- 1Department of Plant Science and Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 2Plant Genomics and Breeding Institute, Seoul National University, Seoul, South Korea
- 3Center for Applied Genetic Technologies, University of Georgia, Athens, GA, USA
- 4Department of Crop Science and Biotechnology, Dankook University, Cheonan, South Korea
The depletion of the ozone layer in the stratosphere has led to a dramatic spike in ultraviolet B (UV-B) intensity and increased UV-B light levels. The direct absorption of high-intensity UV-B induces complex abiotic stresses in plants, including excessive light exposure, heat, and dehydration. However, UV-B stress signaling mechanisms in plants including soybean (Glycine max [L.]) remain poorly understood. Here, we surveyed the overall transcriptional responses of two soybean genotypes, UV-B-sensitive Cheongja 3 and UV-B-resistant Buseok, to continuous UV-B irradiation for 0 (control), 0.5, and 6 h using RNA-seq analysis. Homology analysis using UV-B-related genes from Arabidopsis thaliana revealed differentially expressed genes (DEGs) likely involved in UV-B stress responses. Functional classification of the DEGs showed that the categories of immune response, stress defense signaling, and reactive oxygen species (ROS) metabolism were over-represented. UV-B-resistant Buseok utilized phosphatidic acid-dependent signaling pathways (based on subsequent reactions of phospholipase C and diacylglycerol kinase) rather than phospholipase D in response to UV-B exposure at high fluence rates, and genes involved in its downstream pathways, such as ABA signaling, mitogen-activated protein kinase cascades, and ROS overproduction, were upregulated in this genotype. In addition, the DEGs for TIR-NBS-LRR and heat shock proteins are positively activated. These results suggest that defense mechanisms against UV-B stress at high fluence rates are separate from the photomorphogenic responses utilized by plants to adapt to low-level UV light. Our study provides valuable information for deep understanding of UV-B stress defense mechanisms and for the development of resistant soybean genotypes that survive under high-intensity UV-B stress.
Introduction
The depletion of the ozone layer in the stratosphere has led to increased levels terrestrial ultraviolet B (UV-B, 280–315 nm) radiation until now since the late 1970s (Searles et al., 2001; Austin and Wilson, 2006). Furthermore, it is causing periodic or unpredictable spikes in UV-B intensity in the polar and temperate zones (Kerr and McElroy, 1993). The Montreal Protocol has been effective in stabilizing UV-B exposure since the mid-1990s but the recovery of the ozone layer to pre-1980 levels may require several decades (McKenzie et al., 2011). UV-B photons can function as both environmental stressors and developmental signals in plants (Britt, 1996). Since sunlight is required for photosynthesis, increased UV-B intensity inevitably threatens plant viability, as most vascular plants cannot adapt to such an influx of UV-B (Jordan, 2002; Paul and Gwynn-Jones, 2003). High level UV-B radiation causes physiological damage, such as reduced photosynthetic capacity and impaired pollen fertility, as well as morphological changes including plant stunting, leaf discoloration, and reduced biomass and seed yields (Frohnmeyer and Staiger, 2003; Lytvyn et al., 2010).
Despite the complex effects of UV-B radiation on plants, recent studies have revealed a series of components implicated in UV-B-specific photomorphogenic (non-damaging) regulation in the circadian rhythm pathway (Ulm and Nagy, 2005). These components include the UV-B photoreceptor UV RESISTANCE LOCUS8 (UVR8), the E3 ubiquitin ligase (transducin/WD40 repeat-like superfamily protein) CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), the basic leucine-zipper transcription factor ELONGATED HYPOCOTYL5 (HY5), and its interacting partner LONG HYPOCOTYL5-LIKE (HYH) (Ulm, 2003). In addition, CHALCONE SYNTHASE (CHS) and other genes involved in the biosynthesis of secondary metabolites (e.g., flavonoids and phenylpropanoids) are positively activated in response to UV-B radiation (Heijde and Ulm, 2012). On the other hand, UV-B radiation above ambient level, like other environmental stresses, elicits nonspecific (genotoxic) damage responses that trigger stress defense signaling (Jenkins, 2009). In Arabidopsis thaliana, reactive oxygen species (ROS) production are induced by high dose UV-B irradiation (Heijde and Ulm, 2013; Parihar et al., 2015). Genes activated by stress responses to high level UV-B are distinct from those playing a role in photomorphogenesis specific to low dose UV-B (Müller-Xing et al., 2014). However, the mechanism by which UV-B stress triggers the intracellular defense signaling pathway remains poorly understood (Müller-Xing et al., 2014).
Annual soybean [Glycine max (L.) Merr.] is one of the most important crops that were used as food, energy, and industrial resources worldwide. Only a few studies have compared the morphological, anatomical, and biochemical differences between UV-B-sensitive and -resistant soybean cultivars (Essex vs. Williams) in response to enhanced UV-B radiation (Murali et al., 1988; Jordan, 1993, 2002; Grant et al., 2010). Recently, quantitative trait loci associated with resistance to supplementary UV-B treatment were localized between Satt495 and Satt238 on chromosome 19 using a recombinant inbred line population of Keunol (UV-B sensitive) x Iksan10 (UV-B resistant) (Shim et al., 2015; Lee et al., 2016). Meanwhile, our previous study showed the evaluation of a total of 140 genotypes including 94 G. max and 46 Glycine soja accessions for their sensitivity to supplemental UV-B radiation, resulting in the selection of the most resistant Buseok and the most sensitive Cheongja 3 (Kim et al., 2015). This UV-B resistant genotype Buseok will be a valuable plant material not only for breeding of UV-B resistant cultivars but for molecular genetic study on UV-B resistance mechanisms.
The present study was conducted to figure out how soybean copes with UV-B of high intensity as environmental stress at a transcriptomic level and to verify if UV-B defense pathways are dependent on the UV-B-specific photomorphogenic regulation or not. We surveyed the overall transcriptional responses of two soybean genotypes, UV-B-sensitive Cheongja 3 and UV-B-resistant Buseok, to continuous UV-B irradiation for 0 (control), 0.5, and 6 h at high fluence rates. We compared the transcript abundance between the se genotypes in response to supplementary UV-B irradiation, as well as between control and UV-B treatments in each genotype. The differentially expressed genes (DEGs) were subjected to further filtering using a set of UV-B related genes from A. thaliana to identify DEGs that are most likely involved in UV-B stress defense. Furthermore, the biological functions of four DEGs were confirmed using A. thaliana knock-out mutants. The results of this study provide insights into the molecular basis of the capacity for plants to tolerate UV-B stress at high fluence rates.
Materials and Methods
Plant Materials and Growth Conditions
UV-B-sensitive Cheongja 3 and UV-B-resistant Buseok (IT162669), which were identified as the most sensitive and resistant soybean genotypes to supplementary UV-B irradiation, respectively (Kim et al., 2015), were used in this study. Under 6 h of UV-B treatment, Buseok had only one or two slightly yellowish leaves and still showed vigorous growth, whereas in Cheongja 3, most leaves turned yellow with red spots, ultimately leading to defoliation (Figure S1). Seeds of both soybean genotypes were planted in plastic pots (18 cm diameter, 20 cm deep) in a 1:1:1 mixture of field soil, desalinated sand, and commercial peat soil in a greenhouse at Seoul National University Experimental Farm. One plant per pot was grown under a natural photoperiod of 11.5–14.5 h per day using standard cultivation methods.
UV-B Treatment
For UV-B treatment, soybean plants were divided into three groups; no UV-B treatment (0 h) as a control, 0.5- and 6-h UV-B treatments). Supplementary UV-B irradiation was conducted at V4 growth stage as previously described (Kim et al., 2015). UV-B irradiation began at 9:00 am, and the plants were exposed to prolonged UV-B stress for 0.5 and 6 h. The intensity of 0.5 h UV-B irradiation was equivalent to 11.5 kJ/m2 daily soybean UV-B biological effective dose (UV-BBE), and the 6 h treatment was equivalent to a dose of irradiation that was 12-times higher than daily UV-BBE (Caldwell, 1971). For Illumina RNA sequencing, the uppermost trifoliate leaves were collected from UV-B-treated and non-treated plants. At each sampling time, the leaf tissues from four pots per group of each genotype were pooled together as one biological replicate, frozen immediately in liquid nitrogen, and stored at −80 until use. Thus, a total of six leaf samples (three treatments [0, 0.5, and 6 h] × two genotypes [Cheongja 3 and Buseok] × one biological replicate) were collected for RNA-seq analysis.
RNA Isolation and RNA Sequencing using Illumina HiSeq
Total RNA was extracted from the leaf samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cDNA library for Illumina HiSeq 2000 sequencing was constructed using an mRNA-seq sample preparation kit (TruSeq® RNA Sample Prep Kit v2, Cat.8207217, Illumina Inc., San Diego, CA, USA), including mRNA purification, cDNA synthesis, end-repair of cDNA, adaptor ligation, and cDNA amplification. Six cDNA libraries were pooled at equal molar concentration for loading onto two lanes of an Illumina flow cell and then merged for data analysis. Sequencing runs were performed in paired-end mode for 10l cycles using the Illumina HiSeq 2000 platform.
Read Alignment and RNA-Seq Analysis
The 100 bp paired-end sequence reads were mapped to the soybean reference genome (Glyma v1.1) from Phytozome (http://www.phytozome.net/soybean) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) and TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) in default modes (Langmead et al., 2009; Trapnell et al., 2009). The Cufflinks program was used to assemble gene transcripts and to normalize transcript abundance in terms of fragments per kilobase pair of transcript per million mapped reads (FPKM) (Trapnell et al., 2012). The Cuffdiff program (http://cufflinks.cbcb.umd.edu/index.html) was used to test statistically significant differences in transcript expression in seven pairs of comparisons, which consisted of three comparisons of Cheongja 3 vs. Buseok under control conditions and two UV-B treatments, and four comparisons of control vs. UV-B treatments in the two genotypes (Figure S2). Significant DEGs were detected using the following criteria: (i) absolute fold-change > 1 and (ii) q-value (false discovery rate [FDR]) <0.05 and p-value < 0.004. R studio (https://www.rstudio.com/, version 3. 3. 0) was used to run custom R scripts to perform hierarchical cluster analysis of DEG expression and to construct heat maps.
Survey of A. thaliana UV-B Related Genes
UV-B related genes from A. thaliana were surveyed using Gene Ontology (GO) (http://amigo.geneontology.org/amigo/search/ontology?q=UV-B) and The Arabidopsis Information Resource (TAIR) (https://www.Arabidopsis.org/) database. In addition, a list of A. thaliana genes shown to be involved in the UV-B response was compiled from the literature; 194 UV-B-related A. thaliana genes were ultimately obtained (Figure S2, Supplementary Table 1). To detect genes that function in networks with the UV-B related genes, the set of selected UV-B related genes was analyzed with Aranet (http://www.functionalnet.org/aranet/), a probabilistic functional gene network for A. thaliana, resulting in the identification of 4666 genes predicted to respond directly or indirectly to UV-B (Figure S2, Supplementary Table 2). A total of 15,074 soybean homologs of these A. thaliana genes were identified using Blast analysis. Based on this soybean gene list, DEGs putatively involved in the defense response to UV-B stress were identified (Figure S2).
Functional Classification of DEGs by BINGO and KEGG
To better understand the biological functions of the DEGs in response to UV-B, enrichment of GO categories among the DEGs was assessed using BINGO software (http://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) (Maere et al., 2005). Significantly over-represented GO categories were visualized in Cytoscape (http://www.cytoscape.org). The biochemical pathways involving the DEGs were predicted using Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/).
Quantitative Reverse-Transcription PCR Validation of DEGs
Gene-specific primers were designed based on the nucleotide sequences of the chosen DEGs for qRT-PCR analysis using Primer3 software (http://primer3plus.com/web_3.0.0/primer3web_input.htm) (Supplementary Table 3). Total RNA from six samples (Cheongja 3 and Buseok; control, 0.5, and 6 h UV-B treatments) was used to synthesize cDNA using a Bio-Rad iScript™ cDNA Synthesis Kit (Cat. 170-8891, Hercules, CA, USA). The synthesized cDNAs were used for qRT-PCR with a Bio-Rad iQ™ SYBR Green Supermix Kit (Cat. 170-8882) using a LightCycler® 480 (Roche Diagnostics, Laval, QC, Canada). The qRT-PCR reaction mixtures (total volume of 20 μl) contained 100 ng of cDNA, each primer at 300 μM, 8 μl of sterile water, and 10 μl of Bio-Rad iQ™ SYBR Green Supermix. The amplification conditions were as follow: 5 min denaturation at 95°C followed by 40 cycles of 95°C for 10 s and 60°C for 1 min. The samples were analyzed in triplicate to ensure statistical significance, and the tubulin gene was used as a reference gene for normalization of target gene expression in soybean. Data were analyzed based on the stable expression level of the reference gene according to the method of Livak and Schmittgen (2001). To assess treatment effects on soybean genotypes, significances were analyzed with Fisher's least significant difference tests (P < 0.05) using the Statistical Analysis System (SAS 9.4 for window, SAS Institute Inc., Cary, NC, USA).
Functional Validation of DEGs Using Arabidopsis Knock-Out Mutants
Seeds of knock-out mutants for the target genes were obtained from the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu/) at Ohio State University, USA. Seven mutant lines were utilized, including mutants for five members of TIR-NBS-LRR family (AT1G64070.1; SALK_042846C, AT5G17680.1; SALK_004241C, AT5G36930.1; SALK_124056C, AT5G41540.1; SALK_034471C, and AT4G36150.1; SALK_084909C), one DGK mutant (AT5G07920.1; SALK_033664C), and one PIP5K mutant (AT1G34260.1; SALK_047604C). To identify homozygous mutants for the target genes, seeds incubated at 4°C were sown in commercial peat soil and grown under 16/8 h light conditions in a growth chamber for 14 days. Homozygous mutant plants were confirmed by RT-PCR using primers designed based on the inserted T-DNA and target gene DNA sequences, resulting in the identification of only four knock-out mutants (Figure S3; Supplementary Table 4). Both knock-out mutant and Columbia ecotype (Col-0) seeds were sterilized and incubated at 4°C in the dark for 4 days to synchronize germination and to ensure uniform growth, followed by cultivation under 16 h white fluorescent light for 7 days in a growth chamber. As a pilot UV-B irradiation test to determine suitable irradiation times for A. thaliana, wild-type (Col-0) plants were subjected to different UV-B treatments (1, 2, 4, and 8 h); 4 h UV-B treatment was determined to be suitable (Figure S4). Seven-days-old knock-out mutants were treated with 4 h UV-B irradiation at 22°C. For qRT-PCR analysis, rosette leaves were collected from UV-B-treated and non-treated mutant plants, frozen immediately in liquid nitrogen, and stored at −80°C until use. The specific primers for qRT-PCR were designed using primer 3 (Supplementary Table 5).
Results
RNA-seq Analysis and DEG Identification Related to UV-B Stress in Cheongja 3 and Buseok
To compare transcriptomic variation in the soybean lines in response to UV-B stress, Cheongja3 and Buseok leaves were collected after continuous UV-B exposure at a high fluence rate for 0 (control), 0.5, and 6 h, with a total of six samples. Using Illumina HiSeq 2000 system, 0.3 billion 100 bp paired-end reads were produced, ranging from 47 to 62 million reads per sample (Supplementary Table 6), which were mapped against the soybean reference sequence. The RNA-seq analysis workflow depicted in Figure S2 was implemented to analyze the sequencing data. Of the total reads, 78% were properly mapped to the reference sequence, resulting in approximately 25-fold average coverage (Supplementary Table 6). About 20% unmapped reads appears to be attributable to either misassembled or absent sequences in the reference assembly.
To identify DEGs related to UV-B stress, we compared transcript abundances among the six samples from two soybean genotypes differing in UV-B tolerance based on FPKM values. Two-way comparison data analysis was used to investigate transcriptomic variations, representing the comparison (i) between Cheongja 3 and Buseok by UV-B treatment and (ii) between control and UV-B treatments by genotype (Figure S2). To further identify DEGs in response to supplementary UV-B irradiation, we then utilized a set of 4666 A. thaliana genes predicted to be implicated in UV-B stress defense, either directly or indirectly, via Aranet (Figure S2; Supplementary Tables 1, 2). Homology comparative analysis revealed that 1875 DEGs detected in the Cheongja 3 vs. Buseok comparison were orthologs of UV-B-related Arabidopsis genes (Figure S2). In the control vs. treatment comparison, 228 DEGs in Buseok and 129 in Cheongja 3 were found to be homologous to the UV-B related Arabidopsis genes (Figure S2; Supplementary Table 7–9).
Using the final sets of DEGs, we generated Venn diagrams to identify treatment- or genotype-specific DEGs and common DEGs (Figure 1A). A larger number of DEGs were specific to each genotype and each UV-B treatment time. In the Cheongja 3 vs. Buseok comparison, 499 DEGs specific to the controls were considered to be due to differences in the genetic backgrounds between the genotypes and were thus excluded from further functional classification. Of the 1376 DEGs in Buseok relative to Cheongja 3 (Supplementary Table 9), 511 were upregulated and 345 were downregulated under 0.5 h UV-B treatment (Figure 1B). Under 6 h UV-B treatment, the number of up- and downregulated DEGs was similar. The control vs. treatment comparisons by genotype showed that Buseok had more DEGs than Cheongja 3 both for 0 vs. 5 h and 0 vs. 6 h (Figure 1B). In Cheongja 3, the number of DEG is higher in upregulation than downregulation for 0 vs. 6 h. There were 149 and 50 Buseok- and Cheongja 3-specific DEGs, respectively (Figure 1A). Most DEGs in both genotypes were upregulated by 0.5 h UV-B treatment but more strongly downregulated by 6 h UV-B exposure (Figure 1B).
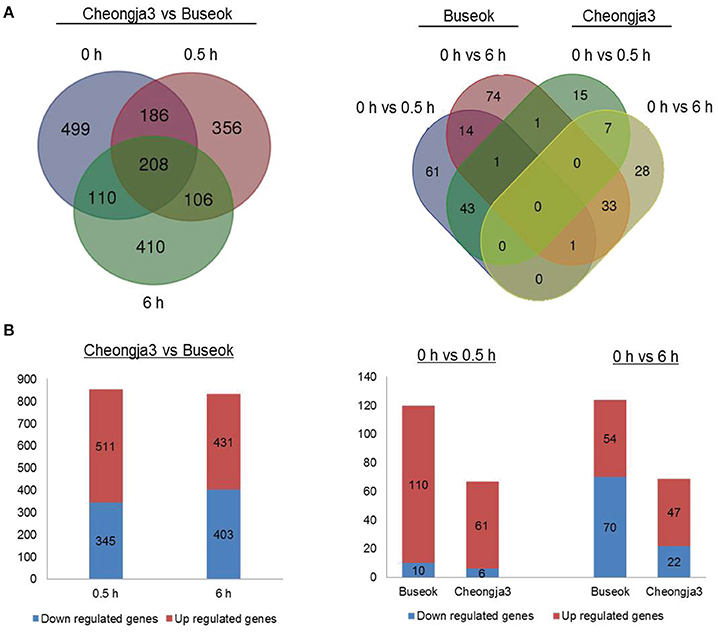
Figure 1. Number of differentially expressed genes (DEGs). (A) Left: DEGs between Cheongja 3 and Buseok under control, 0.5, and 6 h UV-B treatments. Right: DEGs between the control and UV-B treatments in Cheongja 3 and Buseok. (B) Up- and down-regulated DEGs detected in each comparison.
Functional Classification of DEGs by Bingo and KEGG
We investigated the biological significance of the changes in transcript abundance provoked by UV-B irradiation stress in soybean. To address the functional distribution of the identified DEGs, we performed GO term enrichment using Cytoscape plug-in BINGO. GO assignment of 690 DEGs among the 1376 DEGs identified in the Cheongja 3 vs. Buseok comparison by UV-B treatment resulted in significant overrepresentation of 31 GO terms, including 17 in the biological process category and 14 in the metabolic process category (Figure 2; Supplementary Table 10). In the biological process category, over-represented GO terms were mainly classified into four clusters, as shown in Figure 2, including cell morphogenesis, immune response, signaling, and apoptosis. A large portion of the DEGs involved in the GO clusters immune response, signaling, and apoptosis were overlapping, i.e., toll/interleukin-1 nucleotide-binding-site leucine-rich repeat (TIR-NBS-LRR) genes. The GO terms over-represented in metabolic process were primarily divided into five clusters including light harvesting system, fatty acid biosynthetic process, small molecule biosynthetic process, ROS metabolic process, and oxidation reduction. From the control vs. treatment comparisons in UV-B-resistant Buseok, 176 DEGs induced by 0.5 h UV-B treatment were grouped into six GO clusters, including biological regulation, immune response, signaling, apoptosis, ion transport, and polysaccharide metabolic process (Figure S5; Supplementary Table 11). However, no GO term was over-represented by the 124 DEGs in Buseok in response to 6 h UV-B treatment. In the control vs. treatment comparisons in UV-B-sensitive Cheongja 3, 9 DEGs under 0.5 h UV-B treatment were grouped into the GO term metal ion transport (GO:0030001) and 26 DEGs under 6 h UV-B treatment were grouped into oxidation reduction (GO:0055114) (Figure S6; Supplementary Table 12). According to BINGO analysis, the GO terms involved in immune response, cell death, and signaling were the most dominant.
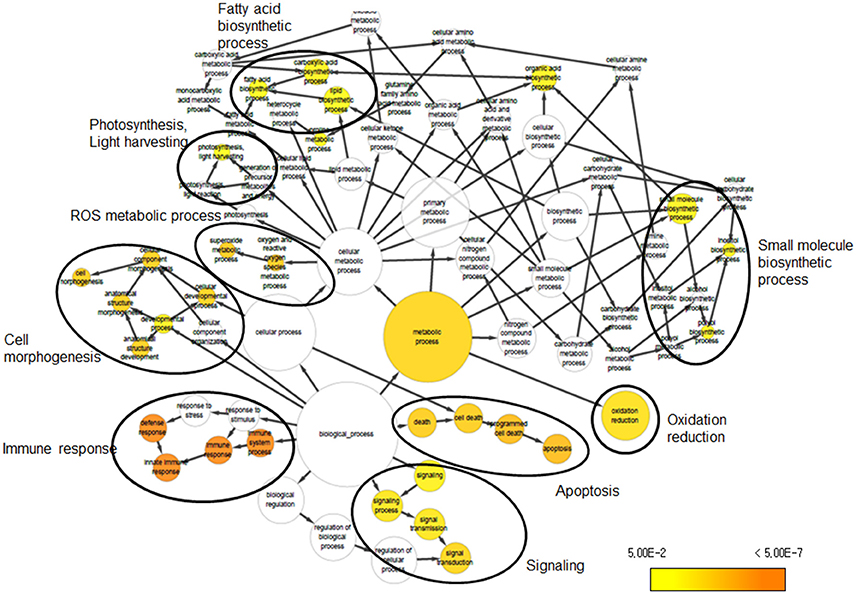
Figure 2. Over-represented GO terms of DEGs that were identified from the comparison between Cheongja 3 and Buseok under UV-B treatments.
KEGG pathway mapping of the 1376 DEGs identified in the Cheongja 3 vs. Buseok comparison under UV-B treatments revealed the involvement of 101 biological pathways (Supplementary Table 13). The 228 and 129 DEGs in the control vs. treatment comparisons in Buseok and Cheongja 3 were assigned to 28 and 52 KEGG pathways, respectively (Supplementary Tables 14, 15). Interestingly, phosphatidylinositol signaling system (gmx04070) and mTOR signaling pathway (map04150), which are involved in environmental information processing and signal transduction, were also identified.
Based on the results of BINGO and KEGG analysis, we specifically focused on DEGs in over-represented GO and KEGG terms such as cell death and immune system, stress defense signaling, and ROS metabolism, as described below. We also investigated the expression patterns of genes in the photosystem and circadian rhythm categories.
Photosystem and Circadian Rhythm
Supplemental UV-B radiation downregulates genes encoding several key photosynthetic proteins, including small subunits of Rubisco, subunits of ATP synthase, and chlorophyll a/b binding protein of the light harvesting antenna complex of photosystem II (Jenkins, 2009). In the present study, there are no photosynthetic DEGs by UV-B treatments in both Buseok and Cheongja 3 compared to the control (Supplementary Tables 7–9, 16). However, differences between the two soybean genotypes were observed in the transcript abundance of chlorophyll a/b binding protein and photosystem light harvesting complex genes under both control and elevated UV-B conditions (Supplementary Table 16). Inherent variations in the expression of two genes encoding light harvesting complex subunits were detected between Buseok and Cheongja 3 under ambient light (control). In addition, the expression patterns of six genes associated with photosystem during UV-B irradiation significantly differed between genotypes, which included two homologs (Glyma09g0826 and Glyma15g19810) for photosystem I light harvesting complex gene 6, one (Glyma04g33360) for light-harvesting chlorophyll-protein complex I subunit A4, one (Glyma16g27995) for light-harvesting chlorophyll-protein complex II subunit B1, and one (Glyma11g35130) for light harvesting complex photosystem II (Supplementary Table 16). These results indicate that the transcriptional regulation of photosynthetic genes in response to supplementary UV-B irradiation is different between soybean genotypes.
Photomorphogenic UV-B responses are specifically mediated by (but are not independent of) other aspects of light signaling, including photoreceptors and circadian rhythms. We observed significant differences in the transcript abundance of genes involved in UV-B specific photomorphogenic pathways between UV-B-resistant Buseok and UV-B-sensitive Cheongja 3. Specifically, HY5 (Glyma08g41450) and COP1 (Glyma14g05430) were upregulated by 6 h UV-B treatment (Supplementary Table 9). Most DEGs encoding CHS proteins, which were expressed at lower levels in Buseok compared to Cheongja 3 under control conditions, were upregulated by UV-B irradiation (Supplementary Table 16). Additional key genes involved in the circadian rhythm pathway, such as PHYTOCHROME A, GIGANTEA PHYTOCHROME INTERACTING FACTOR 3, and PSEUDO-RESPONSE REGULATOR, were also differentially expressed in response to supplementary UV-B exposure (Supplementary Table 16). In the control vs. treatment comparisons by genotype, UVR8, HY5, and COP1 were not detected among DEGs (data not shown).
Cell Death and Immune Response
To investigate which components in defense signaling pathways are activated in response to UV-B stress, we investigated significant differences in the expression of genes participating in programmed cell death and the immune system. The comparison of Cheongja 3 vs. Buseok under UV-B treatments revealed differential expression of 20 TIR-NBS-LRR genes, 11 and 9 of which were upregulated in Buseok and Cheongja 3, respectively (Table 1; Figure 3). Seven additional TIR-NBS-LRR and two coiled-coil-nucleotide-binding-site leucine-rich repeat (CC-NBS-LRR) genes were induced by 0.5 h UV-B irradiation only in Buseok (Table 1), while none of these genes were induced by 6 h treatment.
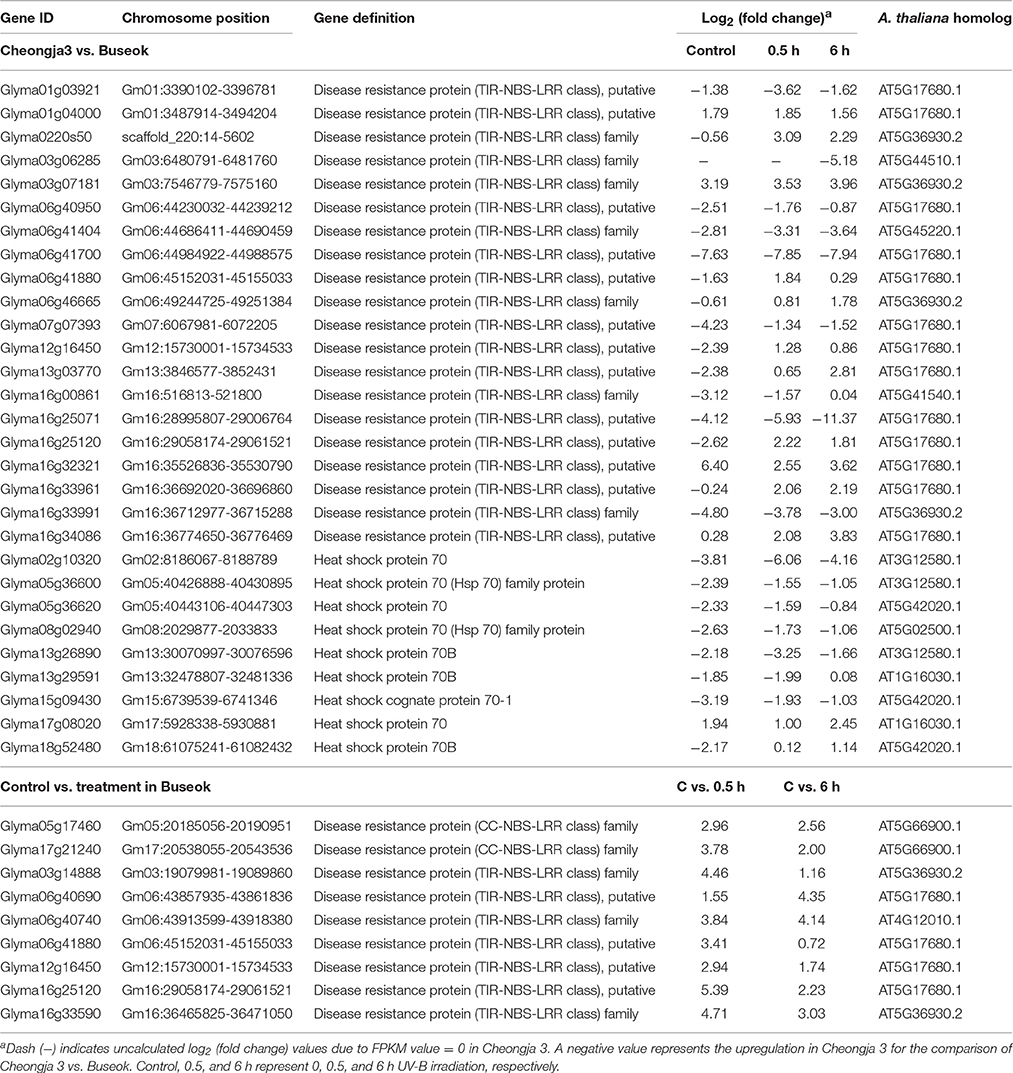
Table 1. Differential expressed genes encoding TIR/CC-NBS-LRRs and heat shock proteins identified by the transcriptomic comparisons of UV-B sensitive Cheongja 3 vs. resistant Buseok and control vs. UV-B treatments in Buseok.
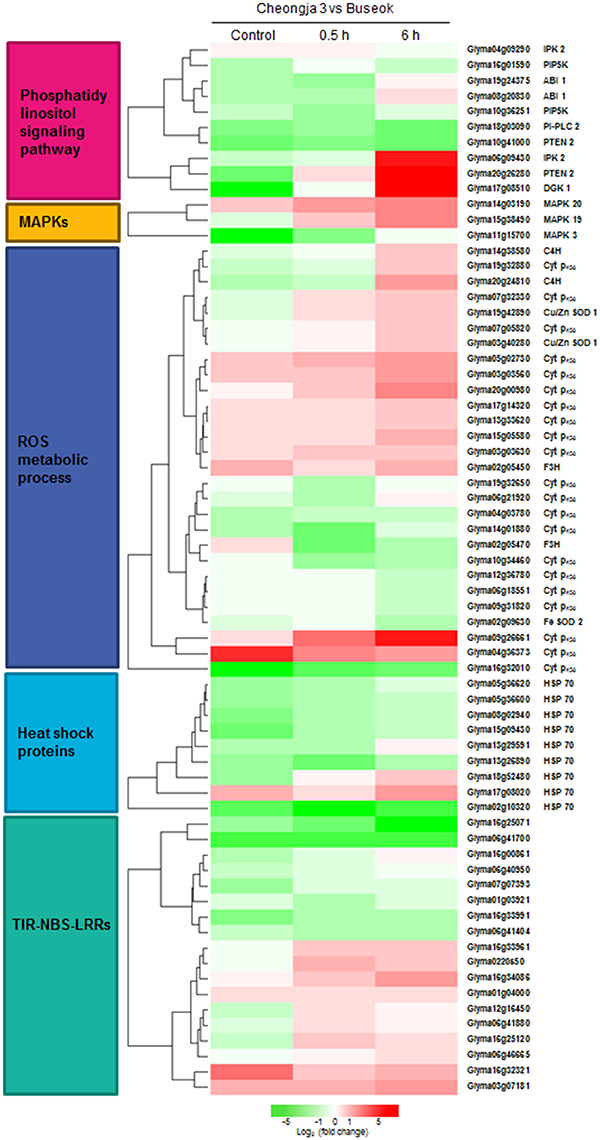
Figure 3. Heatmap showing the differential expression levels of genes encoding components involved in the over-represented GO and KEGG terms between Cheongja 3 and Buseok under control, 0.5, and 6 h UV-B treatments.
Nine members of the heat shock 70 (HSP70) protein family were differentially expressed in response to UV-B irradiation in Buseok relative to Cheongja 3, six of which were down-regulated (Table 1; Figure 3). However, there was no significant difference in the expression patterns of HSP70 genes between control and 0.5 or 6 h UV-B treatment in Buseok or Cheongja 3.
Stress Defense Signaling
Genome-wide transcript analysis in response to supplemental UV-B irradiation revealed DEGs involved in the phosphatidylinositol signaling pathway (Table 2). The phosphatidylinositol signaling pathway generates phosphatidic acid (PA) from structural phospholipids as a secondary messenger. Under UV-B stress, transcripts for phosphatidylinositol-4-phosphate 5-kinase (PIP5K) family protein were more abundant in Buseok than in Cheongja 3 (Table 2); this protein generates phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] as a substrate of phospholipase C (PLC) from phosphatidylinositol 4-phosphate (Ptdins4P) in the plasma membrane (Mueller-Roeber and Pical, 2002). Also, PtdIns(4,5)P2 is produced from the phosphorylation of phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] by phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase (PTEN 2), for which one gene (Glyma20g26280) was upregulated in Buseok under UV-B irradiation (Table 2). PtdIns(4,5)P2 is hydrolyzed into inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG) by phosphatidylinositol-specific PLC (PI-PLC) proteins. One DEG (Glyma18g03090) encoding PI-PLC was upregulated in Cheongja 3 under 6 h UV-B treatment relative to Buseok (Table 2). In UV-B-resistant Buseok, PI-PLC genes were expressed at low levels and independent of UV-B treatment. Meanwhile, two DEGs (Glyma17g08510 and Glyma06g39760) for diacylglycerol kinase (DGK) family proteins were upregulated in Buseok by UV-B stress (Table 2); these proteins convert DAG produced by PI-PLC to PA (Laxalt and Munnik, 2002). The coupling of DGK to the activation of PI-PLC may be induced in response to UV-B stress in Buseok, leading to increased levels of PA in the cells, thereby activating several downstream defense responses.
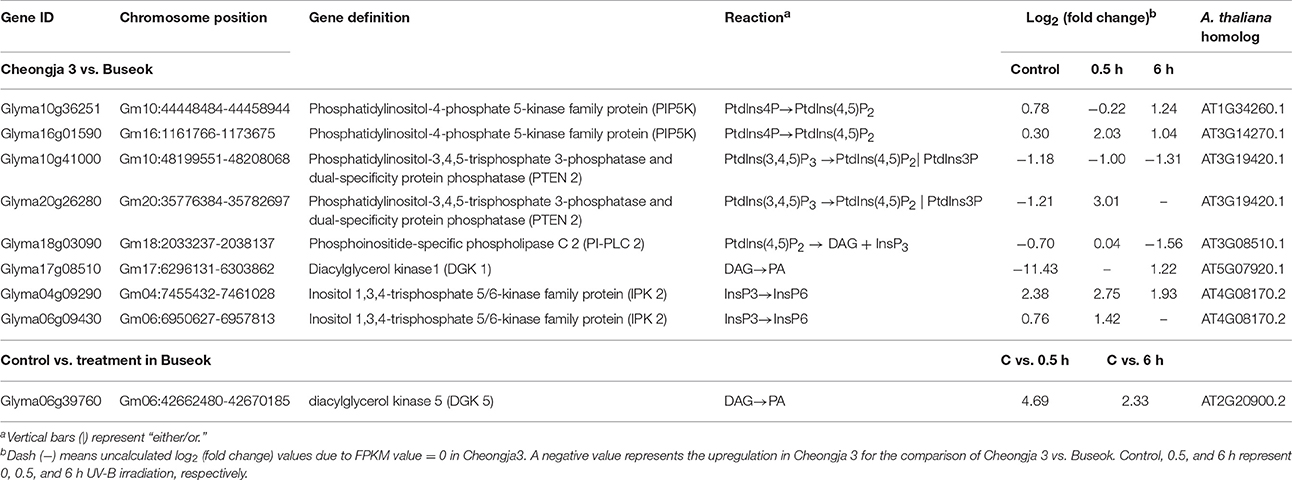
Table 2. Differential expressed genes encoding five enzymes, PIP5K, PTEN 2, PI-PLC 2, DGK, and IPK2 in the phosphatidic acid signaling pathway identified by the transcriptomic comparisons of UV-B sensitive Cheongja 3 vs. resistant Buseok and control vs. UV-B treatments in Buseok.
InsP3 generated via the hydrolysis of PI-PLC diffuses into the cytosol and is involved in the release of Ca2+ from intracellular stores (Ruelland et al., 2015). Owing to a lack of InsP3 receptors in plants, InsP3 is converted into the more phosphorylated forms of inositol, i.e., tetra, penta, and hexaphosphates (InsP4, InsP5, and InsP6), through further phosphorylation steps involving at least two types of inositol polyphosphate 2-kinase (IPK1) and inositol polyphosphate kinase 2 (IPK2, synonym for inositol 1,3,4-trisphosphate 5/6-kinase) (Munnik and Vermeer, 2010; Zhou et al., 2012; Sparvoli and Cominelli, 2015). Among these enzymes, Buseok exhibited enhanced expression of two IPK2 family genes under 0.5 and 6 h UV-B irradiation compared to Cheongja 3 (Table 2). Therefore, in UV-B-resistant Buseok, the genes for four enzymes involved in the PA-dependent signaling pathway were upregulated in response to UV-B stress (Figure 3).
We also identified some DEGs encoding putative target proteins that interact with PA, including two DEGs (Glyma08g20830, Glyma19g24375) encoding protein phosphatase 2 family proteins, which were upregulated in Buseok under 0.5 h UV-B treatment (Figure 3; Supplementary Table 16); these proteins carry a PA binding motif. One example of a functionally characterized protein phosphatase targeted by PA is the protein phosphatase 2C ABI1 (ABA insensitive 1), which is bound by PA to negatively regulate ABA signaling (Zhang et al., 2004). We detected differential transcript accumulation of three genes encoding MAPKs in response to UV-B stress between Cheongja 3 and Buseok (Figure 3; Supplementary Table 16); MAPK 19 (Glyma15g38490) and MAPK 20 (Glyma14g03190) were upregulated in Buseok under 6 h UV-B treatment and MAPK 3 (Glyma11g15700) was upregulated in Cheongja 3 under 0.5 h UV-B treatment (Figure 3; Supplementary Table 16). In both Buseok and Cheongja 3, however, no significant difference in MAPK expression was detected in the control vs. UV-B treatment comparison.
ROS Production and Scavenging
Abiotic stresses including UV-B stress induce ROS production and scavenging. Four genes encoding copper/zinc superoxide dismutase (Cu/Zn SOD) family proteins, which catalyze the dismutation of superoxide anion () to oxygen (O2) and hydrogen peroxide (H2O2), were upregulated in Buseok in response to UV-B stress, while in Cheongja 3, the iron (Fe)-SOD gene was upregulated in response to 6 h UV-B treatment. Among the genes assigned to the GO term “oxidation reduction,” 21 cytochrome P450 (Cyt P450) family genes were identified, 12 and 9 of which were upregulated in Buseok and Cheongja 3, respectively (Figure 3; Supplementary Table 16). Six of eight Cyt P450 DEGs were downregulated in Cheongja 3 under 6 h UV-B treatment compared to the control (Supplementary Table 17). NAD(P)H-dependent electron transport involving cytochrome P450 produces in the endoplasmic reticulum (Sharma et al., 2012). Genes of the Cyt P450 family 76 subfamily C2 and the Cyt P450 family 706 subfamily A are induced by UV-C irradiation (Narusaka et al., 2004). To avoid injury from ROS overproduction, ROS scavenging or detoxification is performed by antioxidative systems consisting of both nonenzymatic and enzymatic antioxidants (Sharma et al., 2012). In Buseok, we identified upregulated DEGs involved in flavonoid biosynthesis to produce phenolic compounds with antioxidant properties, such as anthocyanidin and tannin, including genes encoding flavanone-3-hydroxylase (F3H) and cinnamate-4-hydroxylase (C4H) (Supplementary Table 18). By contrast, a gene encoding the enzymatic oxidant ascorbate peroxidase was upregulated in Cheongja 3. Under 6 h UV-B treatment, genes encoding glutathione peroxidase were also upregulated only in Cheongja 3 compared to the control (Supplementary Table 17).
RNA-seq Validation by qRT-PCR
Using qRT-PCR, we confirmed that five genes (Glyma03g07121, Glyma06g41880, Glyma12g16450, Glyma16g34086, and Glyma19g07650) in the TIR-NBS-LRR family were upregulated in Buseok in response to 0.5 h UV-B irradiation (Figures 4A–E). The TIR-NBS-LRRs' expression patterns obtained by qRT-PCR also showed significantly increased transcript abundance in Buseok by UV-B treatment. Interestingly, transcript accumulation of Glyma03g07121 (TIR-NBS-LRR) was not observed in Cheongja 3 by qRT-PCR or RNA-seq. In addition, the expression profiles of individual DEGs encoding IPK2 (Glyma06g09430), PIP5K (Glyma10g36251), DGK 1 (Glyma17g08510) and DGK 5 (Glyma06g39760) in the PA signaling pathway were obtained by qRT-PCR (Figures 4F–I). Significant upregulation of IPK2 (Glyma06g09430), PIP5K (Glyma10g36251) and DGK 5 (Glyma06g39760) was observed in Buseok compared to Cheongja 3 under UV-B stress. DGK 1 (Glyma17g08510) was significantly activated only in Buseok by 6 h UV-B irradiation relative to control. Finally, we investigated the expression patterns of three DEGs encoding Cyt P450 (Glyma06g21920), Cu/Zn SOD (Glyma11g19840), and C4H (Glyma14g38580), which are involved in ROS production and scavenging, using qRT-PCR (Figures 4J–L). Two genes Cu/Zn SOD (Glyma11g19840) and C4H (Glyma14g38580), except Cyt P450 (Glyma06g21920) showed significant higher expression in Buseok by both RNA-seq and qRT-PCR analyses, There was no significantly differential expression of Cyt P450 (Glyma06g21920) between Cheongja 3 and Buseok but 6 h UV-B treatment induced upregulation of Cyt P450 in both genotypes. All but one of the gene expression patterns measured by qRT-PCR agreed with ones analyzed by RNA-seq.
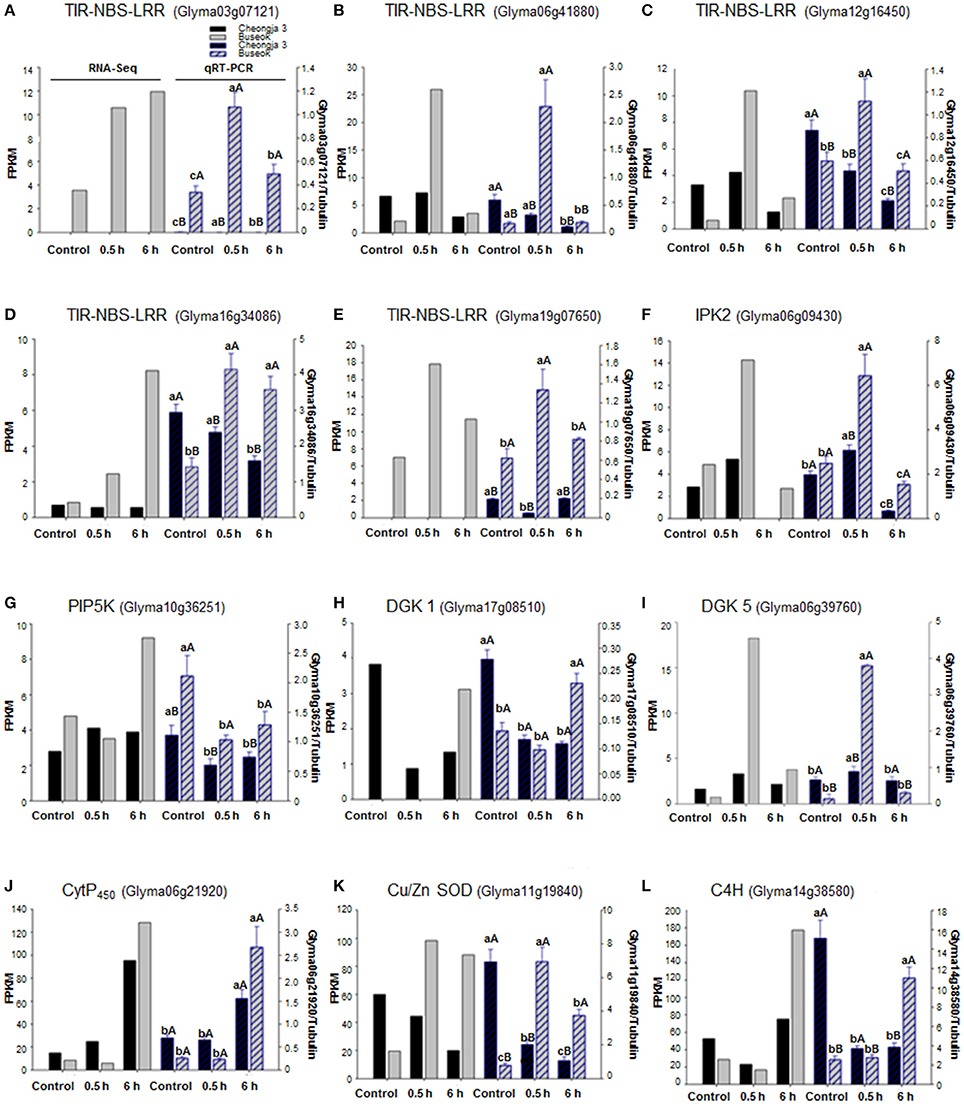
Figure 4. Expression validations via qRT-PCR for UV-B related DEGs identified between UV-B-sensitive Cheongja 3 and UV-B-resistant Buseok. Left and right Y-axes represent relative transcript abundance from RNA-seq and qRT-PCR results, respectively. Black and gray bars indicate Cheongja 3 and Buseok, respectively. Error bars represent the SE for three independent replicates. Bars with the same lower letters within a treatment indicate are not significant differences; Bars with same upper case letters within genotype indicate not significant differences by Fisher's least significant difference (LSD) test at P < 0.05. Control, 0.5, and 6 h on the x-axis refer to 0, 0.5, and 6 h UV-B irradiation, respectively. (A–E) TIR-NBS-LRR genes (Glyma03g07121, Glyma06g41880, Glyma12g16450, Glyma16g34086, and Glyma19g07650); (F) IPK2 (Glyma06g09430); (G) PIP5K (Glyma10g36251); (H) DGK 1 (Glyma17g08510); (I) DGK 5 (Glyma06g39760); (J) Cyt P450 (Glyma06g21920); (K) Cu/Zn SOD (Glyma11g19840); (L) C4H (Glyma14g38580).
Functional Validation of UV-B Stress Defense Signaling Genes Using A. thaliana Knock-Out Mutants
We identified four A. thaliana knock-out mutant lines, including two for TIR-NBS-LRR (AT5G36930:SALK_124056C and AT4G36150: SALK_084909C), one for DGK (AT5G07920:SALK_033664C), and one for PIP5K (AT1G34260:SALK_047604C) (Figure 5). The knock-out mutant lines did not show any noticeable morphological changes compared to wild-type (Col-0) under normal growing condition, suggesting that these genes do not play vital roles in basic growth and development. However, 2 days after 4 h UV-B stress, three of the mutant lines (AT5G36930:SALK_124056C, AT5G07920:SALK_033664C, and AT1G34260:SALK_047604C) showed severe chlorosis and stagnant growth, indicating increased sensitivity to UV-B stress (Figure 5). By contrast, the wild type and the remaining mutant lines (affected in the TIR-NBS-LRR gene) exhibited continued growth despite the presence of leaf curling and slight wilting. We measured the expression patterns of the four target genes in the wild type and mutant lines in response to 4 h UV-B irradiation by qRT-PCR. In the wild type, all of the genes except AT5G36930 (TIR-NBS-LRR) were induced by 4 h UV-B irradiation. As expected, the expression levels of all target genes were highly reduced in the corresponding mutant lines under both control and UV-B treatment. Functional validation using the A. thaliana knock-out mutants revealed that some genes implicated in the phosphatidic acid signaling pathway and immune response play key roles in UV-B stress defense.
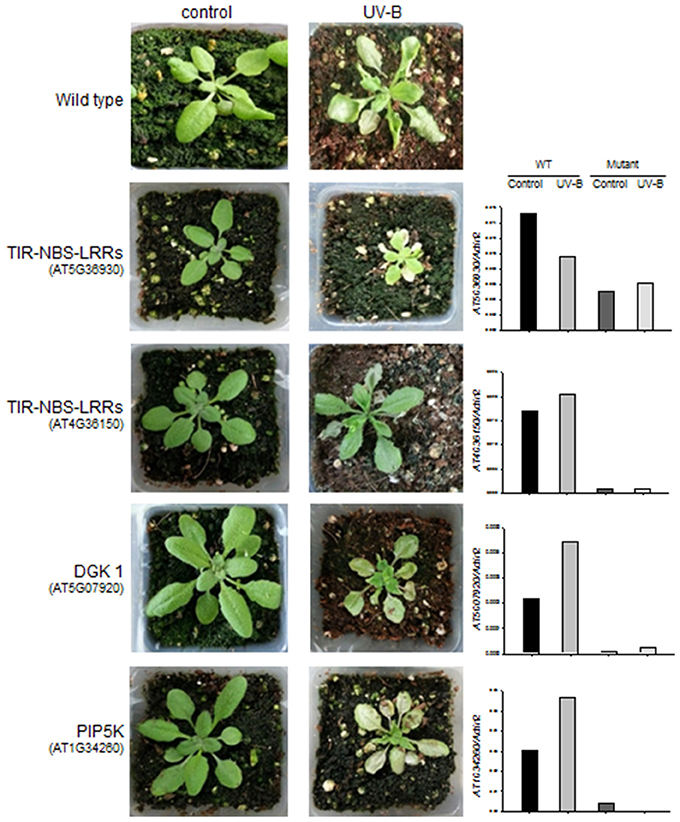
Figure 5. Changes in morphology and gene expression levels of wild type and the A. thaliana knock-out mutants after 4 h UV-B treatment. Left: morphological changes of the wild type (in the 1st row) and the four knock-out mutants (in the 2nd to 5th rows) for TIR-NBS-LRRs (AT5G36930 and AT4G36150), DGK 1 (AT5G07920), and PIP5K (AT1G34260) between control and 4 h UV-B treatment. Right: expression levels of the target genes (TIR-NBS-LRRs, DGK 1, and PIP5K) that were measured by qRT-PCR from the wild type and the knock-out mutants under control and 6 h UV-B treatment.
Discussion
Two approaches are often used to investigate the responses of plants to UV-B radiation. One approach is to observe the damage and subsequent recovery of plants at specific times after UV-B irradiation (Kilian et al., 2007; Safrany et al., 2008; Biedermann and Hellmann, 2010), and the other is to investigate the accumulated responses of plants exposed to continuous UV-B treatment (Casati and Walbot, 2004; Gruber et al., 2010); both were utilized in the present study. A series of studies on the intracellular responses of maize have been conducted at different irradiation times from 5 to 90 min up to 6 h (Casati et al., 2011a,b,c), which were primarily focused on understanding plant acclimation to UV-B. In the current study, we surveyed differences in whole transcript abundance in response to supplementary UV-B exposure for 0.5 and 6 h using RNA-seq. Our transcriptome analysis provided evidence for the notion that intracellular photomorphogenic responses for adaptation to low UV-B levels are separate from defense mechanisms against UV-B stress at high fluence rates (Figure 6). While the responses to UV-B stress are considered to be mediated by signaling pathways not specific to UV-B and are also induced by other stresses, our understanding of how plants activate components of defense signaling pathways against UV-B stress remains limited.
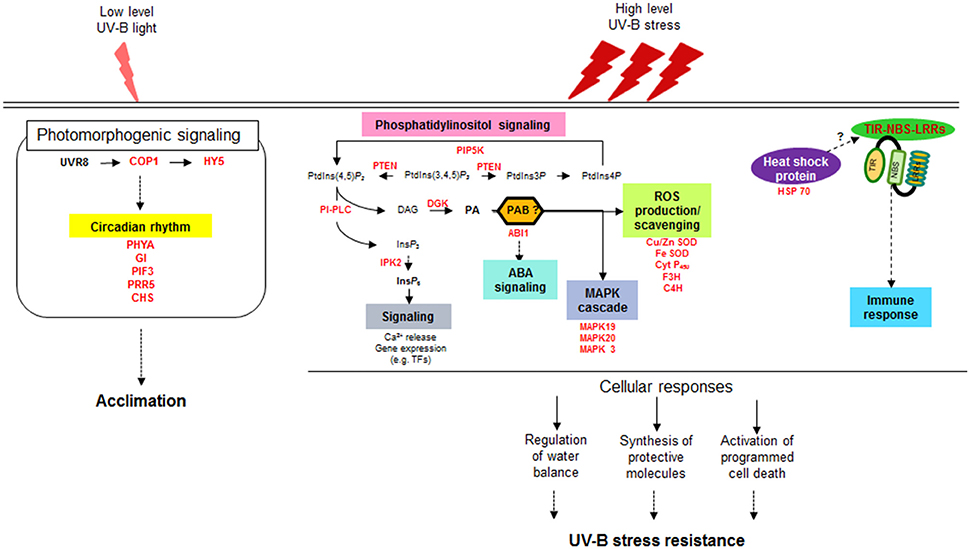
Figure 6. Schematic diagram of distinct signaling pathways depending on UV-B intensity. UVR8, UV Resistance Locus 8; COP1, Constitutively Photomorphogenic 1; HY5, Elongated Hypocotyl 5; PHYA, Phytochrome A; GI, Gigantea; PIF3, Phytochrome interacting factor 3; PRR5, Pseudo-response regulator 5; CHS, Chalcone and stilbene synthase family protein; PIP5K, phosphatidylinositol-4-phosphate 5-kinase family protein; PTEN, Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase; PLC, Phospholipase C; DG, Diacylglycerol; DGK, Diacylglycerol kinase; PA, Phosphatidic acid; PAB, Phosphatidic acid binding protein; ABI1, ABA insensitive phosphatase 1; IPK2, Inositol 1,3,4-trisphosphate 5/6-kinase family protein; IP6, inositol hexaphosphate; Cu/Zn SOD, Copper/Zinc superoxide dismutase; Fe- SOD, Iron-superoxide dismutase; Cyt P450, Cytochrome P450 family protein; F3H, Flavanone-3-hydroxylase; C4H, Cinnamate-4-hydroxylase; MAPK, Mitogen-activated-protein kinase; HSP 70, Heat shock protein 70; TIR-NBS-LRRs, Toll/interleukin-1 receptor nucleotide binding site leucine-rich repeat.
The UV-B photoreceptor UVR8, which was isolated in A. thaliana (Kliebenstein et al., 2002; Favory et al., 2009), was not identified as a DEG in response to 0.5 and 6 h UV-B treatment in the current study, but other UV-B-specific photomorphogenic signaling components, including HY5, COP1, and CHS, were upregulated in UV-B-resistant Buseok compared to Cheongja 3. In addition, key genes involved in the circadian rhythm, such as PHYTOCHROM A, GIGANTEA, and PHYTOCHROM INTERACTING FACTOR 3, were upregulated by UV-B irradiation (Figure 6). Low levels of UV-B are perceived by UVR8 followed by several downstream signaling pathways (Frohnmeyer and Staiger, 2003; Heijde and Ulm, 2012). The threshold UV-B doses that initiate photomorphogenic responses are much lower than those that induce stress defense gene expression (Boccalandro et al., 2001; Brown and Jenkins, 2008; Jenkins, 2009). In the current study, it is unclear if UVR8 expression was upregulated instantly (≤ 1–2 min) after the start of UV-B irradiation to activate downstream photomorphogenic signaling and other light signaling pathways, followed by a return to the ground state (Brown et al., 2005; Heijde and Ulm, 2012; Jenkins, 2014). However, soybean is not presumed to have another UV-B photoreceptor in addition to UVR8 for photomorphogenic responses.
Once plants recognize that irradiated UV-B fluence rates are beyond their adaptive capacity and are stressors, they immediately operate stress defense mechanisms for survival. The UV-B dosages used in our study are not low, and they represent dramatic spikes in UV-B intensity within short periods of time. Our results suggest that defense mechanisms against UV-B stress are provoked through PA-dependent signaling pathways via the sequential actions of PI-PLC and DGK, and that cell death and immunity systems also operate during this response (Figure 6). Significant advances have been made in elucidating stress-induced PA signal transduction during the past several years, as described in recent reviews (Ruelland et al., 2015; Singh et al., 2015; Hong et al., 2016; Hou et al., 2016). PA is the common product of the phospholipase D (PLD) and PLC/DGK pathways, which use different phospholipids as substrates. The PLD pathway is involved in a wide range of responses to external stresses, such as cold, salinity, drought, and pathogen attack (Hong et al., 2016). Nonetheless, we found that UV-B stress triggered only a PLC/DGK-mediated PA signaling pathway in the current study. PLC is induced by the osmotic stress caused by salt and dehydration (Darwish et al., 2009). Since UV-B stress is a complex environmental stress comprising heat and dehydration as well as excessive light exposure, our finding is in line with previous results.
UV-B-resistant Buseok showed higher transcript abundance for four key enzymes implicated in PI-PLC/DGK-mediated signaling cascades in plants (Table 2; Figures 3, 6), resulting in the production of polyphosphoinositols (membrane lipids) and inositol polyphosphates (water-soluble, IPP). These enzymes include PIP5K, PTEN2, DGK, and IPK2, exclusive of PI-PLC. In plants, the cellular levels of PtdIns(4,5)P2, which is produced from PtdIns4P by PIP5K and functions as a subtract for PI-PLC, are extremely low (30–100-fold lower than in mammalian cells) (Munnik and Vermeer, 2010). Nevertheless, Buseok had significantly higher levels of PIP5K transcript under both normal conditions (UV-B untreated) and UV-B exposure compared to Cheongja 3 (Table 2). Increased PtdIns(4,5)P2 levels by concurrent activation of PIP5K might lead to earlier upregulation of PI-PLC in Buseok vs. Cheongja 3 (under 0.5 h UV-B treatment), which is in agreement with the results obtained for plant tissues exposed to salt or osmotic stress (Darwish et al., 2009) as well as heat stress (Mishkind et al., 2009). In the current study, the Arabidopsis mutant defective in PIP5K showed increased sensitivity to UV-B stress compared to wild type (Figure 5). Upregulation of PI-PLC was observed only in Cheongja 3 under 6 h UV-B treatment (Table 2) and Buseok showed stable expression patterns of PI-PLC independent of UV-B treatment. However, the activation of DGK, which catalyzes ATP-dependent DAG phosphorylation for PA biosynthesis, is more specifically regulated in response to UV-B stress and more dependent on genotype. In Buseok, DGK was downregulated under control conditions and upregulated by UV-B treatment (Table 2). By contrast, in Cheongja 3, DGK was not significantly upregulated by UV-B treatment. Thus, the regulation of DGK is likely more critical in the PI-PLC/DGK-dependent PA signaling pathways in response to UV-B stress and appears to be transcriptionally mediated, even though the type of TF that binds to its promoter region is currently unknown. A study performed more than a decade ago revealed that a UV-B light insensitive (uli) T-DNA insertional Arabidopsis mutant, which displays hyposensitivity to low-fluence UV-B irradiation, is defective in DGK (Suesslin and Frohnmeyer, 2003). However, in the current study, an Arabidopsis DGK knock-out mutant showed increased damage to high UV-B intensity (Figure 5). Although whether DGK mediates different signaling pathways according to UV-B fluence rates is currently unclear, DGK is likely a specific component involved in UV-B-induced signal transduction in plants.
Increases in cytosolic Ca2+ levels in plant cells are the hallmark of stress defense responses (Singh et al., 2015). In mammalian systems, InsP3, the other product of PI-PLC hydrolysis of PtdIns(4,5)P2,releases Ca2+ from intracellular reservoirs by binding to ligand gated-calcium channels (Munnik and Nielsen, 2011). However, no InsP3 receptor has been identified in plants; instead, its multiple phosphorylated form InsP6, through stepwise phosphorylation by the IPP multikinases IPK2 and IPK1, is thought to stimulate increases in Ca2+ levels and to function as a signaling molecule itself (Munnik and Vermeer, 2010; Hou et al., 2016). In the current study, the transcript levels of IPK2, which phosphorylates Ins(1,4,5)P3 to Ins(1,3,4,5,6)P5, were higher in Buseok than in Cheongja 3 (Table 2).
PA binds to various target proteins to mediate downstream signal transduction during diverse defense responses such as ABA-mediated pathways, MAPK signaling cascades, and ROS over-accumulation (Hou et al., 2016). Among the dozens of identified effector proteins of PA in these cellular responses in plants (Ruelland et al., 2015; Hou et al., 2016), our RNA-seq analysis showed that ABI1, MAPK, and SOD were upregulated in Buseok under UV-B exposure (Figure 3). PA binding to ABI1 helps tether it to the plasma membrane where it interact with ATHB6, a negative regulator of ABA signaling involved in stomatal closure in response to drought and salinity stress (Zhang et al., 2004). PA also can bind to and activate MAPK in the response of A. thaliana and soybean to salt stress (Yu et al., 2010; Im et al., 2012). Based on the current results, ROS generation induced by UV-B stress is more likely dependent on SOD than on NADPH oxidase, even though PLD-derived PA binds to and activates NADPH oxidases under environmental stress (Park et al., 2004; Zhang et al., 2009).
Another interesting result of the present study is that TIR-NBS-LRR genes are upregulated in resistant Buseok under UV-B stress (Figure 3, Table 1; Supplementary Table 16). Plant NBS-LRR proteins can be divided into two major subfamilies based on the presence of TIR or CC motifs in their N-terminal domains (Tameling and Joosten, 2007; Göhre and Robatzek, 2008). Similarly, dozens of TIR-NBS-LRR genes, but no CC-NBS-LRRs, are differentially expressed in A. thaliana under heat and drought stress (Prasch and Sonnewald, 2013). A surprising role for TIR-NBS-LRRs has been proposed in the sensing of red light (Faigón-Soverna et al., 2006). A mutant of the constitutive shade avoidance (CSA1) gene in Arabidopsis is defective in red light-induced responses and produces a truncated protein with a structure similar to TIR.
We also observed increased accumulation of HSP70 transcripts in response to UV-B stress in Buseok (Figure 3, Table 1; Supplementary Table 16). Indeed, in Arabidopsis, HSPs and heat shock factors are upregulated in response to pathogen infection, abiotic stresses including UV, and wounding (Swindell et al., 2007). HSPs including HSP90 are thought to regulate the function of NBS-LRR (Belkhadir et al., 2004). Similarly, in soybean, HSP70 is upregulated under high temperature stress (Ahsan et al., 2010). Therefore, the highly expressed TIR-NBS-LRR proteins and HSPs under UV-B stress likely play important roles in the UV-B resistance response. Further studies on which and how genetic factors activate the transcription of TIR-NBS-LRR and HSP genes will be valuable for breeding of UV-B resistant soybean cultivars.
Taken with the previous reports together, a valid assumption as shown in Figure 6 can be drawn from our transcriptomic results, which is that high fluence rates of UV-B triggers the multiple defense pathways that are overlap with defense signaling pathways against other stress but independent of UVR8-mediated UV-B pathways at ambient levels. The second signal messenger PA, generated by the reaction of PI-PLC and DGK, induces the expression of genes characteristic of stress responses such as ABA signaling, MAPK signaling cascades and ROS accumulation. Meantime (or subsequently), the genes for TIR-NBS-LRRs and HSPs are also activated under high dose UV-B, involved in programmed cell death and immune response. Orchestration of these multiple defense pathways leads to the regulation of water balance and synthesis of protective molecules, resulting in showing UV-B stress resistance in plants.
The present study provides comprehensive insights into defense signaling pathways against high-intensity UV-B stress, from signal transduction by second messengers to downstream defense-related gene expression (Figure 6). It is currently likely indisputable that the generation of PPI-based signaling molecules such as PA and IP6 is the primary event in the signaling cascades from stress awareness to defensive metabolism. Based on our findings, further studies should be performed investigating how the key enzymes involved in PA signaling pathways are upregulated under UV-B stress and how the derived signaling molecules are integrated into downstream pathways, together with the identification of TFs specific to UV-B stress. Such studies will provide essential information for breeding resistant soybean genotypes that survive under high-intensity UV-B stress and can adapt to other adverse conditions for sustainable productivity in the future.
Author Contributions
MY performed all experiments and prepared the manuscript. MK participated in work design and data interpretation. SS performed RNA-seq bioinformatic analyses. KK provided the plant materials. JH performed statistical analysis of qRT-PCR data. JS and SK revised the manuscript. SL initiated and coordinated the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the Next Generation BioGreen 21 Program (Code No. PJ01102701), Rural Development Administration, Republic of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01917/full#supplementary-material
References
Ahsan, N., Donnart, T., Nouri, M.-Z., and Komatsu, S. (2010). Tissue-specific defense and thermo-adaptive mechanisms of soybean seedlings under heat stress revealed by proteomic approach. J. Proteome Res. 9, 4189–4204. doi: 10.1021/pr100504j
Austin, J., and Wilson, R. J. (2006). Ensemble simulations of the decline and recovery of stratospheric ozone. J. Geophys. Res. 111, 1–16. doi: 10.1029/2005jd006907
Belkhadir, Y., Subramaniam, R., and Dangl, J. L. (2004). Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. doi: 10.1016/j.pbi.2004.05.009
Biedermann, S., and Hellmann, H. (2010). The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 62, 404–415. doi: 10.1111/j.1365-313X.2010.04157.x
Boccalandro, H. E., Mazza, C. A., Mazzella, M. A., Casal, J. J., and Ballaré, C. L. (2001). Ultraviolet B radiation enhances a phytochrome-B-mediated photomorphogenic response in Arabidopsis. Plant Physiol. 126, 780–788. doi: 10.1104/pp.126.2.780
Britt, A. B. (1996). DNA damage and repair in plants. Annu. Rev. Plant Biol. 47, 75–100. doi: 10.1146/annurev.arplant.47.1.75
Brown, B. A., Cloix, C., Jiang, G. H., Kaiserli, E., Herzyk, P., Kliebenstein, D. J., et al. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230. doi: 10.1073/pnas.0507187102
Brown, B. A., and Jenkins, G. I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588. doi: 10.1104/pp.107.108456
Caldwell, M. M. (1971). Solar UV irradiation and the growth and development of higher plants. Photophysiology 6, 131–177. doi: 10.1016/B978-0-12-282606-1.50010-6
Casati, P., Campi, M., Morrow, D. J., Fernandes, J. F., and Walbot, V. (2011b). Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12:321. doi: 10.1186/1471-2164-12-321
Casati, P., Campi, M., Morrow, D. J., Fernandes, J., and Walbot, V. (2011a). Transcriptomic, proteomic and metabolomic analysis of maize responses to UV-B: comparison of greenhouse and field growth conditions. Plant Signal. Behav. 6, 1146–1153. doi: 10.4161/psb.6.8.15751
Casati, P., Morrow, D. J., Fernandes, J., and Walbot, V. (2011c). UV-B signaling in maize: transcriptomic and metabolomic studies at different irradiation times. Plant Signal. Behav. 6, 1926–1931. doi: 10.4161/psb.6.12.18164
Casati, P., and Walbot, V. (2004). Rapid transcriptome responses of maize (Zea mays) to UV-B in irradiated and shielded tissues. Genome Biol. 5:1. doi: 10.1186/gb-2004-5-3-r16
Darwish, E., Testerink, C., Khalil, M., El-Shihy, O., and Munnik, T. (2009). Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 50, 986–997. doi: 10.1093/pcp/pcp051
Faigón-Soverna, A., Harmon, F. G., Storani, L., Karayekov, E., Staneloni, R. J., Gassmann, W., et al. (2006). A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell 18, 2919–2928. doi: 10.1105/tpc.105.038810
Favory, J. J., Stec, A., Gruber, H., Rizzini, L., Oravecz, A., Funk, M., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601. doi: 10.1038/emboj.2009.4
Frohnmeyer, H., and Staiger, D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133, 1420–1428. doi: 10.1104/pp.103.030049
Göhre, V., and Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
Grant, R. H., Apostol, K. G., and Schmitz, H. F. (2010). “Physiological impacts of short-term UV irradiance exposures on cultivars of glycine max,” in UV Radiation in Global Climate Change: Measurements, Modeling and Effects on Ecosystems, eds W. Gao, J. R. Slusser, and D. L. Schmoldt (Berlin; Heidelberg: Springer), 458–487.
Gruber, H., Heijde, M., Heller, W., Albert, A., Seidlitz, H. K., and Ulm, R. (2010). Negative feedback regulation of UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 20132–20137. doi: 10.1073/pnas.0914532107
Heijde, M., and Ulm, R. (2012). UV-B photoreceptor-mediated signaling in plants. Trends Plant Sci. 17, 230–237. doi: 10.1016/j.tplants.2012.01.007
Heijde, M., and Ulm, R. (2013). Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. U.S.A. 110, 1113–1118. doi: 10.1073/pnas.1214237110
Hong, Y., Zhao, J., Guo, L., Kim, S. -C., Deng, X., Wang, G., et al. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signaling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Im, J. H., Lee, H., Kim, J., Kim, H. B., and An, C. S. (2012). Soybean MAPK, GMK1 is dually regulated by phosphatidic acid and hydrogen peroxide and translocated to nucleus during salt stress. Mol. Cells 34, 271–278. doi: 10.1007/s10059-012-0092-4
Jenkins, G. I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60, 407–431. doi: 10.1146/annurev.arplant.59.032607.092953
Jenkins, G. I. (2014). The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26, 21–37. doi: 10.1105/tpc.113.119446
Jordan, B. R. (1993). “The molecular biology of plants exposed to Ultraviolet-B radiation and the interaction with other stresses,” in Interacting Stresses on Plants in a Changing Climate, eds M. B. Jackson and C. R. Black (Berlin; Heidelberg: Springer), 153–170. doi: 10.1007/978-3-642-78533-7_9
Jordan, B. R. (2002). Review: molecular response of plant cells to UV-B stress. Funct. Plant Biol. 29, 909–916. doi: 10.1071/FP02062
Kerr, J., and McElroy, C. (1993). Evidence for large upward trends of ultraviolet-B radiation linked to ozone depletion. Science 262, 1032–1034. doi: 10.1126/science.262.5136.1032
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, K. D., Yun, M. Y., Shin, J. H., Kang, Y. J., Kim, M. Y., and Lee, S.-H. (2015). Underlying genetic variation in the response of cultivated and wild soybean to enhanced ultraviolet-B radiation. Euphytica 202, 207–217. doi: 10.1007/s10681-014-1271-5
Kliebenstein, D. J., Lim, J. E., Landry, L. G., and Last, R. L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130, 234–243. doi: 10.1104/pp.005041
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:1. doi: 10.1186/gb-2009-10-3-r25
Laxalt, A. M., and Munnik, T. (2002). Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5, 332–338. doi: 10.1016/S1369-5266(02)00268-6
Lee, J. S., Kim, S., Ha, B. -K., and Kang, S. (2016). Positional mapping and identification of novel quantitative trait locus responsible for UV-B radiation tolerance in soybean [Glycine max (L.) Merr.]. Mol. Breed. 36, 1–10. doi: 10.1007/s11032-016-0471-1
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lytvyn, D. I., Yemets, A. I., and Blume, Y. B. (2010). UV-B overexposure induces programmed cell death in a BY-2 tobacco cell line. Environ. Exp. Bot. 68, 51–57. doi: 10.1016/j.envexpbot.2009.11.004
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
McKenzie, R. L., Aucamp, P. J., Bais, A. F., Björn, L. O., Ilyas, M., and Madronich, S. (2011). Ozone depletion and climate change: impacts on UV radiation. Photochem. Photobiol. Sci. 10, 182–198. doi: 10.1039/c0pp90034f
Mishkind, M., Vermeer, J. E., Darwish, E., and Munnik, T. (2009). Heat stress activates phospholipase D and triggers PIP2 accumulation at the plasma membrane and nucleus. Plant J. 60, 10–21. doi: 10.1111/j.1365-313X.2009.03933.x
Mueller-Roeber, B., and Pical, C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46. doi: 10.1104/pp.004770
Müller-Xing, R., Xing, Q., and Goodrich, J. (2014). Footprints of the sun: memory of UV and light stress in plants. Front. Plant Sci. 5:474. doi: 10.3389/fpls.2014.00474
Munnik, T., and Nielsen, E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497. doi: 10.1016/j.pbi.2011.06.007
Munnik, T., and Vermeer, J. E. (2010). Osmotic stress-induced phosphoinositide and inositol phosphate signaling in plants. Plant Cell Environ. 33, 655–669. doi: 10.1111/j.1365-3040.2009.02097.x
Murali, N. S., Teramura, A. H., and Randall, S. K. (1988). Response differences between two soybean cultivars with contrasting UV-B radiation sensitivities. Photochem. Photobiol. 48, 653–657. doi: 10.1111/j.1751-1097.1988.tb02877.x
Narusaka, M., Seki, M., Umezawa, T., Ishida, J., Nakajima, M., Enju, A., et al. (2004). Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol. Biol. 55, 327–342. doi: 10.1007/s11103-004-0685-1
Parihar, P., Singh, S., Singh, R., Singh, V. P., and Prasad, S. M. (2015). Changing scenario in plant UV-B research: UV-B from a generic stressor to a specific regulator. J. Photochem. Photobiol. B Biol. 153, 334–343. doi: 10.1016/j.jphotobiol.2015.10.004
Park, J., Gu, Y., Lee, Y., Yang, Z., and Lee, Y. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134, 129–136. doi: 10.1104/pp.103.031393
Paul, N. D., and Gwynn-Jones, D. (2003). Ecological roles of solar UV radiation: towards an integrated approach. Trends Ecol. Evol. 18, 48–55. doi: 10.1016/S0169-5347(02)00014-9
Prasch, C. M., and Sonnewald, U. (2013). Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 162, 1849–1866. doi: 10.1104/pp.113.221044
Ruelland, E., Kravets, V., Derevyanchuk, M., Martinec, J., Zachowski, A., and Pokotylo, I. (2015). Role of phospholipid signalling in plant environmental responses. Environ. Exp. Bot. 114, 129–143. doi: 10.1016/j.envexpbot.2014.08.009
Safrany, J., Haasz, V., Mate, Z., Ciolfi, A., Feher, B., Oravecz, A., et al. (2008). Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. Plant J. 54, 402–414. doi: 10.1111/j.1365-313X.2008.03435.x
Searles, P. S., Flint, S. D., and Caldwell, M. M. (2001). A meta-analysis of plant field studies simulating stratospheric ozone depletion. Oecologia 127, 1–10. doi: 10.1007/s004420000592
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Shim, H. -C., Ha, B. -K., Yoo, M., and Kang, S. -T. (2015). Detection of quantitative trait loci controlling UV-B resistance in soybean. Euphytica 202, 109–118. doi: 10.1007/s10681-014-1233-y
Singh, A., Bhatnagar, N., Pandey, A., and Pandey, G. K. (2015). Plant phospholipase C family: regulation and functional role in lipid signaling. Cell Calcium 58, 139–146. doi: 10.1016/j.ceca.2015.04.003
Sparvoli, F., and Cominelli, E. (2015). Seed biofortification and phytic acid reduction: a conflict of interest for the plant? Plants 4, 728–755. doi: 10.3390/plants4040728
Suesslin, C., and Frohnmeyer, H. (2003). An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J. 33, 591–601. doi: 10.1046/j.1365-313X.2003.01649.x
Swindell, W. R., Huebner, M., and Weber, A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. doi: 10.1186/1471-2164-8-125
Tameling, W. I., and Joosten, M. H. (2007). The diverse roles of NB-LRR proteins in plants. Physiol. Mol. Plant Pathol. 71, 126–134. doi: 10.1016/j.pmpp.2007.12.006
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Ulm, R. (2003). “Molecular genetics of genotoxic stress signalling in plants,” in Plant Responses to Abiotic Stress, eds H. Hirt and K. Shinozaki (Berlin; Heidelberg: Springer-Verlag), 217–240. doi: 10.1007/978-3-540-39402-0_9
Ulm, R., and Nagy, F. (2005). Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8, 477–482. doi: 10.1016/j.pbi.2005.07.004
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Zhang, W., Qin, C., Zhao, J., and Wang, X. (2004). Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 9508–9513. doi: 10.1073/pnas.0402112101
Zhang, Y., Zhu, H., Zhang, Q., Li, M., Yan, M., Wang, R., et al. (2009). Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377. doi: 10.1105/tpc.108.062992
Keywords: UV-B stress resistance, phosphatidic acid, diacylglycerol kinase, TIR-NBS-LRR, transcriptomic profiling, soybean
Citation: Yoon MY, Kim MY, Shim S, Kim KD, Ha J, Shin JH, Kang S and Lee S-H (2016) Transcriptomic Profiling of Soybean in Response to High-Intensity UV-B Irradiation Reveals Stress Defense Signaling. Front. Plant Sci. 7:1917. doi: 10.3389/fpls.2016.01917
Received: 05 August 2016; Accepted: 02 December 2016;
Published: 19 December 2016.
Edited by:
Chengdao Li, Murdoch University, AustraliaReviewed by:
Seonghee Lee, University of Florida, USATae-Hwan Jun, Pusan National University, South Korea
Copyright © 2016 Yoon, Kim, Shim, Kim, Ha, Shin, Kang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suk-Ha Lee, sukhalee@snu.ac.kr
 Min Young Yoon
Min Young Yoon Moon Young Kim1,2
Moon Young Kim1,2 Kyung Do Kim
Kyung Do Kim Suk-Ha Lee
Suk-Ha Lee