- Crop Improvement Division, ICAR-Directorate of Groundnut Research, Junagadh, India
Peanut, an important oilseed crop, is gaining priority for the development of drought tolerant genotypes in recent times, since the area under drought is constantly on the rise. To achieve this, one of the important strategies is to genetically engineer the ruling peanut varieties using transcription factor regulating the expression of several downstream, abiotic-stress responsive gene(s). In this study, eight independent transgenic peanut (cv. GG20) lines were developed using AtDREB1A gene, encoding for a transcription factor, through Agrobacterium-mediated genetic transformation. The transgene insertion was confirmed in (T0) using PCR and Dot-blot analysis, while copy-number(s) was ascertained using Southern-blot analysis. The inheritance of AtDREB1A gene in individual transgenic plants (T1 and T2) was confirmed using PCR. In homozygous transgenic plants (T2), under soil-moisture deficit stress, elevated level of AtDREB1A transgene expression was observed by RT-PCR assay. The transgenic plants at 45-d or reproductive growth stage showed tolerance to severe soil-moisture deficit stress. Physio-biochemical parameters such as proline content, osmotic potential, relative water content, electrolytic leakage, and total-chlorophyll content were found positively correlated with growth-related traits without any morphological abnormality, when compared to wild-type. qPCR analysis revealed consistent increase in expression of AtDREB1A gene under progressive soil-moisture deficit stress in two homozygous transgenic plants. The transgene expression showed significant correlation with improved physio-biochemical traits. The improvement of drought-stress tolerance in combination with improved growth-related traits is very essential criterion for a premium peanut cultivar like GG20, so that marginal farmers of India can incur the economic benefits during seasonal drought and water scarcity.
Introduction
Peanut or groundnut (Arachis hypogaea L.) is an important, leguminous oilseed crop, which is cultivated mostly in the tropical and sub-tropical regions of the world, under rainfed environments and low-input agricultural system (Bhauso et al., 2014b). Across the globe, its cultivation occupies ~20–25 million ha of land, yielding 35–40 million tons of pods annually. India stood second in peanut production after China, having ~5–6 million ha of cultivated area and 6–7 million tons of production (Bhauso et al., 2014a; Mishra et al., 2015). Its productivity suffers from recurrent drought due to insufficient, untimely, and erratic rainfall in those agro-climate areas where it is cultivated (Bosamia et al., 2015). Globally, about 20% of the land surface is under drought at any point of time (Burke et al., 2006); thus, considered as the major environmental constraint to productivity. Annually, peanut productivity suffers a loss of around 6 million tons due to drought alone, in different parts of the World (Bhatnagar-Mathur et al., 2014).
Till now, only minor QTLs were identified for water use efficiency (WUE) in peanut, moreover association of these QTLs with other trait is still unknown. Therefore, introgression of such QTLs, using marker assisted selection (MAS) may result in linkage drag (Bhatnagar-Mathur et al., 2008, 2014). Further, introgression of traits from diploid- or wild- specie to cultivated peanut is very difficult due to cross incompatibilities (Holbrook et al., 2011), linkage-drag (Tiwari et al., 2008), and time consuming breeding practices (Mishra et al., 2015). Thus, genetic engineering of peanut using heterologus drought-tolerance gene, under the control of stress inducible promoter, could be a prudent option to address this problem.
In order to restore cellular function and make plant more tolerant to multiple abiotic stresses, transferring of a single action gene may not be sufficient to confer the required level of tolerance (Bhatnagar-Mathur et al., 2008; Brasileiro et al., 2014). Thus, for enhancing the abiotic stress tolerance like drought, it is more appropriate to transfer a single-gene, encoding for transcription factors (TFs). The TFs regulate the expression of stress inducible, multiple downstream genes through signal transduction pathways, which in turn modulate a series of changes in the plant for adaptation (Datta et al., 2012; Anbazhagan et al., 2015).
The dehydration responsive element binding proteins (DREBs) are important TFs which comprise of conserved ERF/AP2 DNA-binding domain and regulate expression of a set of abiotic stress inducible, downstream, endogenous genes by binding to cis acting element (DRE/CRT) of their promoter and impart abiotic-stress tolerance (Pruthvi et al., 2014). The DREB transcription factors could be grouped into DREB1/CBF (CRT binding factor) and DREB2, which are involved in two separate signal transduction pathways under low temperature; salt and dehydration respectively (Shinozaki and Yamaguchi-Shinozaki, 2000; Pandey et al., 2014). AtDREB1A transgenic peanut genotype, showed tolerance to soil-moisture deficit stress, developed using cv. JL24 (Bhatnagar-Mathur et al., 2007); a variety which is out of cultivation in India. However, overexpressing AtDREB1A gene in transgenic peanut under the control of rd29A promoter showed tolerance to water-deficit stress with improved physiological and agronomic traits such as transpiration efficiency (TE) (Vadez et al., 2007); root-traits (Vadez et al., 2013); higher yield; and improved harvest-index (HI) (Bhatnagar-Mathur et al., 2014).
Constitutive co-expression of three stress-responsive TFs (AtDREB2A, AtHB7, and AtABF3) in transgenic peanut conferred tolerance, and produced improved total biomass under various abiotic stresses (Pruthvi et al., 2014). However, it could be better option to genetically engineer peanut by stress inducible expression of single TF such as AtDREB1A to achieve the desired level of stress tolerance and yield-gain under drought stress, thereby minimizing excessive metabolic load of the transgenics. An earlier report from our lab on the AtDREB1A transgenic peanut, revealed insights about the physio-biochemical parameters and its relationship with the improvement in productivity along with root-traits under both drought- and salinity- stresses (Sarkar et al., 2014). Furthermore, the present study illustrates the development of AtDREB1A transgenic lines and the relationship between transgene expressions with the improved physio-biochemical traits in transgenics, at reproductive-stage, under progressive soil-moisture deficit (drought) stress.
The ultimate aim of this investigation is to develop transgenic peanut cv. GG20; a high yielding, premium, bold-seeded, Virginia bunch type variety; most popular in Western part of India; for improved tolerance to soil-moisture deficit stress, by expressing AtDREB1A gene under the control of rd29A promoter, and to identify best transgenic plants with tolerance to terminal drought.
Materials and Methods
Gene Construct
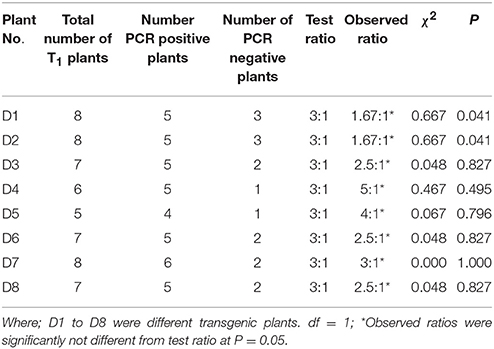
The Agrobacterium tumefaciens strain GV3101 harboring binary vector pCAMBIA2300 containing Prd29A:AtDREB1A:3′ nos and PCaMV35S:nptII:35S polyA expression cassettes, obtained from IARI, New Delhi (India) was used for the genetic transformation. The binary vector comprised of kanamycin resistance gene for bacterial selection (Hajdukiewicz et al., 1994) and harbored nptII gene for selection of genetically transformed plant cells. The nptII expression cassette was fused with transgene expression cassette, isolated from pBI121 binary vector, containing AtDREB1A/CBF3 cDNA (GenBank accession no. AB007787.1) driven by stress inducible rd29A promoter of Arabidopsis thaliana (GenBank accession no. D13044) and nos terminator.
Agrobacterium suspension culture was prepared by inoculating a single bacterial colony in liquid YEM medium (50 mL) containing 25 mgL−1 each of kanamycin, gentamycin, and rifampicin and grown overnight in a rotary shaker (120 rpm; 28°C). The cultures (OD600 = 0.6) were then harvested by centrifugation (5000 × g for 10 min) at room temperature (RT), and cells were re-suspended in 50 mL of co-cultivation MS medium (liquid) (Murashige and Skoog, 1962).
Plant Materials
The seeds of peanut cv. GG20 were obtained from the Genetic Resources Section of the Directorate of Groundnut Research (DGR), Junagadh, India.
Agrobacterium Mediated Transformation and Regeneration of Transgenic Peanut
Mature peanut seeds were surface sterilized in 70% alcohol for 30 s, and then in 0.1% mercuric chloride solution for 4 min, followed by six times rinsing using Milli-Q water and 15 min soaking in Milli-Q water. The embryo was then removed and cotyledonary explants were allowed to get infected with suspension culture of A. tumefaciens harboring binary vector (Figure 1) for 20 min at RT. The genetic transformation protocol as proposed by Mehta et al. (2013) was used for the regeneration of genetically transformed shoots, after which kanamycin based selection of putatively-transformed shoots was done (Bhatnagar-Mathur et al., 2007).
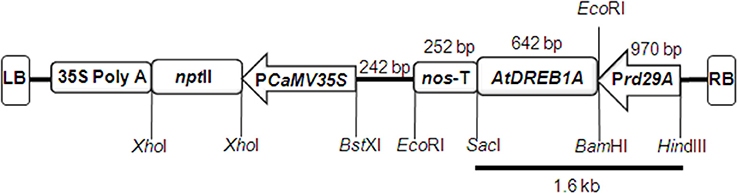
Figure 1. A diagrammatic representation of pCAMBIA2300 plant transformation vector used for transforming tobacco and peanut explants. The vector is harboring rd29A promoter, AtDREB1A transgene, and 3′ nos terminator expression cassette between SacI and BamHI cloning sites.
All the cultures were maintained at 26 ± 2°C in 88 μmol photon m−2s−1 light intensity using white fluorescent tube-lights, under 12 h light and 12 h dark photoperiod in a culture room throughout the experiment. Putative transgenic plants were hardened and allowed to set seeds under controlled polyhouse facility with set humidity (45%) and temperature (30 ± 2°C).
DNA Extraction and Screening of Transgenic Plant by PCR
Genomic DNA was extracted from healthy young leaves of putative peanut transgenics using CTAB method (Doyle and Doyle, 1987), and quantified using Nanodrop spectrophotometer (ND-1000, Thermo Scientific, USA). PCR was performed using thermal cycler (Eppendorf, GmbH, Germany) to screen the putative transgenics for the presence of transgene using AtDREB1A gene specific primer pair (F:5-GCT CCG ATT ACG AGT CTT CGG-3′; R:5′-CTT CTG CCA TAT TAG CCA AC-3′) amplifying 528 bp product. The PCR was carried in 25 μl reaction volume containing 1 × PCR buffer, 2 mM MgCl2, 1.6 μL of dNTP mix (2 mM each), 25 pmole of each primers (Forward and Reverse), 1 U Taq DNA polymerase (Ferments, USA), and 200 ng of genomic DNA as template. A touch down PCR amplification profile was programmed at 94°C for 3 min for a initial denaturation, followed by first 8 cycles of 94°C for 45 s, 63–55°C for 45 s, and 72°C for 1 min, with a 1°C decrement in annealing temperature per cycle; then 27 cycles of 94°C for 45 s with constant annealing temperature of 55°C for 45 s and 72°C for 1 min followed by a final extension for 5 min at 72°C. The genomic DNA of AtDREB1A transgenic tobacco and plasmid DNA were used as positive control, while that of wild-type (WT) or genetically non-transformed peanut plants as negative control. PCR products were analyzed using gel electrophoresis in 1.2% (w/v) agarose gels at 80 V for 60 min.
Segregation Analysis of Transgenic Plants
Independent, PCR screened, T0 transgenic plants were allowed to set seeds in polyhouse containment facility. Subsequently, the T1 generation plants were screened by PCR to analyze the expected transgene segregation ratio (3:1) based on presence or absence of band. Chi-square analysis for transgene inheritance was performed to analyze its segregation pattern. Subsequently, the identification of homozygous transgenic plants was carried out by transgene segregation analysis in T2 generation.
Molecular Characterization of Transgenic Plants
Dot Blot Analysis
The HindIII (Fermentas, USA) digested genomic DNA (15 μg) of PCR positive transgenic (T0) and WT peanut lines along with plasmid DNA were denatured by boiling (10 min) in 3 M NaOH, followed by the addition of 6X SSC buffer. The denatured DNA was loaded onto nylon membrane (Hybond N+, Amersham Pharmacia Biotech, Ireland), wetted in 6X SSC buffer, under low vacuum using DHM-96 dot and slot blotter (SCIE-PLAS, UK). The 528 bp PCR amplified product (using gene-specific primers used for PCR) of plasmid vector was used for the preparation of probe and labeling was done using Biotin DecaLabel DNA Labeling kit (Ferments Life Sciences, USA). Pre-hybridization and hybridization was performed using standard protocols (Sambrook and Russell, 2001) while detection was carried out using the Biotin Chromogenic Detection kit (Ferments Life Sciences, USA).
Southern Blotting
High quality genomic DNA (30 μg) of transgenic (T0) and WT plants was individually isolated using Nucleospin Plant II kit (MN, GmbH). These were then digested with HindIII and the genomic DNA of AtDREB1A transgenic tobacco (isolated using Nucleospin Plant II kit) was double digested with HindIII and SacI (Fermentas, USA) in 1X buffer, separated on 0.8% agarose gel, and transferred to Biodyne plus (0.45 μm) nylon membrane (Pall Life Sciences-Pall corporation, New York, USA) using alkaline transfer buffer (0.4 N NaOH and 1 M NaCl) as per standard protocols (Sambrook and Russell, 2001). The PCR amplified product (528 bp) of AtDREB1A gene from transgenic tobacco was used for the preparation of probe, which was then labeled with thermostable alkaline phosphatase, using Amersham Gene Images AlkaPhos Direct Labeling and Detection System (Amersham, GE Healthcare, UK). Hybridization was carried out overnight at 55°C in hybridization buffer and membrane was washed (at 55°C for 15 min) first in primary wash buffer (25 mL) and then in secondary wash buffer at RT. Hybridized membrane was detected using CDP-Star chemiluminescent as substrate, and signals were visualized on Amersham Hyperfilm ECL (Amersham, GE Healthcare, UK) after 1 h.
RNA Isolation and Reverse Transcription PCR (RT-PCR)
Total mRNA was isolated from the leaves of eight Southern positive plants (D1–D8 in T2 generation) and the WT (exposed to drought stress) using RNeasy kit (Qiagen, GmbH, Germany). This was subjected to RNase-free DNaseI (Fermentas, USA) digestion and purification (Bhauso et al., 2014b). The RNA was quantified using Nanodrop spectrophotometer (ND-1000, Thermo Scientific, USA) and equal amount of RNA from each sample was used for the two-step RT-PCR reaction. The first-strand of cDNA was synthesized from 500 ng RNA per sample using first strand cDNA synthesis kit (Fermentas, USA) and the product obtained was further used for second-strand amplification using AtDREB1A gene specific primers (F: 5′-AAG AAG TTT CGT GAG ACT CG-3′; R: 5′-CTT CTG CCA TAT TAG CCA AC-3′). For internal control, expression of 18S rRNA gene (F: 5′-GGC TCA AGC CGA TGG AAG T-3′, R: 5′-AGC ACG ACA GGG TTT AAC AAG A-3′).
Physio-Biochemical Characterization of Transgenic Peanuts Exposed to Progressive Soil-Moisture Deficit Stress
The plants of eight transgenic (T2) events viz. D1 to D8 were used for the characterization of physio-biochemical and growth-related traits under progressive soil-moisture deficit stress during flowering and pegging stages. The plants were grown in plastic pots (21 × 23 cm) containing 4.5 kg soil mix (1 soil: 1 sand) under containment facility for 45 d until the onset of flowering. The WT plants were divided into two sets as well-watered control (WW), where plants were maintained at about 95% field capacity; and water-stressed control (WS). WS along with the transgenic plants were subjected to drought for a period of 12 d by withdrawal of irrigation, until showing severe wilting symptoms. The experiments were conducted in three replicates. WW plants were used only for phenotypic and growth-related characterization.
The soil-moisture (%) was calculated daily using gravimetric method. The soil-moisture (%) is expressed as the ratio of the mass of water present in the soil sample to the dry-weight of the same soil sample multiplied by 100. The soil-moisture content is calculated from the soil-weight before and after oven drying. Physio-biochemical characterization of all the transgenics and WS were carried out by quantitative estimation of abiotic-stress related parameters such as proline-content, osmotic-potential (OP), relative water content (RWC), electrolytic-leakage (EL), and total chlorophyll content during decreasing soil-moisture regimes at 31, 13, 9, 7, 5% soil-moisture content. Leaves samples, collected from the transgenic and WS plants at the aforesaid soil-moisture contents during drought-stress, were used for further analysis. During the experiment, phenotypic parameters such as wilting of leaves; inhibition of flowering, peg-formation, shoot-length elongation, and development of new-leaf were recorded. At the end of the experiment, 200 ml water was added at an interval of 24 h for recovery of stressed plants up to 3 d and the rate of recovery was visually recorded.
Proline Content
The proline content was estimated according to Bates et al. (1973). The leave samples of both WT and transgenic plants (100 mg) were homogenized using a mortar and pestle in 5 mL sulfosalicylic acid (3%), and centrifuged at 5000 g (10–15 min). The supernatant was collected and diluted to 5 mL with sulfosalicylic acid (3%). Two milliliters each of glacial acetic acid and acid ninhydrin were added to the supernatant (2 mL) and mixed well. The mixture was then boiled for 1 h in a water bath and cooled. Then, for color development, 4 mL toluene was added, mixed well and allowed to stand for 2–3 min. The absorbance of the solution was recorded spectrophotometrically at 520 nm (Epoch microplate spectrophotometer, BioTech, USA). Simultaneously, a blank, containing 2 mL of 3% sulfosalicylic acid was also run. The proline content was calculated as per the proline standard (100 mg/mL in 3% sulfosalicylic acid).
Osmotic Potential
Leaves tissues (100 mg) of transgenics and WS were frozen in liquid nitrogen, followed by thawing in Eppendorf tubes (1.5 mL) at RT for 2 h. It is then centrifuged (at 12,000 rpm for 10 min) to collect the cell-sap. Finally the osmotic potential (osmolality) of cell-sap was measured using Vapor Pressure Osmometer (Model No. 5500; Wescor, USA) (Sarkar et al., 2014).
Electrolytic Leakage (EL)
Fresh leaf discs (1 cm diameter) were washed with distilled water, blotted dried, and incubated in distilled water (25 mL) with continuous shaking for 2 h. Initial electrical conductivity (EC1) was read using pH/EC/TDS Meter (HI991301, HANNA, USA). The leaf discs were then boiled for 30 min in water-bath, followed by cooling at 25°C; and final EC2 was recorded. The percent EL was calculated using the formula (EC1/EC2) × 100% (Wang et al., 2008).
Relative Water Content (RWC)
Initial fresh weight (FW) of leaf discs (1 cm diameter) of both transgenics and WS plants were measured. Then the discs were floated in petri-plates containing water for 8 h, and turgid weight (TW) of hydrated leaf-discs were measured. It was then dried in a hot air oven (80°C for 72 h) and weighed till a consistent dry weight (DW) was obtained. RWC was determined using the formula; RWC = (TW–DW)/(FW–DW) × 100 (Barrs and Weatherley, 1962).
Chlorophyll Content
Fifty mg of leaf tissue was kept in 3 mL vial with screw cap (HIMEDIA, India) containing 2 mL dimethylsulfoxide (DMSO), protected from light and incubated in water bath (65°C for 12 h). Absorbance of the extract was read at 645 and 663 nm spectrophotometrically. Chlorophyll content was determined using the following formula:
Where, V: volume of extract and W: weight of tissue in g (Hiscox and Israelstam, 1979).
Quantitative Expression of Transgene under Progressive Soil-Moisture Deficit Stress
Quantitative PCR (qPCR) analysis was performed to determine differential expression pattern of AtDREB1A gene, after exposing the transgenics at various levels of soil-moisture content (31, 13, 9, 7, and 5%). Total RNA was extracted from the leaf samples and the first strand cDNA was synthesized (as performed for RT-PCR) and qPCR analysis was carried out using a StepOne Real-Time PCR system (Applied Biosystem, USA).
Following first strand cDNA synthesis, the qPCR was carried out in triplicate using gene specific primers (F: 5′-CCT CAG GCG GTG ATT ATA TTC C-3′, R: 5′-ACG ACC CGC CGG TTT C-3′) using QuantiFast SYBR Green PCR Kit (Qiagen, GmbH). The relative quantification of AtDREB1A gene was normalized with respect to 18S rRNA gene as an internal control on qPCR system. Comparative fold expression of transgene was calculated in terms of 2−ΔΔCT method (Livak and Schmittgen, 2001). The ΔCT was determined by subtracting 18S rRNA CT from AtDREB1A CT in a given sample. The ΔCT value of “transgenics exposed to 31% soil-moisture content” was used as calibrators. The ΔΔCT value was determined by subtracting the ΔCT of calibrator from ΔCT of transgenics at different soil-moisture regimes. Each reaction was carried out in 20 μL total volume and consist of 1 × SYBR Green Master mix, 20 pmol each primer and 100 ng of diluted cDNA template.
Evaluation of Transgenics for their Growth-Related Traits
At maturity, the transgenics, WS and WW plants were harvested and carefully divided into shoot and root components. The shoot-length, root-length, root-volumes, and number of pods were measured. The dry-weight of roots, shoots, pod, kernel, and total-biomass were determined. Subsequently root: shoot ratio and HI were calculated.
Statistical Analysis
Statistical analysis was carried out with mean value and standard error (SE) of three replicates per analysis and significance of the treatment effects was determined by one-way ANOVA of SPSS 11.0 (Statistical Package For Social Sciences, SPSS Inc., Illinois) at 5% probability level using Tukey test. Correlation coefficient was determined using PAST (PAlaeontological STatistics, ver. 1.89). The qPCR data was analyzed using two-tailed student's t-test.
Results
Genetic Transformation of Peanut
Out of 682 Agrobacterium-infected explants, 485 responded (71.11% regeneration frequency) and gave rise to 1924 shoots. Further, in the kanamycin (50, 100, and 100 mgL−1) amended selection medium, 683 putative transformed shoots (35.5%) were selected (for three cycles of 15 d each) of which, 83.46% produced roots, and 50.52% plantlets were hardened in the containment facility. Co-cultivation of cotyledonary explants with A. tumefaciens (strain GV3101) for 3–5 d resulted in 2.49% transformation efficiency (Table 1, Figure 2).

Figure 2. Genetic transformation and regeneration of peanut from cotyledons. (A–D) Multiple shoot-buds initiating from co-cultured cotyledonary explants; (E) Shoots regeneration; (F) Selection in antibiotic amended medium; (G) Shoot elongation; (H) Rooting; (I) Hardening of plantlets in Hoagland's solution; (J) Hardened plants grown in pots under containment facility.
Molecular Characterization
Screening of Transgenic Plants by PCR and Dot-Blot Analysis
Screening of 243 putative transgenic peanut plants, grown in containment facility, was carried out by PCR amplification of AtDREB1A gene using gene-specific primers (Figure 3A) and 17 putative transgenic (T0) plants were identified. Further, Dot-blot confirmed the integration of AtDREB1A gene in the genome of ten transgenic (T0) plants only (Figure 3B).
Figure 3. (A) PCR analysis of AtDREB1A transgenics (T0) using gene-specific primers having 528 bp as expected product size. Where lanes, M, marker; PC, positive control (AtDREB1A transgenic tobacco); P, plasmid positive control (pCAMBIA2300 plasmid containing AtDREB1A gene); 1–8, transgenic plants (D1–D8); WT, genetically non-transformed peanut. (B) Dot blot assay of AtDREB1A transgenics (T0), probed with 528 bp PCR amplified product of AtDREB1A coding sequence from plasmid vector pCAMBIA2300 containing Prd29A:AtDREB1A:3′ nos terminator expression cassette. Where, labeled dots, for WT represents HindIII digested DNA from genetically non-transformed plant; P, represents plasmid DNA containing AtDREB1A expression cassette; numbers 1–10, represents HindIII digested DNA from 10 putative transgenic lines. (C) Southern blot analysis of eight transgenic lines (T0). Genomic DNA (30 μg) from individual transgenic peanut plants was digested with Hind III, size fractionated on 0.8% agarose gel, transferred to nylon membrane and hybridized with a probe prepared by direct labeling of PCR amplified product (528 bp) of AtDREB1A gene with alkaline phosphatase. Where, PC is HindIII and SacI digested genomic DNA of AtDREB1A transgenic tobacco, WT is HindIII digested peanut genomic DNA. (D) RT-PCR analysis for AtDREB1A gene in eight transgenic and WT plants exposed to 31% (a,c) and 5% (b,d) soil moisture content using AtDREB1A (a,b) and 18S rRNA (c,d) genes specific primers. Where, in WT, no expression was observed for AtDREB1A gene but expression is recorded for 18S RNA gene; in all the transgenic plants (D1–D8) expression was recorded for both AtDREB1A (442 bp) and 18S RNA (148 bp) genes.
Southern Hybridization
Southern hybridization analysis determined the integration pattern and copy number(s) of AtDREB1A gene into the different parts of genome of eight peanut transgenic events. Of 10 dot-blot positive plants, the presence of hybridization signal was recorded in eight T0 transgenic plants (D1–D8) (Figure 3C). This analysis confirmed the random integration of single copy AtDREB1A transgene in the plants of six transgenic events (D1–D3, D5–D7), while two transgenic plants (D4 and D8) exhibited two copies of transgene in its genome. However, no hybridization signal was observed in the WT.
Reverse Transcription (RT)-PCR
All the Southern-positive transgenic plants (D1–D8) showed transgene expression in T2 generation, as evidenced by RT-PCR. An increase in AtDREB1A gene expression was detected at 5% soil-moisture content (drought stress) as compared to 31% soil-moisture content (controlled condition) in transgenic plants, grown in plastic pots under containment facility. Two bands of 442 bp and 148 bp were amplified from the cDNA of different transgenic plants with AtDREB1A and 18S rRNA gene specific primers, respectively (Figure 3D). No transgene expression was observed in the WT.
All RT-PCR positive transgenic plants were subjected to progressive soil-moisture deficit stress for further evaluation of physio-biochemical and growth-related traits. The transcription level of AtDREB1A gene was relatively high in D3 and D6 lines compared to that of all other transgenic plants as revealed by RT-PCR analysis, and plants of those two events were used for further qPCR analysis.
Transgene Inheritance Analysis
The transgenic plants having Chi-square value < 3.84 (p < 0.05) at 1° of freedom indicated good Mendelian segregation pattern for transgene. In PCR screening, all transgenic plants (D1–D8) in T1 generation, showed Mendelian segregation ratio (3:1) of AtDREB1A gene (Table 2). These transgenic plants were further advanced to T2 generation and based on PCR analysis; homozygous lines in each transgenic event were identified.
Physio-Biochemical Analysis of Transgenic Peanut Exposed to Progressive Soil-Moisture Deficit Stress
Proline Content
Up to 13% of the soil-moisture content, no significant difference in proline content in WS and transgenic plants were recorded. However, at 7% soil-moisture, plants of five transgenic events (D1, D3, D4, D5, and D6) showed very high accumulation of proline; while plants of three transgenic events (D2, D7, and D8) expressed proline content even less than WS. Moreover, at 5% soil moisture, all transgenic plants accumulated more proline than WS, of which D3 and D6 showed maximum proline accumulation to the tune of 2.75 and 2.93 folds respectively (Figure 4A).
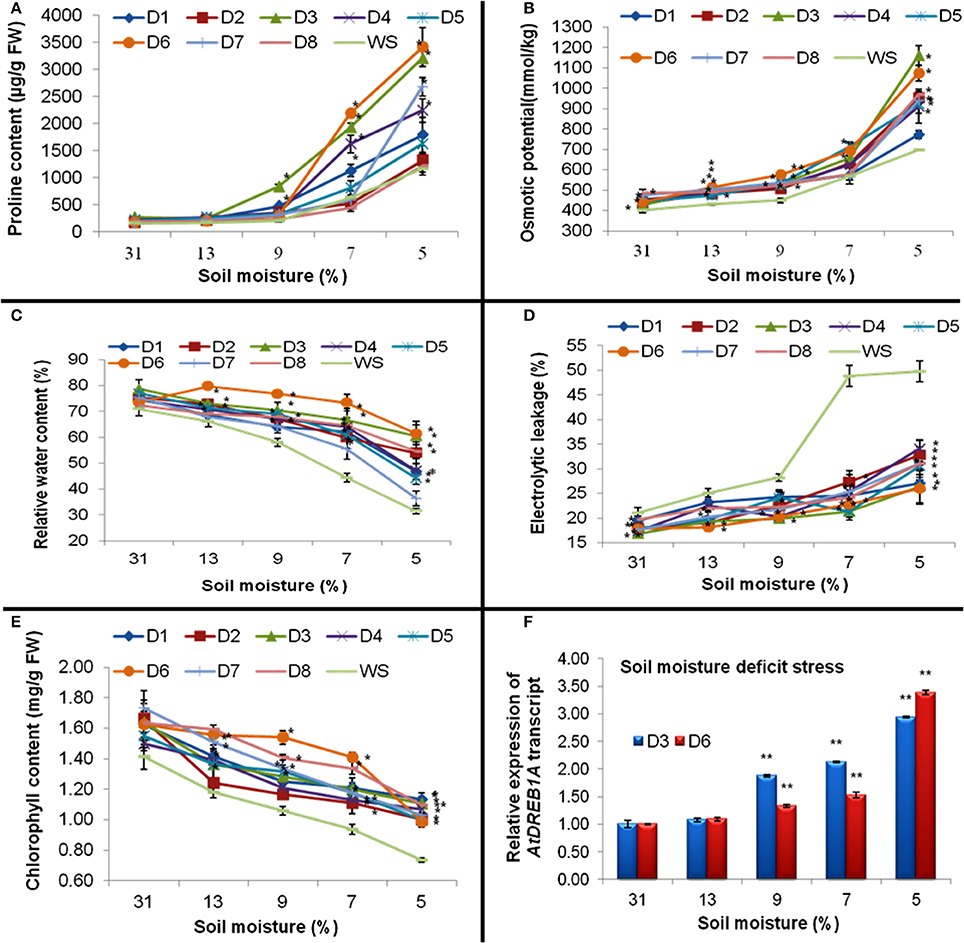
Figure 4. Effect of progressive decrease in soil-moisture percentage on (A) Proline content, (B) osmotic potential, (C) relative water content (D) electrolytic leakage, and (E) chlorophyll content in the leaves of eight transgenic and WS plants. Where; D1–D8 were transgenic lines and WS were water-stressed WT plants. Values represent mean activities (n = 3) ±SE at P = 0.05. (*) represent values significantly different at P ≤ 0.05. (F) Expression of AtDREB1A transcript as analyzed by quantitative PCR in two transgenic plants at different soil moisture regimes. The level of AtDREB1A transcript in transgenic plants was normalized with reference to 18S rRNA taken as an internal control. Data represent means of three replicates. Bars denote fold expression as compared to the expression level at 0 day ± SD. Two-tailed Student's t-test was used to determine highly significant (**P ≤ 0.01).
Osmotic Potential
At 31% soil-moisture, not much difference in osmotic potential was recorded in WS and all transgenic plants. However, across other soil-moisture regimes (starting from 13%), all transgenic plants exhibited significantly better osmotic potential compared to WS. When observed at 5% soil-moisture, two transgenics D3 and D6 prominently showed better osmotic potential over other transgenic plants and WS (Figure 4B). However, a positive correlation was detected between proline content and osmotic potential (r = 0.58; Table 3) in all transgenics and WS under drought stress.
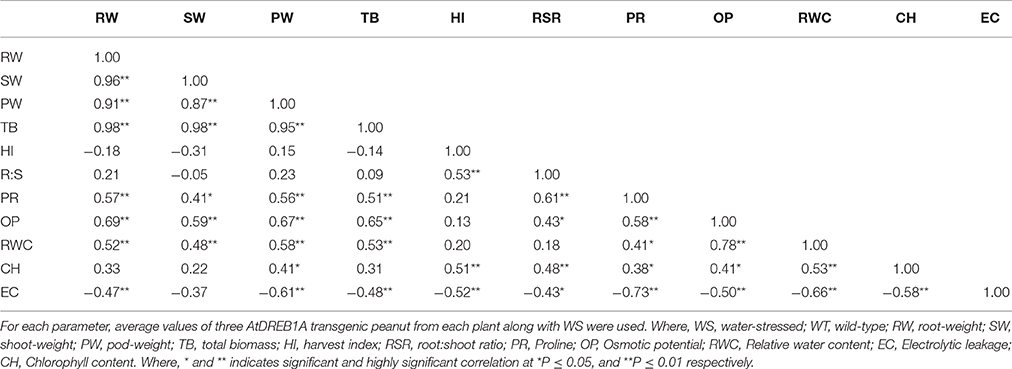
Table 3. Correlation coefficient (r) between growth-related and physio-biochemical parameters of transgenic and WT plants exposed to 5% soil-moisture content.
Relative Water Content
From 13 to 7% of soil-moisture regimes, significantly higher RWC was recorded for all the transgenic plants over WS. But, at 5% soil moisture, plants of D3 and D6 transgenic events witnessed nearly 40% more RWC than WS and were found better than other transgenics (Figure 4C). A positive correlation was noticed between osmotic potential and RWC (r = 0.78) in transgenic plants and WS under drought-stress (Table 3).
Electrolytic Leakage
Although at 31 and 13% soil-moisture content, there were some differences in EL between WS and transgenic plants; but at 5 and 7% of soil-moisture, all transgenic plants witnessed significantly less EL than WS. Moreover, at 5% soil-moisture, D3 and D6 plants showed significantly less EL over other transgenic plants and WS (Figure 4D). However, a significant negative correlation was observed between RWC and EC (r = −0.66) in all transgenic plants and WS under drought stress (Table 3). Furthermore, higher proline accumulation was found negatively correlated with membrane injury or EL in transgenics and WS under drought stress (r = −0.73, Table 3).
Total Chlorophyll Content
Although, at various levels of moisture-deficit stress; all the transgenic plants were found to have significantly more chlorophyll content than WS. Moreover, at 5% soil-moisture, plants of D1, D3, and D8 transgenic events retained relatively more chlorophyll content than WS and other transgenic plants (Figure 4E).
qPCR Analysis of Transgenic Plants Exposed to Progressive Soil-Moisture Deficit Stress
Of eight transgenic events, homozygous D3 and D6 plants showed maximum transgene expression in RT-PCR assay. Further, improved physio-biochemical traits under drought stress were used for the qPCR analysis. At 9% soil-moisture, an increase in the transgene expression to the tune of 1.88 and 1.33 folds and at 5% of soil-moisture, an increase of 2.94 and 3.39 folds were recorded in plants of D3 and D6 events respectively (Figure 4F). Under progressive soil-moisture deficit stress, AtDREB1A gene expression in both the transgenic events were found positively correlated with the improved physio-biochemical parameters like proline content (r = 0.66), osmotic potential (r = 0.63), EL (r = 0.52); while negative correlation was observed with chlorophyll content (r = −0.73), RWC (r = −0.60) (Table 4).
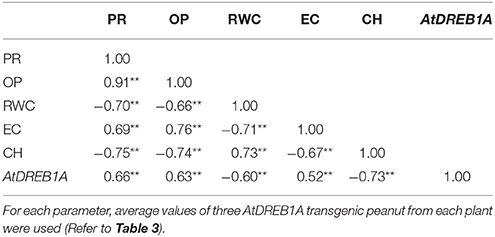
Table 4. Correlation coefficient (r) between AtDREB1A gene expression and different physio-biochemical parameters in two transgenic plants (D3 and D6) under progressive soil-moisture deficit stress.
Visual Observations under Progressive Soil-Moisture Deficit Stress
All the transgenic plants showed improved phenotypic traits over WS during soil-moisture deficit stress and WS exhibited wilting of leaves at 9% soil-moisture, while transgenic plants showed wilting, only when soil-moisture becomes < 7%. Moreover, at 5% soil-moisture, WS showed severe wilting compared to transgenics (data not shown). Upon withdrawal of stress, transgenic plants showed speedy recovery than WS.
Evaluation Growth-Related Parameters
Growth-related parameters like shoot, root, pod and kernel traits were recorded for all the transgenics and WT (WS and WW). Among different transgenics and WS, D3 line was found significantly better for shoot-length and shoot-weight; however, it was significantly higher for WW than all transgenics and WS (P < 0.05) (Table 5). Transgenic plants of D3 and D6 events, exhibited significantly better root-length, root-weight, and root-volume than WS and WW; which might had helped in improving its WUE under drought-stress. Moreover, negative impact of drought-stress was recorded for pod-biomass more prominently in WS compared to transgenic and WW lines. Likewise, kernel-weight, total-biomass and HI of all transgenics and WW were higher or at par with WS lines.
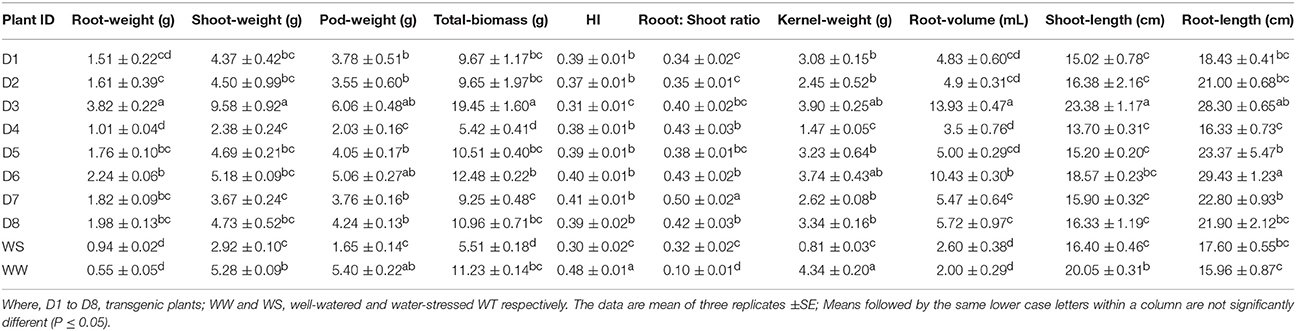
Table 5. Growth-related parameters of transgenic and WT (WS and WW) lines grown under soil-moisture deficit stress.
Similarly, root: shoot ratio of all the transgenics were higher or at par with WS lines under soil-moisture deficit stress (Table 5). Chlorophyll content showed positive correlation with pod-weight (r = 0.41) and HI (r = 0.51); and it also witnessed positive correlation with improved root: shoot ratio in transgenic and WS (r = 0.48) under drought stress (Table 3). Both transgenics and WS showed improved root: shoot ratio which was found positively correlated with HI (r = 0.53; Table 3) under drought stress. Overall, transgenic plants D3 and D6 showed better root-length, root-volume, root-weight, pod-weight, total-biomass, kernel-weight and root: shoot ratio than WS plants, indicating the best agronomic performers across all eight transgenic plants under drought stress (Table 5).
Discussion
This investigation deals with the development of eight AtDREB1A transgenic events (D1–D8) and its characterization at reproductive growth stage, for drought-stress using progressive soil-moisture deficit stress, having advantage of mimicking the real-field conditions. Whereas, the results published earlier (Sarkar et al., 2014) is part of the ongoing work in our lab, which reports the characterization of only three transgenic events (D1–D3) at seedling and vegetative-growth stages for both drought and salinity stresses using different levels of Polyethylene glycol (PEG) and NaCl respectively, under hydroponic conditions.
In this investigation, Agrobacterium-mediated genetic transformation, successfully transformed the cotyledonary explants of peanut cv. GG20 with AtDREB1A gene and shoots were regenerated following the protocols previously reported in peanut by our group (Radhakrishnan et al., 2000; Mehta et al., 2013; Bhauso et al., 2014b). Further, cotyledon have been used as explants, as it showed better transformation efficiency, compared to immature peanut leaves as reported by Mehta et al. (2013). The selection scheme has identified the putatively transformed shoots in kanamycin amended selection medium based on visual observations, and genetically non-transformed shoots have been eliminated (Bhatnagar-Mathur et al., 2007).
This investigation produced transgenic peanut (cv. GG20) with AtDREB1A transgene, under the regulatory mechanism of stress inducible rd29A promoter, with 2.49% transformation efficiency. The regeneration of multiple shoots from cotyledonary explants was 71.11%, which validated the reports on genetic transformation of peanut, where it ranged from 60.4 to 84.3% (Bhatnagar-Mathur et al., 2007; Mehta et al., 2013). However, certain other reports showed varying transformation efficiency in different genotypes like 5.18% in cv. GG20 (Bhauso et al., 2014b); 16.5% in cv. K134, 34% in cv. K6 (Mehta et al., 2013) and 70% in cv. JL24 (Bhatnagar-Mathur et al., 2007). It means transformation efficiency is genotype dependent in peanut, as also suggested by Krishna et al. (2015).
Most of the tissue culture raised transgenic plants (T0) produced less number of seeds (5–8), as also reported on cry1EC transgenic peanut (Tiwari et al., 2008), and all the transgenic plants (T1) showed Mendelian segregation ratio (3:1). Further, PCR based screening was used for the progeny advancement, thereby helping in analyzing the transgene inheritance pattern appropriately. Southern analysis showed both, stable integration and copy number(s) of transgene in the genome of individual transgenic plants (T0). No phenotypic alteration in any transgenic plant was recorded, compared to WT under controlled condition; indicating the absence of any metabolic load in transgenics, no disruption of any functional endogenous gene(s), and tightly regulated expression of AtDREB1A transgene under drought-stress as also reported in earlier studies (Vivek et al., 2013; Sarkar et al., 2014).
RT-PCR analysis showed elevated levels of transgene-expression in eight homozygous transgenic plants (T2) under drought-stress (Figure 3D), indicating a positive relationship between transgene expression and stress-tolerance as also observed in case of mtlD transgenic peanut (Bhauso et al., 2014b). Furthermore, even under controlled conditions, RT-PCR showed some basal level of transgene expression in all transgenic plants, which might be responsible for the relatively better physio-biochemical parameters of transgenics than WT. Similarly, basal level of transgene expression were also observed in AtDREB1A transgenic Arabidopsis (Kasuga et al., 1999) and tomato lines (Rai et al., 2013), under unstressed condition. Although, very minute activity of transgene expression was detected under unstressed conditions, but rd29A promoter is still considered stress-inducible due to the rapid induction and higher level of AtDREB1A gene expression under stress, compared to the 35S promoter (Sarkar et al., 2014).
The PCR, dot-blot, southern-blot and RT-PCR results were effectively utilized for the identification and selection of transformed plants of eight transgenic events. Further, confirmed transgenic plants (T2), were then used for the evaluation of physio-biochemical and growth-related parameters. The drought-tolerance evaluation of transgenic plants, during appropriate developmental stage, like initiation of flowering (under field like conditions) is crucial for the identification of better transgenic lines (Bhatnagar-Mathur et al., 2010; Ravikumar et al., 2014). The scheme of progressive soil-moisture deficit stress, followed in this study, for the imposition of drought-stress, has the advantage of mimicking the real-field conditions (Bhatnagar-Mathur et al., 2008; Anbazhagan et al., 2015).
Accumulation of various osmolytes in different concentrations, under drought stress is known to result in the osmotic adjustments, which in turn impart tolerance to the plants (Macková et al., 2013; Ravikumar et al., 2014; Nahar et al., 2015). Proline is one such important osmolytes which gets accumulated as the first line of response, when plants were exposed to drought stress (Ghobadi et al., 2013; dosSantos et al., 2013; Bhauso et al., 2014a). In this study too, proline levels has been increased profoundly as a function of decreasing soil-moisture content. Similar observation were also reported for AtNDPK2 transgenic alfalfa (Wang et al., 2014), mtlD transgenic peanut (Bhauso et al., 2014a,b) and AtDREB1A transgenic peanut under both drought and salinity stresses (Sarkar et al., 2014).
A gradual increase in osmotic-potential was recorded, after imposition of progressive soil-moisture deficit stress, across all the transgenic, and WS peanut lines. However, transgenics exhibited better osmotic adjustment than WS in both the investigations (Sarkar et al., 2014). Increase in osmotic potential, by way of proline accumulation is known to be a vital plant defense mechanism to face environmental constraints (Pruthvi et al., 2014). Positive correlation between proline content and osmotic potential (Table 3) reiterated the fact that free proline accumulation might be responsible for better osmotic adjustments (Hayat et al., 2012).
RWC, often used to determine the water retention capacity and drought tolerance, reflects metabolic activities of plant tissues (Ravikumar et al., 2014). The transgenic peanut plants showed more RWC, compared to WS, under drought stress. It is assumed that the improved membrane integrity in transgenics over WS resulted in controlled water-efflux as indicated by better RWC under drought stress. Similar reports were also observed in LEA Rab28 transgenic maize (Amara et al., 2013) and AtNDPK2 transgenic alfalfa (Wang et al., 2014) under drought stress. Positive and negative correlation of RWC with osmotic potential and EC respectively, in all the transgenic and WS (Table 3) peanut lines supports the analogy that improved osmotic adjustment reflects higher water retention capacity, lower rate of water loss, as well as higher WUE (Mao et al., 2012).
Abiotic stress imparts detrimental effects on membrane integrity, as also reflected by higher EL. Moreover, the leaking ions are equally vital for the proper functioning of cell (Khare et al., 2010). A gradual increase in EL along with progressive decline in soil-moisture content, across all the transgenic and WS peanut lines was recorded. But, transgenic peanut lines showed relatively less EL compared to WS, as also reported by Gao et al. (2013) on transgenic TaLEA poplar under drought-stress. Similarly, Xu et al. (2014) also reported that MaPIP1;1 transgene insertion in Arabidopsis genome conferred enhanced drought-stress tolerance by reducing membrane injury and maintaining osmotic balance in transgenics. A negative correlation was observed between the proline content and EL (Table 3), which means, proline accumulation might be responsible for lesser EL in both transgenics and WS; but, EL was quite less in transgenics than WS (Hayat et al., 2012).
The activation of stress-response pathway of any plant is highly energy-demanding mechanism; therefore, protection of photosynthetic machinery contributes to its ability to withstand the stress. Extensive reactive oxygen species (ROS) were generated by the drought stress which is detrimental to the photosynthetic machinery including D1 protein of photosystem II (PSII) and total-chlorophyll content (Wang et al., 2008; Macková et al., 2013). Under decreasing soil-moisture regime, transgenics peanut plants showed significantly less reduction in total chlorophyll content than WS, which is in agreement with various other reports (Amara et al., 2013; Liu et al., 2014). The chlorophyll retention capacity might have resulted in improved pod- and kernel- weight of transgenic than WS peanut lines, as also reported for AtDREB1A transgenic rice (Ravikumar et al., 2014).
qPCR analysis showed a consistent increase of around three-fold in the expression of AtDREB1A gene in transgenic peanut (D3 and D6) with progressive decrease in soil-moisture content (Figure 4F). Similarly, in our previous study, more than two-fold increase in AtDREB1A gene expression was observed within 2 h of drought (20% PEG) and salinity (200 mM NaCl) stress imposition (Sarkar et al., 2014). Furthermore, soil-moisture contents had direct impacts on the level of transgene expression in the leaves of transgenic plants. It means these transgenic plants could be used for the peanut genetic improvement programmes for drought tolerance.
Under stressed conditions, transgenic plants showed delay in the wilting of leaves; and speedy recovery of transgenics over WS after withdrawal of stress. This could be supported by the fact that there is a consistent increased in expression of AtDREB1A gene in transgenics across various soil-moisture regimes, as also reported in other transgenic crops expressing various TFs such as LbDREB (Ban et al., 2011); AtDREB1A (Bhatnagar-Mathur et al., 2014); AtDREB2A, AtHB7, and AtABF3 (Pruthvi et al., 2014).
Our earlier studies on three AtDREB1A transgenic peanut lines (D1, D2, and D3) also showed better tolerance of transgenics to both PEG induced drought and NaCl induced salinity stresses, and its rapid recovery upon withdrawal of stress, compared to WT (Sarkar et al., 2014). All the transgenics exhibited improved water retention capacity compared to WS peanut lines, which could be due to the regulation of stomatal behavior and transpiration under stress (Datta et al., 2012; Anbazhagan et al., 2015). However, it needs further experimental confirmation for our transgenic plants.
Correlations were elucidated between AtDREB1A gene-expression and various physio-biochemical parameters in response to drought-stress (Table 4). Significant positive correlation between AtDREB1A gene-expression with proline content and osmotic potential; signifies the fact that transgene expression could be one of the reasons for improved osmotic adjustments (Figures 4A,B; Table 4). A negative correlation between transgene expression with RWC and chlorophyll content could be attributed in part to the enhanced water and chlorophyll retention capacity of transgenics (Table 4). Positive relationship was observed between transgene expression and EC under drought stress (r = 0.52) (Figure 4D; Table 4). This means, enhanced tolerance in transgenic peanut was associated with stress inducible expression of AtDREB1A gene, as also reported in AtDREB1A transgenic tomato (Rai et al., 2013).
Growth-related parameters indicated that, transgenics performed better than WS and showed full potential growth and yield, at severe drought-stress (up to 5% soil moisture) during reproductive growth stages (Table 5). Based on phenotypic response, physio-biochemical characterization, and transgene expression analysis, it could be inferred that at ≤ 9% soil-moisture content, transgenics made better use of stress-responsive mechanism than WS, which could be due to heterologous expression of AtDREB1A gene. Differential improvement in various growth-related traits including shoot-, root-, pod-, and kernel-traits at maturity was also observed in our earlier report (Sarkar et al., 2014).
Significantly, improved root-traits like root: shoot ratio, root-volume of transgenic plants over WS under drought stress, could be due to better AtDREB1A gene expression in root system compared to leaf tissues as also reported by Ban et al. (2011). Improved root-architecture like profuse and deeper rooting system in transgenics might be the reason for enhanced water uptake from deeper soil layer, resulting in improved yield, as also observed in AtDREB1A transgenic peanut by Vadez et al. (2013). Furthermore, most of the transgenic plants showed improved total-biomass, pod-weight, kernel-weight and HI over WS under drought stress.
Transgenics also showed better partitioning of total-biomass, more toward roots and pods, than shoots. This might have resulted in higher root: shoot ratio, pod-weight, and HI in transgenics, compared to WS. Moreover, higher water-uptake capacity of transgenic peanut lines was considered as a function of increased root: shoot ratio under drought stress (Vadez et al., 2007). Positive correlation was found between root: shoot ratio and HI under drought stress and transgenics showed better improvement for these traits than WS (Tables 3, 5). It means better root: shoot ratio might have contributed to the enhancement in HI more prominently in transgenics than WS. Similarly, positive relationship between improved root-traits and productivity was observed in AtDREB1A transgenic peanut under soil-moisture deficit stress (Vadez et al., 2007; Jagana et al., 2012; Bhatnagar-Mathur et al., 2014).
Moreover, transgenic plants showed improved root: shoot ratio and root-traits over WS which might have resulted in better access of roots to the deeper soil-layers for improved water uptake (Bhatnagar-Mathur et al., 2010; Anbazhagan et al., 2015). Based on correlation analysis (Table 3), it could be inferred that an enhanced (denser and deeper) root-system in transgenics might be responsible for improved accumulation of ions (especially Mg2+) from soil which seems to also have a positive role in the chlorophyll retention capacity under drought stress (Werner et al., 2010; Macková et al., 2013).
Furthermore, AtDREB1A transgenic rice lines derived from a single plant also showed difference in stress tolerance, physiological, and growth parameters under drought stress (Ravikumar et al., 2014). Improved physio-biochemical and growth-related parameters of the transgenic peanut plants, might be either due to the activation of an array of drought inducible genes or devising some sort of conservative growth pattern as drought tolerance/avoidance mechanism compared to WS (Bhatnagar-Mathur et al., 2008; Turyagyenda et al., 2013; Ravikumar et al., 2014). Thus, both investigations showed that, AtDREB1A transgenic peanut events developed in our lab revealed abiotic-stress tolerance.
Conclusions
A premium peanut variety (GG20) of India, with high marketability, but not tolerant to drought was used for the development of drought tolerant AtDREB1A transgenics. Both, present investigation and our earlier report (Sarkar et al., 2014) demonstrated better tolerance of AtDREB1A transgenic plants to drought, not only at seedling and reproductive growth stages, but also at maturity level. Out of eight transgenic events (viz. D1–D8), two D3 and D6, clearly showed much improved drought tolerance under progressive soil-moisture deficit stress. A strong association was recorded between AtDREB1A gene induction and drought-stress tolerance in the transgenic plants. It seems that the enhancement of various growth-related traits could be due to the prompt and tightly regulated expression of AtDREB1A gene, driven by stress inducible rd29A promoter, and its subsequent participation in signaling cascade followed by up-regulation of various stress-inducible, downstream endogenous genes (Kasuga et al., 1999; Ravikumar et al., 2014), which eventually have resulted in drought-stress tolerance. This study could be a valuable attempt for application of transgenic peanut in agriculture, and subsequently in trade and commerce. Although, a few transgenic peanut plants having enhanced drought tolerance have been developed by various groups of researchers; but so far, no commercial varieties have been released (Bhauso et al., 2014b) from any part of the World. Thus, the transgenic plants characterized in this study, can be used further for the contained multi-year, multi-location trials under real-field conditions. Subsequently, the best one(s) can be used in the breeding programmes either as valuable pre-breeding resources or directly as enhanced drought-tolerant genotype for commercial cultivation.
Author Contributions
RT: Conceived the experiment and planned. TS: Executed the experiments and recorded data. AK: Assisted in carrying out the laboratory work. GM: Prepared the manuscript. JD: Assisted in carrying out the laboratory work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research work was funded by Indian Council of Agricultural Research, New Delhi, India under the project entitled “ICAR Network Project on Transgenic Crops.”
Abbreviations
ABF3, ABRE binding factor; HB7, Homeodomain-leucine zipper; DREB2A, Dehydration responsive element-binding factor.
References
Amara, I., Capellades, M., Ludevid, M. D., Pagès, M., and Goday, A. (2013). Enhanced water stress tolerance of transgenic maize plants over-expressing LEA Rab28 gene. J. Plant Physiol. 170, 864–873. doi: 10.1016/j.jplph.2013.01.004
Anbazhagan, K., Bhatnagar-Mathur, P., Vadez, V., Dumbala, S. R., Kavi Kishor, P. B., and Sharma, K. K. (2015). DREB1A overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 34, 199–210. doi: 10.1007/s00299-014-1699-z
Ban, Q., Liu, G., and Wang, Y. A. (2011). DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco. J. Plant Physiol. 168, 449–458. doi: 10.1016/j.jplph.2010.08.013
Barrs, H. D., and Weatherley, P. E. (1962). A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 24, 413–428.
Bates, R., Waldren, R. P., and Teare, I. D. (1973). A rapid determination of free proline for water stress studies. Plant Soil. 39, 205–207. doi: 10.1007/BF00018060
Bhatnagar-Mathur, P., Devi, M. J., Reddy, D. S., Lavanya, M., Vadez, V., Serraj, R., et al. (2007). Stress-inducible expression of AtDREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 26, 2071–2082. doi: 10.1007/s00299-007-0406-8
Bhatnagar-Mathur, P., Rao, J. S., Vadez, V., and Sharma, K. K. (2010). Transgenic strategies for improved drought tolerance in legumes of semi-arid tropics. J. Crop Improv. 24, 92–111. doi: 10.1080/15427520903337095
Bhatnagar-Mathur, P., Rao, J. S., Vadez, V., Reddy, S. D., Rathore, A., Yamaguchi-Shinozaki, K., et al. (2014). Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33, 327–340. doi: 10.1007/s11032-013-9952-7
Bhatnagar-Mathur, P., Vadez, V., and Sharma, K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 27, 411–424. doi: 10.1007/s00299-007-0474-9
Bhauso, T. D., Radhakrishnan, T., Kumar, A., Mishra, G. P., Dobaria, J. R., and Rajam, M. V. (2014a). Over-expression of bacterial mtlD gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic peanut (Arachis hypogaea) through accumulation of mannitol. Aust. J. Crop Sci. 8, 413–421. Available online at: http://www.cropj.com/thankappan_8_3_2014_413_421.pdf
Bhauso, T. D., Radhakrishnan, T., Kumar, A., Mishra, G. P., Dobaria, J. R., Patel, K. K., et al. (2014b). Overexpression of bacterial mtlD gene in peanut improves drought tolerance through accumulation of mannitol. Scientific World J. 2014:125967. doi: 10.1155/2014/125967
Bosamia, T. C., Mishra, G. P., Radhakrishnan, T., and Dobaria, J. R. (2015). Novel and stress relevant EST derived SSR markers developed and validated in peanut. PLoS ONE 10:e0129127. doi: 10.1371/journal.pone.0129127
Brasileiro, A. C. M., Araujo, A. C. G., Leal-Bertioli, S. C., and Guimarães, P. M. (2014). Genomics and genetic transformation in Arachis. Int. J. Plant Biol. Res. 2, 1017. Available online at: http://www.jscimedcentral.com/PlantBiology/plantbiology-2-1017.pdf
Burke, E. J., Brown, S. J., and Christidis, N. (2006). Modelling the recent evolution of global drought and projections for the twenty-first century with the Hadley Centre climate model. J. Hydrometeorol. 7, 1113–1125. doi: 10.1175/JHM544.1
Datta, K., Baisakh, N., Ganguly, M., Krishnan, S., Shinozaki, K. Y., and Datta, S. K. (2012). Overexpression of Arabidopsis and rice stress genes' inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. doi: 10.1111/j.1467-7652.2012.00688.x
dosSantos, C. M., Verissimo, V., de Lins Wanderley Filho, H. C., Ferreira, V. M., da Silva Cavalcante, P. G., Rolim, E. V., et al. (2013). Seasonal variations of photosynthesis, gas exchange, quantum efficiency of photosystem II and biochemical responses of Jatropha curcas L. grown in semi-humid and semi-arid areas subject to water stress. Indus. Crop Produc. 41, 203–213. doi: 10.1016/j.indcrop.2012.04.003
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Photochem. Bull. 19, 11–15.
Gao, W., Bai, S., Li, Q., Gao, C., Liu, G., and Feili, T. (2013). Overexpression of TaLEA Gene from Tamarix androssowii improves salt and drought tolerance in transgenic poplar (Populus simonii × P. nigra). PLoS ONE 8:e67462. doi: 10.1371/journal.pone.0067462
Ghobadi, M., Taherabadi, S., Ghobadi, M. E., Mohammadi, G. R., and Jalali-Honarmand, S. (2013). Antioxidant capacity, photosynthetic characteristics and water relations of sunflower (Helianthus annuus L.) cultivars in response to drought stress. Indus. Crop Product. 50, 29–38. doi: 10.1016/j.indcrop.2013.07.009
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994.
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 7, 1–11. doi: 10.4161/psb.21949
Hiscox, J. D., and Israelstam, G. F. (1979). A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334. doi: 10.1139/b79-163
Holbrook, C. C., Ozias-Akins, P., Chu, Y., and Guo, B. (2011). Impact of molecular genetic research on peanut cultivar development. Agronomy 1, 3–17. doi: 10.3390/agronomy1010003
Jagana, S. R., Vadez, V., Bhatnagar-Mathur, P., Narasu, M. L., and Sharma, K. K. (2012). Better root:shoot ratio conferred enhanced harvest index in transgenic groundnut overexpressing the rd29A:DREB1A gene under intermittent drought stress in an outdoor lysimetric dry-down trial. J. SAT Agric. Res. 10, 1–7. Available online at: http://ejournal.icrisat.org/Volume10/Groundnut/KKS.pdf
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Khare, N., Goyary, D., Singh, N. K., Shah, P., Rathore, M., Anandhan, S., et al. (2010). Transgenic tomato cv. Pusa Uphar expressing a bacterial mannitol-1-phosphate dehydrogenase gene confers abiotic stress tolerance. Plant Cell Tiss. Organ Cult. 103, 267–277. doi: 10.1007/s11240-010-9776-7
Krishna, G., Singh, B. K., Kim, E.-K., Morya, V. K., and Ramteke, P. R. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)- A review. Plant Biotechnol. J. 13, 147–162. doi: 10.1111/pbi.12339
Liu, Y., Wang, L., Jiang, S., Pan, J., Cai, G., and Li, D. (2014). Group 5 LEA protein, ZmLEA5C, enhance tolerance to osmotic and low temperature stresses in transgenic tobacco and yeast. Plant Physiol. Biochem. 84, 22–31. doi: 10.1016/j.plaphy.2014.08.016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the method. Method 25, 402–408. doi: 10.1006/meth.2001.1262
Macková, H., Hronková, M., Dobrá, J., Turečková, V., Novák, O., Lubovská, Z., et al. (2013). Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/ dehydrogenase gene expression. J. Exp. Bot. 64, 2805–2815. doi: 10.1093/jxb/ert131
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, J. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Mehta, R., Radhakrishnan, T., Kumar, A., Yadav, R., Dobaria, J. R., Thirumalaisamy, P., et al. (2013). Coat protein-mediated transgenic resistance of peanut (Arachis hypogaea L.) to peanut stem necrosis disease through Agrobacterium-mediated genetic transformation. Ind. J. Virol. 24, 205–213. doi: 10.1007/s13337-013-0157-9
Mishra, G. P., Radhakrishnan, T., Kumar, A., Thirumalaisamy, P. P., Kumar, N., Bosamia, T. C., et al. (2015). Advancements in molecular marker development and their applications in the management of biotic stresses in peanuts. Crop Protect. 77, 74–86. doi: 10.1016/j.cropro.2015.07.019
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nahar, K., Hasanuzzaman, M., Alam, M. M., and Fujita, M. (2015). Glutathione-induced drought stress tolerance in mung bean: coordinated roles of the antioxidant defence and methylglyoxal detoxification systems. AoB Plants 7:plv069. doi: 10.1093/aobpla/plv069
Pandey, B., Sharma, P., Saini, M., Pandey, D. M., and Sharma, I. (2014). Isolation and characterization of dehydration-responsive element-binding factor 2 (DREB2) from Indian wheat (Triticum aestivum L.) cultivars. Aust. J. Crop Sci. 8, 44–54. Available online at: http://www.cropj.com/pandey_8_1_2014_44_54.pdf
Pruthvi, V., Narasimhan, R., and Nataraja, K. N. (2014). Simultaneous Expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS ONE 12:e111152. doi: 10.1371/journal.pone.0111152
Radhakrishnan, T., Murthy, T. G. K., Chandran, K., and Bandyopadhyay, A. (2000). Micro propagation in peanut (A. hypogaea L.). Biol. Plant. 43, 447–450. doi: 10.1023/A:1026743822546
Rai, G. K., Rai, N. P., Rathaur, S., Kumar, S., and Major, S. (2013). Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol. Biochem. 69, 90–100. doi: 10.1016/j.plaphy.2013.05.002
Ravikumar, G., Manimaran, P., Voleti, S. R., Subrahmanyam, D., Sundaram, R. M., Bansal, K. C., et al. (2014). Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res. 23, 421–439. doi: 10.1007/s11248-013-9776-6
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Sarkar, T., Radhakrishnan, T., Kumar, A., Mishra, G. P., and Dobaria, J. R. (2014). Heterologus expression of AtDREB1A gene in transgenic peanut conferred tolerance to drought and salinity stresses. PLoS ONE 9:e110507. doi: 10.1371/journal.pone.0110507
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Tiwari, S., Devesh, K., Mishra, A. S., Singh, P. K., and Tuli, R. (2008). Expression of a synthetic cry1EC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.). Plant Cell Rep. 27, 1017–1025. doi: 10.1007/s00299-008-0525-x
Turyagyenda, L. F., Kizito, E. B., Ferguson, M., Baguma, Y., Agaba, M., Harvey, J. J. W., et al. (2013). Physiological and molecular characterization of drought responses and identification of candidate tolerance genes in cassava. AoB Plants 5:plt007. doi: 10.1093/aobpla/plt007
Vadez, V., Rao, J. S., Bhatnagar-Mathur, P., and Sharma, K. K. (2013). DREB1A promotes root development in deep soil layers and increases water extraction under water stress in groundnut. Plant Biol. 15, 45–52. doi: 10.1111/j.1438-8677.2012.00588.x
Vadez, V., Rao, J. S., Sharma, K. K., Bhatnagar-Mathur, P., and Devi, M. J. (2007). DREB1A allows for more water uptake in groundnut by a large modification in the root/shoot ratio under water deficit. J. SAT Agric. Res. 5, 1–5.
Vivek, R. J., Tuteja, N., and Soniya, E. V. (2013). CDPK1 from ginger promotes salinity and drought stress tolerance without yield penalty by improving growth and photosynthesis in Nicotiana tabacum. PLoS ONE 8:e76392. doi: 10.1371/journal.pone.0076392
Wang, Z., Li, H., Ke, Q., Jeong, J. C., Lee, H. S., Xu, B., et al. (2014). Transgenic alfalfa plants expressing AtNDPK2 exhibit increased growth and tolerance to abiotic stresses. Plant Physiol. Biochem. 84, 67–77. doi: 10.1016/j.plaphy.2014.08.025
Wang, Z., Zhang, Y., Huang, Z., and Huang, L. (2008). Antioxidative response of metal-accumulator and non-accumulator plants under cadmium stress. Plant Soil. 310, 137–149. doi: 10.1007/s11104-008-9641-1
Werner, T., Nehnevajova, E., Kollmer, I., Novák, O., Stmad, M., Kramer, U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Keywords: abiotic stress tolerance, drought stress, growth-related traits, groundnut, genetic transformation, regeneration
Citation: Sarkar T, Thankappan R, Kumar A, Mishra GP and Dobaria JR (2016) Stress Inducible Expression of AtDREB1A Transcription Factor in Transgenic Peanut (Arachis hypogaea L.) Conferred Tolerance to Soil-Moisture Deficit Stress. Front. Plant Sci. 7:935. doi: 10.3389/fpls.2016.00935
Received: 15 March 2016; Accepted: 13 June 2016;
Published: 28 June 2016.
Edited by:
Guo-Liang Jiang, Virginia State University, USAReviewed by:
Taras P. Pasternak, University of Freiburg, GermanyCharles Y. Chen, Auburn University, USA
Copyright © 2016 Sarkar, Thankappan, Kumar, Mishra and Dobaria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhakrishnan Thankappan, radhakrishnan.nrcg@gmail.com
 Tanmoy Sarkar
Tanmoy Sarkar Radhakrishnan Thankappan
Radhakrishnan Thankappan Abhay Kumar
Abhay Kumar Gyan P. Mishra
Gyan P. Mishra Jentilal R. Dobaria
Jentilal R. Dobaria