipocytes via P38 pathways. J. Mol. Endocrinol. 61, 37–45. doi: 10.1530/JME-17-0267
Kalenik, B. M., Góra-Sochacka, A., Stachyra, A., Pietrzak, M., Kopera, E., and Fogtman, A. (2018). Transcriptional response to a prime/boost vaccination of chickens with three vaccine variants based on HA DNA and Pichia-produced HA protein. Dev. Comp. Immunol. 88, 8–18. doi: 10.1016/j.dci.2018.07.001
Kelly, D., King, T., and Aminov, R. (2007). Importance of microbial colonization of the gut in early life to the development of immunity. Mutat. Res. Mol. Mech. Mutagen 622, 58–69. doi: 10.1016/j.mrfmmm.2007.03.011
Kogut, M. H., Genovese, K. J., and Lowry, V. K. (2001). Differential activation of signal transduction pathways mediating phagocytosis, oxidative burst, and degranulation by chicken heterophils in response to stimulation with opsonized Salmonella Enteritidis. Inflammation. 25, 7–15. doi: 10.1023/a:1007067426499
Kong, B. W., Lassiter, K., Piekarski-Welsher, A., Dridi, S., Reverter-Gomez, A., Hudson, N. J., and Bottje, W. G. (2016). Proteomics of breast muscle tissue associated with the phenotypic expression of feed efficiency within a pedigree male broiler line: I. Highlight on mitochondria. PLoS ONE. 11:e0155679. doi: 10.1371/journal.pone.0155679
Kumar, S., Ingle, H., Prasad, D. V. R., and Kumar, H. (2013). Recognition of bacterial infection by innate immune sensors. Crit. Rev. Microbiol. 39, 229–246. doi: 10.3109/1040841X.2012.706249
Kuttappan, V. A., Bottje, W., Ramnathan, R., Hartson, S. D., Coon, C. N., and Kong, B. W. (2017). Proteomic analysis reveals changes in carbohydrate and protein metabolism associated with broiler breast myopathy. Poult. Sci. 96, 2992–2999. doi: 10.3382/ps/pex069
Liu, X., Chang, X., Wu, H., Xiao, J., Gao, Y., and Zhang, Y. (2014). Role of intestinal inflammation in predisposition of Edwardsiella tarda infection in zebrafish (Danio rerio). Fish Shellfish Immunol. 41, 271–278. doi: 10.1016/j.fsi.2014.09.009
Nedjic, J., Aichinger, M., Emmerich, J., Mizushima, N., and Klein, L. (2008). Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 455, 396–400. doi: 10.1038/nature07208
Nutrient Requirements of Poultry. (1994). Nutrient Requirements of Poultry. Washington, DC: National Academies Press. doi: 10.17226/2114
Pedroso, A. A., Batal, A. B., and Lee, M. D. (2016). Effect of in ovo administration of an adult-derived microbiota on establishment of the intestinal microbiome in chickens. Am. J. Vet. Res. 77, 514–526. doi: 10.2460/ajvr.77.5.514
Piccinini, A. M., and Midwood, K. S. (2010). DAMPening inflammation by modulating TLR signalling. Mediators Inflamm. Special issue, ID 705612. doi: 10.1155/2010/672395
Rodrigues, D. R., Wilson, K. M., Trombetta, M., Briggs, W. N., Duff, A. F., Chasser, K. M., et al. (2020a). A proteomic view of the cross-talk between early intestinal microbiota and poultry immune system. Front. Physiol. 11:20. doi: 10.3389/fphys.2020.00020
Rodrigues, D. R., Winson, E., Wilson, K. M., Briggs, W. N., Duff, A. F., Chasser, K. M., et al. (2020b). Intestinal pioneer colonizers as drivers of ileal microbial composition and diversity of broiler chickens. Front. Microbiol. 10:2858. doi: 10.3389/fmicb.2019.02858
Roto, S. M., Kwon, Y. M., and Ricke, S. C. (2016). Applications of in ovo technique for the optimal development of the gastrointestinal tract and the potential influence on the establishment of its microbiome in poultry. Front Vet. Sci. 3:63. doi: 10.3389/fvets.2016.00063
Saeed, M., Babazadeh, D., Naveed, M., Alagawany, M., Abd El-Hack, M. E., Arain, M. A., et al. (2019). In ovo delivery of various biological supplements, vaccines and drugs in poultry: current knowledge. J. Sci. Food Agric. 99, 3727–3739. doi: 10.1002/jsfa.9593
Sawant, P. M., Dhama, K., Rawool, D. B., Wani, Y., Tiwari, R., Singh, S. D., et al. (2015). Development of a DNA vaccine for chicken infectious anemia and its immunogenicity studies using high mobility group box 1 protein as a novel immunoadjuvant indicated induction of promising protective immune responses. Vaccine 33, 333–340. doi: 10.1016/j.vaccine.2014.11.020
Schmid, D., Pypaert, M., and Münz, C. (2007). MHC class II antigen loading compartments continuously receive input from autophagosomes. Immunity 26, 79–92. doi: 10.1016/j.immuni.2006.10.018
Schokker, D., Peters, T., Hoekman, A., and Rebel, J. (2010). Effects of Salmonella on spatial-temporal processes of jejunal development in chickens. Dev. Comp. Immunol. 34, 1090–1100. doi: 10.1016/j.dci.2010.05.013
Sekelova, Z., Stepanova, H., Polansky, O., Varmuzova, K., Faldynova, M., Fedr, R., et al. (2017). Differential protein expression in chicken macrophages and heterophils in vivo following infection with Salmonella Enteritidis. Vet. Res. 48:35. doi: 10.1186/s13567-017-0439-0
Shira, E. B., and Friedman, A. (2018). Innate immune functions of avian intestinal epithelial cells: Response to bacterial stimuli and localization of responding cells in the developing avian digestive tract. PLoS ONE 13:e0200393. doi: 10.1371/journal.pone.0200393
Simon K Verwoolde MB, Zhang J, Smidt H, de Vries Reilingh G, Kemp B, Lammers A. Long-term effects of early life microbiota disturbance on adaptive immunity in laying hens. Poult Sci. 95, 1543–1554. doi: 10.3382/ps/pew088.
Slowik, V., and Apte, U. (2017). Leukocyte cell-derived chemotaxin-2: it's role in pathophysiology and future in clinical medicine. Clin. Transl. Sci. 10, 249–259. doi: 10.1111/cts.12469
Teague, K. D., Graham, L. E., Dunn, J. R., Cheng, H. H., Anthony, N., and Latorre, J. D. (2017). In ovo evaluation of FloraMax -B11 on Marek's disease HVT vaccine protective efficacy, hatchability, microbiota composition, morphometric analysis, and Salmonella enteritidis infection in broiler chickens. Poult. Sci. 96, 2074–2082. doi: 10.3382/ps/pew494
Vuong, C. N., Kuttappan, V. A., Faulkner, O. B., Berghman, L. R., Wolfenden, A. D., Tellez-Isaias, G., et al. (2020). Comparison of oil emulsion, mannosylated chitosan, and Bacillus vector adjuvants for vaccination against influenza in chickens. J. Appl. Poult. Res. 29, 653–664. doi: 10.1016/j.japr.2020.04.003
Wilson, K. M., Rodrigues, D. R., Briggs, W. N., Duff, A. F., Chasser, K. M., Bielke, L. R., et al. (2019). Evaluation of the impact of in ovo administered bacteria on microbiome of chicks through 10 days of age. Poult. Sci. 98, 5949–5960. doi: 10.3382/ps/pez388
Wilson, K. M., Rodrigues, D. R., Briggs, W. N., Duff, A. F., Chasser, K. M., Bottje, W. G., et al. (2020). Impact of in ovo administered pioneer colonizers on intestinal proteome on day of hatch. Poult. Sci. 99, 1254–1266. doi: 10.1016/j.psj.2019.10.017
Yanai, H., Chiba, S., Ban, T., Nakaima, Y., Onoe, T., Honda, K., et al. (2011). Suppression of immune responses by nonimmunogenic oligodeoxynucleotides with high affinity for high-mobility group box proteins (HMGBs). Proc. Natl. Acad. Sci. USA. 108, 11542–11547. doi: 10.1073/pnas.1108535108
Yang, H., Antoine, D. J., Andersson, U., and Tracey, K. J. (2013). The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 93, 865–873. doi: 10.1189/jlb.1212662
Yang, Y., Wolfenden, A., Mandal, R. K., Faulkner, O., Hargis, B., Kwon, Y. M., et al. (2017). Evaluation of recombinant Salmonella vaccines to provide cross-serovar and cross-serogroup protection. Poult. Sci. 96, 4352–4360. doi: 10.3382/ps/pex144
Keywords: immune programming, probiotics, Enterobacteriaceae, lactic acid bacteria, intestinal microbiome, innate immune system, proteome, pathways
Citation: Rodrigues DR, Wilson KM and Bielke LR (2021) Proper Immune Response Depends on Early Exposure to Gut Microbiota in Broiler Chicks. Front. Physiol. 12:758183. doi: 10.3389/fphys.2021.758183
Received: 13 August 2021; Accepted: 13 September 2021;
Published: 15 October 2021.
Edited by:
Michael Kogut, United States Department of Agriculture, United StatesReviewed by:
Shaaban Saad Elnesr, Fayoum University, EgyptMonika Proszkowiec-Weglarz, United States Department of Agriculture, United States
Melha Mellata, Iowa State University, United States
Copyright © 2021 Rodrigues, Wilson and Bielke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa R. Bielke, bielke.1@osu.edu
 Denise R. Rodrigues
Denise R. Rodrigues Kim M. Wilson
Kim M. Wilson Lisa R. Bielke
Lisa R. Bielke