Marsupials and Multi-Omics: Establishing New Comparative Models of Neural Crest Patterning and Craniofacial Development
- The School of BioSciences, University of Melbourne, Parkville, VIC, Australia
Studies across vertebrates have revealed significant insights into the processes that drive craniofacial morphogenesis, yet we still know little about how distinct facial morphologies are patterned during development. Studies largely point to evolution in GRNs of cranial progenitor cell types such as neural crest cells, as the major driver underlying adaptive cranial shapes. However, this hypothesis requires further validation, particularly within suitable models amenable to manipulation. By utilizing comparative models between related species, we can begin to disentangle complex developmental systems and identify the origin of species-specific patterning. Mammals present excellent evolutionary examples to scrutinize how these differences arise, as sister clades of eutherians and marsupials possess suitable divergence times, conserved cranial anatomies, modular evolutionary patterns, and distinct developmental heterochrony in their NCC behaviours and craniofacial patterning. In this review, I lend perspectives into the current state of mammalian craniofacial biology and discuss the importance of establishing a new marsupial model, the fat-tailed dunnart, for comparative research. Through detailed comparisons with the mouse, we can begin to decipher mammalian conserved, and species-specific processes and their contribution to craniofacial patterning and shape disparity. Recent advances in single-cell multi-omics allow high-resolution investigations into the cellular and molecular basis of key developmental processes. As such, I discuss how comparative evolutionary application of these tools can provide detailed insights into complex cellular behaviours and expression dynamics underlying adaptive craniofacial evolution. Though in its infancy, the field of “comparative evo-devo-omics” presents unparalleled opportunities to precisely uncover how phenotypic differences arise during development.
1 Introduction
One of the most remarkable, yet enigmatic aspects of the vertebrate skull is the broad diversity of craniofacial shapes observed between species. While our understanding of craniofacial biology has been significantly enhanced through investigations across several vertebrate models, we still know very little about the processes that drive the development of distinct craniofacial adaptations. Comparative embryology and developmental biology in jawed and jawless vertebrates have revealed that craniofacial morphogenesis is driven by a transient population of embryonic progenitors called the neural crest (Kuratani et al., 2018; Fish, 2019). Multipotent neural crest cells (NCCs) direct patterning and development of the head and neck, amongst other structures, and are controlled by deeply conserved gene regulatory networks (GRNs) constituting a species-generic program (Green et al., 2015; Martik and Bronner, 2021). The combination of these developmental and evolutionary observations, with forward genetics and human clinical models of craniofacial disease, have provided a holistic understanding of how the craniofacial prominences are patterned and skull bones develop (Wilkie and Morriss-Kay, 2001; Szabo-rogers et al., 2010; Murillo-Rincón and Kaucka, 2020). However, despite this fundamental understanding of craniofacial biology across vertebrates, we still know remarkably little about how species-specific diversity arises and is patterned during development.
One way we can begin to address this phenomenon is by utilizing comparative models to quantitatively examine how disparities or similarities arise during development. These models need to be suitably chosen depending on the hypothesis being tested. i.e., examining closely related species with unique skull morphologies (disparity), versus distantly related species with similar skull morphologies (convergence). Mammals provide excellent examples to address these hypotheses, owing to their conserved anatomy yet remarkable craniofacial disparity or convergence, shared developmental patterns, heterochrony and lineage-specific constraints, and appropriate divergence times, e.g., within orders or across clades. Through application of these models, we can begin to tease apart how facial morphogenesis and shape diversity is regulated at the cellular and molecular level (Newton et al., 2017; Usui and Tokita, 2018; Newton and Pask, 2020), informing new models of development.
In this article, I outline my perspectives on establishing new comparative mammalian models for investigations into the developmental basis of craniofacial patterning. I discuss the underlying biology of craniofacial morphogenesis, including NCC biology, its influence on patterning, and heterochrony between therian mammals. I emphasize the importance of establishing an appropriate marsupial model for comparative investigations with the eutherian laboratory mouse, including the establishment and utilization of transgenic approaches. Finally, I discuss how single-cell multi-omic approaches, regularly utilized in developmental biology, should be applied to comparative craniofacial models to scrutinize differential cell and molecular behaviours underlying mammalian craniofacial patterning and shape diversity. Establishing a marsupial model for comparative mammalian biology will strengthen our understanding of craniofacial development and how morphological diversity is generated throughout evolution.
2 Neural Crest Cells and Patterning of the Head
Development of the vertebrate head and craniofacial skeleton is achieved largely through the contribution of migratory NCCs. NCC specification is regulated through a deeply-conserved GRN comprised of shared suites of core transcriptional regulators, constituting a species-generic program (Green et al., 2015; Martik and Bronner, 2021). During early embryogenesis, NCCs arise within the neuroectoderm at the neural plate border (Figure 1A). Initially, WNT, FGF, and BMP signalling pathways define the border and initiate pre-migratory NCC specification in response to activation of SOX9 (Cheung and Briscoe, 2003). Committed NCCs undergo activation of epithelial-to-mesenchymal transcription factors, SOX10, SNAIL and SLUG, and other NCC-specific transcription factors such as MSX1 and TFAP2A (Martik and Bronner, 2021), causing the cells to delaminate and migrate away from the forming neural tube (Figure 1A). The spatial location of NCCs along the anterior-posterior axis of the embryo predefine their paths of migration. The anterior-most cranial NCCs of the forebrain and hindbrain populate the frontonasal process and maxillary arch (Figures 1B,C), contributing to development of the facial skeleton, whereas more posterior cranial NCCs populate the pharyngeal arches to form the musculoskeletal elements of the lower jaw and neck (Figures 1B,C). NCC migration into their target primordia occur in response to cues within the local extracellular environment. Here, as NCCs populate the developing prominences, reciprocal FGF, BMP, SHH, and retinoic acid signalling interactions between mesenchymal NCCs and the epithelial ectoderm and endoderm direct their spatial organization and activate GRNs responsible for proliferation, outgrowth and differentiation of the craniofacial skeleton [(Creuzet et al., 2004; Minoux and Rijli, 2010; Dash and Trainor, 2020; Murillo-Rincón and Kaucka, 2020) and references within].
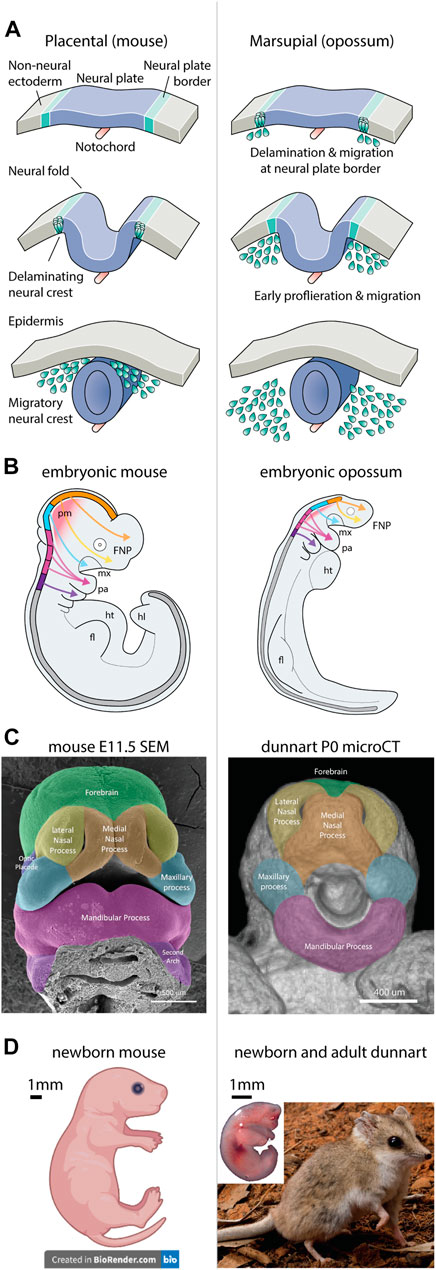
FIGURE 1. Neural crest and craniofacial development between therian mammals. Craniofacial heterochrony between therian mammals arises from altered neural crest cell behaviours. (A) In placental mammals, the neural crest forms in the neural folds and delaminates from the neural tube to migrate throughout the embryo. In marsupials however, the neural crest forms and delaminates from the neural plate border, leading to accelerated migration in the early embryo—redrawn from (Martik and Bronner, 2021). (B) Neural crest migration pathways are shared between therian mammals, though are accelerated in marsupials relative to developmental stage. Note marsupials display rapid development of the facial complex and forelimbs, while the CNS and hindlimbs are rudimentary. (C) The facial prominences of newborn marsupials resemble those observed in an embryonic mouse (credit FaceBase.org) (Samuels, B. D., 2020). (D) Comparative images of newborn mouse and dunnart, demonstrating the altriciality of the dunnart at birth. The adult dunnart superficially resembles a mouse. Image credits: dunnart newborn—Laura Cook; Dunnart Adult—David Paul—Museums Victoria; mouse pup—created with BioRender.com. Abbreviations: fl, forelimb; FNP, frontonasal process; hl, hindlimb; ht, heart; md, mandibular process; mx, maxillary process; pa, pharyngeal arch; pm, paraxial (head) mesoderm.
2.1 The Origin of Species-Specific Pattern
The specific influence of NCCs patterning the vertebrate head has been showcased through cross-species transplantations and xenografts. These experiments have revealed that NCCs possess intrinsic programming and autonomous behaviours which drive species-specific patterning (Schneider, 2018). NCC transplantation chimeras in avian embryos see recipient species develop donor-specific patterning, bone formation and craniofacial morphology (Schneider and Helms, 2003; Chen et al., 2012; Hall et al., 2014; Ealba et al., 2015). Such morphological outcomes are driven via intrinsic NCC behaviours, including donor-specific regulation of the cell-cycle and distinct expression of transcriptional regulators and signalling factors (Hall et al., 2014). These unique NCC behaviours are further suggested to influence their local environment to produce distinct morphological outcomes. Here, NCC-derived signals modulate activation of reciprocal signalling pathways to the surrounding ectoderm and endoderm which determine gene expression and spatiotemporal patterning of the facial primordia (Schneider, 2018). In agreement with this, while shared (species-generic) genes and patterning factors are active in the developing facial prominences, each species displays distinct expression profiles during beak outgrowth and development (Wu et al., 2004; Wu et al., 2006; Brugmann et al., 2010). Together, these data suggest that intrinsic species-specific NCC programming influences interactions with their local environment, regulating differentiation of craniofacial cells and tissues and the development of distinct morphological identities.
2.2 Marsupial Heterochrony, Accelerated Neural Crest Cell Specification and Migration
The mechanisms underlying mammalian neural crest patterning and craniofacial development have been largely ascertained from studies in mouse. However, while these findings may be relevant for eutherian mammals, comparative studies in marsupials have revealed pronounced heterochrony in their NCC behaviours. During specification at the neural plate border, marsupial NCCs undergo rapid delamination and migration prior to neural plate folding (Figure 1A) (Smith, 1997; Smith, 2001; Smith, 2020; Vaglia and Smith, 2003; Wakamatsu et al., 2014), leading to large accumulations of NCCs within the forming facial prominences at an earlier equivalent developmental stage to that seen in eutherians (Figure 1B) (Smith, 2001). Remarkably, very little is known about the molecular regulation of marsupial NCCs during development. Two related studies revealed that in the opossum embryo, NCC specification and delamination is accelerated as a result of sequence alteration in a SOX9 enhancer which drives early activation of SOX9 in the neural plate border (Wakamatsu et al., 2014; Wakamatsu and Suzuki, 2019). This accelerated activation likely influences the heterochronic migration, proliferation, and ossification observed in marsupials, though no other studies have interrogated these processes or drawn comparisons with eutherians. This represents a large gap in our understanding of how NCC behaviours influence development and differ between distinct mammalian clades. Future studies should address the genetic underpinning of these behavioural differences and their contribution to craniofacial patterning. Curiously, it remains to be seen whether transplantation of marsupial NCCs into recipient mouse embryos (or vice versa) would retain their heterochronic behaviours and promote differential establishment of the facial prominences and skeleton. As such, observations into marsupial NCC biology and comparative heterochrony between eutherians are required for a complete understanding of mammalian NCC patterning and craniofacial development.
3 Craniofacial Patterning, Disparity, and Convergence in Therian Mammals
Mammals have evolved unique cranial adaptations which distinguish them from other vertebrates. Evolutionary novelties such as a hinged jaw, middle ear bones and muzzle or semi-motile snout (Higashiyama et al., 2021) have allowed mammals to adapt to a diverse range of ecological niches. However, since diverging ∼160 million years ago (Bininda-Emonds et al., 2007), therian mammals (marsupials and eutherians) have evolved distinct reproductive and developmental strategies, resulting in heterochrony and lineage-specific constraints (Smith, 1997). The marsupial mode of reproduction requires well-developed jaws and forelimbs at a comparatively early stage to allow the altricial neonate to crawl from the birth canal into pouch and attach to the teat for an extended period of suckling. These distinct functional requirements require differences in the onset of development of the olfactory and central nervous system, and musculoskeletal element of the head body and limbs (Smith, 1997; Nunn and Smith, 1998; Weisbecker et al., 2008; Sears, 2009; Keyte and Smith, 2010; Keyte and Smith, 2012; Chew et al., 2014). Particularly, during craniofacial development, ossification and suture closure of the facial bones are advanced to meet the functional requirements associated with suckling (Sánchez-Villagra et al., 2008; Rager et al., 2014; Spiekman and Werneburg, 2017; Cook et al., 2021). Overall, these constraints imposed on the marsupial orofacial bones are suggested to limit evolvability of their cranial anatomy.
Marsupials have evolved altered patterns of cranial modularity (Goswami, 2006; Goswami, 2007; Goswami et al., 2009; Goswami et al., 2012; Goswami et al., 2016) which are thought to reduce their overall skull shape diversity compared to eutherians (Bennett and Goswami, 2013; Fabre et al., 2021). For example, the marsupial jaws form a functionally constrained module, while the frontonasal bones and neurocranium are under relaxed constraint and can evolve more freely (Goswami et al., 2016). On the other hand, eutherian mammals largely lack these constraints during development, thus their cranial bone groups are free to evolve independently producing a greater range of morphological adaptations. Importantly, these frontonasal, jaw and neurocranium bone groups (anatomical modules) possess distinct embryonic origins, arising from the cranial NC, first arch NC, or head mesoderm, respectively (developmental modules) (Couly et al., 1993; Jiang et al., 2002; Yoshida et al., 2008). These semi-independent origins, known as mosaicism, allow flexibility in how different cranial morphologies can evolve and change (Felice and Goswami, 2018), even in the presence of functional constraints. Importantly, the combination of cranial mosaicism with cell-autonomous programming of NCCs provide clues as to how particular cranial adaptations can arise during evolution. Specifically, it can be hypothesized that evolution within GRNs associated with cranial progenitor cell types can produce adaptive morphological outcomes.
These evolutionary hypotheses have been recently applied, investigating the origins of the remarkable craniofacial convergence observed between the marsupial thylacine and eutherian wolf (Goswami et al., 2011; Feigin et al., 2018). During postnatal ontogeny, the thylacine and wolf frontonasal and neurocranial bones develop with strong shape convergence, whilst the thylacine’s maxillary bones (upper jaw) possess constrained shape shared with other marsupials and disparate patterns to that seen in the wolf (Newton et al., 2021). This supports the notion that adaptive evolution (similarity and disparity) of the mammalian skull is modular (Goswami, 2006; Goswami and Polly, 2010), facilitated by mosaic evolution of select bone groups. Furthermore, the distinct embryological origins of the convergent bone groups observed between the thylacine and wolf suggest their underlying GRNs may be convergently targeted by selection. Indeed, comparative genomic investigations of the loci underlying the thylacine and wolf’s cranial convergence revealed enrichment of homoplasy in GRNs associated with cranial mesenchyme migration, differentiation, and ossification (Feigin et al., 2019). Taken together, these studies support the hypothesis that evolution within GRNs of embryonic cranial precursors may specify species-specific patterning of the facial primordia, ultimately influencing craniofacial shape. However, this hypothesis requires further validation, particularly into the role of mammalian NCC heterochrony during early facial development and patterning. As such, establishing a marsupial model of NCC patterning and craniofacial biology that is amenable to manipulation is essential to our understanding of how evolutionary adaptations are produced during development.
4 A Marsupial Model to Investigate Mammalian Heterochrony
Modern studies of NCC development and craniofacial patterning in mammals have leveraged the mouse Cre-Lox system, with several transgenic reporter lines established to target various stages of the NCC or skull developmental pathway (Zhang et al., 2002; Rodda and McMahon, 2006; Yoshida et al., 2008; Stine et al., 2009; Rauch et al., 2010; Lewis et al., 2013). Of these, the Wnt1-cre strain has been widely utilized for NCC developmental biology to uncover the spatiotemporal decisions underlying NCC differentiation (Soldatov et al., 2019), defining tissue boundaries between NCC and non NCC-derived cranial structures (Jiang et al., 2002; Hu et al., 2004; Lewis et al., 2013), as well as a multipotent NCC line to define models of differentiation (Ishii et al., 2012; Nguyen et al., 2018). In addition, chromatin profiling of mouse NCCs and craniofacial prominences have annotated the regulatory landscape of craniofacial enhancers and putative GRNs (Visel et al., 2009a; Visel et al., 2009b; Attanasio et al., 2013; Pennacchio et al., 2018). Yet while these tools provide powerful and valuable outcomes, they are scarcely utilized outside murine models, limiting comparative mammalian research. Recently however, several new marsupial resources are actively being established, including a pioneering study to generate the first genetically modified marsupials—founder lines of tyrosinase knockout opossums (Kiyonari et al., 2021). Though marsupial transgenic resources are still in their infancy, these advances have primed the generation of new marsupial Cre-Lox resources for developmental investigations. Of note, the generation of a marsupial orthologous WNT1-cre line would allow targeted labelling of neural crest cells and their craniofacial derivatives, opening the door for comparative developmental studies and investigations into mammalian heterochrony.
4.1 The Dunnart as the Gold-Standard Marsupial Model
In the past, several marsupial species have provided insights to various aspects of mammalian biology, reproduction, and development (Selwood and Coulson, 2006), with the American opossum (Monodelphis domestica) informing models of NCC and limb development (Martin and Mackay, 2003; Vaglia and Smith, 2003; Goswami et al., 2012; Beiriger and Sears, 2014). However, Monodelphis are basal American marsupials (superorder Ameridelphia) possessing an 80-million-year divergence from Australian marsupials, similar to times shared between human and mouse. Therefore, an Australian laboratory-based marsupial model with similar easy husbandry, year-round breeding and experimental manipulation is still required for a more complete understanding of mammalian (and marsupial) biology. Dunnarts (Sminthopsis sp.; superorder Australidelphia) are small, carnivorous, mouse-like marsupials that are easy to maintain, possess simple husbandry and are polyovular, poly-oestrous and spontaneous ovulators which produce multiple litters of up to 10 pouch young year round (Frigo and Woolley, 1996; Frigo and Woolley, 1997; Suárez et al., 2017; Cook et al., 2021). Owing to this, several new resources are being established for the fat-tailed dunnart (S. crassicaudata, hereafter referred to as the dunnart) as the gold-standard model for next-gen marsupial biology. These include a chromosome level assembly, transcriptomic and gene regulatory datasets, induced pluripotent cells, inbred strains, and transgenic laboratory lines (Eldridge et al., 2020). Furthermore, like other marsupials, the dunnart possesses significant heterochrony in development of its head, brain and limbs compared with eutherian species, making it an excellent model for comparative mammalian research.
One of the most remarkable features of dunnart biology is its rapid gestation and ultra-altricial state at birth (Suárez et al., 2017; Cook et al., 2021). Dasyurid marsupials, including the dunnart, represent some of the most altricial of all extant mammals. Dunnart neonates are born after a rapid 13.5-day gestation, compared to ∼20 days in mouse (Figure 1D), and superficially resemble a eutherian foetus. At birth, the dunnart orofacial region appears as rudimentary facial prominences akin to an embryonic day 11.5–12 mouse (Figure 1C), despite being functional to accommodate suckling. The newborn dunnart lacks a developed brain and has paddle-like hindlimbs, but possesses highly developed, muscularized forelimbs with claws to accommodate crawling (Figure 1D) (Suárez et al., 2017; Cook et al., 2021). Remarkably, newborn dunnarts lack mineralized bone in the facial skeleton and forelimbs, which rapidly ossify within the first 24 h, while the hindlimbs do not start to ossify until ∼D5 (Cook et al., 2021). This extreme heterochrony and altriciality at birth allows direct manipulations of these developmental systems ex utero, at equivalent eutherian embryonic stages (Paolino et al., 2018). Critically, the ultra-altricial birth of dasyurids demand additional acceleration of the onset of NCC specification, migration and proliferation, compared to the opossum (Smith, 2020). These features distinguish the dunnart as an exceptional mammalian model to investigate NCC-derived craniofacial patterning and ossification. However, detailed analyses which substantiate these early NCC behaviours in Sminthopsis have yet to be performed, representing an important first step to understand their NCC biology and thus heterochrony in mammals. Nevertheless, the dunnart is well positioned to determine how altered developmental timing influences ontogeny and craniofacial morphogenesis, providing new insights into the origin of species-specific pattern.
5 A Look to the Future: Comparative Evo-Devo-Omics
The age of comparative and functional genomics has accelerated investigations into the molecular basis of mammalian trait evolution. Comparative genomics has allowed identification of genes and regulatory regions under selection within and between lineages (Capra et al., 2013; Parker et al., 2013; Foote et al., 2015; Feigin et al., 2018, 2019); comparative bulk RNA-seq has revealed differentially expressed genes between tissues or developing structures (Eckalbar et al., 2016; Cooper et al., 2020); and chromatin pulldown or accessibility assays (ChIP, HiC, or ATAC-seq) define the gene regulatory landscape associated with these tissues or developing structures (Visel et al., 2009a; Attanasio et al., 2013). However, though powerful, individually these analyses are static and may overlook dynamic processes that contribute to development of complex traits. For example, while identification of differentially expressed genes or enhancers active in the embryonic orofacial region may constitute components that contribute to mammalian facial shape diversity (species-generic), such analyses are unable to capture dynamic regulation of these and the GRNs that influence development of unique anatomical features (species-specific) (Schneider, 2018)—as exemplified in avian models (Schneider and Helms, 2003; Chen et al., 2012; Hall et al., 2014; Ealba et al., 2015). As such, alternative approaches are required to disentangle the complex landscape of craniofacial development between disparate species.
The advent of single-cell omics has revolutionized developmental biology, producing high-resolution atlases of diverse developmental processes (Cao et al., 2019). To date, single-cell studies have been applied to multiple aspects of neural crest patterning and craniofacial development (Li et al., 2019; Soldatov et al., 2019; Farmer et al., 2021; Morrison et al., 2021; Pagella et al., 2021; Tatarakis et al., 2021), providing unique insights into how these complex developmental processes are regulated. Single-cell transcriptomics (scRNA-seq) allow detailed characterization of transcriptional profiles and cell-types present within developing structures, fate decisions and gene expression dynamics underlying differentiation of progenitors into mature cell types (Trapnell et al., 2014), and cell-cell signalling interactions between adjacent tissues (Jin et al., 2021). Importantly, scRNA-seq data can be integrated with genome-wide assays for Transposase-Accessible Chromatin (ATAC-seq) to define regulatory elements active within their underlying cell types (Buenrostro et al., 2015; Cao et al., 2018; Stuart et al., 2019), or spatial transcriptomics to resolve cellular gene expression profiles in individual cells (Xia et al., 2019) or developing tissues in situ (Marx, 2021). Used in combination, these techniques provide powerful methods to integrate developmental biology with gene expression dynamics and construction of species-specific GRNs.
Despite their potential, single-cell multi-omic approaches have been scarcely applied in comparative evolutionary biology. Such studies of “comparative evo-devo-omics” between taxa are becoming rapidly viable to investigate the molecular mechanisms underlying convergence, constraint, or innovation in specific developmental processes (Shafer, 2019; Mahadevaiah et al., 2020). However, the lack of these applied studies are largely in response to significant technical limitations surrounding integration and batch correction of disparate datasets, specificity and stage-matching of tissues and homologous cell types between disparate species, and quality of the underlying genome and transcriptome—reviewed by (Shafer, 2019). Nevertheless, these limitations can be mitigated through application of tools aiding dataset integration, sequencing depth and batch correction, as well as issues with transcriptome quality and gene orthology (Butler et al., 2018; Haghverdi et al., 2018). Furthermore, new applied methodologies are being produced to better identify and match homologous cell and tissue types (Tosches et al., 2018; Welch et al., 2019; Feregrino and Tschopp, 2021) and their proportions between distantly related species and datasets (Phipson et al., 2021). Given the rapid rate by which these limitations are being resolved by the community, comparative evo-devo-omics presents a powerful platform to interrogate the cell, molecular and developmental mechanisms underlying heterochronic NCC specification and facial patterning between marsupial and eutherian mammals.
5.1 Mammalian Craniofacial Heterochrony at Single Cell Resolution
Using the above examples, I present a hypothetical workflow for detailed investigations into mammalian craniofacial heterochrony and evolution through a comparative lens. First, sampling of single-cell RNA, chromatin and spatial profiles of stage-matched dunnart and mouse embryos (Figure 2A) will allow generation of species-specific transcriptional atlases, building on existing datasets (Soldatov et al., 2019) and producing novel mammalian resources. The resulting transcriptomic, epigenetic and spatial profiles can be clustered and integrated for detection of homologous and novel cell types (Tosches et al., 2018; Welch et al., 2019), conserved and disparate gene co-expression modules (Feregrino and Tschopp, 2021), cell type proportions (Phipson et al., 2021) and spatial quantification of genes in situ (Figure 2B). From here, evolutionary hypotheses can be tested through identification of dynamic transcriptional, signalling, epigenetic and spatial relationships, revealing shared (species-generic) and unique (species-specific) processes underlying facial development and evolution (Figure 2C). Through application of these approaches, we can begin to determine how NCC gene regulatory architecture differs between therian mammals, and their influence on heterochronic craniofacial patterning. Ultimately, this will not only provide valuable data into how diverse facial shapes are produced during development, but also provide novel insights into how mammalian craniofacial diversity arises during evolution.
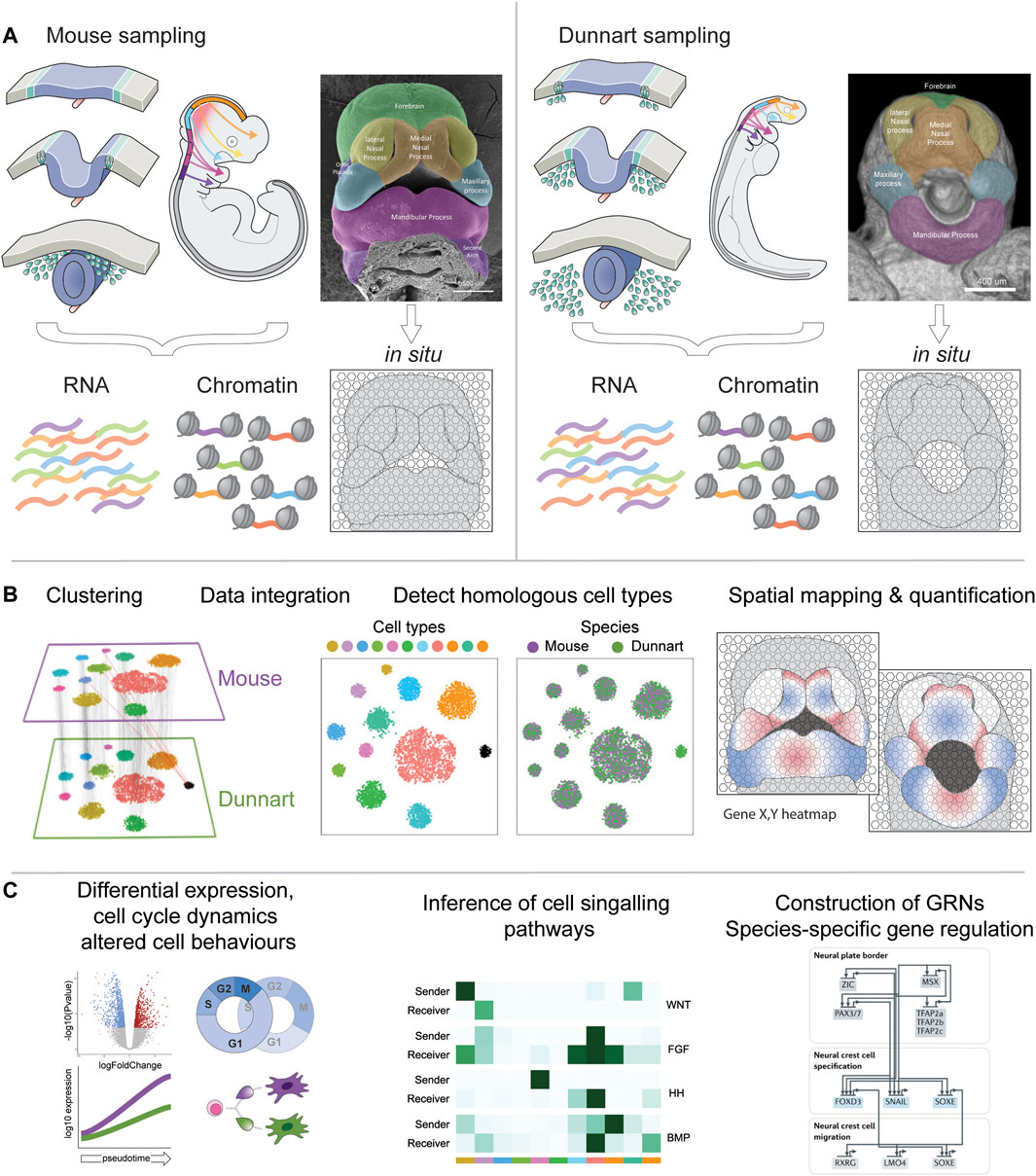
FIGURE 2. Workflow for comparative craniofacial single-cell multiomics. (A) Single cells can be isolated from dunnart and mouse NCCs and developing craniofacial prominences for multiplexed isolation and sequencing of RNA and open chromatin. Facial tissue sections can be processed for in situ spatial transcriptomics or MERFISH (Xia et al., 2019). Single-cell RNA and ATAC-seq data can be readily integrated using pipelines such as Seurat (Butler et al., 2018). (B) Dunnart and mouse datasets can be individually clustered or integrated to generate an atlas of homologous cell type populations—adapted from (Stuart et al., 2019). Cell transcriptomic and epigenetic profiles can be further mapped back to their spatial organization in the embryo. (C) The combination of these methods allows sophisticated downstream workflows to examine differential expression between species-specific clusters (pseudobulk) or cell lineage differentiation (pseudotime), inference of cell-cell signalling relationships (Jin et al., 2021), or construction of GRNs and species-specific patterns of gene regulation (Martik and Bronner, 2021).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AN conceived the study and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
I thank my mentors Andrew Pask, Christy Hipsley, Stephen Frankenberg, and Craig Smith for constructive discussions and guidance which led to the preparation of this manuscript.
References
Attanasio, C., Nord, A. S., Zhu, Y., Blow, M. J., Li, Z., Liberton, D. K., et al. (2013). Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers. Science 342, 1–20. doi:10.1126/science.1241006
Beiriger, A., and Sears, K. E. (2014). Cellular Basis of Differential Limb Growth in Postnatal Gray Short-Tailed Opossums (Monodelphis Domestica). J. Exp. Zool. Mol. Dev. Evol.) 322, 221–229. doi:10.1002/jez.b.22556
Bennett, C. V., and Goswami, A. (2013). Statistical Support for the Hypothesis of Developmental Constraint in Marsupial Skull Evolution. BMC Biol. 11. doi:10.1186/1741-7007-11-52
Bininda-Emonds, O. R. P., Cardillo, M., Jones, K. E., MacPhee, R. D. E., Beck, R. M. D., Grenyer, R., et al. (2007). The Delayed Rise of Present-Day Mammals. Nature 446, 507–512. doi:10.1038/nature05634
Brugmann, S. A., Powder, K. E., Young, N. M., Goodnough, L. H., Hahn, S. M., James, A. W., et al. (2010). Comparative Gene Expression Analysis of Avian Embryonic Facial Structures Reveals New Candidates for Human Craniofacial Disorders. Hum. Mol. Genet. 19, 920–930. doi:10.1093/hmg/ddp559
Buenrostro, J. D., Wu, B., Litzenburger, U. M., Ruff, D., Gonzales, M. L., Snyder, M. P., et al. (2015). Single-cell Chromatin Accessibility Reveals Principles of Regulatory Variation. Nature 523, 486–490. doi:10.1038/nature14590
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 36, 411–420. doi:10.1038/nbt.4096
Cao, J., Cusanovich, D. A., Ramani, V., Aghamirzaie, D., Pliner, H. A., Hill, A. J., et al. (20182018). Joint Profiling of Chromatin Accessibility and Gene Expression in Thousands of Single Cells. Science 361, 1380–1385. doi:10.1126/science.aau0730
Cao, J., Spielmann, M., Qiu, X., Huang, X., Ibrahim, D. M., Hill, A. J., et al. (2019). The Single-Cell Transcriptional Landscape of Mammalian Organogenesis. Nature 566, 496–502. doi:10.1038/s41586-019-0969-x
Capra, J. A., Erwin, G. D., Mckinsey, G., Rubenstein, J. L. R., and Pollard, K. S. (2013). Many Human Accelerated Regions Are Developmental Enhancers. Phil. Trans. R. Soc. B 368, 20130025. doi:10.1098/rstb.2013.0025
Chen, C.-C., Balaban, E., and Jarvis, E. D. (2012). Interspecies Avian Brain Chimeras Reveal that Large Brain Size Differences Are Influenced by Cell-Interdependent Processes. PLoS One 7, e42477. doi:10.1371/journal.pone.0042477
Cheung, M., and Briscoe, J. (2003). Neural Crest Development Is Regulated by the Transcription Factor Sox9. Development 130, 5681–5693. doi:10.1242/dev.00808
Chew, K. Y., Shaw, G., Yu, H., Pask, A. J., and Renfree, M. B. (2014). Heterochrony in the Regulation of the Developing Marsupial Limb. Dev. Dyn. 243, 324–338. doi:10.1002/dvdy.24062
Cook, L. E., Newton, A. H., Hipsley, C. A., and Pask, A. J. (2021). Postnatal Development in a Marsupial Model, the Fat-Tailed Dunnart Sminthopsis crassicaudata; Dasyuromorphia: Dasyuridae. Commun. Biol. 4, 1–14. doi:10.1038/s42003-021-02506-2
Cooper, K., Saxena, A., Sharma, V., Neufeld, S., Tran, M., Gutierrez, H., et al. (2020). Interspecies Transcriptome Analyses Identify Genes that Control the Development and Evolution of Limb Skeletal Proportion. FASEB J. 34, 1–1. doi:10.1096/fasebj.2020.34.s1.00363
Couly, G. F., Coltey, P. M., and Le Douarin, N. M. (1993). The Triple Origin of Skull in Higher Vertebrates: a Study in Quail-Chick Chimeras. Development 117, 409–429. doi:10.1242/dev.117.2.409
Creuzet, S., Schuler, B., Couly, G., and Le Douarin, N. M. (2004). Reciprocal Relationships between Fgf8 and Neural Crest Cells in Facial and Forebrain Development. Proc. Natl. Acad. Sci. U.S.A. 101, 4843–4847. doi:10.1073/pnas.0400869101
Dash, S., and Trainor, P. A. (2020). The Development, Patterning and Evolution of Neural Crest Cell Differentiation into Cartilage and Bone. Bone 137, 115409. doi:10.1016/j.bone.2020.115409
Dickel, D. E., Ypsilanti, A. R., Pla, R., Zhu, Y., Barozzi, I., Mannion, B. J., et al. (2018). Ultraconserved Enhancers Are Required for Normal Development. Cell 172, 491–499. e15. doi:10.1016/j.cell.2017.12.017
Ealba, E. L., Jheon, A. H., Hall, J., Curantz, C., Butcher, K. D., and Schneider, R. A. (2015). Neural Crest-Mediated Bone Resorption Is a Determinant of Species-specific Jaw Length. Dev. Biol. 408, 151–163. doi:10.1016/j.ydbio.2015.10.001
Eckalbar, W. L., Schlebusch, S. A., Mason, M. K., Gill, Z., Parker, A. V., Booker, B. M., et al. (2016). Transcriptomic and Epigenomic Characterization of the Developing Bat Wing. Nat. Genet. 48, 528–536. doi:10.1038/ng.3537
Eldridge, M. D. B., Deakin, J. E., MacDonald, A. J., Byrne, M., Fitzgerald, A., Johnson, R. N., et al. (2020). The Oz Mammals Genomics (OMG) Initiative: Developing Genomic Resources for Mammal Conservation at a Continental Scale. Aust. Zool. 40, 505–509. doi:10.7882/AZ.2020.003
Fabre, A.-C., Dowling, C., Portela Miguez, R., Fernandez, V., Noirault, E., and Goswami, A. (2021). Functional Constraints during Development Limit Jaw Shape Evolution in Marsupials. Proc. R. Soc. B 288, 1–8. doi:10.1098/rspb.2021.0319
Farmer, D. J. T., Mlcochova, H., Zhou, Y., Koelling, N., Wang, G., Ashley, N., et al. (2021). The Developing Mouse Coronal Suture at Single-Cell Resolution. Nat. Commun. 12, 1–14. doi:10.1038/s41467-021-24917-9
Feigin, C. Y., Newton, A. H., Doronina, L., Schmitz, J., Hipsley, C. A., Mitchell, K. J., et al. (2018). Genome of the Tasmanian Tiger Provides Insights into the Evolution and Demography of an Extinct Marsupial Carnivore. Nat. Ecol. Evol. 2, 182–192. doi:10.1038/s41559-017-0417-y
Feigin, C. Y., Newton, A. H., and Pask, A. J. (2019). Widespread Cis-Regulatory Convergence between the Extinct Tasmanian Tiger and Gray Wolf. Genome Res. 29, 1648–1658. doi:10.1101/gr.244251.118
Felice, R. N., and Goswami, A. (2018). Developmental Origins of Mosaic Evolution in the Avian Cranium. Proc. Natl. Acad. Sci. U.S.A. 115, 555–560. doi:10.1073/pnas.1716437115
Feregrino, C., and Tschopp, P. (2021). Assessing Evolutionary and Developmental Transcriptome Dynamics in Homologous Cell Types. Dev. Dyn., 1–18. doi:10.1002/dvdy.384
Fish, J. L. (2019). Evolvability of the Vertebrate Craniofacial Skeleton. Seminars Cell & Dev. Biol. 91, 13–22. doi:10.1016/j.semcdb.2017.12.004
Foote, A. D., Liu, Y., Thomas, G. W. C., Vinař, T., Alföldi, J., Deng, J., et al. (2015). Convergent Evolution of the Genomes of Marine Mammals. Nat. Genet. 47, 272–275. doi:10.1038/ng.3198
Frigo, L., and Woolley, P. A. (1997). Growth and Development of Pouch Young of the Stripe-Faced Dunnart, Sminthopsis Macroura (Marsupialia : Dasyuridae), in Captivity. Aust. J. Zool. 45, 157–170. doi:10.1071/ZO97002
Frigo, L., and Woolley, P. (1996). Development of the Skeleton of the Stripe-Faced Dunnart, Sminthopsis Macroura (Marsupialia: Dasyuridae). Aust. J. Zool. 44, 155–164. doi:10.1071/ZO9960155
Goswami, A. (2007). Cranial Modularity and Sequence Heterochrony in Mammals. Evol. Dev. 9, 290–298. doi:10.1111/j.1525-142X.2007.00161.x
Goswami, A. (2006). Cranial Modularity Shifts during Mammalian Evolution. Am. Nat. 168, 270–280. doi:10.1086/505758
Goswami, A., Milne, N., and Wroe, S. (2011). Biting through Constraints: Cranial Morphology, Disparity and Convergence across Living and Fossil Carnivorous Mammals. Proc. R. Soc. B 278, 1831–1839. doi:10.1098/rspb.2010.2031
Goswami, A., Polly, P. D., Mock, O. B., and Sánchez-villagra, M. R. (2012). Shape, Variance and Integration during Craniogenesis: Contrasting Marsupial and Placental Mammals. J. Evol. Biol. 25, 862–872. doi:10.1111/j.1420-9101.2012.02477.x
Goswami, A., and Polly, P. D. (2010). The Influence of Modularity on Cranial Morphological Disparity in Carnivora and Primates (Mammalia). PLoS One 5, e9517–8. doi:10.1371/journal.pone.0009517
Goswami, A., Randau, M., Polly, P. D., Weisbecker, V., Bennett, C. V., Hautier, L., et al. (2016). Do developmental Constraints and High Integration Limit the Evolution of the Marsupial Oral Apparatus? Integr. Comp. Biol. 56, 404–415. doi:10.1093/icb/icw039
Goswami, A., Weisbecker, V., and Sánchez-Villagra, M. R. (2009). Developmental Modularity and the Marsupial-Placental Dichotomy. J. Exp. Zool. 312B, 186–195. doi:10.1002/jez.b.21283
Green, S. A., Simoes-costa, M., and Bronner, M. E. (2015). Evolution of Vertebrates as Viewed from the Crest. Nature 520, 474–482. doi:10.1038/nature14436
Haghverdi, L., Lun, A. T. L., Morgan, M. D., and Marioni, J. C. (2018). Batch Effects in Single-Cell RNA-Sequencing Data Are Corrected by Matching Mutual Nearest Neighbors. Nat. Biotechnol. 36, 421–427. doi:10.1038/nbt.4091
Hall, J., Jheon, A. H., Ealba, E. L., Eames, B. F., Butcher, K. D., Mak, S.-S., et al. (2014). Evolution of a Developmental Mechanism: Species-specific Regulation of the Cell Cycle and the Timing of Events during Craniofacial Osteogenesis. Dev. Biol. 385, 380–395. doi:10.1016/j.ydbio.2013.11.011
Higashiyama, H., Koyabu, D., Hirasawa, T., Werneburg, I., Kuratani, S., and Kurihara, H. (2021). Mammalian Face as an Evolutionary Novelty. Proc. Natl. Acad. Sci. U.S.A. 118, 1–8. doi:10.1073/pnas.2111876118
Hu, Q., Ueno, N., and Behringer, R. R. (2004). Restriction of BMP4 Activity Domains in the Developing Neural Tube of the Mouse Embryo. EMBO Rep. 5, 734–739. doi:10.1038/sj.embor.7400184
Ishii, M., Arias, A. C., Liu, L., Chen, Y.-B., Bronner, M. E., and Maxson, R. E. (2012). A Stable Cranial Neural Crest Cell Line from Mouse. Stem Cells Dev. 21, 3069–3080. doi:10.1089/scd.2012.0155
Jiang, X., Iseki, S., Maxson, R. E., Sucov, H. M., and Morriss-Kay, G. M. (2002). Tissue Origins and Interactions in the Mammalian Skull Vault. Dev. Biol. 241, 106–116. doi:10.1006/dbio.2001.0487
Jin, S., Guerrero-Juarez, C. F., Zhang, L., Chang, I., Ramos, R., Kuan, C.-H., et al. (2021). Inference and Analysis of Cell-Cell Communication Using CellChat. Nat. Commun. 12, 1–20. doi:10.1038/s41467-021-21246-9
Keyte, A. L., and Smith, K. K. (2010). Developmental Origins of Precocial Forelimbs in Marsupial Neonates. Development 137, 4283–4294. doi:10.1242/dev.049445
Keyte, A., and Smith, K. K. (2012). Heterochrony in Somitogenesis Rate in a Model Marsupial,Monodelphis Domestica. Evol. Dev. 14, 93–103. doi:10.1111/j.1525-142X.2011.00524.x
Kiyonari, H., Kaneko, M., Abe, T., Shiraishi, A., Yoshimi, R., Inoue, K.-i., et al. (2021). Targeted Gene Disruption in a Marsupial, Monodelphis Domestica, by CRISPR/Cas9 Genome Editing. Curr. Biol. 31, 3956–3963. doi:10.1016/j.cub.2021.06.056
Kuratani, S., Kusakabe, R., and Hirasawa, T. (2018). The Neural Crest and Evolution of the Head/trunk Interface in Vertebrates. Dev. Biol. 444, S60–S66. doi:10.1016/j.ydbio.2018.01.017
Lewis, A. E., Vasudevan, H. N., O’Neill, A. K., Soriano, P., and Bush, J. O. (2013). The Widely Used Wnt1-Cre Transgene Causes Developmental Phenotypes by Ectopic Activation of Wnt Signaling. Dev. Biol. 379, 229–234. doi:10.1016/j.ydbio.2013.04.026
Li, H., Jones, K. L., Hooper, J. E., and Williams, T. (2019). The Molecular Anatomy of Mammalian Upper Lip and Primary Palate Fusion at Single Cell Resolution. Dev 146, dev174888. doi:10.1242/dev.174888
Mahadevaiah, S. K., Sangrithi, M. N., Hirota, T., and Turner, J. M. A. (2020). A Single-Cell Transcriptome Atlas of Marsupial Embryogenesis and X Inactivation. Nature 586, 612–617. doi:10.1038/s41586-020-2629-6
Martik, M. L., and Bronner, M. E. (2021). Riding the Crest to Get a Head: Neural Crest Evolution in Vertebrates. Nat. Rev. Neurosci. 22, 616–626. doi:10.1038/s41583-021-00503-2
Martin, K. E. A., and Mackay, S. (2003). Postnatal Development of the Fore- and Hindlimbs in the Grey Short-Tailed Opossum, Monodelphis Domestica. J. Anat. 202, 143–152. doi:10.1046/j.1469-7580.2003.00149.x
Marx, V. (2021). Method of the Year: Spatially Resolved Transcriptomics. Nat. Methods 18, 9–14. doi:10.1038/s41592-020-01033-y
Minoux, M., and Rijli, F. M. (2010). Molecular Mechanisms of Cranial Neural Crest Cell Migration and Patterning in Craniofacial Development. Development 137, 2605–2621. doi:10.1242/dev.040048
Morrison, J. A., McLennan, R., Teddy, J. M., Scott, A. R., Kasemeier-Kulesa, J. C., Gogol, M. M., et al. (2021). Single-cell Reconstruction with Spatial Context of Migrating Neural Crest Cells and Their Microenvironments during Vertebrate Head and Neck Formation. Dev 148, dev199468. doi:10.1242/dev.199468
Murillo-Rincón, A. P., and Kaucka, M. (2020). Insights Into the Complexity of Craniofacial Development From a Cellular Perspective. Front. Cell Dev. Biol. 8, 1–8. doi:10.3389/fcell.2020.620735
Newton, A. H., Feigin, C. Y., and Pask, A. J. (2017). RUNX2 Repeat Variation Does Not Drive Craniofacial Diversity in Marsupials. BMC Evol. Biol. 17, 1–9. doi:10.1186/s12862-017-0955-6–
Newton, A. H., and Pask, A. J. (2020). Evolution and Expansion of the RUNX2 QA Repeat Corresponds with the Emergence of Vertebrate Complexity. Commun. Biol. 3, 771. doi:10.1038/s42003-020-01501-3
Newton, A. H., Weisbecker, V., Pask, A. J., and Hipsley, C. A. (2021). Ontogenetic Origins of Cranial Convergence between the Extinct Marsupial Thylacine and Placental Gray Wolf. Commun. Biol. 4, 51. doi:10.1038/s42003-020-01569-x
Nguyen, B. H., Ishii, M., Maxson, R. E., and Wang, J. (2018). Culturing and Manipulation of O9-1 Neural Crest Cells. J. Vis. Exp. 9, 58346. doi:10.3791/58346
Nunn, C. L., and Smith, K. K. (1998). Statistical Analyses of Developmental Sequences: The Craniofacial Region in Marsupial and Placental Mammals. Am. Nat. 152, 82–101. doi:10.1086/286151
Pagella, P., de Vargas Roditi, L., Stadlinger, B., Moor, A. E., and Mitsiadis, T. A. (2021). A Single-Cell Atlas of Human Teeth. iScience 24, 102405. doi:10.1016/j.isci.2021.102405
Paolino, A., Fenlon, L. R., Kozulin, P., Richards, L. J., and Suárez, R. (2018). Multiple Events of Gene Manipulation via in Pouch Electroporation in a Marsupial Model of Mammalian Forebrain Development. J. Neurosci. Methods 293, 45–52. doi:10.1016/j.jneumeth.2017.09.004
Parker, J., Tsagkogeorga, G., Cotton, J. A., Liu, Y., Provero, P., Stupka, E., et al. (2013). Genome-wide Signatures of Convergent Evolution in Echolocating Mammals. Nature 502, 228–231. doi:10.1038/nature12511
Phipson, B., Sim, C. B., Porrello, E., Hewitt, A. W., Powell, J., and Oshlack, A. (2021). Propeller: Testing for Differences in Cell Type Proportions in Single Cell Data. bioRxiv 2021, 470236. doi:10.1101/2021.11.28.470236
Rager, L., Hautier, L., Forasiepi, A., Goswami, A., and Sánchez-Villagra, M. R. (2014). Timing of Cranial Suture Closure in Placental Mammals: Phylogenetic Patterns, Intraspecific Variation, and Comparison with Marsupials. J. Morphol. 275, 125–140. doi:10.1002/jmor.20203
Rauch, A., Seitz, S., Baschant, U., Schilling, A. F., Illing, A., Stride, B., et al. (2010). Glucocorticoids Suppress Bone Formation by Attenuating Osteoblast Differentiation via the Monomeric Glucocorticoid Receptor. Cell Metab. 11, 517–531. doi:10.1016/j.cmet.2010.05.005
Rodda, S. J., and McMahon, A. P. (2006). Distinct Roles for Hedgehog and Canonical Wnt Signaling in Specification,differentiation and Maintenance of Osteoblast Progenitors. Development 133, 3231–3244. doi:10.1242/dev.02480
Samuels, B. D., Aho, R., Brinkley, J. F., Bugacov, A., Feingold, E., Fisher, S., et al. (2020). FaceBase 3: Analytical Tools and FAIR Resources for Craniofacial and Dental Research. Development (Cambridge, England), 147(18), dev191213. doi:10.1242/dev.191213
Sánchez-Villagra, M. R., Goswami, A., Weisbecker, V., Mock, O., and Kuratani, S. (2008). Conserved Relative Timing of Cranial Ossification Patterns in Early Mammalian Evolution. Evol. Dev. 10, 519–530. doi:10.1111/j.1525-142X.2008.00267.x
Schneider, R. A., and Helms, J. A. (2003). The Cellular and Molecular Origins of Beak Morphology. Science 299, 565–568. doi:10.1126/science.1077827
Schneider, R. A. (2018). Neural Crest and the Origin of Species-specific Pattern. Genesis 56, e23219–33. doi:10.1002/dvg.23219
Sears, K. E. (2009). Differences in the Timing of Prechondrogenic Limb Development in Mammals: The Marsupial-Placental Dichotomy Resolved. Evol. (N. Y). 63, 2193–2200. doi:10.1111/j.1558-5646.2009.00690.x
Selwood, L., and Coulson, G. (2006). Marsupials as Models for Research. Aust. J. Zool. 54, 137. doi:10.1071/ZOv54n3_IN
Shafer, M. E. R. (2019). Cross-Species Analysis of Single-Cell Transcriptomic Data. Front. Cell Dev. Biol. 7, 1–9. doi:10.3389/fcell.2019.00175
Smith, K. K. (1997). Comparative Patterns of Craniofacial Development in Eutherian and Metatherian Mammals. Evolution 51, 1663. doi:10.2307/2411218
Smith, K. K. (2001). Early Development of the Neural Plate, Neural Crest and Facial Region of Marsupials. J. Anat. 199, 121–131. doi:10.1017/S002187820100820210.1046/j.1469-7580.2001.19910121.x
Smith, K. K. (2020). J P Hill and Katherine Watson's Studies of the Neural Crest in Marsupials. J. Morphol. 281, 1567–1587. doi:10.1002/jmor.21270
Soldatov, R., Kaucka, M., Kastriti, M. E., Petersen, J., Chontorotzea, T., Englmaier, L., et al. (2019). Spatiotemporal Structure of Cell Fate Decisions in Murine Neural Crest. Science 364, eaas9536. doi:10.1126/science.aas9536
Spiekman, S. N. F., and Werneburg, I. (2017). Patterns in the Bony Skull Development of Marsupials: High Variation in Onset of Ossification and Conserved Regions of Bone Contact. Sci. Rep. 7, 1–11. doi:10.1038/srep43197
Stine, Z. E., Huynh, J. L., Loftus, S. K., Gorkin, D. U., Salmasi, A. H., Novak, T., et al. (2009). Oligodendroglial and Pan-Neural Crest Expression of Cre Recombinase Directed bySox10enhancer. Genesis 47, 765–770. doi:10.1002/dvg.20559
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., et al. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902. e21. doi:10.1016/j.cell.2019.05.031
Suárez, R., Paolino, A., Kozulin, P., Fenlon, L. R., Morcom, L. R., Englebright, R., et al. (2017). Development of Body, Head and Brain Features in the Australian Fat-Tailed Dunnart (Sminthopsis crassicaudata; Marsupialia: Dasyuridae); A Postnatal Model of Forebrain Formation. PLoS One 12, e0184450–18. doi:10.1371/journal.pone.0184450
Szabo-rogers, H. L., Smithers, L. E., Yakob, W., and Liu, K. J. (2010). New Directions in Craniofacial Morphogenesis. Dev. Biol. 341, 84–94. doi:10.1016/j.ydbio.2009.11.021
Tatarakis, D., Cang, Z., Wu, X., Sharma, P. P., Karikomi, M., MacLean, A. L., et al. (2021). Single-cell Transcriptomic Analysis of Zebrafish Cranial Neural Crest Reveals Spatiotemporal Regulation of Lineage Decisions during Development. Cell Rep. 37, 110140. doi:10.1016/j.celrep.2021.110140
Tosches, M. A., Yamawaki, T. M., Naumann, R. K., Jacobi, A. A., Tushev, G., and Laurent, G. (2018). Evolution of Pallium, hippocampus, and Cortical Cell Types Revealed by Single-Cell Transcriptomics in Reptiles. Science. 360, 881–888. doi:10.1126/science.aar4237
Trapnell, C., Cacchiarelli, D., Grimsby, J., Pokharel, P., Li, S., Morse, M., et al. (2014). The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 32, 381–386. doi:10.1038/nbt.2859
Usui, K., and Tokita, M. (2018). Creating Diversity in Mammalian Facial Morphology: A Review of Potential Developmental Mechanisms. Evodevo 9, 1–17. doi:10.1186/s13227-018-0103-4
Vaglia, J. L., and Smith, K. K. (2003). Early Differentiation and Migration of Cranial Neural Crest in the Opossum, Monodelphis Domestica. Evol. Dev. 5, 121–135. doi:10.1046/j.1525-142X.2003.03019.x
Visel, A., Blow, M. J., Li, Z., Zhang, T., Akiyama, J. A., Holt, A., et al. (2009a). ChIP-seq Accurately Predicts Tissue-specific Activity of Enhancers. Nature 457, 854–858. doi:10.1038/nature07730
Visel, A., Rubin, E. M., and Pennacchio, L. A. (2009b). Genomic Views of Distant-Acting Enhancers. Nature 461, 199–205. doi:10.1038/nature08451
Wakamatsu, Y., Nomura, T., Osumi, N., and Suzuki, K. (2014). Comparative Gene Expression Analyses Reveal Heterochrony forSox9expression in the Cranial Neural Crest during Marsupial Development. Evol. Dev. 16, 197–206. doi:10.1111/ede.12083
Wakamatsu, Y., and Suzuki, K. (2019). Sequence Alteration in the Enhancer Contributes to the Heterochronic Sox9 Expression in Marsupial Cranial Neural Crest. Dev. Biol. 456, 31–39. doi:10.1016/j.ydbio.2019.08.010
Weisbecker, V., Goswami, A., Wroe, S., and Sánchez-Villagra, M. R. (2008). Ossification Heterochrony in the Therian Postcranial Skeleton and the Marsupial-Placental Dichotomy. Evol. (N. Y). 62, 2027–2041. doi:10.1111/j.1558-5646.2008.00424.x
Welch, J. D., Kozareva, V., Ferreira, A., Vanderburg, C., Martin, C., and Macosko, E. Z. (2019). Single-Cell Multi-Omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887. e17. doi:10.1016/j.cell.2019.05.006
Wilkie, A. O. M., and Morriss-Kay, G. M. (2001). Genetics of Craniofacial Development and Malformation. Nat. Rev. Genet. 2, 458–468. doi:10.1038/35076601
Wu, P., Jiang, T.-X., Shen, J.-Y., Widelitz, R. B., and Chuong, C.-M. (2006). Morphoregulation of Avian Beaks: Comparative Mapping of Growth Zone Activities and Morphological Evolution. Dev. Dyn. 235, 1400–1412. doi:10.1002/dvdy.20825
Wu, P., Jiang, T.-X., Suksaweang, S., Widelitz, R. B., and Chuong, C.-M. (2004). Molecular Shaping of the Beak. Science 305, 1465–1466. doi:10.1126/science.1098109
Xia, C., Fan, J., Emanuel, G., Hao, J., and Zhuang, X. (2019). Spatial Transcriptome Profiling by MERFISH Reveals Subcellular RNA Compartmentalization and Cell Cycle-dependent Gene Expression. Proc. Natl. Acad. Sci. U.S.A. 116, 19490–19499. doi:10.1073/pnas.1912459116
Yoshida, T., Vivatbutsiri, P., Morriss-Kay, G., Saga, Y., and Iseki, S. (2008). Cell Lineage in Mammalian Craniofacial Mesenchyme. Mech. Dev. 125, 797–808. doi:10.1016/j.mod.2008.06.007
Zhang, M., Xuan, S., Bouxsein, M. L., Von Stechow, D., Akeno, N., Faugere, M. C., et al. (2002). Osteoblast-specific Knockout of the Insulin-like Growth Factor (IGF) Receptor Gene Reveals an Essential Role of IGF Signaling in Bone Matrix Mineralization. J. Biol. Chem. 277, 44005–44012. doi:10.1074/jbc.M208265200
Keywords: NCC, mammal, heterochrony, constraint, evolution, GRN, skull
Citation: Newton AH (2022) Marsupials and Multi-Omics: Establishing New Comparative Models of Neural Crest Patterning and Craniofacial Development. Front. Cell Dev. Biol. 10:941168. doi: 10.3389/fcell.2022.941168
Received: 11 May 2022; Accepted: 06 June 2022;
Published: 23 June 2022.
Edited by:
Neva P. Meyer, Clark University, United StatesReviewed by:
Shunsuke Suzuki, Shinshu University, JapanAnna Keyte, The Rockefeller University, United States
Copyright © 2022 Newton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Axel H. Newton, axel.newton@unimelb.edu.au
†ORCID: Axel H. Newton, orcid.org/0000-0001-7175-5978
 Axel H. Newton
Axel H. Newton