- 1Division of Plant and Microbial Biotechnology, Institute of Life Sciences, Bhubaneswar, India
- 2Department of Molecular Plant Virology and Plant Genetic Engineering, KTRDC, College of Agriculture, Food and Environment, University of Kentucky, Lexington, KY, United States
Development of disease-resistant plant varieties achieved by engineering anti-microbial transgenes under the control of strong promoters can suffice the inhibition of pathogen growth and simultaneously ensure enhanced crop production. For evaluating the prospect of such strong promoters, we comprehensively characterized the full-length transcript promoter of Cassava Vein Mosaic Virus (CsVMV; -565 to +166) and identified CsVMV8 (-215 to +166) as the highest expressing fragment in both transient and transgenic assays. Further, we designed a new chimeric promoter ‘MUASCsV8CP’ through inter-molecular hybridization among the upstream activation sequence (UAS) of Mirabilis Mosaic Virus (MMV; -297 to -38) and CsVMV8, as the core promoter (CP). The MUASCsV8CP was found to be ∼2.2 and ∼2.4 times stronger than the CsVMV8 and CaMV35S promoters, respectively, while its activity was found to be equivalent to that of the CaMV35S2 promoter. Furthermore, we generated transgenic tobacco plants expressing the totiviral ‘Killer protein KP4’ (KP4) under the control of the MUASCsV8CP promoter. Recombinant KP4 was found to accumulate both in the cytoplasm and apoplast of plant cells. The agar-based killing zone assays revealed enhanced resistance of plant-derived KP4 against two deuteromycetous foliar pathogenic fungi viz. Alternaria alternata and Phoma exigua var. exigua. Also, transgenic plants expressing KP4 inhibited the growth progression of these fungi and conferred significant fungal resistance in detached-leaf and whole plant assays. Taken together, we establish the potential of engineering “in-built” fungal stress-tolerance in plants by expressing KP4 under a novel chimeric caulimoviral promoter in a transgenic approach.
Introduction
Designing of pathogen-resistant plants can help to alleviate global food crisis. An annual crop loss of 10% occurs worldwide due to fungal diseases; fungicides are the most commonly used means to prevent the colonization, sporulation, and growth of phytopathogenic fungi (Hahn, 2014; Drenth and Guest, 2016). But, the situation is worsened by the appearance of resistant isolates of various pathogens, which render the regular use of these fungicides, impractical (Nicot et al., 2016). In addition, the use of potent fungicides has already been pronounced as ‘obsolete’ due to the presence of high genetic diversity of phytopathogens as well as undesirable after-effects of these chemicals on the environment (Burdon, 1993; Damalas and Eleftherohorinos, 2011; Lorenzini and Zapparoli, 2014; Czislowski et al., 2017). There are numerous resistant varieties of plant fungal pathogens that have been documented viz. Colletotrichum graminicola causing anthracnose of turf grasses (Avila-Adame et al., 2003), Plasmopara viticola causing downy mildew of grapes (Genet et al., 2006), Venturia inaequalis causing apple scab (Köller et al., 2004), Monilinia fructicola causing brown rot of stone fruits (Chen et al., 2013), Botrytis cinerea causing post-harvest gray mold of apple (Hahn, 2014) and many other. In continuation, plant-pathogenic species of Alternaria cause devastating effects on crops of nutritional and economic value by attacking several members of Cucurbitaceae, Brassicaceae, and Solanaceae plant families (Mamgain et al., 2013). Phoma, on the other hand, causes epidemic disease development in cabbage, Tasmanian pyrethrum and oilseed rape (West et al., 2001; Pethybridge et al., 2005; Dilmaghani et al., 2009). Besides, it has devastating effects on field peas when associated with Ascochyta (Liu et al., 2016). Recently, both Alternaria and Phoma were found to develop resistance against QoI or strobilurins and benzimidazole group of fungicides, respectively (Van de Graaf et al., 2003; Karaoglanidis et al., 2011; Kim et al., 2017). Alternaria directly penetrates and infects annual plants including several vegetables, ornamentals and fruit trees (Sharma et al., 2013). On the other-hand, Phoma being a hemibiotrophic pathogen enters through wounds and attacks several herbaceous plants including vegetables like common bean, artichoke, romaine lettuce, and field pea (Kubota and Abiko, 2002; Fitt et al., 2006; Koike et al., 2006; Khani et al., 2016). Furthermore, Phoma-Didymella complex in association with Peronospora sparsa causes epidemic wilting symptom in Arctic bramble (Rubus arcticus L.) (Lindqvist-Kreuze et al., 2003) in Finland. On one-hand, where such pathogens cause heavy crop losses, there are serious constraints associated with the emerging resistant strains of the same. Therefore, there is an urgent need to explore new alternatives that can mount a broader resistance spectrum as well as stable pattern of inheritance in the resistant plants (Russell, 2013).
Till date there have been numerous advances toward the development of disease-resistant plant varieties via conventional breeding (Collard and Mackill, 2008). However, raising biotic-stress tolerant plants by traditional breeding approaches is fairly slow and may lead to unwanted linkage-drag due to the presence of quantitative traits at multiple loci (Jauhar, 2006). In contrast, the transgenic approach offers an excellent asexual means to insert foreign genes into plant cells (Sanghera et al., 2011).
The need to develop proficient plant-expression systems coupled to novel regulatory elements (promoter) that can efficiently escalate the disease resistance by expressing anti-microbial transgenes (at moderate/high level) in the engineered plants is gradually increasing (Rushton et al., 2002). Usually, the naturally occurring promoters have certain limitations and have low transcriptional activities. On the contrary, recombinant synthetic promoters are generated through shuffling of unique domains namely the distal upstream activation sequence (UAS) and proximal core promoter (CP) region of the same or different naturally occurring promoters. Such tailor-made chimeric promoters with altered cis-architecture usually appear to be efficient in driving transgenes in a plant cell (Dey et al., 2015). For example, the M24 promoter (Dey and Maiti, 1999), a chimera containing the duplicated enhancer domains derived from the Mirabilis Mosaic Virus (MMV) is capable of driving constitutive expression of heterologous genes in engineered plants such as, the SAC domain of PAR-4 (Prostate Apoptosis Response 4) (Sarkar et al., 2015) and Glucocerebrosidase (GCB) (Chatterjee et al., 2017). The chimeric promoter ‘MUAS35SCP’ (Patro et al., 2012) has been established to drive constitutive and stable expression of human beta-defensins (hβD-1 and hβD-2) in transgenic tobacco lines (Patro et al., 2015). Likewise, in the present study, we generated a chimeric promoter “MUASCsV8CP” by coupling the UAS of MMV (MUAS) with the newly identified CsVMV8 promoter fragment of Cassava vein mosaic virus (CsVMV), as CP and tested its efficacy in plant modification. The CsVMV, a putative member of the caulimoviral group infects cassava (Manihot esculenta L.) plants in Brazil. It has a unique genome organization with a promoter sequence that contains several organ-specific cis- elements and has been partially characterized in rice and cassava (Verdaguer et al., 1998; Oyelakin et al., 2015). In this context, the transcription start site (TSS) of CsVMV promoter was earlier reported as an adenine residue located 35 bp downstream of the TATA-box (Verdaguer et al., 1996).
Transgenic plants expressing anti-fungal proteins have previously been tested for disease tolerance. The first fungus-resistant transgenic plant expressing bean-chitinase in tobacco and Brassica napus conferred resistance toward Rhizoctonia solani (Broglie et al., 1991). Since then, various transgenic plants such as those expressing Glucose oxidase gene from Talaromyces flavus (Murray et al., 1997), family 19 chitinase of Streptomyces griseus (Itoh et al., 2003), endochitinase gene from Trichoderma virens (Emani et al., 2003), bean chitinase from Phaseolus vulgaris (Tohidfar et al., 2005) and Rpsl-k from Soyabean (Gao et al., 2005) have provided increased resistance to various fungal pathogens. The corn smut fungus, Ustilago maydis secretes a virally encoded fungal toxin “Killer Protein 4” (KP4) when infected by the P4 strain of UMV4 totivirus (Allen et al., 2011). KP4 killer toxin is a α/β sandwich protein having 105 amino acids and is the only member of Killer protein family that is not processed by Kex2p (Ganesa et al., 1991). KP4 functions as a calcium channel chelator that blocks a cAMP-regulated growth pathway (Gage et al., 2001). In continuation, Schlaich et al. (2006) reported that KP4 specifically blocks the calcium channels of fungi. However, it has no effect on human and plants. Furthermore, transgenic maize expressing KP4 was found to be highly resistant to corn smut, a devastating fungal disease where galls start appearing on aboveground parts of plants particularly on leaves. This calls for a comprehensive evaluation of the anti-fungal properties of KP4 wherein we can find other candidate fungi that can be targeted using this molecule in a transgenic approach.
In this study, we fully characterized the full-length transcript promoter of CsVMV. We were able to identify the ‘CsVMV8’ promoter having equivalent activity to the most widely used CaMV35S promoter. To further enhance the activity of CsVMV8, we performed inter-molecular hybridization between the MUAS and CsVMV8-CP (CsV8CP) to generate a chimera ‘MUASCsV8CP’. The activity of this newly designed chimeric promoter (coupled to a GUS reporter) was evaluated in the transient (protoplast and agro-infiltration) and transgenic assays. Further, this recombinant promoter was employed for raising transgenic tobacco lines expressing KP4. Next, we examined the fungal resistance of transgenic lines expressing KP4 against two foliar pathogenic fungi namely Alternaria alternata and Phoma exigua var. exigua. The in vitro agar-based killing zone and leaf detachment assays were performed to evaluate whether the recombinant KP4 could substantially restrain the growth of both the above fungi. To further inspect the anti-fungal activity of KP4 transgenic plants, we performed the disease resistance assays using whole plants against them. Overall, this study describes the effectiveness of the totiviral KP4 driven by a newly designed chimeric caulimoviral promoter MUASCsV8CP to restrict the growth of devastating foliar fungi, which may open new avenues in the field of plant protection against phyto-pathogens.
Materials and Methods
Construction of Binary Vectors for Expression in Plants
A total of thirteen 5′- (CsVMV1-CsVMV13) and three 3′- (CsVMVR1-CsVMVR3) end deletion fragments were PCR amplified (using primers listed in Supplementary Table S1) adding EcoRI and HindIII sites at the 5′- and 3′- ends, respectively. The PCR amplified products were subsequently cloned in the EcoRI and HindIII sites of pUC119 sequencing vector and subsequently sequenced and analyzed to check their integrity. The fragments were finally sub-cloned into protoplast and plant expression vectors pUCPMAGUS and pKYLX71GUS (Schardl et al., 1987; Dey and Maiti, 1999), respectively.
For the generation of chimeric promoter construct MUASCsV8CP, the UAS of MMV (MUAS; 335 bp, -297 to -38) and the proximal domain of CsVMV8-Flt (CsV8CP; 381 bp, -215 to +166) promoter were PCR amplified using appropriate primers (Supplementary Table S1) containing overhangs of EcoRI and HincII at the 5′- end and SmaI and HindIII at the 3′- end. The PCR amplified fragments were cloned into the corresponding sites of pUC119 following a previously published protocol (Acharya et al., 2014). Subsequently, the resultant promoter fragment was sub-cloned into pUCPMAGUS and pKYLX71GUS to generate the pUPMUASCsV8CPGUS and pKMUASCsV8CPGUS plant expression vectors.
To generate the recombinant KP4 chimeric construct, the totiviral KP4 gene [UniProtKB-Q90121(KP4T_UMV4)] was fused with a 35-nucleotide untranslated region of AlMV RNA4 (5′ AMV; translational enhancer) and apoplast targeting sequence (aTP) of Arabidopsis 2S2 protein gene (Kroumova et al., 2013) at its 5′- end; and a hexa (6x) His-tag at its 3′- end. The above recombinant sequence was then cloned into the XhoI and SstI sites of the binary vector pKMUASCsV8CPGUS and pKCaMV35S2GUS (replacing GUS), to design the plasmids pKMUASCsV8CP-KP4His and CaMV35S2-KP4His. The pKMUASCsV8CP construct was also designed to serve as a vector control (VC).
Transient and Transgenic Assays of Promoter Clones
The native and chimeric promoters along with CaMV35S and CaMV35S2 (cloned in pUCPMAGUS) were electroporated into viable tobacco protoplasts following standard protocols (Acharya et al., 2014). Transient GUS activities from respective promoter constructs were calculated biochemically by fluorimetric GUS assay (Jefferson, 1987). For transient Agro-infiltration assays of the promoter constructs in tobacco, petunia, and tomato, Agrobacterium tumefaciens strain LBA4404 was transformed with respective promoter constructs following the freeze-thaw method as described previously (Chen et al., 1994). Leaves of whole plants of tobacco, petunia and tomato were mechanically infiltrated with individual Agrobacterium constructs (Yang et al., 2000). Quantitative fluorimetric GUS assay for each promoter construct was carried out 3–4 days’ post-infiltration (Jefferson, 1987; Ranjan et al., 2011; Acharya et al., 2014).
For comparative GUS activity analysis in agro-infiltrated leaves expressing CsVMV native deletion fragments cloned in pKYLXGUS, pKMUASCsV8CPGUS, pKMUAS35SCPGUS, pK35SGUS, and pK35S2GUS fragments; total leaf protein from transgenic seedlings (21 days old) were isolated and GUS activity analysis was performed according to standard protocols (Bradford, 1976; Jefferson, 1987). Histochemical GUS staining of 21 days old transgenic tobacco seedlings (T2 generation) was performed according to a previously published protocol (Jefferson et al., 1987).
Production of Transgenic Tobacco Plants by Tobacco Leaf Disk Transformation
The native and chimeric promoter fragments along with KP4-His and VC constructs were used to transform tobacco plants by Agrobacterium-mediated gene transfer (Dey and Maiti, 1999) using LBA4404 strain. On an average, seventeen to eighteen T0 transgenic tobacco lines were generated for each of the constructs. All transgenic plants showed normal growth and development. Subsequently, the T1 generation seeds were subjected to segregation analysis (Patro et al., 2015). Transgenic lines with a chi-square value of <1.5 (p ≤ 0.05) were considered to be true transgenic lines. Independently re-generated transgenic lines showing proper segregation ratios (KanR:KanS::3:1) were selected and respective T2 generation transgenic plants were propagated in greenhouse conditions [photoperiod: 16/8 h (light/dark), light intensity: 220 μmol/m2/s, temperature: 28 ± 2°C and humidity: 70–75%]. The transgenic tobacco plants expressing KP4-His along with VC lines were also compared for their agronomical parameters such as plant height, leaf architecture, seed fertility and flowering time. Likewise, the GUS activities of the transgenic plants expressing pKYLXGUS, pKMUASCsV8CPGUS, pKMUAS35SCPGUS, pK35SGUS, and pK35S2GUS was performed following the standard protocol (Bradford, 1976; Jefferson et al., 1987).
Gene Integration Assays
Total genomic DNA was extracted from 21-days old T2 transgenic seedlings of individual lines expressing VC and pKMUASCsV8CP-KP4His, respectively (Dellaporta et al., 1983). Gene integration assays for uidA, KP4, rbcSE9 and nptII were performed in respective transgenic lines using gene-specific primer sets (Supplementary Table S1). Transgenic lines expressing native and chimeric promoter constructs along with KP4-His and VC plants were subjected to PCR amplification (of the genes mentioned above) following the standard protocol using Taq DNA Polymerase (Thermo Fischer Scientific; Cat No. #EP0402) having proofreading activity (Kumar et al., 2011) and all the above amplicons were sequenced by Sanger sequencing method employing a sequencer (Applied Biosystems 3500 Genetic Analyzer).
Southern Blotting
Briefly, 20 μg of genomic DNA isolated from selected T1 progenies of transgenic and VC lines was digested using the XhoI restriction enzyme (Fermentas ER0691) overnight (16 h) at 37°C, electrophoresed on 0.8% agarose gel, blotted on nylon membrane (nylon-N+, Amersham) and finally probed with PCR amplified αP32-labeled GUS and KP4-His probes using specific primers (Supplementary Table S1). The entire procedure of Southern Blotting was performed following a previously described protocol (Sambrook et al., 1989; Sarkar et al., 2015).
Quantitative Real-Time PCR Analysis
Total RNA from transgenic tobacco seedlings expressing the respective constructs were isolated using the RNeasy plant mini kit (Qiagen, Chatsworth, United States) and cDNA synthesis was performed. After that, Real-time PCR analysis was performed using Premix Ex TaqTM II (Perfect Real Time, Takara Bio Inc., Japan) employing the Opticon-2Real-time PCR (MJ Research, Bio-Rad; Model; CFD-3220). The transcript levels for individual constructs were calculated by the 2-ΔΔCT (CT-comparative threshold cycle) method (Livak and Schmittgen, 2001) and expressed as fold change in comparison to respective controls where 18S was used as the housekeeping gene (Kumar et al., 2012).
Western Blot Analysis of T2 Generation KP4 Transgenic Plants
The uppermost fully expanded leaves of 6-week-old greenhouse grown plants (T2 generation) of KP4L2#2, KP4L2#3, KP4L2#4, KP4L2#7, KP4L2#11, and VC were extracted and crushed in liquid nitrogen to make a fine powder. Next, the total soluble protein (TSP) was extracted from the crushed samples using an extraction buffer containing 1X PBS with 0.05% β-mercaptoethanol and plant protease inhibitor cocktail (Sigma- P9599-5mL), pH 7.0. An aliquot of 10 μg of total soluble protein obtained from transgenic lines was resolved on 15% SDS-PAGE for Western blot analysis. For detection of the recombinant proteins, primary antibody specific to the hexa- (6x) His-tag [(Mouse monoclonal His-probe (SC-57598) and Rabbit polyclonal His-probe (SC-803)] was used following the standard protocol (Sarkar et al., 2015).
Purification of KP4 From Transgenic Plants
KP4 was purified from 21-days-old transgenic KP4-His seedling (T2) using Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions with slight modifications as described earlier (Ma et al., 2005). Briefly, the seedlings of KP4L2#2 transgenic lines were homogenized (in 1X PBS, 1 mM phenylmethylsulfonyl fluoride and 1.5% PVP-40), filtered through layers of miracloth and centrifuged at 14,000 g for 20 min at 4°C. The supernatant obtained was dialyzed against 50 mM Tris-HCl (pH 7.0) buffer for 24 h at 4°C and adjusted to 40% ammonium sulfate saturation. After centrifugation at 30,000 g for 40 min, the enriched fraction obtained as pellet was solubilized, dialyzed (overnight) and filtered through a 0.45 μM membrane filter, and loaded on to a Ni-NTA agarose column. Finally, the column was washed with wash buffer (2 mM imidazole, 20 mM Na2HPO4, 100 mM NaCl). Ni-NTA agarose-bound KP4 was eluted with elution buffer (500 mM imidazole, 20 mM Na2HPO4, 250 mM NaCl). The eluted fractions were dialyzed extensively in 1x PBS and concentrated with a centrifugal filter device (Amicon 3 kDa cut-off).
Purification of KP4 From Interstitial Fluid (IF) of Transgenic Plants
The extraction of IF from transgenic leaves (T2) expressing KP4 was performed following the infiltration- centrifugation technique (Chatterjee et al., 2017). The IF obtained post-extraction was enriched by 40% ammonium sulfate saturation and purified using Ni-NTA affinity purification. Finally, the eluted fractions were dialyzed and concentrated to detect KP4.
In Vitro Agar-Based Killing Zone Assay
For agar-based killing zone assay, we followed a previously published protocol with slight modifications (Allen et al., 2011). The plant material for plate-diffusion assay was prepared by crushing 50 mg of leaf material in 1 ml of sterile 1X Phosphate buffered saline (PBS, pH 7.0). A. alternata (MTCC No.10833) and P. exigua var. exigua (MTCC No. 2321) cultures procured from MTCC (Microbial Type Culture Collection, Chandigarh, India) were grown on Potato Carrot Agar (PCA) medium (HiMedia- M696-500G) for 3 and 5 days, respectively. Fine wells were cut into the agar with the help of a cork borer (diameter = 0.5 cm) and 100 μl of the enriched test plant material prepared from both transgenic and VC lines were added to each well on either side of the fungal growth. Two other wells containing 100 μl each of sterile water and PBS were used as controls. The plates were then incubated at 25°C under constant light for A. alternata and, 12:12 h light:dark cycle for P. exigua var. exigua. The plates were photographed when clear zones of inhibition started to appear around the point of application of test samples.
To compare the protection efficacies of MUASCsV8CP and CaMV35S2, we cloned the KP4-His downstream of the above promoters to generate pKMUASCsV8CP-KP4His and pKCaMV35S2-KP4His constructs. The recombinant constructs were then agro-infiltrated in healthy tobacco leaves. After 3–4 days of incubation, the infiltrated leaf segments were excised and total protein was isolated as described above. Equal concentrations of protein from both the constructs were used for the agar-based killing zone assays against A. alternata and P. exigua var. exigua.
In Vivo Disease Resistance Assays on Detached Leaves
Fungal resistance assays on detached leaves (employing A. alternata and P. exigua var. exigua) were performed according to a standard protocol with minor modifications (Emani et al., 2003). Briefly, young and healthy leaves were collected from 3 to 4 weeks old greenhouse grown plants and kept on wet paper towels in a petri-dish. On the other hand, A. alternata and P. exigua var. exigua were grown for 2 weeks on Potato Carrot Agar and Oat Meal Agar medium, respectively. Finally, an agar plug containing full mycelial growth was removed with the help of a cork borer (diameter = 0.5 cm) and agar plugs containing A. alternata and P. exigua var. exigua were placed on the adaxial surface of the leaves. In case of P. exigua var. exigua assay, the leaves were wounded with the help of a sterile needle at multiple points (6–7). Alongside, a mock leaf was kept to evaluate the effects of wounding inflicted by the needle. The petri-plates were sealed and kept in dark at 28°C. The leaves were photographed on the 14th and 18th day post-inoculation (dpi) of A. alternata and P. exigua var. exigua, respectively. The lesion area percentage was calculated with the help of millimeter graph paper method (Pandey and Singh, 2011).
Whole Plant Assays Against A. alternata and P. exigua var. exigua
The whole plant assays were performed following a previously published protocol. Healthy leaves of 8-weeks-old transgenic KP4L2#2, KP4L2#3, and VC plants were sprayed with conidial suspension of A.alternata and P. exigua var. exigua (Bithell and Stewart, 1998) using a sprayer. In case of P. exigua var. exigua, however, the leaves were wounded before applying the conidial suspension. The greenhouse conditions [photoperiod: 12:12 h (light:dark), light intensity: 220 μmol/m2/s, temperature: 25 ± 2°C and humidity: 90–95%] were kept extremely humid and the symptoms were recorded/photographed 20 dpi. The lesion area percentage was recorded as described above.
Statistical Analysis
All experiment procedures were carried out in at least three independent replicates and the statistical analyses was performed using one-way analysis of variance (ANOVA) where a P-value of <0.05 was considered as significant.
Results
Molecular Characterization of the Full-Length Transcript Promoter(-565 to +166) of CsVMV
The transient activities of each of the sixteen 5′ and 3′-end deletion constructs coupled to GUS reporter (pUCPMAGUS-CsVMV1 to pUCPMAGUS-CsVMVR3) (Figure 1A) were evaluated in tobacco protoplasts (cv. Xanthi Brad). Figure 1B depicts the average GUS activities obtained for each of the deletion constructs along with the CaMV35S promoter and an empty vector (EV) control with respective standard deviations (SDs). The data obtained clearly suggests that the relative GUS activity of the CsVMV8 (-215 to +166) was almost equivalent (∼1.1 times) to that obtained for the CaMV35S promoter; while the CsVMV7 (-256 to +166) ranked second. Furthermore, the TATA-less fragments CsVMVR2 (-565 to +16) and CsVMVR3 (-565 to -34) showed diminished GUS activities.
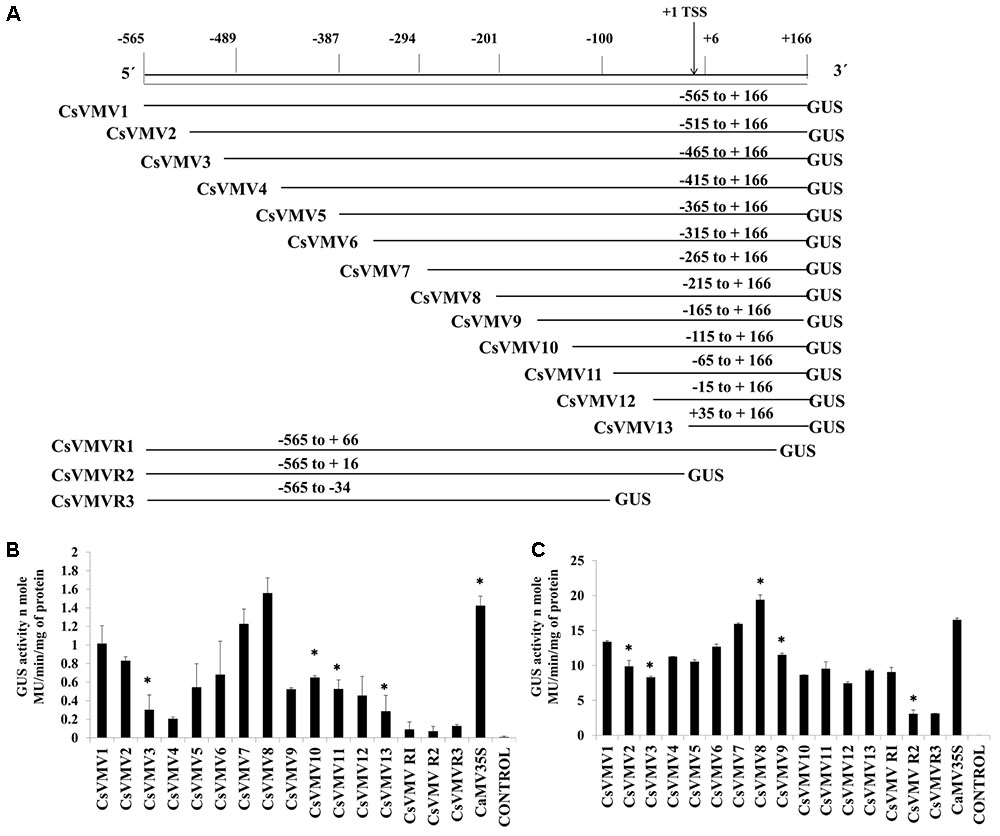
FIGURE 1. 5′–3′ and 3′–5′ end deletion analysis of CsVMV promoter fragment. (A) Schematic representation of deletion constructs (16 numbers) coupled to GUS reporter gene and their respective co-ordinates. The relative positions of TATA-box and TSS are shown. (B) Transient protoplast GUS expression analysis of the above mentioned deletion constructs in tobacco protoplasts. (C) Transient GUS activities of the promoter deletion constructs in whole plant of Nicotiana tabacum Samsun NN following Agro-infiltration assays. Average GUS activities (nmole MU/min/mg protein) of three independent experiments obtained from transformed protoplasts and agro-infiltration assays for above deletion constructs were presented with respective SD. Asterisks indicate level of significance where “∗∗” is more significant than “∗”.
Additionally, we evaluated the transient activities of the above constructs in whole plants of Nicotiana tabacum Samsun NN by agro-infiltration assays. The average GUS activities along with respective SD were presented in Figure 1C. The data obtained was in accordance with that of the protoplast assays.
Finally, we raised transgenic tobacco plants by Agrobacterium mediated leaf disk transformation protocol as described in Methods. We successfully raised 7–10 independent transgenic lines expressing uidA gene under the control of CsVMV1, CsVMV7, CsVMV8, and CaMV35S promoter fragments, respectively. Next, we performed segregation and Southern-blot analysis of T1 progeny transgenic plants. We observed, single copy integration of the transgene uidA in the transgenic lines (T1 generation) CsVMV1L4, CsVMV7L7, CsVMV8L6, CsVMV8L13, CaMV35SL9, and CaMV35S2L11. Finally, from the Southern-blot analysis, those transgenic plants having single copy insertion of the transgene (uidA) and exhibiting appropriate segregation ratio (3:1), namely CsVMV1L4, CsVMV7L7, CsVMV8L13, and CaMV35SL9 were chosen for further analyses (Figure 2A).
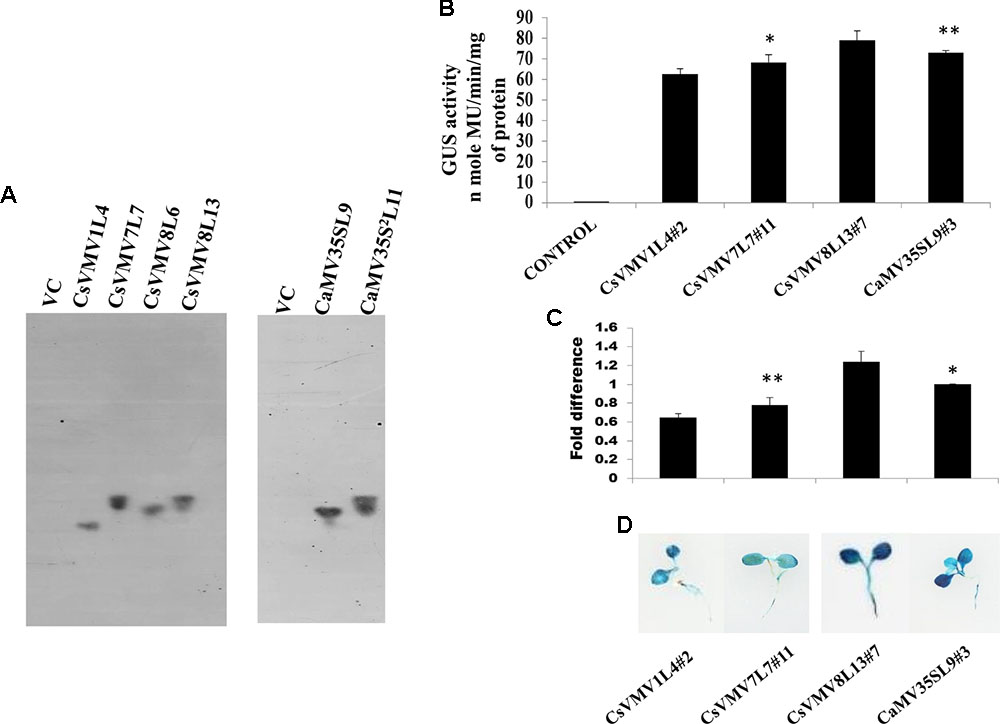
FIGURE 2. Transgenic activity analyses of CsVMV1L4, CsVMV7L7, CsVMV8L13, and CaMV35SL9. (A) Southern blot analyses to investigate the copy number of uidA gene in transgenic tobacco lines (T1 generation). (B) Stable transgenic GUS expression analyses of the above promoter constructs were presented with their corresponding SD. (C) Relative fold difference of uidA transcript levels in 21-day old transgenic seedlings under the control of respective promoter fragments. (D) Images of X-Gluc stained whole seedlings (T2 generation) expressing CsVMV1L4#2, CsVMV7L7#11, CsVMV8L13#7, and CaMV35SL9#3 promoters coupled to GUS reporter gene. Asterisks indicate level of significance where “∗∗” is more significant than “∗”.
We evaluated the stable GUS activities of 3-week old transgenic tobacco seedlings (T2 generation). On analysis of the results, we observed that the CsVMV8 showed ∼1.1 times stronger activity than that of the CaMV35S promoter (Figure 2B). Additionally, we performed qRT-PCR analysis of the transgenic plants as described in section “Materials and Methods.” The relative accumulation levels of uidA-mRNA level in transgenic plants (T2 generation) harboring the respective promoter constructs (viz. CsVMV1L4#2, CsVMV7L7#11, CsVMV8L13#7, and CaMV35SL9#3) confirmed ∼1.1-fold more expression of uidA under the control of CsVMV8 promoter in comparison to the CaMV35S promoter (Figure 2C). Finally, we obtained a fairly strong intensity of X-Gluc staining of 21 days-old transgenic seedlings expressing the CsVMV8 promoter (Figure 2D), in comparison to that obtained under other constructs.
Characterization of MUASCsV8 Chimeric Promoter
Figure 3A represents the schematic map with essential components of the chimeric promoter MUASCsV8CP along with CaMV35S, CaMV35S2 and MUAS35SCP. Upon transient activity analysis in tobacco protoplasts (Figure 3B) and in whole plants of tobacco, petunia, and tomato (Supplementary Figure S1), we detected that the GUS activity of MUASCsV8CP was ∼2.4 times stronger as compared to the CaMV35S promoter.
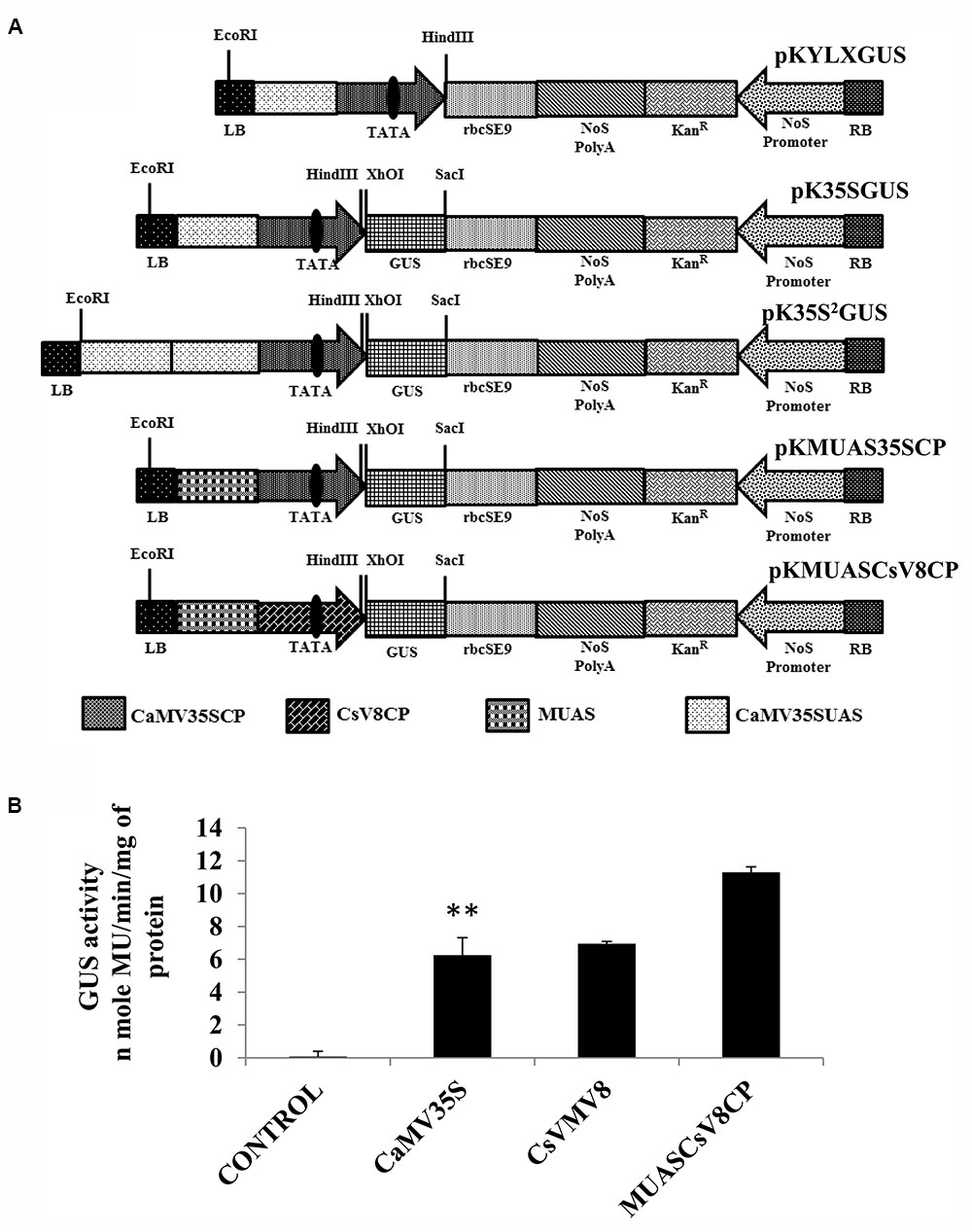
FIGURE 3. (A) Graphical representation of plant expression vectors harboring promoter constructs namely pKYLX (containing 35S promoter), pKYLXGUS, pK35S2GUS, pK35SGUS, pKMUAS35SCP, and pKMUASCsV8CP coupled to GUS reporter gene. The relative position of UAS, CP with TATA element and different essential components of the expression cassette were illustrated. (B) Transient activity analyses of the above mentioned recombinant promoters in tobacco protoplasts. Asterisks indicate level of significance where “∗∗” is more significant than “∗”.
We raised transgenic plants harboring the MUASCsV8CP-GUS and CaMV35S2-GUS (a modified version of CaMV35S with double enhancer domain) (Kay et al., 1987) constructs individually as already described. Southern blot analysis was performed to determine the copy number of the uidA transgene in above plant lines (T1 generation). The appearance of distinct southern-positive bands in MUASCsV8CPL4 and MUASCsV8CPL6 indicate integration of the transgene construct as a single copy insertion in the T1 progeny plant genome (Figure 4A). The progenies (T2 generation) of line MUASCsV8CPL6 exhibited expected segregation ratio and was chosen for further analyses.

FIGURE 4. Transgenic activity analyses of CsVMV8L13, MUASCsV8CPL6, MUAS35SCPL1, CaMV35SL9, and CaMV35S2L11. (A) Southern blot analyses performed to investigate uidA gene copy number in transgenic tobacco lines. (B) GUS expression analyses of the above promoter constructs in 21-day old transgenic seedlings. (C) Each bar represents the relative fold difference of uidA transcript levels of the respective constructs. The mean relative fold differences obtained from three independent experiments with respective SD for each promoter construct was presented. (D) Images of X-Gluc treated whole seedlings (T2 generation) expressing GUS under the control of CsVMV8L13#7, MUASCsV8CPL6#2, MUAS35SCPL1#1, CaMV35SL9#3 and CaMV35S2L11#6 promoter constructs. Asterisks indicate level of significance where “∗∗” is more significant than “∗”.
We measured the GUS activities of the respective constructs using 21 days-old T2 transgenic seedlings having the CaMV35S, CaMV35S2, MUASCsV8CP, and MUAS35SCP promoters. The data obtained clearly showed that the chimeric promoter MUASCsV8CP, reported in this study has ∼2.4 and ∼1.1 times enhanced GUS activity as compared to CaMV35S and CaMV35S2 promoters, respectively. The data also indicates toward the equivalence in activities of MUASCsV8CP and MUAS35SCP promoters (Figure 4B).
Additionally, we performed the Real-time PCR analysis and histochemical X-Gluc staining of the transgenic seedlings expressing GUS under the control of CaMV35S, CaMV35S2, MUASCsV8CP, and MUAS35SCP promoters. The fold change as shown in Figure 4C indicates that the uidA-mRNA accumulation in MUASCsV8CP was nearly 1.8 times and 1.1 times as compared to CaMV35S and CaMV35S2 promoters, respectively, while MUASCsV8CP and MUAS35SCP showed the previous trend of being equivalent. Finally, the histochemical staining also suggested strongest intensity using MUASCsV8CP and MUAS35SCP chimeric promoters (Figure 4D).
Generation of Tobacco Lines Expressing Totiviral Killer Protein (KP4) Under the Control of MUASCsV8CP Chimeric Promoter
Using Agrobacterium-mediated gene transformation method we raised 18 independent T1 generation transgenic tobacco plants (KanR) expressing totiviral KP4 driven by MUASCsV8CP. Figure 5A represents the schematic map of both KP4-His and VC constructs. We performed segregation analysis of T1 generation transgenic plants as described in section “Materials and Methods.” Out of eighteen transgenic lines, the progenies (T1 generation) of eight independent lines showed proper segregation ratio (KanR: KanS::3:1) and found to be Kanamycin (300 mg/L) resistant on half strength Murashige Skoog (MS) medium (Zhu et al., 1999) (Supplementary Table S2). Seeds from these selected plants (T1 generation) showing proper segregation ratios were propagated to get homozygous T2 transgenic plants.
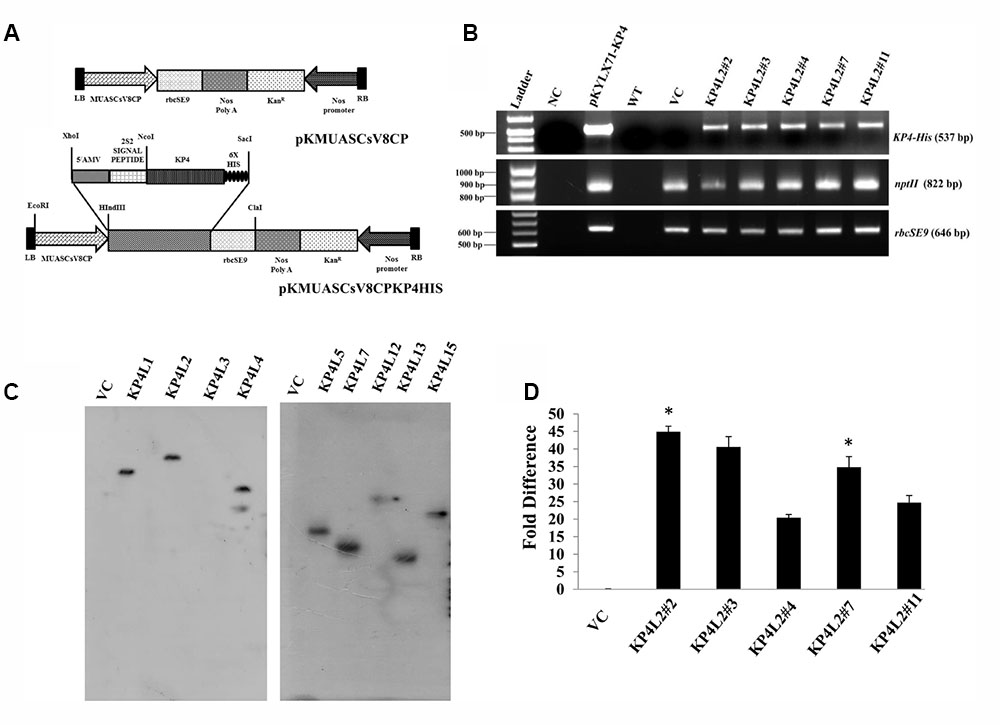
FIGURE 5. Gene-integration analyses of transgenic plants expressing KP4. (A) Schematic representation of the plant transformation vectors for the expression of KP4. (B) PCR amplification of different genes viz. (i) KP4 (ii) rbcSE9, and (iii) nptII from transgenic plant lines KP4L2#2, KP4L2#3, KP4L2#4, KP4L2#7, and KP4L2#11. The KP4 amplicon was electrophoresed along with a 1 kb DNA ladder (BioLit ProxiO 1 kb DNA Ladder plus BLL007) while the amplicons of rbcSE9 and nptII were electrophoresed along with a 100 bp Ladder (Thermo Scientific GeneRuler, 100 bp DNA Ladder #SM0241). (C) Southern blot analyses to check the copy number of the KP4 transgene in the above transgenic plant lines. (D) Real-time PCR analyses to check the abundance of KP4 m-RNA in different transgenic lines. Asterisks indicate level of significance where “∗∗” is more significant than “∗”.
Molecular Analysis of the Transgenic Plants Expressing MUASCsV8CP-KP4His
The integration of transgenes was confirmed by PCR amplification using primers specific to KP4 (537 bp), nptII (822 bp), and rbcSE9 (646 bp) (Supplementary Table S1). We confirmed the integration of different components of KP4 expression construct in T2 progenies of transgenic plant lines KP4L2#2, KP4L2#3, KP4L2#4, KP4L2#7, and KP4L2#11 whereas no amplification for KP4 was observed in the VC plant line (Figure 5B). We analyzed the sequence integrity of the amplicons and did not detect any mutations in the nucleotide sequences of KP4, nptII, and rbcSE9.
Next, Southern-blot analysis was performed with gDNA isolated from different KP4-His transgenic lines and a VC plant (T1 generation progenies). Southern analysis confirmed the single copy insertion of KP4-His transgene construct in KP4L1, KP4L2, KP4L5, KP4L7, KP4L12, KP4L13, and KP4L15 transgenic tobacco plants (Figure 5C). KP4L4 however, showed two copies of the transgene. Further, the transcript analysis of KP4 in KP4L2 transgenic lines (selected on the basis of segregation analysis) was performed by Real-time PCR. The result obtained from independent transgenic lines KP4L2#2, KP4L2#3, KP4L2#4, KP4L2#7, and KP4L2#11 clearly showed the accumulation of KP4 in respective transgenic lines in the following order: KP4L2#2 > KP4L2#3 > KP4L2#7 > KP4L2#11 > KP4L2#4 as shown in Figure 5D.
Isolation of Total Soluble Protein (TSP) and Western Blot Analysis of KP4-His
In order to detect the KP4-His protein in transgenic plants, we isolated and concentrated TSP from individual transgenic seedlings (T2 generation). After isolation and quantification of TSP, we performed western blot analysis using anti-His polyclonal (Figure 6A) primary antibody as described in section “Materials and Methods.” The KP4-His expressed in transgenic tobacco plants representing 2S2aTP-KP4-His was seen as a prominent band, the molecular mass being ∼18 kDa along with a few faint non-specific bands. To avoid the appearance of any contaminating bands, we performed western blot with anti-His monoclonal primary antibody (Figure 6B). In this case, a discrete single band of ∼18 kDa indicating the presence of 2S2aTP-KP4-His in TSP was confirmed.
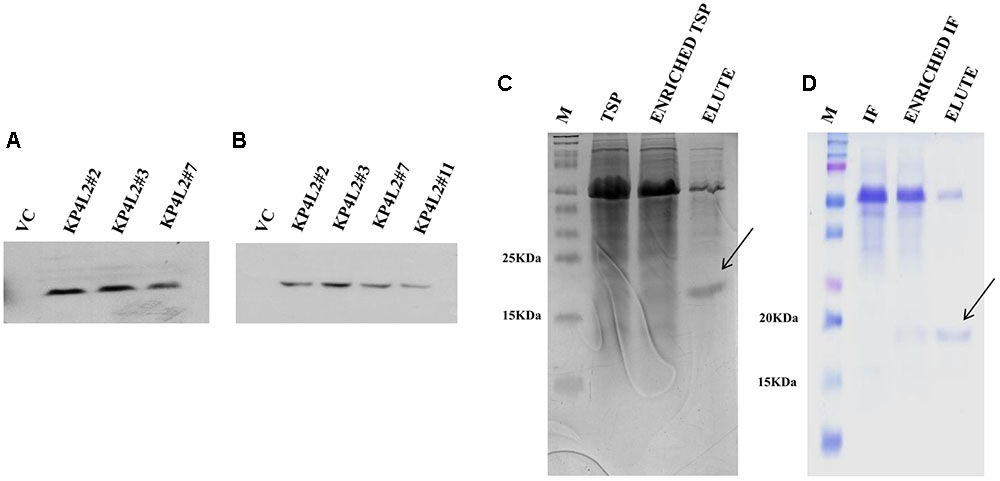
FIGURE 6. Western blot analysis of KP4 protein expression in transgenic tobacco plants using (A) anti-His polyclonal and (B) anti-His monoclonal primary antibody. Coomassie-stained 15% SDS-PAGE showing the partial purification of KP4 from (C) TSP and (D) IF fractions from pKMUASCsV8CP-KP4His transformed line KPL2#2.
Partial Purification of KP4 From Transgenic Tobacco Plants
To further detect the presence of KP4 in transgenic plants we performed nickel-nitrilotriacetic acid (Ni-NTA) affinity purification using TSP obtained from 21 days-old KP4L2#2 T2 transgenic plants as described in section “Materials and Methods.” The C-terminal 6xHis tag present in MUASCsV8CP-KP4-His (plant expression construct) facilitated the purification of recombinant KP4 using Ni-NTA resin. The SDS-PAGE analysis clearly showed a band of ∼18 kDa after 40% ammonium sulfate enrichment coupled to Ni-NTA affinity chromatography process (Figure 6C). We also observed a prominent contaminating band of ∼55 kDa which we assume might be of the RuBiSCO large subunit.
Detection of KP4 from Interstitial Fluid (IF) of Transgenic Tobacco Plants
In order to confirm the accumulation of KP4 in the IF of transgenic plants, we isolated and partially purified the intercellular fluid (as described in section “Materials and Methods”) from the transgenic line KP4L2#2. The presence of 2S2aTP from Arabidopsis 2S2 protein gene facilitated the accumulation of KP4 in the IF. The 40% ammonium sulfate enrichment and Ni-NTA affinity purification module was adopted for purifying KP4 from the IF of transgenic plants. Appearance of distinct bands of ∼18 kDa in the eluted fraction (as shown in Figure 6D) indicated that the KP4 is properly targeted to the plant apoplast or intercellular space. In this case also we observed the band of ∼55 kDa as a contaminant.
In Vitro Agar-Based Killing Zone Assay Against Foliar Fungal Pathogens
As KP4 blocks voltage-gated calcium channels of fungi, we sought to check the efficacy of plant-derived KP4 against two deuteromycetous fungal phyto-pathogens: A. alternata and P. exigua var. exigua using the ‘plate-diffusion’ assay (Allen et al., 2011). Enriched protein extracts for KP4L2#2, KP4L2#3, and KP4L2#7 were evaluated for anti-fungal activity as shown in Figures 7A–C (A. alternata) and Figures 7D–F (P. exigua var. exigua). It was evident that the KP4 activity represents a clear zone of inhibition around the point of application. This inhibition might be due to the blockage in calcium uptake by the fungal cells causing precocious hyphal disintegration.
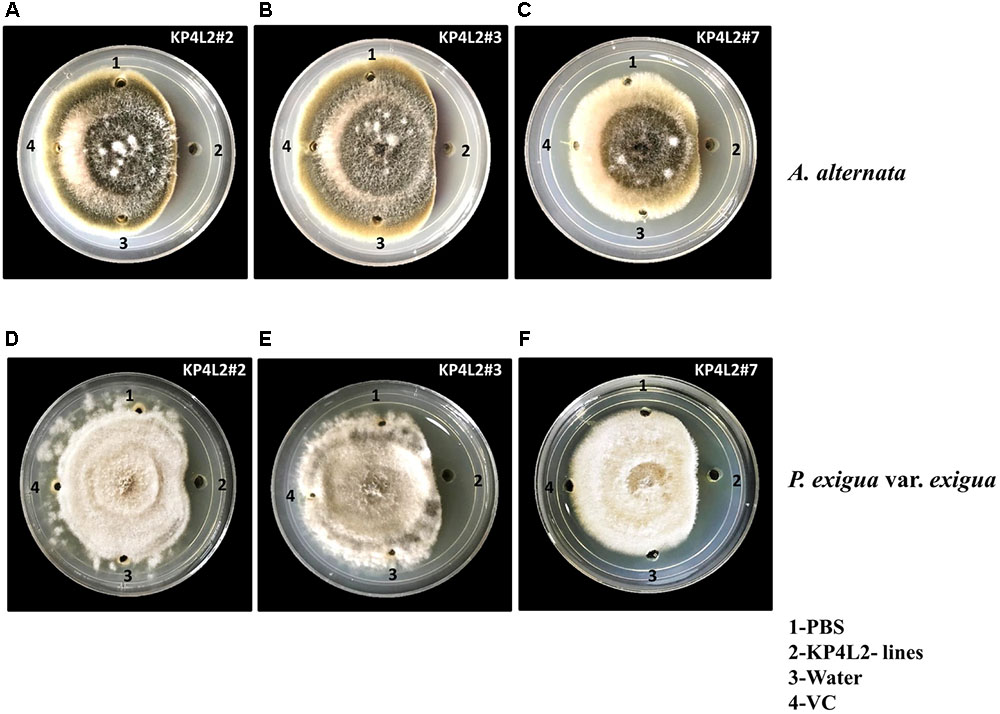
FIGURE 7. In vitro agar plate diffusion assays against (A–C) Alternaria alternata and (D–F) Phoma exigua var. exigua. Here, wells on each side of the fungal growth represent the VC, KP4L2#2, KP4L2#3, and KP4L2#7 plant materials grounded in 1X sterile PBS along with sterile water and buffer (PBS) taken as controls.
Comparative in Vitro Agar-Based Assays for MUASCsV8CP-KP4His and CaMV35S2-KP4His
Using KP4 as the gene of interest, we checked the efficacies of the MUASCsV8CP and CaMV35S2 promoters in the frame of fungal inhibition against A. alternata and P. exigua var. exigua. For this assay, we used transiently expressed KP4 from tobacco leaves infiltrated with pKMUASCsV8CP-KP4-His and pKCaMV35S2-KP4-His constructs (Supplementary Figure S3). Although we observed a slightly enhanced inhibition in case of pKMUASCsV8CP-KP4-His, we assume equivalence in activities of both promoter constructs.
Disease Resistance Assays With Alternaria alternata and Phoma exigua var. exigua
Deuteromycetous necrotrophic fungal pathogen Alternaria and hemi-biotrophic pycnidial pathogen Phoma causes foliar diseases in a wide variety of vegetables and annual plants (Momol et al., 2004; Mamgain et al., 2013). A. alternata causes brown spot disease on tobacco which is characterized by the appearance of roughly circular necrotic spots with concentric rings surrounded by yellow halo. On the other-hand, P. exigua var. exigua causes Phoma blight or ragged leaf spot in tobacco which is characterized by the appearance of irregular brown to ashy gray spots. In order to evaluate the anti-fungal efficacy of transgenic plants expressing KP4, both A. alternata and P. exigua var. exigua were used as test pathogens. Prior to performing the bio-assays with the above fungal strains (procured from MTCC), we checked for their virulence toward infecting tobacco plants. We found that both the pathogens could successfully infect mature tobacco leaves and develop visible symptoms (data not shown).
The disease resistance assays for both the pathogens were carried out with detached leaves kept on wet paper towels on petri dishes as described in section “Materials and Methods.” The detached-leaf technique is a routinely used method to determine the disease resistance of a particular transgenic plant against necrotrophic pathogens (Emani et al., 2003). In case of A. alternata, yellowish halos began to appear 7–8 days dpi while in P. exigua var. exigua, irregular brown spots appeared 10–11 dpi. The lesion area was measured 2 weeks post-inoculation for A. alternata (Figures 8A,B) while 18 dpi for P. exigua var. exigua (Figures 9A,B). The results revealed a high degree of protection in transgenic plants expressing KP4 as compared to the VC, as supported by the observation that the leaves of transgenic plants were found to be unaffected or developed smaller lesions than the VC plants.
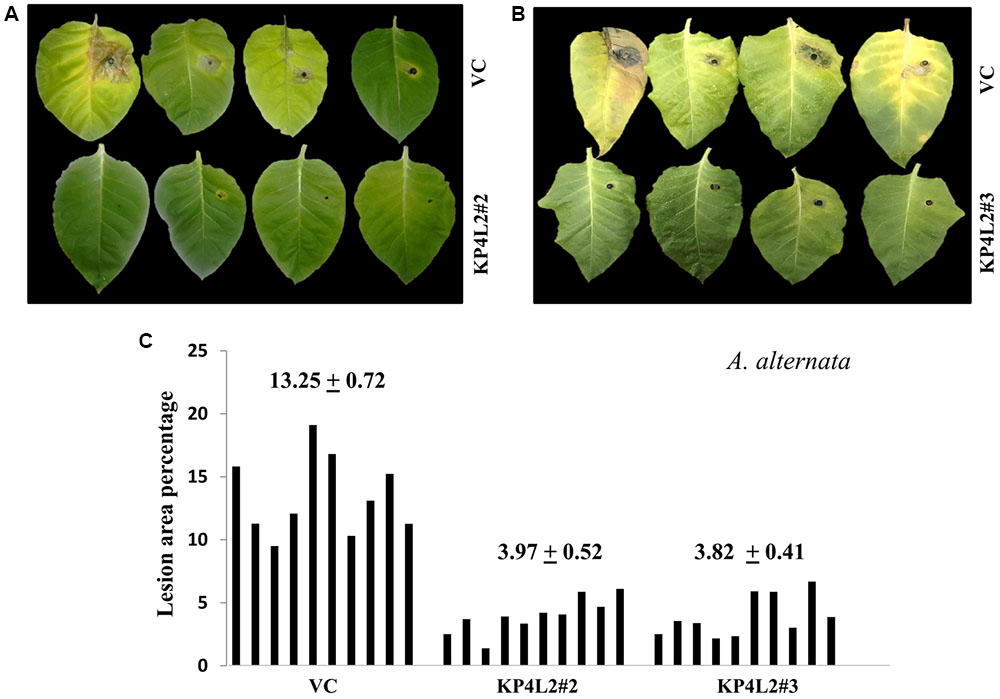
FIGURE 8. (A,B) Alternaria alternata disease resistance assays on detached leaves of VC, KP4L2#2 and KP4L2#3. An agar plug containing the full growth of the pathogen was placed on adaxial surface of the leaves. The images indicate lesion development 2 weeks post-infection. (C) Leaves from ten plant each of KP4L2#2, KP4L2#3, and VC were infected with one agar plug and subsequently lesion area percentage was measured by millimeter graph paper method. The numbers above each set of bars indicate the mean of percent lesion area ± standard error (SE) of the mean.
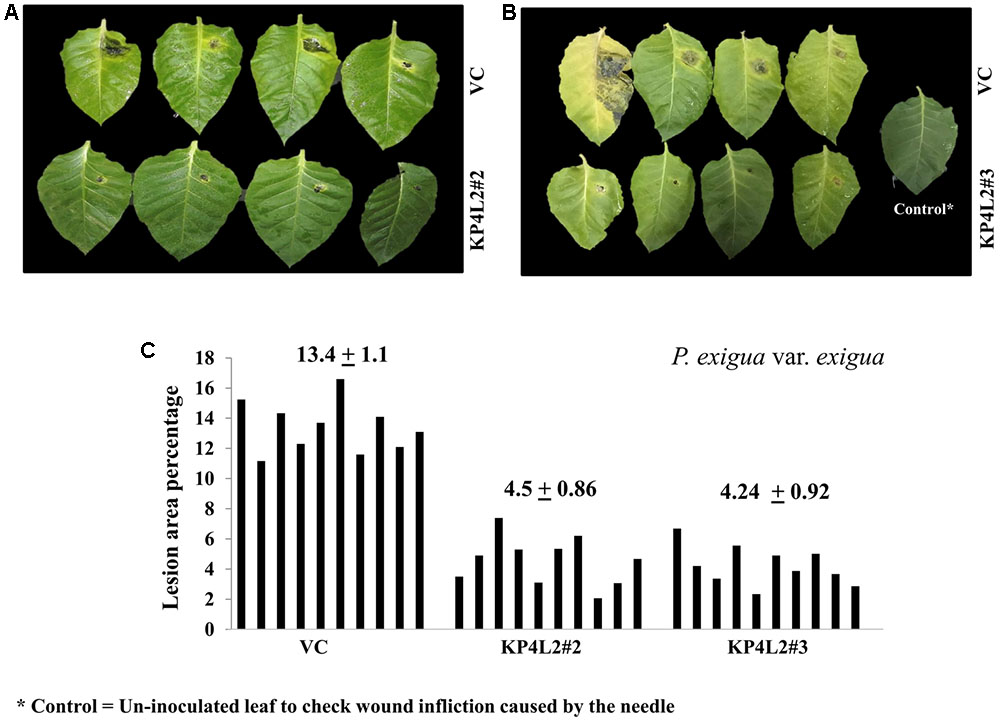
FIGURE 9. (A,B) P. exigua var. exigua disease resistance assays on detached leaves of VC, KP4L2#2, and KP4L2#3 indicating lesion development two and half weeks post-infection. (C) Graph showing lesion area percentage along with respective means and SE.
Following detached leaf assay, the lesion area percentage was determined using ‘Millimeter graph paper method’ as described in section “Materials and Methods.” We found, lesser lesion area in case of KP4 transgenic leaves as compared to the control plants (Figures 8C, 9C). These results suggested an enhanced anti-fungal activity of KP4 against these foliar fungal pathogens, which can be considered as significant.
Disease Resistance Assays Using Whole Plants Against A. alternata and P. exigua var. exigua
As described in section “Materials and Methods,” three independent experiments were carried out with whole plants of transgenic and VC lines. In all sets of experiments, the VC leaves developed distinct lesions much earlier than the KP4 transgenic plants. Significant lesions of A. alternata infection were observed 9–10 dpi in VC plants (Figure 10A). On an average, we observed that the VC plants showed 16.4% lesion area, while KP4L2#2 and KP4L2#3 showed 5.5 and 5.9% lesion areas, respectively (Figure 10B). Likewise, we observed higher degree of lesions caused by P. exigua var. exigua in VC plants as compared to the KP4 transgenic lines KP4L2#2 and KP4L2#3 (Figure 11A). We observed 12.4% lesion area (as a mean value) in VC plants, in comparison to 6.7 and 5.7% lesion areas in KP4L2#2 and KP4L2#3 transgenic lines, respectively (Figure 11B).
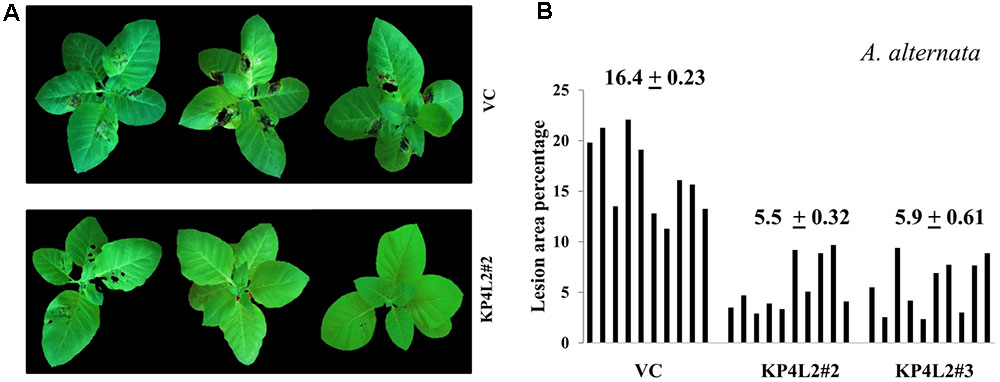
FIGURE 10. Whole plant assays against A. alternata. (A) Representative images of transgenic plant line KP4L2#2 along with VC 13 dpi. Ten plants each of KP4L#2, KP4L2#3, and VC were sprayed with conidial suspension of the fungus (107 CFU/mL) to initiate infection. (B) The graph represents lesion area percentage showing symptoms ± SE calculated on the 13th dpi.
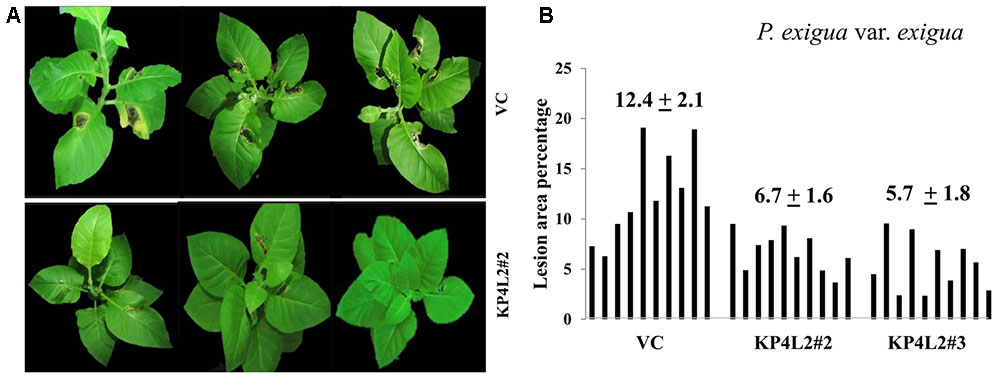
FIGURE 11. Whole plant assays against P. exigua var. exigua. (A) Representative images of transgenic plant line KP4L2#2 along with VC 20 dpi. Ten plants each of KP4L2#2, KP4L2#3, and VC were wounded with a sterile needle and conidial suspension of the fungus (107 CFU/mL) was applied to initiate infection. (B) The graph represents percent lesion area ± SE calculated on the 20th dpi.
Consistent with our results of detached-leaf assays for A. alternata and P. exigua var. exigua, we found significantly lesser leaf-area exhibiting symptoms in case of KP4 transgenic leaves as compared to the VC plants. Although, some of the leaves showed inconsistent results, the percent lesion area clearly suggested inhibition and/or delayed lesion development in transgenic plants expressing KP4. The inhibition found in detached-leaf and whole plant assays further proved the anti-fungal efficacy of KP4 against both A. alternata and P. exigua var. exigua.
Discussion
The emergence of various fungicide-resistant fungal-isolates deeply necessitates the evaluation of novel candidate genes coding for anti-microbial compounds. In the current scenario, where great genetic diversity and resistant isolates of pathogens thrive in nature, production of resistant crop varieties by transgenic approach is considered one of the easiest and effective device to control recalcitrant plant pathogens (Mundt, 2014). There are recent reports where in expression of transgenes in plants has conferred increased resistance against fungal pathogens, such as transgenic plants engineered with glucanase gene from alfalfa (Singh et al., 2014); ERF1-V gene from Haynaldia villosa (Xing et al., 2017) and PSS1 from Arabidopsis thaliana (Wang et al., 2017). Likewise, this study is a nascent report of significant anti-fungal activity exerted by recombinant KP4 on two deuteromycetous foliar pathogenic fungi A. alternata and Phoma exigua var. exigua.
Targeted gene expression in a transgenic platform lies under the control of the key genetic regulator ‘promoter’ which determines its spatio-temporal regulation. Designing of synthetic promoters can be of immense importance in plant modification through genetic engineering that can withstand biotic stress while also ensuring maximum productivity in the field of translational biology (Dey et al., 2015). In this context, we have developed a near constitutive synthetic promoter “MUASCsV8CP” having altered cis-architecture (as described in section “Materials and Methods”) with higher activity. It showed different levels of constitutive expression in different plant parts as follows: leaf > stem > root (Supplementary Figure S2). Owing to the above features, it may appear as an important tool for plant modification. It can be efficiently used in combination with other caulimoviral promoters like CaMV35S and/or CaMV35S2 for co-expressing multiple genes (gene pyramiding) in a plant cell to avoid unwanted genetic recombination. Furthermore, the MUASCsV8CP can stand as a substitute for CaMV35S2 promoter in plant biotechnology-based applications as its activity is nearly comparable to that of the latter. Our in vitro agar-based well diffusion assays revealed that the recombinant KP4 expressed under both CaMV35S2 and MUASCsV8CP promoters showed almost equal zone of inhibitions against A. alternata and P. exigua var. exigua, suggesting that the newly designed promoter can also be capable of imparting protection against phytopathogens.
Keeping in mind the above facts, we attempted to express a gene displaying anti-fungal properties under the control of our newly designed chimeric promoter MUASCsV8CP and evaluated its ability to inhibit the growth of phyto-pathogenic fungi. Here, KP4 was chosen as the gene of interest based upon the inter-strain hindrance found in double-stranded DNA totiviruses. Having structural similarity to the scorpion toxin AaHII, it was proposed and eventually proved that KP4 prevents uptake of calcium by fungal cells. As a result, it prevents hyphal tip growth, budding and viability of the fungi. KP4 has no sequence similarity to any other member of the ‘killer protein’ gene family and is extremely basic in nature with a pI of 9.0. The KP4 is known to be composed of five-stranded antiparallel β-sheet and two antiparallel α-helices that are stabilized by five disulfide bonds. It has been reported that KP4 has no apparent effect on the viability or subcellular structures of human, plant or insect cell lines as well as found to be harmless to soil inhabitants. Moreover, KP4 degrades in the stomach fluid and has no sequence resemblance with any known allergens (Schlaich et al., 2006; Allen et al., 2011).
A total of eight transgenic plant lines harboring MUASCsV8CP-KP4His (KP4L1, KP4L2, KP4L5, KP4L7, KP4L12, KP4L13, KP4L15, and KP4L17) showed normal agronomical parameters with proper segregation ratio but slight delay in root growth (Supplementary Table S2). However, this delay was not as evident as observed by Allen et al. (2008) probably because purified KP4 was applied exogenously in their experiments. The distinct southern positive bands in KP4L1, KP4L2, KP4L5, KP4L7, KP4L12, KP4L13, and KP4L15 (T1 progenies) indicated the single copy insertion of the transgene construct. We chose KP4L2 for our further studies as it exhibited good growth and maximum number of homozygous lines. Molecular analysis of the transgenic plants expressing KP4 confirmed the integration of KP4 in T2 progenies of KP4L2 lines viz. KP4L2#2, KP4L2#3, KP4L2#4, KP4L2#7, and KP4L2#11. Subsequently, Real-time PCR analysis of the above transgenic plants indicated toward the maximum transcript accumulation in KP4L2#2.
Plants monitor the apoplastic fluid, secrete hydrolytic enzymes and anti-microbial metabolites in response to fungal colonization which re-models the apoplast to become the first battleground for any plant-fungal interaction (Du et al., 2016; Ma et al., 2017). Hence, we targeted the chimeric construct harboring KP4 toward the plant apoplast using an ‘apoplast targeting sequence’ from Arabidopsis 2S2 protein gene (Sarkar et al., 2015). Also, our chimeric construct had a 6X-His tag at the 3′-end which aided the detection and purification of the same. We successfully purified recombinant KP4 from both TSP and IF of KP4L2#2 via Ni-NTA affinity purification. The SDS-PAGE analysis clearly showed the presence of 18 kDa band which appeared to be unglycosylated (Figures 6B,C).
We found strong antifungal activity of the KP4 extract against the virulent phytopathogens namely A. alternata and P. exigua var. exigua in the in vitro agar plate diffusion assays. This report is in accordance with the earlier published results of Spelbrink et al. (2004) where KP4 was extrinsically applied and found to be active against Fusarium graminearum, the causal agent of devastating head blight of small grain cereals (Yang et al., 2013). Interestingly, KP4 was found to inhibit the growth and progression of Phoma which has been previously described to form pseudosclerotia that perennate in soil during unfavorable environmental conditions.
The ‘detached-leaf’ assay is one of the most extensively used approaches for scoring disease symptoms. There are previous reports where this assay has been successfully employed to determine the anti-fungal potency of various compounds against Sclerotinia sclerotiorum, Cercospora arachidicola, and Blumeria graminis (Chenault et al., 2005; Roy-Barman et al., 2006; Anuradha et al., 2008). Likewise, we scored the disease symptoms against A. alternata and P. exigua var. exigua using this technique and observed that the transgenic lines expressing KP4 displayed substantial resistance against these foliar pathogens. The whole plant assays performed against A. alternata and P. exigua var. exigua also suggested effective anti-fungal protection offered by KP4 in planta.
Furthermore, we sought to investigate the anti-fungal activity of KP4 against Verticillium dahliae (MTCC No. 9998), which causes vascular wilt in a wide range of plant species (Fradin and Thomma, 2006). Interestingly, we did not observe any contemplating inhibition against this pathogen (Supplementary Figure S4) which may be due to its ability to form long-lasting resistant melanised clumps called microsclerotia in soil/media (Goud et al., 2003; Tian et al., 2016). Regardless, to our knowledge this is the first report of significant anti-fungal activity exhibited by transgenic plants expressing KP4 against two deuteromycetous phytopathogens.
Conclusion
Our study has a tripartite vision: firstly, the characterization of CsVMV promoter where CsVMV8 (-215 to +66) was found to be the highest expressing fragment; secondly, hybridization of the CsVMV8 fragment with UAS of MMV (-297 to -38) to develop an inter-molecularly shuffled recombinant promoter “MUASCsV8CP” and finally, expression of the totiviral KP4 under this chimeric promoter to develop transgenic tobacco resistant against two foliar pathogenic fungi – A. alternata and P. exigua var. exigua. This study provides insights into the development of potential candidates having in-built biotic-stress tolerance coupled to efficient caulimoviral promoters with discrete cis-organization in a transgenic approach for plant biotechnology.
Author Contributions
IM and ND conceptualized the experiments. DD, AS, and ND designed the experiments and critically analyzed the results and wrote the paper. DD performed all the experiments.
Funding
This research was supported by grants from core funds of Institute of Life Sciences, Department of Biotechnology, Government of India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Director, Institute of Life Sciences for his interest and support in this study. We also thank Mr. Abhimanyu Das for his technical help and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00278/full#supplementary-material
FIGURE S1 | Transient agro-infiltration assays to evaluate the GUS expression in whole plants of (A) Nicotiana tabacum (B) Petunia hybrida, and (C) Solanum lycopersicum expressing GUS under respective promoter constructs along with VC leaves.
FIGURE S2 | Spatial distributions of GUS activities under VC, CaMV35S, CaMV35S2, and MUASCsV8CP promoters. GUS activities (in nmole MU/min/mg protein) from the (A) root, (B) stem, and (C) leaf of transgenic tobacco seedlings (21 days old) expressing GUS under the control of the above promoter constructs were measured. Average GUS activities from three independent experiments for each of the promoter construct were presented with corresponding SDs.
FIGURE S3 |In vitro agar plate diffusion assays using transiently expressed KP4-His. Equal amounts of TSP extracted from tobacco leaves infiltrated with constructs MUASCsv8CP-KP4His and CaMV35S2-KP4His were used to compare the inhibitions conferred against (A,B) Alternaria alternata and (C,D) Phoma exigua var. exigua, respectively. Sterile PBS and water were used as controls.
FIGURE S4 |In vitro agar plate diffusion assays against Verticillium dahliae. Wells on each side of the fungal growth represent the VC and KP4L2 lines viz. KP4L2#2 (A) and KP4L2#3 (B) plant materials grounded in 1X sterile PBS.
TABLE S1 | List of oligonucleotides (primers) used in this study. ∗ The annealing temperature for all the above PCR/Real Time PCR primer sets was 58°C.
TABLE S2 | Segregation analysis of T1 generation plants expressing KP4-His.
References
Acharya, S., Ranjan, R., Pattanaik, S., Maiti, I. B., and Dey, N. (2014). Efficient chimeric plant promoters derived from plant infecting viral promoter sequences. Planta 239, 381–396. doi: 10.1007/s00425-013-1973-2
Allen, A., Islamovic, E., Kaur, J., Gold, S., Shah, D., and Smith, T. J. (2011). Transgenic maize plants expressing the Totivirus antifungal protein, KP4, are highly resistant to corn smut. Plant Biotechnol. J. 9, 857–864. doi: 10.1111/j.1467-7652.2011.00590.x
Allen, A., Snyder, A. K., Preuss, M., Nielsen, E. E., Shah, D. M., and Smith, T. J. (2008). Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth. Planta 227, 331–339. doi: 10.1007/s00425-007-0620-1
Anuradha, T. S., Divya, K., Jami, S., and Kirti, P. (2008). Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep. 27, 1777–1786. doi: 10.1007/s00299-008-0596-8
Avila-Adame, C., Olaya, G., and Köller, W. (2003). Characterization of Colletotrichum graminicola isolates resistant to strobilurin-related QoI fungicides. Plant Dis. 87, 1426–1432. doi: 10.1094/PDIS.2003.87.12.1426
Bithell, S., and Stewart, A. (1998). “Evaluation of the pathogenicity of Phoma exigua var. exigua on Californian Thistle,” in Proceedings of the New Zealand Plant Protection Conference: New Zealand Plant Protection Society, Palmerston North, 179–183.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Broglie, K., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., et al. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. doi: 10.1126/science.254.5035.1194
Burdon, J. (1993). “Genetic variation in pathogen populations and its implications for adaptation to host resistance,” in Durability of Disease Resistance, eds T. H. Jacobs and J. E. Parlevliet (Dordrecht: Kluwer), 41–56.
Chatterjee, A., Das, N. C., Raha, S., Maiti, I. B., Shrestha, A., Khan, A., et al. (2017). Enrichment of apoplastic fluid with therapeutic recombinant protein for efficient biofarming. Biotechnol. Prog. 33, 726–736. doi: 10.1002/btpr.2461
Chen, F., Liu, X., and Schnabel, G. (2013). Field strains of Monilinia fructicola resistant to both MBC and DMI fungicides isolated from stone fruit orchards in the eastern United States. Plant Dis. 97, 1063–1068. doi: 10.1094/PDIS-12-12-1177-RE
Chen, H., Nelson, R., and Sherwood, J. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16, 664–668, 670.
Chenault, K., Melouk, H., and Payton, M. (2005). Field reaction to Sclerotinia blight among transgenic peanut lines containing antifungal genes. Crop Sci. 45, 511–515. doi: 10.2135/cropsci2005.0511
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. London B Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Czislowski, E., Fraser-Smith, S., Zander, M., O’Neill, W. T., Meldrum, R. A., Tran-Nguyen, L. T., et al. (2017). Investigating the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. doi: 10.1111/mpp.12594 [Epub ahead of print].
Damalas, C. A., and Eleftherohorinos, I. G. (2011). Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 8, 1402–1419. doi: 10.3390/ijerph8051402
Dellaporta, S. L., Wood, J., and Hicks, J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/BF02712670
Dey, N., and Maiti, I. B. (1999). Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol. 40, 771–782. doi: 10.1023/A:1006285426523
Dey, N., Sarkar, S., Acharya, S., and Maiti, I. B. (2015). Synthetic promoters in planta. Planta 242, 1077–1094. doi: 10.1007/s00425-015-2377-2
Dilmaghani, A., Balesdent, M., Didier, J., Wu, C., Davey, J., Barbetti, M., et al. (2009). The Leptosphaeria maculans–Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 58, 1044–1058. doi: 10.1111/j.1365-3059.2009.02149.x
Drenth, A., and Guest, D. I. (2016). Fungal and oomycete diseases of tropical tree fruit crops. Annu. Rev. Phytopathol. 54, 373–395. doi: 10.1146/annurev-phyto-080615-095944
Du, Y., Stegmann, M., and Misas Villamil, J. C. (2016). The apoplast as battleground for plant–microbe interactions. New Phytol. 209, 34–38. doi: 10.1111/nph.13777
Emani, C., Garcia, J. M., Lopata-Finch, E., Pozo, M. J., Uribe, P., Kim, D. J., et al. (2003). Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol. J. 1, 321–336. doi: 10.1046/j.1467-7652.2003.00029.x
Fitt, B. D., Brun, H., Barbetti, M., and Rimmer, S. (2006). “World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus),” in Sustainable Strategies for Managing Brassica napus (Oilseed Rape) Resistance to Leptosphaeria maculans (Phoma Stem Canker), eds B. D. L. Fitt, N. Evans, B. J. Howlett, and B. M. Cooke (Berlin: Springer), 3–15. doi: 10.1007/1-4020-4525-5_1
Fradin, E. F., and Thomma, B. P. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Gage, M. J., Bruenn, J., Fischer, M., Sanders, D., and Smith, T. J. (2001). KP4 fungal toxin inhibits growth in Ustilago maydis by blocking calcium uptake. Mol. Microbiol. 41, 775–785. doi: 10.1046/j.1365-2958.2001.02554.x
Ganesa, C., Flurkey, W., Randhawa, Z., and Bozarth, R. (1991). Ustilago maydis virus P4 killer toxin: characterization, partial amino terminus sequence, and evidence for glycosylation. Arch. Biochem. Biophys. 286, 195–200. doi: 10.1016/0003-9861(91)90027-G
Gao, H., Narayanan, N. N., Ellison, L., and Bhattacharyya, M. K. (2005). Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microbe Interact. 18, 1035–1045. doi: 10.1094/MPMI-18-1035
Genet, J. L., Jaworska, G., and Deparis, F. (2006). Effect of dose rate and mixtures of fungicides on selection for QoI resistance in populations of Plasmopara viticola. Pest Manag. Sci. 62, 188–194. doi: 10.1002/ps.1146
Goud, J.-K. C., Termorshuizen, A. J., and Gams, W. (2003). Morphology of Verticillium dahliae and V. tricorpus on semi-selective media used for the detection of V. dahliae in soil. Mycol. Res. 107, 822–830. doi: 10.1017/S0953756203008050
Hahn, M. (2014). The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 7, 133–141. doi: 10.1007/s12154-014-0113-1
Itoh, Y., Takahashi, K., Takizawa, H., Nikaidou, N., Tanaka, H., Nishihashi, H., et al. (2003). Family 19 chitinase of Streptomyces griseus HUT6037 increases plant resistance to the fungal disease. Biosci. Biotechnol. Biochem. 67, 847–855. doi: 10.1271/bbb.67.847
Jauhar, P. P. (2006). Modern biotechnology as an integral supplement to conventional plant breeding: the prospects and challenges. Crop Sci. 46, 1841–1859. doi: 10.2135/cropsci2005.07-0223
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Karaoglanidis, G., Luo, Y., and Michailides, T. (2011). Competitive ability and fitness of Alternaria alternata isolates resistant to QoI fungicides. Plant Dis. 95, 178–182. doi: 10.1094/PDIS-07-10-0510
Kay, R., Chan, A., Daly, M., and McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236, 1299–1302. doi: 10.1126/science.236.4806.1299
Khani, M., Davidson, J., Sosnowski, M., and Scott, E. (2016). Survival of Phoma koolunga, a causal agent of ascochyta blight, on field pea stubble or as pseudosclerotia in soil. Plant Pathol. 65, 1246–1253. doi: 10.1111/ppa.12506
Kim, E., Lee, H. M., and Kim, Y. H. (2017). Morphogenetic alterations of Alternaria alternata exposed to dicarboximide fungicide, iprodione. Plant Pathol. J. 33, 95–100. doi: 10.5423/PPJ.NT.06.2016.0145
Koike, S. T., Subbarao, K. V., Verkley, G. J., Fogle, D., and O’Neill, T. M. (2006). Phoma basal rot of romaine lettuce in California caused by Phoma exigua: occurrence, characterization, and control. Plant Dis. 90, 1268–1275. doi: 10.1094/PD-90-1268
Köller, W., Parker, D., Turechek, W., Avila-Adame, C., and Cronshaw, K. (2004). A two-phase resistance response of Venturia inaequalis populations to the QoI fungicides kresoxim-methyl and trifloxystrobin. Plant Dis. 88, 537–544. doi: 10.1094/PDIS.2004.88.5.537
Kroumova, A. B., Sahoo, D. K., Raha, S., Goodin, M., Maiti, I. B., and Wagner, G. J. (2013). Expression of an apoplast-directed, T-phylloplanin-GFP fusion gene confers resistance against Peronospora tabacina disease in a susceptible tobacco. Plant Cell Rep. 32, 1771–1782. doi: 10.1007/s00299-013-1490-6
Kubota, M., and Abiko, K. (2002). Black rot of artichoke leaves caused by two Phoma species in Japan. J. Gen. Plant Pathol. 68, 208–211. doi: 10.1007/PL00013078
Kumar, D., Patro, S., Ghosh, J., Das, A., Maiti, I. B., and Dey, N. (2012). Development of a salicylic acid inducible minimal sub-genomic transcript promoter from Figwort mosaic virus with enhanced root-and leaf-activity using TGACG motif rearrangement. Gene 503, 36–47. doi: 10.1016/j.gene.2012.04.053
Kumar, D., Patro, S., Ranjan, R., Sahoo, D. K., Maiti, I. B., and Dey, N. (2011). Development of useful recombinant promoter and its expression analysis in different plant cells using confocal laser scanning microscopy. PLoS One 6:e24627. doi: 10.1371/journal.pone.0024627
Lindqvist-Kreuze, H., Hellqvist, S., Koponen, H., and Valkonen, J. (2003). Phoma–Didymella complex on hybrid arctic bramble with wilting symptoms. Plant Pathol. 52, 567–578. doi: 10.1046/j.1365-3059.2003.00885.x
Liu, N., Xu, S., Yao, X., Zhang, G., Mao, W., Hu, Q., et al. (2016). Studies on the control of ascochyta blight in field peas (Pisum sativum L.) Caused by Ascochyta pinodes in Zhejiang Province, China. Front. Microbiol. 7:481. doi: 10.3389/fmicb.2016.00481
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenzini, M., and Zapparoli, G. (2014). Characterization and pathogenicity of Alternaria spp. strains associated with grape bunch rot during post-harvest withering. Int. J. Food Microbiol. 186, 1–5. doi: 10.1016/j.ijfoodmicro.2014.06.008
Ma, S., Huang, Y., Davis, A., Yin, Z., Mi, Q., Menassa, R., et al. (2005). Production of biologically active human interleukin-4 in transgenic tobacco and potato. Plant Biotechnol. J. 3, 309–318. doi: 10.1111/j.1467-7652.2005.00125.x
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Mamgain, A., Roychowdhury, R., and Tah, J. (2013). Alternaria pathogenicity and its strategic controls. Res. J. Biol. 1, 1–9.
Momol, T., Kucharek, T., and Dankers, H. (2004). Fungal and Bacterial Disease Diagnoses for Distance Diagnostic and Identification System (DDIS). Gainesville, FL: University of Florida.
Mundt, C. C. (2014). Durable resistance: a key to sustainable management of pathogens and pests. Infect. Genet. Evol. 27, 446–455. doi: 10.1016/j.meegid.2014.01.011
Murray, F., Llewellyn, D., Peacock, W., and Dennis, E. (1997). Isolation of the glucose oxidase gene from Talaromyces flavus and characterisation of its role in the biocontrol of Verticillium dahliae. Curr. Genet. 32, 367–375. doi: 10.1007/s002940050290
Nicot, P. C., Stewart, A., Bardin, M., and Elad, Y. (2016). “Biological control and biopesticide suppression of Botrytis-incited diseases,” in Botrytis–the Fungus, the Pathogen and its Management in Agricultural Systems, eds S. Fillinger and Y. Elad (Cham: Springer), 165–187. doi: 10.1007/978-3-319-23371-0_9
Oyelakin, O. O., Opabode, J. T., Raji, A. A., and Ingelbrecht, I. L. (2015). A Cassava vein mosaic virus promoter cassette induces high and stable gene expression in clonally propagated transgenic cassava (Manihot esculenta Crantz). S. Afr. J. Bot. 97, 184–190. doi: 10.1016/j.sajb.2014.11.011
Pandey, S., and Singh, H. (2011). A simple, cost-effective method for leaf area estimation. J. Bot. 2011:6. doi: 10.1155/2011/658240
Patro, S., Kumar, D., Ranjan, R., Maiti, I. B., and Dey, N. (2012). The development of efficient plant promoters for transgene expression employing plant virus promoters. Mol. Plant 5, 941–944. doi: 10.1093/mp/sss028
Patro, S., Maiti, S., Panda, S. K., and Dey, N. (2015). Utilization of plant-derived recombinant human β-defensins (hBD-1 and hBD-2) for averting salmonellosis. Transgenic Res. 24, 353–364. doi: 10.1007/s11248-014-9847-3
Pethybridge, S. J., Esker, P., Hay, F., Wilson, C., and Nutter, F. W. Jr. (2005). Spatiotemporal description of epidemics caused by Phoma ligulicola in Tasmanian pyrethrum fields. Phytopathology 95, 648–658. doi: 10.1094/PHYTO-95-0648
Ranjan, R., Patro, S., Kumari, S., Kumar, D., Dey, N., and Maiti, I. B. (2011). Efficient chimeric promoters derived from full-length and sub-genomic transcript promoters of Figwort mosaic virus (FMV). J. Biotechnol. 152, 58–62. doi: 10.1016/j.jbiotec.2011.01.015
Roy-Barman, S., Sautter, C., and Chattoo, B. B. (2006). Expression of the lipid transfer protein Ace-AMP1 in transgenic wheat enhances antifungal activity and defense responses. Transgenic Res. 15, 435–446. doi: 10.1007/s11248-006-0016-1
Rushton, P. J., Reinstädler, A., Lipka, V., Lippok, B., and Somssich, I. E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 14, 749–762. doi: 10.1105/tpc.010412
Russell, G. E. (2013). Plant Breeding for Pest and Disease Resistance: Studies in the Agricultural and Food Sciences. Oxford: Butterworth-Heinemann.
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold spring harbor laboratory press.
Sanghera, G. S., Wani, S. H., Hussain, W., and Singh, N. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12, 30–43. doi: 10.2174/138920211794520178
Sarkar, S., Jain, S., Rai, V., Sahoo, D. K., Raha, S., Suklabaidya, S., et al. (2015). Plant-derived SAC domain of PAR-4 (Prostate Apoptosis Response 4) exhibits growth inhibitory effects in prostate cancer cells. Front. Plant Sci. 6:822. doi: 10.3389/fpls.2015.00822
Schardl, C. L., Byrd, A. D., Benzion, G., Altschuler, M. A., Hildebrand, D. F., and Hunt, A. G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61, 1–11. doi: 10.1016/0378-1119(87)90359-3
Schlaich, T., Urbaniak, B. M., Malgras, N., Ehler, E., Birrer, C., Meier, L., et al. (2006). Increased field resistance to Tilletia caries provided by a specific antifungal virus gene in genetically engineered wheat. Plant Biotechnol. J. 4, 63–75. doi: 10.1111/j.1467-7652.2005.00158.x
Sharma, P., Deep, S., Sharma, M., and Bhati, D. S. (2013). Genetic variation of Alternaria brassicae (Berk.) Sacc., causal agent of dark leaf spot of cauliflower and mustard in India. J. Gen. Plant Pathol. 79, 41–45. doi: 10.1007/s10327-012-0417-3
Singh, D., Ambroise, A., Haicour, R., Sihachakr, D., and Rajam, M. V. (2014). Increased resistance to fungal wilts in transgenic eggplant expressing alfalfa glucanase gene. Physiol. Mol. Biol. Plants 20, 143–150. doi: 10.1007/s12298-014-0225-7
Spelbrink, R. G., Dilmac, N., Allen, A., Smith, T. J., Shah, D. M., and Hockerman, G. H. (2004). Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol. 135, 2055–2067. doi: 10.1104/pp.104.040873
Tian, L., Wang, Y., Yu, J., Xiong, D., Zhao, H., and Tian, C. (2016). The mitogen-activated protein kinase kinase VdPbs2 of Verticillium dahliae regulates microsclerotia formation, stress response, and plant infection. Front. Microbiol. 7:1532. doi: 10.3389/fmicb.2016.01532
Tohidfar, M., Mohammadi, M., and Ghareyazie, B. (2005). Agrobacterium-mediated transformation of cotton (Gossypium hirsutum) using a heterologous bean chitinase gene. Plant Cell Tissue Organ Cult. 83, 83–96. doi: 10.1007/s11240-004-6155-2
Van de Graaf, P., O’Neill, T., Chartier-Hollis, J., and Joseph, M. (2003). Aspects of the biology and control of benzimidazole resistant isolates of Phoma clematidina, cause of leaf spot and wilt in clematis. J. Phytopathol. 151, 442–450. doi: 10.1046/j.1439-0434.2003.00748.x
Verdaguer, B., De Kochko, A., Beachy, R. N., and Fauquet, C. (1996). Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol. Biol. 31, 1129–1139. doi: 10.1007/BF00040830
Verdaguer, B., de Kochko, A., Fux, C. I., Beachy, R. N., and Fauquet, C. (1998). Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 37, 1055–1067. doi: 10.1023/A:1006004819398
Wang, B., Sumit, R., Sahu, B. B., Ngaki, M. N., Srivastava, S. K., Yang, Y., et al. (2017). An Arabidopsis glycine-rich plasma membrane protein enhances disease resistance in soybean. Plant Physiol. doi: 10.1104/pp.16.01982 [Epub ahead of print].
West, J. S., Kharbanda, P., Barbetti, M., and Fitt, B. D. (2001). Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 50, 10–27. doi: 10.1046/j.1365-3059.2001.00546.x
Xing, L., Di, Z., Yang, W., Liu, J., Li, M., Wang, X., et al. (2017). Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8:1948. doi: 10.3389/fpls.2017.01948
Yang, F., Jacobsen, S., Jørgensen, H. J., Collinge, D. B., Svensson, B., and Finnie, C. (2013). Fusarium graminearum and its interactions with cereal heads: studies in the proteomics era. Front. Plant Sci. 4:37. doi: 10.3389/fpls.2013.00037
Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi: 10.1046/j.1365-313x.2000.00760.x
Keywords: killer protein, Alternaria alternata, Phoma exigua, caulimovirus, recombinant promoter
Citation: Deb D, Shrestha A, Maiti IB and Dey N (2018) Recombinant Promoter (MUASCsV8CP) Driven Totiviral Killer Protein 4 (KP4) Imparts Resistance Against Fungal Pathogens in Transgenic Tobacco. Front. Plant Sci. 9:278. doi: 10.3389/fpls.2018.00278
Received: 17 October 2017; Accepted: 16 February 2018;
Published: 05 March 2018.
Edited by:
Fernando Ponz, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainReviewed by:
Rosa Rao, University of Naples Federico II, ItalyKarabi Datta, University of Calcutta, India
Maria Raffaella Ercolano, University of Naples Federico II, Italy
Copyright © 2018 Deb, Shrestha, Maiti and Dey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nrisingha Dey, nrisinghad@gmail.com; ndey@ils.res.in
 Debasish Deb
Debasish Deb Ankita Shrestha
Ankita Shrestha Indu B. Maiti
Indu B. Maiti Nrisingha Dey
Nrisingha Dey